Abstract
Background: Several members of the SIRT family (SIRT1-7), a highly conserved family of NAD+-dependent enzymes, play an important role in tumor formation. Recently, several studies have suggested that SIRT4 may function as both a tumor oncogene and a tumor suppressor. However, its relationship with breast cancer remains unclear. Methods: We investigated SIRT4 protein levels in breast cancer and its possible association with selected clinicopathological parameters by immunohistochemical staining of a tissue microarray that included samples from 94 breast cancer patients. We further invested the effect of SIRT4 on the proliferation, migration and invasion of breast cancer cells. Results: SIRT4 protein levels in breast were markedly higherthan their non-neoplastic tissue counterparts (P<0.001). Additionally, SIRT4 promoted the proliferation, migration and invasion of breast cancer cells. Conclusions: Our results show that SIRT4 possess oncogenic properties at the human cancer cell level and indicate that SIRT4 may participate in the development of breast cancer.
Keywords: Breast cancer, pathology, oncogene, sirtuin, SIRT4
Introduction
Breast cancer, the most common malignancy in women and one of the three most common cancers worldwide, is associated with more than half a million deaths per year [1]. The etiology and pathogenesis of breast cancer is highly complex and involves many risk factors and a variety of genetic and epigenetic alterations. Over the past few decades, many key genes and signaling pathways were found to play key roles in the pathogenesis of breast cancer. These included human epidermal growth factor receptor 2 (HER2), fibroblast growth factor receptor 1 (FGFR1), inhibitor of nuclear factor kappa-B kinase subunit beta (IKBKB), epidermal growth factor receptor (EGFR), and phenylethanolamine N-methyltransferase (PNMT) [2].
The SIRT family (SIRT1-7) is a group of NAD+-dependent deacetylases and ADP-ribosyltransferases that regulate pressure resistance, genomic stability, energy metabolism and aging [3]. SIRT4, an NAD+-dependent ADP-ribosyltransferase localized in the mitochondria, catalyzes the transfer of ADP ribosyl to GDH [4] and regulates insulin secretion and fatty acid oxidation [4-7]. Recent studies indicate that SIRT4 can suppress tumors by regulating glutamine metabolism [7-11]. On the contrary, some studies have demonstrated SIRT4 may also function as an oncogene [12,13]. Shi Q et al. showed that SIRT4 protein levels in breast cancer tissues were higher than in corresponding non-neoplastic tissue by immunohistochemical staining of 241 breast cancer samples. Because the effect of SIRT4 on the function of breast cancer cells has not been studied, the role of SIRT4 in breast cancer remains unclear.
By employing high-throughput tissue microarray and immunohistochemical approaches, we investigated the expression of SIRT4 in breast cancer and analyzed the relationship between SIRT4 protein levels and clinically relevant pathological parameters in breast cancer. Using the human breast cancer cell line MDA-MB-453S, we further demonstrated that SIRT4 overexpression enhanced proliferation, migration and invasion of this cell line. Together, these results support an oncogenic role for SIRT4 in breast cancer.
Materials and methods
This study was approval by the First Affiliated Hospital of Wenzhou Medical University (Wen Zhou, China) and was conducted in accordance with the principles of the Declaration of Helsinki.
Patient and tissue samples
For this study, patient samples from 94 cases of breast cancer were collected. The age range of the patients was 22 to 82 years of age with a mean age of 54 years.The patients did not receive preoperative chemotherapy or radiotherapy before breast cancer resection.The clinicopathologic parameter information included the following: patient age, gender, tumor size, pathological grade, depth of tumor invasion (T stage), lymph node status (N stage) and the American Joint Committee on Cancer (AJCC, 7th edition) staging information. Information for pathological grade, T stage, N stage, and AJCC stage were unavailable for 5, 2, 16, and 5 patients respectively. The major clinicopathological parameters are shown in Table 2.
Table 2.
Correlation between the clinicopathologic variables and SIRT4 expression in breast cancer
| Clinicopathologic parameters | SIRT4 expression | Χ2 | P-valuea | ||
|---|---|---|---|---|---|
|
|
|||||
| All cases | Low | High | |||
| Age (years) | 0.122 | 0.727 | |||
| ≤60 | 76 | 43 | 33 | ||
| >60 | 18 | 11 | 6 | ||
| Tumor size (cm) | 0.237 | 0.626 | |||
| ≤2 | 43 | 24 | 19 | ||
| >2 | 51 | 31 | 20 | ||
| Differentiation | 1.197 | 0.274 | |||
| G2 | 47 | 30 | 17 | ||
| G3 | 42 | 22 | 20 | ||
| Stage (T) | 0.942 | 0.624 | |||
| T1 | 34 | 18 | 16 | ||
| T2 | 48 | 28 | 20 | ||
| T3 | 10 | 7 | 3 | ||
| Stage (N) | 0.644 | 0.725 | |||
| N0 | 47 | 25 | 22 | ||
| N1 | 31 | 19 | 12 | ||
| N2-3 | 10 | 5 | 5 | ||
| AJCC stage | 0.990 | 0.610 | |||
| I | 24 | 12 | 12 | ||
| II | 46 | 28 | 18 | ||
| III | 19 | 12 | 7 | ||
Chi-square test.
Tissue gene array chips were obtained commercially (Superchip Inc., Shanghai, China). There were 94 breast cancer resection specimens of which 86 had the corresponding adjacent non-neoplastic tissues specimen available. Thus, there were 180 points on one tissue microarray. The diameter of tissue pieces on the tissue microarray was 1.5 mm, and all points were overlaid with paraffin wax.
Immunohistochemistry
The tissue microarray was prepared by incubating at 60°C for 2 hrs and then incubated twice in xylene for 5 min at room temperature to deparaffinize the specimen. The tissue microarray was then transferred to successively graded concentrations of ethanol washes at 100%, 100%, 95%, 85%, and 70% every 5 min to rehydrate the specimen. Antigen retrieval was performed in a pressure cooker with citrate buffer (10 mM citrate and 0.05% Tween 20, pH 6.0). The microarray chip was then incubated in 0.3% H2O2 in Tris-HCl buffer for 15 min at room temperature to suppress endogenous peroxidase activity. The tissue microarray was then incubated with polyclonal rabbit anti-SIRT4 (HPA029692, 1:400, Sigma, USA) at 4°C overnight. Secondary antibody was applied using the GTVision Kit (Gene Tech Inc., Shanghai, China). The microarray chip section was then stained with diaminobenzidine (DAB) and counterstained with hematoxylin, followed by the chip being dehydrated and sealed with a coverslip. Tissue that was treated with buffer alone served as a negative control.
Two pathologists blinded to the patient information evaluated the SIRT4 immunostaining intensity under a light microscope. Each tissue point was assigned a score based on the staining intensity multiplied by the area of the stain [14]. The staining intensity was divided into four categories and included the following criteria: 0 = no staining; 1 = weak staining; 2 = moderate staining; and 3 = strong staining. Staining area assessment wasas follows: 0 = 5% or none of the cells were stained; 1 = 5%-25% of the cells werestained positive; 2 = 26%-50% of the cells werestained positive; 3 = 51-75% of the cells werestained positive; and 4 = more than 75% of the cells had stained positive. Finally, the eventual degree of staining was divided into two categories: 0-5 = low expression; and 6-12 = high expression. When evaluation of the staining pattern differed, both pathologists came to a consensus opinion.
Vector and virus production
Lentivirus for overexpression of SIRT4 was purchased from Hanbio, Shanghai, China. The viral vector is pHBLV-CMVIE-ZsGreen-T2A-Puro. The final virus titer of the over expressing lentivirus and the negative control virus was 2×108 PFU/ml.
Western blot
Cells were lysed with Ripa lysis buffer (Beyotime, China) supplemented with protease inhibitor cocktail (Beyotime, China). Protein concentrations were determined by BCA protein concentration reagent kit (Beyotime, China). Proteins from cell lysates were separated by SDS-PAGE and transferred to PVDF membrane. The following antibodies were used: rabbit anti-human SIRT4 polyclonal antibody (HPA029692, Sigma, USA) andrabbit anti-human beta-actin polyclonal antibody (ab11971, Abcam, England).
Cell lines and culture conditions
The human breast cancer cell line MDA-MB-435S was purchased from Shanghai Institute of cell biology, Chinese Academy of sciences. The cells were maintained in Dulbecco’s modified Eagle’s media (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and penicillin/streptomycin (Gibco, USA) at 37°C and 5% CO2. For stable SIRT4 overexpression, MDA-MB-435S cells were transduced with lentivirus and screened with 2 µg/ml puromycin for two weeks.
Cell proliferation activity
Cells were seeded at 1000/well in a 96-well plate to assess proliferation. To measure relative cell number, 10 µl CCK-8 (Dojindo, Japan) solution was added to each well and then the absorbance was read at 450 nm after culture in a CO2 incubator for 2 hours. The calculation for cell proliferation was as follows: cell viability (%) = A450 of treated cells/A450 of untreated cells.
Colony formation
Cells were seeded at 150/well in 6-well plates, and the media was changed every other day. After 2 weeks of culture, cells were fixed with methanol, stained with Giemsa, and the number of clones was counted with the naked eye.
Wound healing assay
MDA-MB-435S cells and SIRT4 overexpressing MDA-MB-435S cells were seeded at 5×105 per well in 6-well plates and cultured overnight. When the cell density reached about 90%, a 200 µL tip held perpendicular to the bottom of the 6-well plate was used to scratch a line through the cell monolayer. The cells were then washed 3 times with PBS, and then cultured in serum-free medium. The cell migration distances were quantified.
Cell migration and invasion assays
Migration and invasion assays were performed as previously described [15]. For the migration assay, the transfected cells (6×104 cells) were suspended in 0.2 ml of serum-free medium and seeded in the upper chambers of 24-well transwell plates (Corning Inc., Corning, NY, USA). The lower chambers were filled with 0.6 ml of growth medium containing 10% FBS. Cells were cultured at 37°C and allowed to migrate for 18 h. After migration, cells in the top chambers were removed by a cotton swab, and cells in the bottom chambers were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet (Sigma). The stained cells were counted under a microscope (Olympus). For the invasion assay, a similar protocol was followed except that the top chamber of the transwell plate was coated with Matrigel (BD Biosciences).
Statistical analysis
Statistical analysis was performed using the SPSS software package version 20.0 (SPSS, Inc., IBM, USA). A paired student’s t-test was used to analyze the final score of the tumor and non-tumor tissues. Chi-squared analysis was used to analyze the relationships between SIRT4 expression and the clinicopathological parameters. Other experiments, unless otherwise noted, were analyzed by the non-paired t test. P<0.05 (two-tailed) was considered statistically significant.
Results
SIRT4 expression is elevated in breast cancer tissues
To confirm that SIRT4 is overexpressed in breast cancer, we evaluated SIRT4 expression in tissue samples via immunohistochemistry. As expected, SIRT4 was predominantly expressed in the cytoplasm (Figure 1). Importantly, the staining intensity of SIRT4 was significantly higher in breast canceras compared to adjacent non-neoplastic tissue. We divided the samples into a low or high staining group. We found that 41.5% of breast cancer tissues had high expression of SIRT4, while only 15.1% of adjacent non-neoplastic tissues had high expression of SIRT4 (Table 1).
Figure 1.
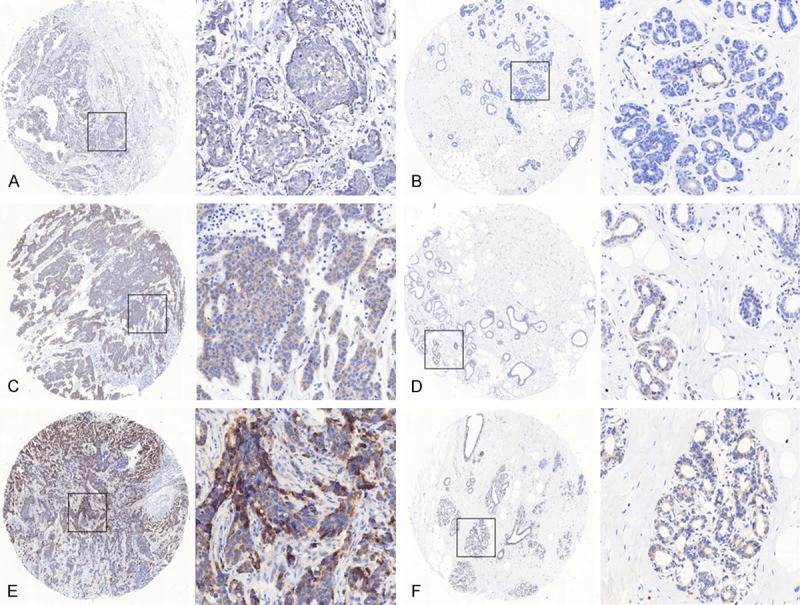
Representative immunohistochemical staining of SIRT4 in tumor cells. SIRT4 was expressed in the cytoplasm, and was significantly higher in tumor tissues as compared with adjacent non-neoplastic breast tissues. The micrographs showed weak (A), medium (C) and strong (E) expression of SIRT4 in breast cancer tissues. The relevant expression of SIRT4 in corresponding adjacent non-neoplastic breast tissues for (A, C and E) were shown in (B, D and F), respectively. (Magnification: left panel 40×, right panel 200×).
Table 1.
SIRT4 protein levels in breast cancer and adjacent non-tumor breast tissues
| SIRT4 expression | Χ2 | P-valuea | |||
|---|---|---|---|---|---|
|
|
|||||
| All cases | Low (%) | High (%) | |||
| Tissue type | 15.206 | 0.000 | |||
| Normal | 86 | 73 (84.9%) | 13 (15.1%) | ||
| Cancer | 94 | 55 (58.5%) | 39 (41.5%) | ||
Bold values are statistically significant (P<0.05).
Chi-square test.
Relationship between SIRT4 levels and clinicopathological parameters in breast cancer patients
Associations between SIRT4 levels and clinicopathological features were evaluated using immunohistochemistry. We did not find any significant associations between SIRT4 levels and parameters that included age, pathological grade, tumor size, T and N and AJCC staging (P>0.05) (Table 2).
Effect of SIRT4 inhibition on breast cancer cell line proliferation, migration and invasion
We constructed a stable MDA-MB-453S cell line overexpressing SIRT4 by lentivirus. The over expression was verified with Western blot (Figure 2A). SIRT4 overexpression significantly enhanced the proliferation of MDA-MB-453S cells (Figure 2B). Moreover, SIRT4 overexpression significantly reduced the number and size of the colonies formed by MDA-MB-453S cells (Figure 2C). We also found that the wound healing rate was accelerated after overexpression of SIRT4 (Figure 2D). Next, we found that overexpression of SIRT4 significantly increased the migration and invasion of breast cancer cells (Figure 2E and 2F). Together, these results indicate SIRT4 can promote the proliferation, migration and invasion of breast cancer cells.
Figure 2.
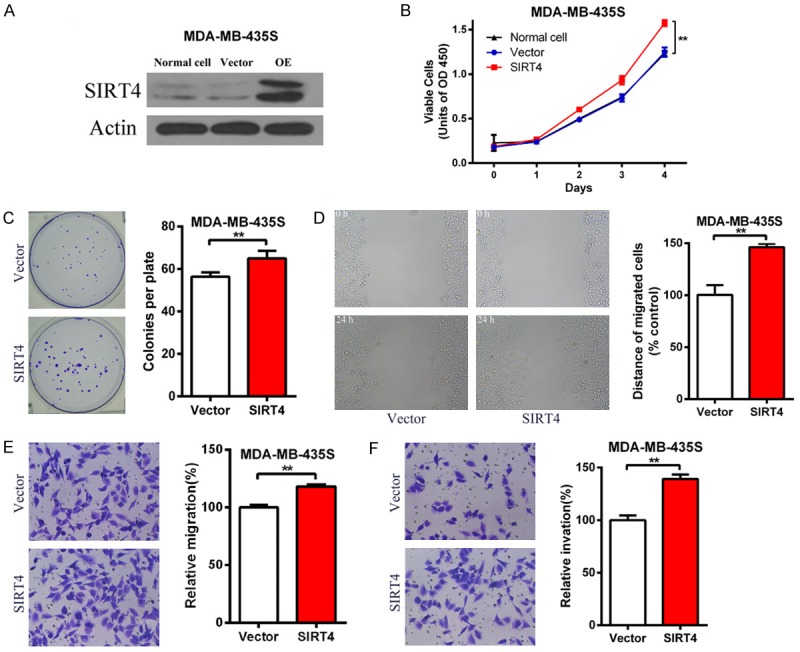
SIRT4 promotes cell proliferation, migration and invasion of breast cancer cells. (A) Western blot verification SIRT4 overexpression after two week treatment with 2 µg/ml puromycin. β-actin is an internal control. (B) Proliferation curve of vector and SIRT4-OE MDA-MB-435S cells. Cell Proliferation was measured every 24 h for 4 consecutive days. (C) Representative picture of colony formation experiment of vector and SIRT4-OE MDA-MB-435S cells (left). Cells stained with Giemsa after 14 day culture (right). (D) Representative picture of wound healing assay of vector and SIRT4-OE MDA-MB-435S cells (left). Ratio of cell migration in SIRT4-OE cells to that of vector cells (right). (E and F) The migration (E) and invasion (F) of MDA-MB-435S cell were determined by transwell assays. Vector and SIRT4-OE MDA-MB-435S cells were subjected to transwell assays. After being cultured for 18 h in transwell plates, the migrated or invaded cells were stained and counted under a microscope. **P<0.01.
Discussion
Based on the literature, multiple SIRT family members play a variety of roles in different tumors depending on the specific tissue and tumor type [16]. For example, the presence of SIRT1 in stomach [17], colon [18], prostate [19], skin [20] cancers, and several other tumors suggests that SIRT1 might promote tumor development in these cancers. However, other studies have found reduced SIRT1 expression in human breast cancer [21]. Moreover, SIRT1 expression in the mouse APCmin/+ model inhibits the formation of intestinal tumors [22]. This particular observation is similar to that found for SIRT2, which was down-regulated in human breast [23], glioma [24] and skin cancers [25]. However, SIRT2 expression was enhanced in acute myeloid leukemia [26] and prostate cancer [27]. Thus, the observations and conclusions made for one tumortype cannot be extrapolated to that of another tumor type.
Some studies have determined that SIRT4 acts as a tumor suppressor. For example, Jeong et al. [7] found SIRT4 can suppress tumor formation by inhibiting glutamine metabolism. Overexpression of SIRT4 can inhibit the growth of HeLa cells; SIRT4 knockout MEF cells formed larger tumors in nude mice; and SIRT4 knockout mice spontaneously developed lung cancer, liver cancer, breast cancer and lymphoma. Csibi et al. [8] found that overexpression of SIRT4 can inhibit the growth of the human colon cancer cell line DLD-1 and human prostate cancer cell line DU145. We and others found that SIRT4 is downregulated in gastric cancer and colon cancer tissues and associated with pathological grading and other clinicopathological parameters [9-11].
In contrast, some studies have shown that SIRT4 may not only function as a tumor suppressor but may function as a tumor promoter. At the cellular level, SIRT4 was increased in DNA damage conditions including: camptothecin (CPT), ultraviolet (UV), and oncogene-induced transformation. In addition, SIRT4 overexpression increased the survival of HepG2 cells in these DNA damage conditions [12]. Using tissue microarray of 93 esophageal carcinoma and adjacent non-tumor tissue samples, we found that SIRT4 protein levels in esophageal cancer is higher than in adjacent non-tumor tissues of the esophagus [13]. Of 241 breast cancer tissue samples with paired adjacent non-neoplastic tissues, Shi Q et al. found that 19.92% of tumor tissue stained positive for SIRT4, while only 12.03% of tumor adjacent non tumor breast tissue stained positive [28]. However, this study also found that in invasive breast cancer, the median survival time of the SIRT4 high expression group was longer than the SIRT4 low expression group (73.2 vs 67.8 months) although the difference is not very obvious [28]. Though still in need of further research, these studies support the dual role of SIRT4 as both a tumor suppressor and an oncogene in some tumors, especially in breast.
In this study, we analyzed the SIRT4 protein levels in breast cancer and the relationship between SIRT4 levels and the clinicopathological parameters of patients with breast cancer. We found that SIRT4 is significantly upregulated in breast cancer as compared with adjacent non-neoplastic tissues. Using a breast cancer cell line, we found that SIRT4 can promote cell proliferation, migration and invasion. These observations suggest that SIRT4 may possess oncogene properties in the context of breast cancer.
Research shows that SIRT4 can suppress tumors by inhibiting metabolism, especially glutamine metabolism [7,8,29]. The higher the degree of malignancy, the faster the proliferation, the corresponding energy demand is also increased. Although SIRT4 is clearly implicated in cell metabolism, little research on the mechanism of SIRT4 action exists. Like other members of the SIRT family, SIRT4 may also have complex regulatory networks underscoring its role as both a tumor suppressor and oncogenes. Therefore, the regulatory network of SIRT4 in the tumor must be explored to facilitate in-depth understanding of its role in cancer. In summary, our results suggest that SIRT4 may possess oncogenic properties in the context of breast cancer and therefore may be a promising target for the treatment of breast cancer.
Acknowledgements
This research is financially supported by the Project of Wenzhou Science and Technology Bureau (No. Y20160404), Zhejiang Natural Science Foundation (No. LY18H160055).
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 5.Nasrin N, Wu X, Fortier E, Feng Y, Bare’ OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285:31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 7.Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, Xu X, Li C, Wang RH, Lee J, Csibi A, Cerione R, Blenis J, Clish CB, Kimmelman A, Deng CX, Haigis MC. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang G, Cui F, Yu F, Lu H, Zhang M, Tang H, Peng Z. Sirtuin-4 (SIRT4) is downregulated and associated with some clinicopathological features in gastric adenocarcinoma. Biomed Pharmacother. 2015;72:135–139. doi: 10.1016/j.biopha.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K, Ogawa H, Hamabe A, Uemura M, Nishimura J, Hata T, Takemasa I, Mizushima T, Doki Y, Mori M, Ishii H. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer. 2015;113:492–499. doi: 10.1038/bjc.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang G, Cheng J, Yu F, Liu X, Yuan C, Liu C, Chen X, Peng Z. Clinical and therapeutic significance of sirtuin-4 expression in colorectal cancer. Oncol Rep. 2016;35:2801–10. doi: 10.3892/or.2016.4685. [DOI] [PubMed] [Google Scholar]
- 12.Jeong SM, Hwang S, Seong RH. SIRT4 regulates cancer cell survival and growth after stress. Biochem Biophys Res Commun. 2016;470:251–256. doi: 10.1016/j.bbrc.2016.01.078. [DOI] [PubMed] [Google Scholar]
- 13.Lai X, Yu Z, Chen X, Huang G. SIRT4 is upregulated in Chinese patients with esophageal cancer. Int J Clin Exp Pathol. 2016;9:10543–10549. [Google Scholar]
- 14.Huang G, Cheng J, Yu F, Liu X, Yuan C, Liu C, Chen X, Peng Z. Clinical and therapeutic significance of sirtuin-4 expression in colorectal cancer. Oncol Rep. 2016;35:2801–2810. doi: 10.3892/or.2016.4685. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Xu K, Shi L, Zhang L, Zhao Z, Xu H, Liang F, Li H, Zhao Y, Xu X, Tian Y. Overexpression of MicroRNA-216a suppresses proliferation, migration, and invasion of glioma cells by targeting leucine-rich repeat-containing G protein-coupled receptor 5. Oncol Res. 2017;25:1317–1327. doi: 10.3727/096504017X14874323871217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2014;33:1609–20. doi: 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH, Park HS, Lee H, Chung MJ, Kang MJ, Lee DG, Moon WS, Jang KY. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin Cancer Res. 2009;15:4453–4459. doi: 10.1158/1078-0432.CCR-08-3329. [DOI] [PubMed] [Google Scholar]
- 18.Stünkel W, Peh BK, Tan YC, Nayagam VM, Wang X, Salto-Tellez M, Ni B, Entzeroth M, Wood J. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J. 2007;2:1360–1368. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- 19.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 20.Hida Y, Kubo Y, Murao K, Arase S. Strong expression of a longevity-related protein, SIRT1, in Bowen’s disease. Arch Dermatol Res. 2007;299:103–106. doi: 10.1007/s00403-006-0725-6. [DOI] [PubMed] [Google Scholar]
- 21.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, Ji J, Wang XW, Park SH, Cha YI, Gius D, Deng CX. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20:487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, Oshimura M. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 25.Ming M, Qiang L, Zhao B, He YY. Mammalian SIRT2 inhibits keratin 19 expression and is a tumor suppressor in skin. Exp Dermatol. 2014;23:207–209. doi: 10.1111/exd.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dan L, Klimenkova O, Klimiankou M, Klusman JH, van den Heuvel-Eibrink MM, Reinhardt D, Welte K, Skokowa J. The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica. 2012;97:551–559. doi: 10.3324/haematol.2011.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou H, Chen W, Zhao L, Zuo Q, Zhang G, Zhang X, Wang H, Gong H, Li X, Wang M, Wang Y, Li X. Cortactin is associated with tumour progression and poor prognosis in prostate cancer and SIRT2 other than HADC6 may work as facilitator in situ. J Clin Pathol. 2012;65:1088–1096. doi: 10.1136/jclinpath-2012-200940. [DOI] [PubMed] [Google Scholar]
- 28.Shi Q, Liu T, Zhang X, Geng J, He X, Nu M, Pang D. Decreased sirtuin 4 expression is associated with poor prognosis in patients with invasive breast cancer. Oncol Lett. 2016;12:2606–2612. doi: 10.3892/ol.2016.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]


