Abstract
Accumulating evidence has indicated that microRNAs (miRNAs) can act as candidate tumor suppressors. However, the mechanism of miR-27b-3p in colorectal cancer (CRC) is still unknown. In this study, the relative expression levels of miR-27b-3p and LIMK1 were detected by quantitative real-time PCR (qRT-PCR) in CRC and adjacent normal tissues as well as in CRC cell lines. The proliferation, cell cycle and migratory and invasive abilities of SW620 cells were analyzed by CCK-8, flow cytometry, wound healing assays, and Transwell assays, respectively. The regulatory relationships between LIMK1 and miR-27b-3p were detected by qRT-PCR, luciferase reporter gene and Western blot assays. The results showed that the expression levels of miR-27b-3p were downregulated in CRC samples compared with those in adjacent nontumor tissues. Moreover, we found that miR-27b-3p suppressed CRC cell proliferation, cycle and migration and invasion. Additionally, there was an inverse correlation between LIMK1 expression and miR-27b-3p expression in CRC tissues. LIMK1 was identified as a direct target of miR-27b-3p. Our findings indicated that miR-27b-3p inhibited CRC cell proliferation, migration and invasion by targeting LIMK1.
Keywords: miR-27b-3p, LIMK1, colorectal cancer, proliferation, migration, invasion
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed malignancies worldwide and is one of the most common leading causes of cancer-specific mortality [1]. In the past few years, the survival rates of patients with CRC have gradually increased because of earlier diagnosis and improved treatments; however, approximately 30-50% of patients who undergo excision surgery suffer from local tumor recurrence or metastasis [2]. According to available statistics, patients treated with chemotherapy in combination with monoclonal antibody have an average overall survival of 20 months, and the best response rates are 50% [3]. However, this treatment is expensive for patients with CRC, and chemotherapy may lead to overtreatment of patients with toxic drugs that generate severe adverse effects [4]. Therefore, new biomarkers and therapeutic targets should be found to obtain effective treatments for more aggressive stages of CRC.
miRNAs are a type of small, non-coding RNA 20-22 nucleotides in length that regulate the expression of hundreds of target genes by binding to the 3’-untranslated region (3’-UTR) of their target mRNAs [5]. miRNAs play critical roles in many kinds of physiological and pathological processes such as apoptosis, cell proliferation, differentiation, development, stress response and migration [6,7]. A growing amount of evidence has revealed that miRNAs can act as either tumor suppressors or oncogenes in the progression of many kinds of tumors by regulating posttranscriptional gene expression [8-10]. A variety of miRNAs are aberrantly ex-pressed in CRC [8].
miR-27 is a family of miRNA precursors found in humans. miRNAs are typically transcribed into precursors approximately 70 nucleotides in length and subsequently processed by the Dicer enzyme to yield a product approximately 22 nucleotides in length. miR-27b-3p is the mature miRNA of the miR-27 family. It has been reported that miR-27b-3p is differentially expressed in cervical cancer [11], colorectal neoplasia [12], and gastric cancer [13]. Furthermore, studies have shown that dysregulated miR-27b-3p plays a critical role in the proliferation and invasion of many solid malignancies, which makes it a potential target for tumor treatment [13,14]. However, the exact function of miR-27b-3p in CRC has not yet been studied.
LIMK1 is a member of the LIM kinase (LIMK) family, which contains key regulators of the actin cytoskeleton and are related to cell motility and invasion, and has been identified as a therapeutic target for metastatic disease [15]. A large study showed that LIMK1 as an oncogene participates in the process of various types of cancer [16-18]. For example, LIMK1 affected the invasion ability of prostate cancer by regulating membrane type matrix metalloproteinase 1 (MT1-MMP) [17]. LIMK1 was also verified recently to be involved in the development of CRC [19]. However, the regulatory mechanism and the role of LIMK1 in the progression of CRC are poorly understood.
In the current study, we aimed to detect the expression levels of miR-27b-3p in CRC tissues and cell lines and elucidate the roles of miR-27b-3p on CRC cell proliferation, cell cycle, migration and invasion. Furthermore, we confirmed that LIMK1 is a target gene of miR-27b-3p.
Materials and methods
Samples collection
CRC samples and adjacent normal tissues were collected from 83 patients who underwent tumor resection surgery at our hospital between 2012 and 2016. All patients signed informed consent. The tissues were separately frozen in liquid nitrogen for further study. Protocols were approved by the institutional ethics committee at our hospital and conducted in accordance with the guidelines for ethical management.
Cell lines
Human embryonic kidney 293T (HEK293T) cells, human-derived colonic epithelial NCM460 cells and human CRC cell lines (SW480, DLD-1, HT29, HCT116, and SW620) were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. SW620, HT29 and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen). NCM460, SW480, HCT116, and DLD-1 cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA). Each medium contained 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA) and 100 U/ml penicillin and streptomycin (Invitrogen). All the cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Vector construction, lentivirus production and cell transfection
miR-27b-3p mimics were packaged into lentiviral vector (GenePharma) to overexpress miR-27b-3p in CRC cells, and miR-NC was used as a control. Cells were seeded into 6-well plates at a 30% density and cultured overnight before transfection. Cells were transfected with miR-27b-3p mimics or control mimics using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocols.
Human LIMK1 was amplified from SW620 cells by RT-PCR. The PCR products were cloned into a pcDNA vector. SW620 cells (1×104 cells/well) were seeded in 24-well plates and transfected with either LIMK1 plasmid or negative control (NC) using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocols.
Quantitative real-time PCR
Total RNA was extracted from the tissues or cells using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and quantified by measuring the absorbencies at 260 and 280 nm (A260/280). Subsequently, the RNA was reverse-transcri-bed into cDNA using a reverse transcription system (Thermo Scientific, CA, USA). The expression levels of miR-27b-3p were detected by an Applied Biosystems 7500 Real-time PCR system (ABI) using TaqMan MicroRNA assay kits (Applied Biosystems, California, USA). U6 was used as the control for normalization. The gene expression of LIMK1 also analyzed using SYBR Green and was normalized to GAPDH expression. The primer sequences were as follows: GAPDH, 5’-TGT TCG TCA TGG GTG TGA AC-3’ (forward) and 5’-ATG GCA TGG ACT GTG GTC AT-3’ (reverse) (internal control); LIMK1, 5′-AAG CCA GAC ACT TCA GCT GCT GAT-3’ (forward) and 5’-AAG ATC TGC TGG GAG TAG CTG CTA-3’ (reverse); and U6, 5’-CTC GCT TCG GCA GCA CA-3’ (forward) and 5’-AAC GCT TCA CGA ATT TGC GT-3’ (reverse); The primer sequence for hsa-miR-27b-3p is 5’-TTC GGT TCA CAG TGG CTA AG-3’. The specificity of the primer sequences was analyzed by a dissociation curve, and 2-ΔΔCt (cycle threshold) was used to calculate the relative gene expression levels.
Western blotting
Protein from tissues or cells was collected using RIPA buffer containing a protease inhibitor cocktail and phosphatase inhibitors (Sigma, St. Louis, MO, USA) according to the operating instructions and quantified using a bicinchoninic acid (BCA) kit (Thermo Scientific, CA, USA). Equal amounts of protein (30 μg per lane) were separated by SDS-PAGE and then transferred onto polyvinylidene difluoride (PVDF, Millipore, Bedford, MA, USA) membranes. Western blotting was performed using anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-LIMK1 (Abcam, Cambridge, MA, USA) antibodies. The protein levels were detected with an ECL kit (Thermo Scientific, CA, USA) following the manufacturer’s instructions.
Dual luciferase reporter assay
TargetScan software (http://www.targetscan.org/vert_71/) was used to identify the potential target gene of miR-27b-3p. Either putative miR-27b-3p binding sites or mutated binding sites on the 3’UTR of LIMK1 were cloned into a vector to generate LIMK1 3’-UTR WT and LIMK1 3’-UTR Mut, respectively. Either LIMK1 3’-UTR WT or LIMK1 3’-UTR Mut was transfected into HEK-293T cells that stably over-expressed miR-27b-3p; cells transfected with negative control were used as control cells. A Dual-Luciferase Reporter Assay System (Promega, Madison, USA) was used to detect the luciferase activity 36 hrs. after transfection.
CCK-8 assay
The CCK-8 assay was performed to evaluate cell proliferation following the manufacturer’s protocols. Experimental cells were seeded at a density of 5×103 cells per well in 96-well plates and were transfected with miR-NC, miR-27b-3p, miR-27b-3p and negative control (NC), or miR-27b-3p and LIMK1. The absorbance at 450 nm was measured using an electroluminescence immunosorbent assay reader (Thermo Fisher Scientific, Waltham, MA).
Cell cycle assay
Flow cytometry was used to analyze cell cycle distribution. First, cells were cultured in serum-free medium for 24 hours to induce cell cycle synchronization. Cells were then collected and washed twice with ice-cold phosphate-buffered saline (PBS) to remove floating cells. Cells were then fixed with 70% ethanol, rehydrated with PBS, treated with RNase A (10 mg/ml), and stained with propidium iodide (10 μg/ml). The cell cycle was evaluated using flow cytometry. Data were analyzed using FlowJo 7.6 software.
Wound healing assay
Cells were seeded into 6-well plates. Wounds were created in the confluent cell monolayer using a 200 μl sterile pipette tip, and any free-floating cells and debris were removed by washing the scratched monolayers with PBS three times. Medium was then added, and the culture plates were incubated at 37°C for another 48 hrs. Wound healing within the scraped wound line was measured at the 48-hour time point using a microscope (Nikon, Tokyo, Japan).
Transwell assay
Cell migration and invasion assays were performed using 24-well Transwell plates (8.0-μm pores; BD BioCoat, USA). For the migration assay, transfected cells were plated onto the upper chamber of 24-well plate with a membrane insert. For the invasion assay, the upper chamber of the 24-well plate was pre-treated with Matrigel (100 μg per well; BD Biosciences, USA). In the lower portion of the chambers, fresh medium contained 10% FBS was added to each well. After the cells were incubated for 24 hrs at 37°C, the cells in the upper chamber were carefully removed. Invading cells were fixed with 4% formaldehyde, stained with 0.5% crystal violet, and counted under a microscope.
Mouse xenograft assay
Cells were suspended in serum-free medium and then subcutaneously injected into the flanks of mice at a concentration of 2×105 cells/ml. Three weeks later, the mice were treated with miR-27b-3p, control, miR-27b-3p+NC or miR-27b-3p+LIMK1. At 3-7 weeks after RNA injections, the mice were euthanized, and the tumor volumes were calculated.
Statistical analysis
All quantitative data are presented as the mean ± SD from at least three independent experiments. One-way ANOVA test or Student’s t-test was used to analyze the differences between the groups. The statistical significance was set at P < 0.05. SPSS 18.0 and Graph Pad Prism 6.0 software were used for the data analysis.
Results
Expression and prognosis of miR-27b-3p and LIMK1 in CRC tissues
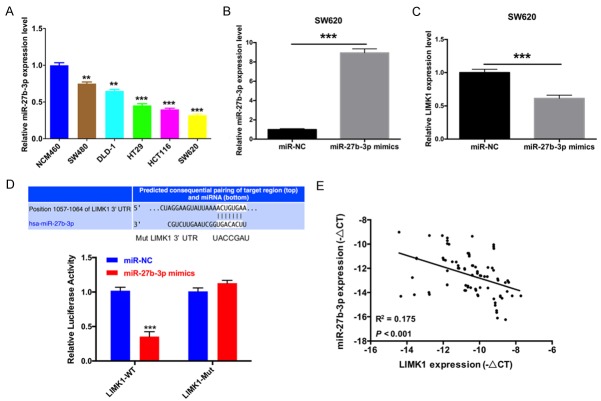
To confirm the underlying molecular mechanisms of CRC, we analyzed the expression levels of miR-27b-3p and LIMK1 in a series of clinical CRC tissues. The results showed that miR-27b-3p expression was markedly decreased in cancer samples compared with that in adjacent normal tissues (Figure 1A). In contrast, the mRNA expression levels of LIMK1 were significantly increased (Figure 1D). Subsequently, we predicted the prevalence rate of CRC using the receiver operating characteristic curves (ROC) of miR-27b-3p and LIMK1 and found that the area under the ROC curves of miR-27b-3p was 0.836 (Figure 1B) and that the area under the ROC curves of LIMK1 was 0.665 (Figure 1E). Furthermore, survival curves were analyzed between high miR-27b-3p expression (n = 41) and low miR-27b-3p expression (n = 42), and the results showed that miR-27b-3p increased the survival rate of CRC patients (Figure 1C) and that LIMK1 reduced the survival rate of CRC patients (Figure 1F). The relationships between miR-27b-3p and LIMK1 expression and the clinicopathological features of the enrolled patients with CRC are summarized in Tables 1 and 2.
Figure 1.
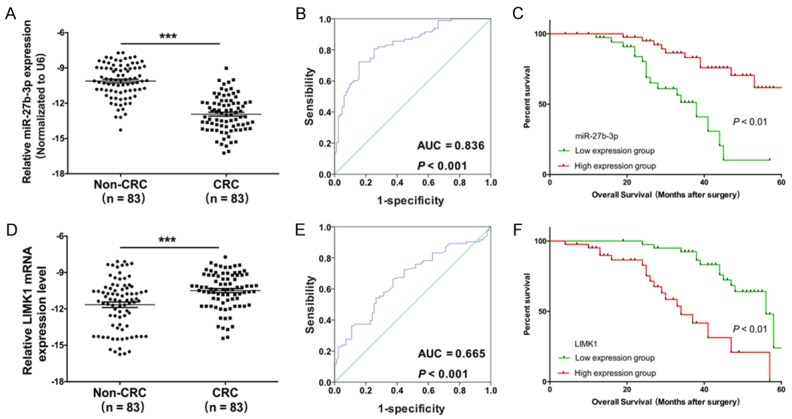
The expression and prognostic power of miR-27b-3p and LIMK1 in CRC tissues. A. The expression levels of miR-27b-were was measured by qRT-PCR in CRC tissues and corresponding noncancerous tissues (n = 83, ***P < 0.001). B. The prevalence rate was predicted by the receiver operating characteristic curve (ROC) of miR-27b-3p (AUC = 0.836, P < 0.001). C. Survival curve comparisons between high (n = 41) and low (n = 42, ***P < 0.01) expression of miR-27b-3p. D. LIMK1 expression was detected by qRT-PCR in CRC tissues (n = 83, ***P < 0.001). E. The prevalence rate was predicted by the ROC of LIMK1 (AUC = 0.665, P < 0.001). F. Survival curve comparisons between high (n = 41) and low (n = 42, ***P < 0.01) levels of LIMK1 expression.
Table 1.
Association between miR-27b-3p expression and the clinicopathological characteristics of enrolled colorectal cancer patients
| Characteristics | No. of patients | Mean ± SD | P value |
|---|---|---|---|
| Total no. of patients | 83 | ||
| Age (years) | |||
| > 60 | 51 (61.4%) | 12.38 ± 1.48 | 0.197 |
| ≤ 60 | 32 (38.6%) | 12.87 ± 1.94 | |
| Gender | |||
| Male | 42 (50.6%) | 11.96 ± 1.64 | 0.279 |
| Female | 41 (49.4%) | 12.32 ± 1.35 | |
| Invasion | |||
| T0-T2 | 56 (67.5%) | 12.68 ± 1.29 | 0.085 |
| T3-T4 | 27 (32.5%) | 12.09 ± 1.73 | |
| Lymphatic metastasis | |||
| N0-N1 | 59 (71.1%) | 11.84 ± 1.73 | 0.001** |
| N2-N3 | 24 (28.9%) | 13.13 ± 1.06 | |
| Distal metastasis | |||
| M0 | 68 (81.9%) | 11.47 ± 2.36 | 0.007** |
| M1 | 15 (18.1%) | 13.26 ± 1.88 | |
| TNM stage | |||
| 0 & I & II | 72 (86.7%) | 11.37 ± 1.94 | 0.003** |
| III & IV | 11 (13.3%) | 13.31 ± 2.16 |
Indicates statistical significance between the two groups (P < 0.01).
Table 2.
Association between LIMK1 expression and clinicopathological characteristics in enrolled colorectal cancer patients
| Characteristics | No. of patients | Mean ± SD | P value |
|---|---|---|---|
| Total no. of patients | 83 | ||
| Age (years) | |||
| > 60 | 51 (61.4%) | 10.46 ± 1.63 | 0.080 |
| ≤ 60 | 32 (38.6%) | 9.79 ± 1.75 | |
| Gender | |||
| Male | 42 (50.6%) | 10.53 ± 1.49 | 0.110 |
| Female | 41 (49.4%) | 9.95 ± 1.77 | |
| Invasion | |||
| T0-T2 | 56 (67.5%) | 9.86 ± 1.85 | 0.217 |
| T3-T4 | 27 (32.5%) | 10.39 ± 1.39 | |
| Lymphatic metastasis | |||
| N0-N1 | 59 (71.1%) | 10.85 ± 2.28 | 0.011* |
| N2-N3 | 24 (28.9%) | 9.47 ± 1.96 | |
| Distal metastasis | |||
| M0 | 68 (81.9%) | 10.75 ± 2.05 | 0.009** |
| M1 | 15 (18.1%) | 9.28 ± 1.27 | |
| TNM stage | |||
| 0 & I & II | 72 (86.7%) | 11.02 ± 1.94 | 0.018* |
| III & IV | 11 (13.3%) | 9.48 ± 2.16 |
Indicates statistical significance between the two groups (P < 0.05);
Indicates statistical significance between the two groups (P < 0.01).
LIMK1 is a direct target of miR-27b-3p
In vitro, we first measured the expression level of miR-27b-3p in human-derived colonic epithelial NCM460 cells and CRC cell lines (SW480, DLD-1, HT29, HCT116, and SW620) using qRT-PCR and found that the expression levels of miR-27b-3p in CRC cells were significantly lower than those in NCM460 cells (Figure 2A). Then, SW620 cells were transfected with either miR-NC or miR-27b-3p. qRT-PCR was used to detect the expression levels of miR-27b-3p and LIMK1. The results revealed that miR-27b-3p was highly expressed in SW620 cells transfected with miR-27b-3p compared with cells transfected with miR-NC (Figure 2B). miR-27b-3p inhibited the expression of LIMK1 (Figure 2C). To illustrate that LIMK1 is a direct target of miR-27b-3p, LIMK1 wild-type (WT) or mutant (Mut) 3’-UTRs were subcloned into a luciferase reporter vector and co-transfected with either miR-27b-3p mimics or negative control (NC) into HEK-293T cells. Then, luciferase activity was detected at 48 hrs after transfection. The results demonstrated that miR-27b-3p markedly inhibited luciferase activity in cells transfected with the LIMK1 WT 3’-UTR but had no influence on cells transfected with the mutant 3’UTR (Figure 2D). To verify whether miR-27b-3p negatively regulates LIMK1, we analyzed the correlation between LIMK1 and miR-27b-3p expression in CRC tissues and found that there was a negative correlation between LIMK1 and miR-27b-3p expression (Figure 2E).
Figure 2.
LIMK1 is a direct target of miR-27b-3p. A. The mRNA expression levels of miR-27b-3p were detected by qRT-PCR in human-derived colonic epithelial NCM460 cells and CRC cell lines (SW480, DLD-1, HT29, HCT116, and SW620, **P < 0.01, ***P < 0.001). B. qRT-PCR was used to detect the expression levels of miR-27b-3p in SW620 cells transfected with miR-NC and miR-27b-3p (***P < 0.001). C. LIMK1 expression in transfected SW620 cells was detected by qRT-PCR (***P < 0.001). D. Sequence of miR-27b-3p and the LIMK1 3’-UTR, which contains a predicted miR-27b-3p binding site. The results of the luciferase assay in CRC cells co-transfected with miR-27b-3p mimics and miR-NC and containing either the LIMK1 3’-UTR (WT) or mutant (Mut) constructs (***P < 0.001). E. The negative correlation between LIMK1 and miR-27b-3p expression in CRC tissues was assayed by qRT-PCR.
miR-27b-3p inhibits CRC cell proliferation via LIMK1
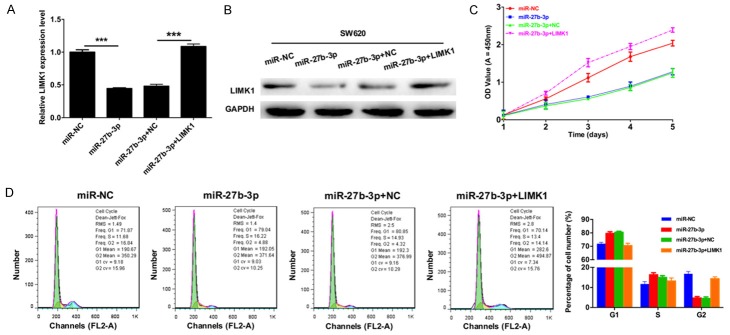
Next, to verify the role of miR-27b-3p on CRC cell proliferation via LIMK1, SW620 cells were transfected with miR-NC, miR-27b-3p, miR-27b-3p and negative control (NC), or miR-27b-3p and LIMK1, and then cells were collected for analysis. The expression levels of LIMK1 in transfected SW620 cells were measured by qRT-PCR and Western blot. The results indicated that the mRNA and protein expression levels of LIMK1 were dramatically suppressed by miR-27b-3p and that LIMK1 expression was increased when we transfected LIMK1 plasmid into SW620 cells transfected with miR-27b-3p (Figure 3A and 3B). The CCK-8 assay demonstrated that miR-27b-3p significantly suppressed SW620 cell proliferation, and the proliferative ability of SW620 cells was significantly restored via overexpression of LIMK1 compared with cells transfected with miR-27b-3p mimics (Figure 3C). Furthermore, to study the cell cycle, we used flow cytometry analysis to detect the cell cycle distribution of the transfected SW620 cells, which is shown in Figure 3D. Then, the cell number of the G1, S and G2 phases was counted, and we found that cell cycle arrest was characterized by a higher percentage of cells in G1/S phase of the cell cycle, and fewer cells were in G2 phase in SW620 cells transfected with miR-27b-3p compared to the cells transfected with miR-NC. Meanwhile, cell cycle arrest was characterized by fewer cells in G1/S phase of the cell cycle, and SW620 cells co-transfected with miR-27b-3p and LIMK1 presented more cells in G2 phase in than cells in the miR-27b-3p group. This suggested that G1 cell cycle progression in SW620 cells was inhibited by miR-27b-3p and that overexpression of LIMK1 can restore this change.
Figure 3.
miR-27b-3p inhibits CRC cell proliferation via LIMK1. SW620 cells were transfected with miR-NC, miR-27b-3p, miR-27b-3p and negative control (NC), or miR-27b-3p and LIMK1. A. The mRNA expression levels of LIMK1 in transfected SW620 cells were measured by qRT-PCR (***P < 0.001). B. Western blotting was used to detect the protein expression levels of LIMK1 in transfected SW620 cells, and GAPDH was used as a protein-loading control. C. The proliferation of CRC cells was detected by the CCK-8 assay. D. Cell cycle distribution was analyzed by flow cytometry.
miR-27b-3p suppresses CRC cell migration and invasion through LIMK1
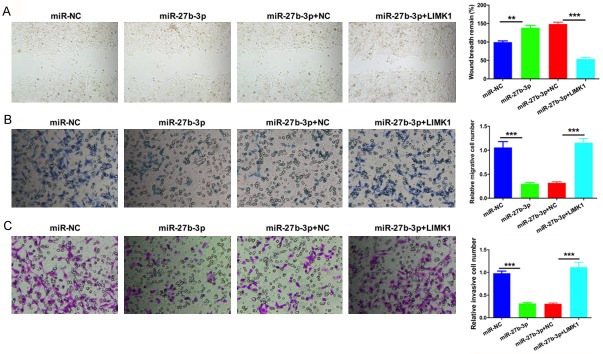
Furthermore, we confirmed the role of miR-27b-3p on CRC migration and invasion through LIMK1. SW620 cells were transfected with miRNC, miR-27b-3p, miR-27b-3p and negative control (NC), or miR-27b-3p and LIMK1. The migratory ability of transfected SW620 cells was measured by wound healing and Transwell migration assays. The results showed that miR-27b-3p suppressed the migratory ability of SW620 cells and that restoration of LIMK1 reversed the anti-migratory activity of miR-27b-3p (Figure 4A and 4B). The invasion abilities of transfected SW620 cells were measured by a Transwell invasion assay. The results showed that miR-27b-3p inhibited the invasive ability of SW620 cells and that the invasion capability of SW620 cells was significantly restored via up-regulation of LIMK1 when compared with cells transfected with miR-27b-3p mimics (Figure 4C).
Figure 4.
miR-27b-3p suppresses CRC cell migration and invasion via LIMK1. SW620 cells were transfected with miR-NC, miR-27b-3p, miR-27b-3p and negative control (NC), or miR-27b-3p and LIMK1. A. The migratory ability of transfected SW620 cells was measured by the wound healing assay (**P < 0.01, ***P < 0.001). B. The migratory ability of transfected SW620 cells was measured by the Transwell assay (***P < 0.001). C. The invasive ability of transfected SW620 cells was measured by Transwell assays (***P < 0.001).
miR-27b-3p inhibited tumor growth in a xenograft nude mouse model
To explore the effects of miR-27b-3p in vivo, a xenograft nude mouse model of SW620 cells was established. SW620 cells were subcutaneously inoculated into the nude mice, which were then treated with miR-27b-3p, control, miR-27b-3p+NC or miR-27b-3p+LIMK1. The results indicated that miR-27b-3p overexpression significantly decreased the volume and weight of the tumor; meanwhile, restoration of LIMK1 reversed the anti-proliferative effects of miR-27b-3p in vivo (Figure 5A and 5B).
Figure 5.
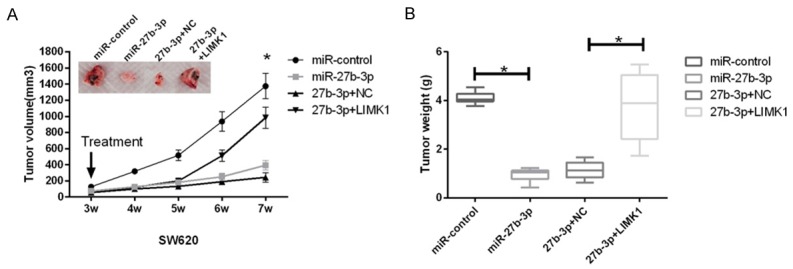
Effects of miR-27b-3p and LIMK1 on tumor growth in an in vivo mouse xenograft model. A and B. SW480 cells were subcutaneously injected into the nude mouse. Three weeks later, the tumors were treated with miR-NC, miR-27b-3p, miR-27b-3p and negative control (NC), or miR-27b-3p and LIMK1. Then, the animals were euthanized, and the volume and weight of the tumors were measured (*P < 0.05).
Discussion
CRC is the most frequently diagnosed malignancy, and the morbidity and mortality due to CRC are the fourth leading cause of malignant tumors in the digestive system after gastric cancer, esophageal cancer and primary liver cancer [20]. Therefore, the early diagnosis of CRC is critical to allow successful treatment. Currently, the main methods for CRC diagnosis are X-ray, endoscopic examination, and serum carcinoembryonic antigen (CEA). However, the therapeutic approaches are relatively limited.
miRNAs are a series of small non-coding RNAs that are widely involved in both physiological and pathological processes [21-23]. For example, miR-135a and miR-21 separately contribute to the cancer pathological process and insulin resistance [24,25]. It has been widely speculated that dysregulating miRNAs may cause cell dysfunction and further promote oncogenesis [26]. Therefore, studying miRNAs may illustrate the understanding of CRC development and be a great help for recognizing potential biomarkers. At present, many studies have reported that miR-27b plays important roles in cancer progression; however, there is much to be confirmed before including miR-27 in the network of cancer progression pathways, and a series of studies have shown that miR-27b serves as a tumor suppressor in numerous malignant tumors [27-29]. However, limited information is certified regarding the clinical potential and underlying mechanisms of miR-27b-3p in CRC.
MiR-27b-3p is one of the most important miRNAs that has been studied in many types of cancers, including breast cancer [14], gastric cancer [13], glioma [30], and esophageal cancer [31]. In our study, we first investigated the expression levels of miR-27b-3p in CRC pa-tients and found that it was markedly decreased in tissues from CRC patients. The expression levels of miR-27b-3p were also decreased in CRC cell lines compared to NCM460 cells. More importantly, miR-27b-3p inhibited CRC cell proliferation, migration and invasion. These results suggested that miR-27b-3p as a tumor suppressor plays an important role in the pathogenesis of CRC.
Increasing evidence has recognized that LIMK1 acts as a novel well-known biomarker for tumors while its effects on CRC oncogenesis have not been defined. There are also studies demonstrating that the expression of LIMK1 is dysregulated in many human cancers and that inhibiting LIMK1 expression can suppress the development of lung cancer [32], gastric cancer [33], breast cancer, and prostate cancer [34]. In the current study, we found that the 3’UTR of LIMK1 includes a highly conserved miR-27b-3p binding site, and direct interaction between these sites an miR-27b-3p downregulated the protein levels of LIMK1. Therefore, the purpose of this study is to demonstrate the antitumor effect of miR-27b-3p and its correlation with LIMK1 in CRC cells. We demonstrated the expression of LIMK1 and miR-27b-3p in CRC cells, analyzed the relationship between LIMK1 and miR-27b-3p, and studied the role of the miR-27b-3p-LIMK1 axis in CRC. Our study is the first report describing the role of miR-27b-3p in CRC cells by regulating LIMK1 expression. Therefore, we suggested that LIMK1 is a direct target gene of miR-27b-3p in CRC cells.
Based on the abovementioned data, we demonstrated that the levels of miR-27b-3p are downregulated in CRC patients and the cell lines. miR-27b-3p markedly suppressed CRC cell proliferation, cell cycle progression and migration and invasion by targeting LIMK1. Our findings introduced a better understanding of miR-27b-3p in CRC progression, which may also provide more precise targets for diagnosing and treating CRC.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No: 81472647). We would like to thank Dr. Chen Guojiang, an associate researcher at the Chinese Academy of Military Sciences.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US multi-society task force on colorectal cancer. Gastroenterology. 2012;143:844–857. doi: 10.1053/j.gastro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Halama N, Herrmann C, Jaeger D, Herrmann T. Treatment with cetuximab, bevacizumab and irinotecan in heavily pretreated patients with metastasized colorectal cancer. Anticancer Res. 2008;28:4111–4115. [PubMed] [Google Scholar]
- 4.Meropol NJ, Schulman KA. Cost of cancer care: issues and implications. J. Clin. Oncol. 2007;25:180–186. doi: 10.1200/JCO.2006.09.6081. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinfeld I, Navon R, Ach R, Yakhini Z. miRNA target enrichment analysis reveals directly active miRNAs in health and disease. Nucleic Acids Res. 2013;41:e45. doi: 10.1093/nar/gks1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 8.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Dalmay T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013;54:29–38. doi: 10.1042/bse0540029. [DOI] [PubMed] [Google Scholar]
- 11.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 12.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 13.Tao J, Zhi X, Zhang X, Fu M, Huang H, Fan Y, Guan W, Zou C. miR-27b-3p suppresses cell proliferation through targeting receptor tyrosine kinase like orphan receptor 1 in gastric cancer. J Exp Clin Cancer Res. 2015;34:139. doi: 10.1186/s13046-015-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen S, Sun Q, Liang Z, Cui X, Ren X, Chen H, Zhang X, Zhou Y. A prognostic model of triple-negative breast cancer based on miR-27b-3p and node status. PLoS One. 2014;9:e100664. doi: 10.1371/journal.pone.0100664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Doherty J, Antonipillai J, Chen S, Devlin M, Visser K, Baell J, Street I, Anderson RL, Bernard O. LIM kinase inhibition reduces breast cancer growth and invasiveness but systemic inhibition does not reduce metastasis in mice. Clin Exp Metastasis. 2013;30:483–495. doi: 10.1007/s10585-012-9553-6. [DOI] [PubMed] [Google Scholar]
- 16.Yoshioka K, Foletta V, Bernard O, Itoh K. A role for LIM kinase in cancer invasion. Proc Natl Acad Sci U S A. 2003;100:7247–7252. doi: 10.1073/pnas.1232344100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tapia T, Ottman R, Chakrabarti R. LIM kinase1 modulates function of membrane type matrix metalloproteinase 1: implication in invasion of prostate cancer cells. Mol Cancer. 2011;10:6. doi: 10.1186/1476-4598-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q, Jiao D, Hu H, Song J, Yan J, Wu L, Xu LQ. Downregulation of LIMK1 level inhibits migration of lung cancer cells and enhances sensitivity to chemotherapy drugs. Oncol Res. 2013;20:491–498. doi: 10.3727/096504013X13657689382699. [DOI] [PubMed] [Google Scholar]
- 19.West NR, McCuaig S, Franchini F, Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. 2015;15:615–629. doi: 10.1038/nri3896. [DOI] [PubMed] [Google Scholar]
- 20.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 21.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Ha TY. MicroRNAs in human diseases: from cancer to cardiovascular disease. Immune Netw. 2011;11:135–154. doi: 10.4110/in.2011.11.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Chen X, Chen X, Wang X, Ji A, Jiang L, Sang F, Li F. miR-135a acts as a tumor suppressor in gastric cancer in part by targeting KIFC1. Onco Targets Ther. 2016;9:3555–3563. doi: 10.2147/OTT.S105736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao XY, Shao K. [Roles of MicroRNA-21 in the pathogenesis of insulin resistance and diabetic mellitus-induced non-alcoholic fatty liver disease] . Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2016;38:144–9. doi: 10.3881/j.issn.1000-503X.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta. 2014;1845:255–265. doi: 10.1016/j.bbcan.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YW, Chen X, Gao JW, Zhang H, Ma RR, Gao ZH, Gao P. High expression of cAMP responsive element binding protein 1 (CREB1) is associated with metastasis, tumor stage and poor outcome in gastric cancer. Oncotarget. 2015;6:10646–10657. doi: 10.18632/oncotarget.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi RU, Miyazaki H, Takeshita F, Yamamoto Y, Minoura K, Ono M, Kodaira M, Tamura K, Mori M, Ochiya T. Loss of microRNA-27b contributes to breast cancer stem cell generation by activating ENPP1. Nat Commun. 2015;6:7318. doi: 10.1038/ncomms8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan L, Zhang L, Fan K, Wang JJ. miR-27b targets LIMK1 to inhibit growth and invasion of NSCLC cells. Mol Cell Biochem. 2014;390:85–91. doi: 10.1007/s11010-013-1959-1. [DOI] [PubMed] [Google Scholar]
- 30.Xu WY, Liu MF, Peng XL, Zhou P, Zhou JW, Xu K, Xu HX, Jiang SS. miR-24-3p and miR-27a-3p promote cell proliferation in glioma cells via cooperative regulation of MXI1. Int J Oncol. 2013;42:757–766. doi: 10.3892/ijo.2012.1742. [DOI] [PubMed] [Google Scholar]
- 31.Hummel R, Sie C, Watson DI, Wang TT, Ansar A, Michael MZ, Van der Hoek M, Haier J, Hussey DJ. MicroRNA signatures in chemotherapy resistant esophageal cancer cell lines. World J Gastroenterol. 2014;20:14904–14912. doi: 10.3748/wjg.v20.i40.14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen QY, Jiao DM, Hu HZ, Song J, Yan J, Wu LJ, Xu LQ. Downregulation of LIMK1 level inhibits migration of lung cancer cells and enhances sensitivity to chemotherapy drugs. Oncol Res. 2013;20:491–498. doi: 10.3727/096504013X13657689382699. [DOI] [PubMed] [Google Scholar]
- 33.Li XD, Ke Q, Li YS, Liu FN, Zhu G, Li F. DGCR6L, a novel PAK4 interaction protein, regulates PAK4-mediated migration of human gastric cancer cell via LIMK1. Int J Biochem Cell Biol. 2010;42:70–79. doi: 10.1016/j.biocel.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Oilman R, Ritchey L, Herchan R, Bossan A, Chakrabarti R. LIMK1 functions as a modulator of expression and targeting of CXCR4 in prostate cancer cells through a negative feedback loop. Cancer Research. 2016;76:209. [Google Scholar]