Abstract
Dermal papilla cells (DPCs) are associated with the development of hair follicles (HFs) and the regulation of the hair growth cycle. Previous studies have shown that Wnt family member 10B (WNT10B) plays an important role in the proliferation and survival of DPCs in vitro, and promotes the growth of HFs. However, the underlying mechanisms have not been fully elucidated. The present study evaluated the role of WNT10B in regulating HF morphogenesis by characterizing the differential gene expression profiles between WNT10B-treated DPCs and control DPCs using RNA-sequencing (RNA-seq). A total of 1,073 and 451 genes were upregulated and downregulated, respectively. The RNA-seq data was subsequently validated by reverse-transcription quantitative PCR. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that 442 GO terms and 21 KEGG pathways were significantly enriched. Further functional analysis revealed that WNT10B decreased translation initiation, elongation and termination, and RNA metabolic processes in cultured DPCs compared with controls in vitro. Human signaling networks were compared using pathway analysis, and treatment of DPCs with WNT10B was revealed to downregulate the ribosome biogenesis pathway and decrease protein synthesis in vitro. KEGG pathway analysis showed that WNT10B upregulated the phosphoinositide 3-kinase/protein kinase B signaling pathway. The present study analyzed the expression of mRNA in WNT10B-treated DPCs using next-generation sequencing and uncovered mechanisms regulating the induction of HFs.
Keywords: mRNA, expression profile analysis, dermal papilla cells, Wnt family member 10B, hair follicles
Introduction
In adults, hair follicles (HFs) undergo cycles of growth, quiescence and regeneration (1). Human dermal papilla cells (DPCs) are mesenchymal cells located in the hair bulb of HFs, and are associated with the development and periodic growth of HFs (2–6). Additionally, DPCs serve an important role in the formation of HFs (7,8). Previous studies have demonstrated that the introduction of exogenous DPCs into the follicles induced the formation of new HFs in mice (9–12). Furthermore, DPCs have been investigated as potential cell therapy for hair loss (13,14). However, previous studies demonstrated that DPCs lose their stemness and inductive ability during in vitro culture (7,12).
According to previous reports, DPCs rapidly lose their inductive ability when cultured in AmnioMAX™-C100 medium (Thermo Fisher Scientific, Inc.) in vitro, and require the addition of chemical factors such as bone morphogenic protein 6 and Wnt3a to maintain their hair-inductive properties (15). The Wnt signaling pathway was demonstrated to be involved in the maintenance of the intrinsic properties in hair follicle morphogenesis and cycling of cultured DPCs in vitro (16). The Wnt signaling pathway is associated with the repair of several tissues, including skin and HFs, and was demonstrated to serve a regulatory role in morphogenesis during embryogenesis, the growth of various tissues, the maintenance of stem cells and the occurrence of tumors (17–19). Among the Wnt family, Wnt family member 10B (WNT10B) was demonstrated to be associated with the proliferation and the maintenance of DPCs in vitro (20–24). Moreover, adenovirus-mediated WNT10B overexpression was shown to induce HF regeneration in vivo (25–29). WNT10B is one of the earliest and most determinate markers during the embryonic stage of HF substrate formation (7,11,12) and promotes the differentiation of epithelial cells and the development of HFs (30). Furthermore, WNT10B promoted the development of HFs during long-term in vitro culture (4,7,11–12,29,31–32). However, the mechanisms linking WNT10B, HF formation and the inductive ability of DPCs have not been fully elucidated. As the downstream target genes of the Wnt signaling pathway, the β-catenin/TCF/LEF transcription family are expressed in epithelial and mesenchymal cells at the early morphogenetic stages of HF development and in post-natal HF stem cells (33). Additionally, β-catenin activity regulates the regeneration of hair (15,20,33–37).
The aim of the current study was to investigate the effect of WNT10B on the DPC transcriptome using mRNA sequencing. Progress in this area of research may facilitate the development of novel and effective therapeutic approaches for HF regeneration dysfunction in alopecia.
Materials and methods
Cells and reagents
HFDPCs isolated from human dermis originating from the lateral scalp were purchased from PromoCell GmbH. Cells were cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.; 10099-14-FBS) and 1% Penicillin-Streptomycin Solution (E607011, Sangon Biotech Co., Ltd.), at 37°C and 5% CO2. Human recombinant WNT10B protein was purchased from R&D Systems (cat. no. 7196-WN). The WNT10B treatment conditions used in the present study were as previously described (38). WNT10B protein was reconstituted in PBS containing 0.1% bovine serum albumin (BSA; Sangon Biotech Co., Ltd.; A602440) to form a solution of 10 µg/ml and used at a final concentration of 1 µg/ml. The expression of the Wnt target gene β-catenin in DPCs was increased following treatment with 1 µg/ml WNT10B protein for 3 days at 37°C, which was in agreement with the results of a previous study (25) indicating that the culture conditions were appropriate.
For RNA-sequencing (RNA-seq), the DPCs were divided into two groups: i) The experimental group, in which DPCs were cultured in DMEM supplemented with 10% FBS and 1 µg/ml recombinant WNT10B for 3 days at 37°C; and ii) the control group, in which DPCs were cultured in DMEM containing 10% FBS and 1 µg/ml BSA for 3 days at 37°C.
RNA extraction and sequencing
Total RNA from WNT10B-treated and control DPCs was extracted using TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. The total RNA quantity and purity were analyzed using Bioanalyzer 2100 (Agilent Technologies, Inc.) and RNA 6000 Nano LabChip Kit (Agilent Technologies, Inc.). RNA samples with an RNA integrity number > 7.0 were used for subsequent experimentation. The Illumina TruSeq RNA Sample Preparation kit (Illumina, Inc.) was used to construct sequencing libraries. Sequencing was performed using an Illumina HiSeq 2500 Sequencing system (Illumina, Inc.) by Shanghai YingBiotech Co., Ltd. Each group was analyzed in triplicate.
Mapping and identification of differentially expressed genes (DEGs)
Prior to read mapping, the raw sequencing data were analyzed using FAST-QC, which assessed the nucleotide quality distribution, PCR duplication rate, position-specific sequencing quality, k-mer frequency and GC content. The clean reads were mapped to the human genome (GRCH37). Then the aligned clean read number was further normalized to reads per kilo of per million mapped reads (RPKM) with RSEM software (version 1.2.3). Bioconductor DESeq2 version 1.12.3 (https://www.rdocumentation.org/packages/DESeq2) was used to identify DEGs using a fold-change (FC) >2 for significant upregulation or downregulation and a false discovery rate (FDR) <0.05. A volcano plot was drawn according to the analysis of the DEGs.
Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
GO (www.geneontology.org) analysis was performed to identify the biologic implications of the DEGs. Fisher's exact test was used to identify the significant GO terms with FDR-adjusted P-values. KEGG pathway analysis was performed to identify biologically important pathways associated with the DEGs. Fisher's exact test was used to select the significant pathways based on P-values (P<0.05) and FDR (FDR<0.27).
Reverse-transcription quantitative PCR (RT-qPCR)
To verify the results obtained from RNA-seq, seven highly expressed and enriched DEGs with high FCs (FC >2 or FC <0.5), including ribosomal protein L17 (RPL17), ribosomal protein L39 (RPL39), Rho associated coiled-coil containing protein kinase 2 (ROCK2), cytoplasmic FMR1 interacting protein 2 (CYFIP2), fibroblast growth factor 14 (FGF14), protein phosphatase 2 regulatory subunit B′ε (PPP2R5E) and vascular endothelial growth factor B (VEGFB), were selected for RT-qPCR. Total RNA was extracted from HFDPCs with TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Approximately 5 µg total RNA from each sample was used for RT, which was performed using PrimeScript™ RT Master mix (Takara Biotechnology Co., Ltd.) following the manufacturer's instructions. qPCR was subsequently performed using the One Step TB Green™ PrimeScript™ PLUS RT-PCR kit (Takara Biotechnology Co., Ltd) and the 7500 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR program was: 95°C for 10 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 60 sec. Gene expression was quantified using was the 2−ΔΔCq method (39) and normalized to the expression of the internal control GAPDH. Each reaction was performed in triplicate. The primers used for qPCR are presented in Table I.
Table I.
Primer sequence for reverse-transcription quantitative PCR.
| Primer sequences | ||
|---|---|---|
| Gene | Forward | Reverse |
| RPL17 | 5′-AGCCTGAGGTGATCTGTGAAAAT-3′ | 5′-CGAGTGTTATTTCGTGGGGTT-3′ |
| RPL39 | 5′-GCCTTCTAAGCTCGTTCTTCCG-3′ | 5′-CGAGCAGCGGAGTCAAGAACA-3′ |
| ROCK2 | 5′-GCAGAAGTGGGTTAGTCGGTTG-3′ | 5′-GGCAGTTAGCTAGGTTTGTTTGG-3′ |
| CYFIP2 | 5′-CCTTAAACCAGCCACTACCTCTC-3′ | 5′-TCTGTATTCTGCACTCATCCGC-3′ |
| FGF14 | 5′-TGCTGGATTGCTTTTCGCC | 5′-GCTGGGGATCAGTTGGGTTCT-3′ |
| PPP2R5E | 5′-TGTCCTCAGCACCAACTACTCCT-3′ | 5′-CAAGATACCTTTTAGCAGCGGC-3′ |
| VEGFB | 5′-GATCCGGTACCCGAGCAGT-3′ | 5′-TTAGGTCTGCATTCACACTGGC-3′ |
RPL17, ribosomal protein L17; RPL39, ribosomal protein L39; ROCK2, Rho associated coiled-coil containing protein kinase 2; CYFIP2, cytoplasmic FMR1 interacting protein 2; FGF14, fibroblast growth factor 14; PPP2R5E, protein phosphatase 2 regulatory subunit B′ε ; VEGFB, vascular endothelial growth factor B.
Statistical analysis
All data are expressed as mean ± standard error of the mean. The statistical analyses were performed using GraphPad Prism software (version 6; GraphPad Software, Inc.). A two-tailed Student's t-test was used to evaluate statistical significant differences. P<0.05 was considered to indicate a statistically significant difference.
Results
Analysis of transcription sequencing of WNT10B-treated DPCs
In order to identify the differential expression of mRNA in DPCs following WNT10B treatment, DPCs were divided into the experimental and control groups. An mRNA library was constructed for each group and subjected to Illumina mRNA deep sequencing. As presented in Table II, 95.37% of 51.5 million reads from the WNT10B-treated group and 93.78% of 52.6 million reads from the control group remained following filtering and quality control.
Table II.
Analysis of the data generated.
| Sample | Total reads | High quality | Reads filter % | Clean reads | Mapped reads | Mapped rate % |
|---|---|---|---|---|---|---|
| DPC-WNT10B-1 | 5.74×107 | 5.44×107 | 9.49×10−1 | 4.05×107 | 3.86×107 | 9.52×10−1 |
| DPC-WNT10B-2 | 5.21×107 | 5.00×107 | 9.58×10−1 | 7.51×107 | 7.16×107 | 9.54×10−1 |
| DPC-WNT10B-3 | 4.5 ×107 | 4.30×107 | 9.54×10−1 | 4.13×107 | 3.95×107 | 9.54×10−1 |
| DPC-NC-1 | 4.08×107 | 3.83×107 | 9.39×10−1 | 5.64×107 | 5.45×107 | 9.65×10−1 |
| DPC-NC-2 | 7.52×107 | 7.10×107 | 9.44×10−1 | 5.18×107 | 5.00×107 | 9.64×10−1 |
| DPC-NC-3 | 4.20×107 | 3.93×107 | 9.38×10−1 | 4.45×107 | 4.31×107 | 9.69×10−1 |
DPC, dermal papilla cells; WNT10B, Wnt family member 10B; NC, negative control. Total reads: All sequencing data getting from next generation sequence. High quality: Reads with high quality of sequencing identity. Reads filter%: The ratio of High quality in Total reads. Clean reads: Remained reads after detecting with FASTQC. Mapped reads: Reads that could be unique mapped to the reference genome. Mapped rate %: The ratio of matched genome in clean reads.
Hierarchical clustering of global gene expression
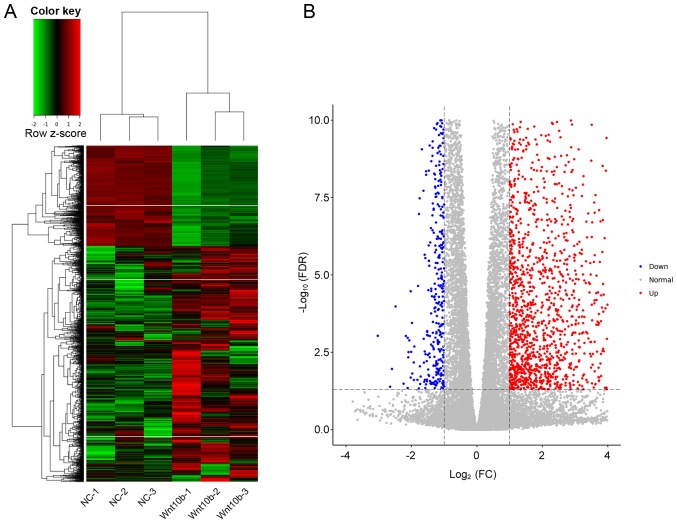
RPKM analysis was performed to evaluate the differential mRNA expression between WNT10B-treated and control DPCs. Using FC >2 and P<0.05 as the cut-off criteria, a total of 1,525 DEGs were identified, of which 1,074 were upregulated and 451 were downregulated following treatment with WNT10B (Table SI). The 3 WNT10B treatment and 3 control samples were used for hierarchical clustering and to construct a volcano plot. The WNT10B-treated DPCs were easily distinguished from the control cells, suggesting that there was a significant difference in gene expression between the two groups (Fig. 1).
Figure 1.
Gene expression in WNT10B-treated and control DPCs. (A) The heat map displays gene expression changes in the 3 WNT10B-treated and 3 control DPCs samples. Red, black, and green represent increased, unchanged, and decreases expression, respectively. (B) Volcano plot of upregulated and downregulated differentially expressed genes between WNT10B-treated and control DPCs. WNT10B, Wnt family member 10B; DPCs, dermal papilla cells; FC, fold-change; FDR, false discovery rate; NC, negative control.
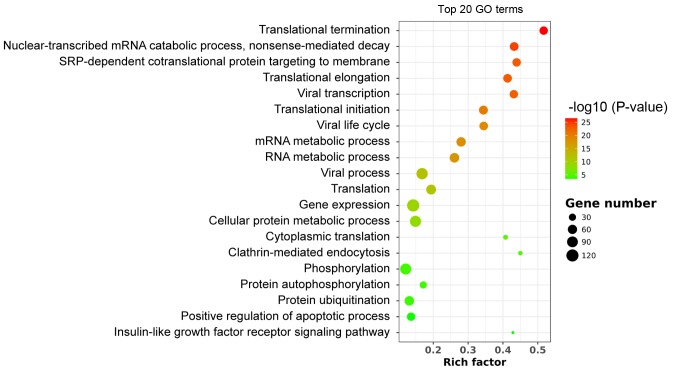
GO analysis
GO enrichment includes biological process, cellular component (CC) and molecular function (MF). Significantly enriched GO terms were associated with translational initiation, elongation and termination (Fig. 2). The ‘structural constituent of ribosome’ in the MF analysis and the ‘cytosolic large ribosomal subunit’ in the CC analysis indicated that WNT10B treatment may influence RNA translation and protein synthesis in DPCs, which subsequently affect HF induction. Several upregulated genes were enriched in the term ‘stem cell maintenance’. These included dicer 1 ribonuclease III, APC regulator of WNT signaling pathway, NIPBL cohesin loading factor, notch receptor 2, cell division cycle 73, replication timing regulatory factor 1, mediator complex subunit 12 and bone morphogenetic protein receptor type 1A (Table SIII).
Figure 2.
Scatter plot of the top enriched 20 GO terms. The size of the circle indicates the number of enriched genes in each pathway. Each color indicates a different P-value as indicated. GO, Gene Ontology. Rich factor is the ratio of enriched genes in selected pathway to the total genes of the pathway.
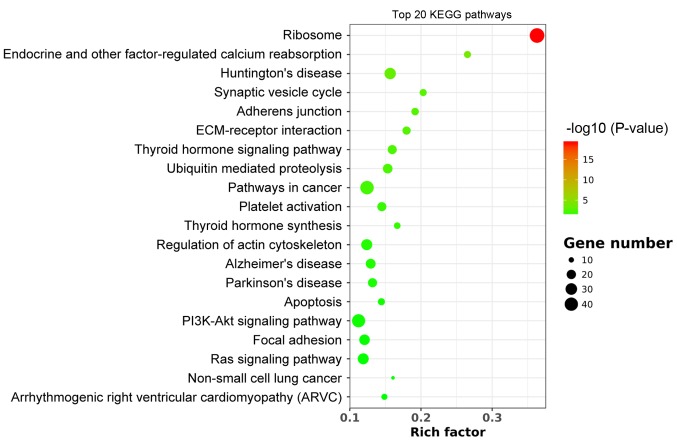
KEGG pathway analysis
KEGG pathway analysis revealed that DEGs were enriched in a total of 21 pathways following WNT10B treatment. Among those pathways, the ‘ribosome’ was identified as the most enriched (Fig. 3), suggesting that WNT10B may influence protein synthesis, which was consistent with the GO analysis.
Figure 3.
Scatter plot of the top enriched 20 KEGG pathways. The rich factor represents the proportion of differentially expressed genes to all the genes that are annotated in a specific pathway term. The size of the black circle indicates the unigene number. Each color indicates a different P-value as indicated. KEGG, Kyoto Encylopedia of Genes and Genomes.
In addition, KEGG pathway analysis revealed that the upregulated DEGs were significantly enriched in the ‘PI3K-Akt signaling pathway’ (Table SII). Therefore, WNT10B may activate the signaling PI3K/Akt pathway and maintain the HF inductive proprieties of DPCs.
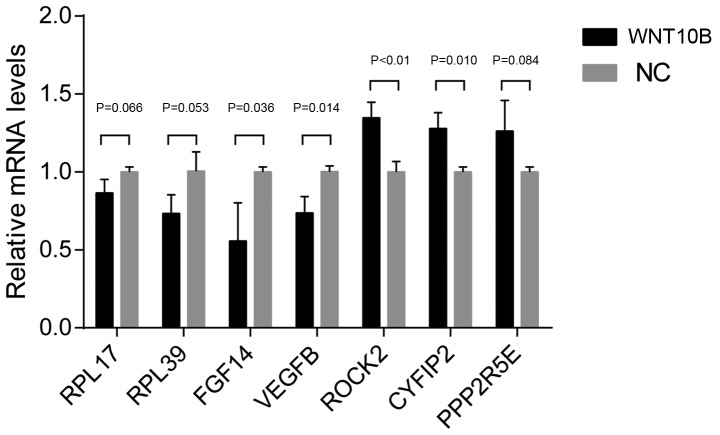
Validation of RNA-seq results
RT-qPCR was performed to verify the expression levels of the DEGs. DEGs with high FPKM and high FCs (FC>2 or FC<0.5) were selected, including RPL17, RPL39, ROCK2, CYFIP2, FGF14, PPP2R5E and VEGFB. The RT-qPCR results were highly consistent with RNA-seq data (Fig. 4).
Figure 4.
RT-qPCR to confirm RNA sequencing results from WNT10B-treated and control dermal papilla cells. A total of 7 genes were selected for the qRT-PCR analysis, including RPL17, RPL39, ROCK2, CYFIP2, FGF14, PPP2R5E and VEGFB. The data are presented as the mean ± SEM of three independent experiments. RT-qPCR, reverse-transcription quantitative PCR; WNT10B, Wnt family member 10B; RPL17, ribosomal protein L17; RPL39, ribosomal protein L39; ROCK2, Rho associated coiled-coil containing protein kinase 2; CYFIP2, cytoplasmic FMR1 interacting protein 2; FGF14, fibroblast growth factor 14; PPP2R5E, protein phosphatase 2 regulatory subunit B′ε; VEGFB, vascular endothelial growth factor B; NC, negative control.
Discussion
Human HFDPCs are a type of mesenchymal cell isolated from the DPCs of HFs (5,6,40–41). According to reports, DPCs serve important roles in the dermal-epidermal interactions regulating hair regeneration and in the hair growth cycle (5,42–43). DPCs have been investigated as potential cell therapy due to their HF inductive ability (44,45). However, this ability rapidly diminishes when DPCs are cultured in vitro (8,12,46), which limits the potential for application in alopecia therapy.
The Wnt signaling pathway is the most important regulator of DPC behavior (47,48). Previous studies have revealed that WNT10B serves a vital role in the proliferation and maintenance of DPCs and promotes the growth of HFs and the differentiation of mouse HF melanocytes (7,11,18,20). However, the mechanisms underlying these processes remain unclear. The present study used GO term and KEGG pathway analyses to explore the potential mechanisms based on mRNA-seq data. Results revealed 1,073 upregulated and 451 downregulated DEGs between WNT10B-treated and control DPCs in vitro. GO and KEGG results suggested that WNT10B significantly downregulated all the three steps (initiation, elongation and termination) of the translation process and as well as the ribosome pathway In the majority of adults, progenitor cells such as hematopoietic progenitor cell are quiescent and undergo lower levels of protein synthesis (49), suggesting that the level of protein synthesis may be related to stemness and properties of HFDPCs. Additionally, KEGG pathway analysis revealed that WNT10B upregulated the PI3K/Akt signaling pathway, which is known to regulate various cellular processes, including proliferation, metabolism, transcription, protein synthesis, growth and survival (49). A previous study revealed that deletion of the PI3K/Akt signaling pathway antagonist phosphatase and tensin homolog increased protein synthesis and depleted hematopoietic stem cells (50), suggesting that translational regulation and lower rates of ribosome biogenesis may maintain the properties of HFDPCs. The results of the present study suggested that WNT10B treatment may downregulate the protein synthesis rate.
The PI3K/Akt signaling pathway promotes the development, proliferation and differentiation of adult stem cells, particularly neural stem cells (51). As the downstream effector of Wnt signaling pathway, β-catenin regulates the proliferation of HFSCs through the PI3K/Akt signaling pathway (52,53). Furthermore, the activation of the PI3K/Akt signaling pathway triggers the expression of growth factors and promotes DPC-mediated hair growth (54–56). The activation of the PI3K/Akt signaling pathway may therefore be essential for DPCs to perform their normal functions.
It has previously been reported that Wnt signaling increases MTOR complex 1 (mTORC1) activity. mTORC1 plays a key role in protein synthesis and positively regulates cellular metabolism, ATP production and lipid synthesis (57–59). Wnt signaling may therefore promote protein synthesis by upregulating mTORC1. However, the regulation of the Wnt signaling pathway is an area requiring further study and may involve other signaling pathways (60,61). Moreover, the Wnt signaling pathway maintains stem cell pluripotency and balances progenitor self-renewal and differentiation (62). A previous study identified several Wnt proteins that were expressed in mouse embryonic and postnatal skin, including WNT3, WNT3A WNT4, WNT5A, WNT6, WNT7A, WNT7B, WNT10A, WNT10B and WNT16. The earliest and most highly expressed Wnt ligand in mouse hair follicle development and hair cycle induction was shown to be WNT10B (47). In agreement with the GO term enrichment results in the present study, WNT10B is involved in signaling pathways controlling stemness, pluripotency and cell fate (63). Recent studies have shown that embryonic and somatic stem cells rely on low translation rates to maintain an undifferentiated state. By contrast, differentiation requires increased protein synthesis. The transition from self-renewal to differentiation relies on enhanced ribosome biogenesis accompanied by increased protein synthesis (64). Therefore, the regulation of protein synthesis is important for cellular differentiation. WNT10B may maintain the stemness of human dermal papilla cells via decreasing the rate of protein synthesis.
The results obtained in the current study may aid in the elucidation of mechanisms of DPC activation and contribute to the development of therapies to treat dysfunction of HF regeneration in diseases such as alopecia.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81573057, 81703135 and 81472889).
Authors' contributions
QZ, YS and HC designed the experiments. QZ, YS, QLZ, and RH carried out the experiments and interpreted the data. QZ and YS prepared the sequenced samples and performed the qPCR. HC supervised all experiments. All the authors contributed to the manuscript.
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Ethical approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ma X, Tian Y, Song Y, Shi J, Xu J, Xiong K, Li J, Xu W, Zhao Y, Shuai J, et al. Msi2 maintains quiescent state of hair follicle stem cells by directly repressing the Hh signaling pathway. J Invest Dermatol. 2017;137:1015–1024. doi: 10.1016/j.jid.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harel S, Higgins CA, Cerise JE, Dai Z, Chen JC, Clynes R, Christiano AM. Pharmacologic inhibition of JAK-STAT signaling promotes hair growth. Sci Adv. 2015;1:e1500973. doi: 10.1126/sciadv.1500973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahoda CA, Reynolds AJ. Hair follicle dermal sheath cells: unsung participants in wound healing. Lancet. 2001;358:1445–1448. doi: 10.1016/S0140-6736(01)06532-1. [DOI] [PubMed] [Google Scholar]
- 4.Yu Z, Jiang K, Xu Z, Huang H, Qian N, Lu Z, Chen D, Di R, Yuan T, Du Z, et al. Hoxc-dependent mesenchymal niche heterogeneity drives regional hair follicle regeneration. Cell Stem Cell. 2018;23:487–500.e486. doi: 10.1016/j.stem.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 5.Driskell RR, Clavel C, Rendl M, Watt FM. Hair follicle dermal papilla cells at a glance. J Cell Sci. 2011;124:1179–1182. doi: 10.1242/jcs.082446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balañá ME, Charreau HE, Leirós GJ. Epidermal stem cells and skin tissue engineering in hair follicle regeneration. World J Stem Cells. 2015;7:711–727. doi: 10.4252/wjsc.v7.i4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouji Y, Yoshikawa M, Shiroi A, Ishizaka S. Wnt-10b promotes differentiation of skin epithelial cells in vitro. Biochem Biophys Res Commun. 2006;342:28–35. doi: 10.1016/j.bbrc.2006.01.104. [DOI] [PubMed] [Google Scholar]
- 8.Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development. 2009;136:2815–2823. doi: 10.1242/dev.038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abaci HE, Coffman A, Doucet Y, Chen J, Jacków J, Wang E, Guo Z, Shin JU, Jahoda CA, Christiano AM. Tissue engineering of human hair follicles using a biomimetic developmental approach. Nat Commun. 2018;9:5301. doi: 10.1038/s41467-018-07579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osada A, Kobayashi K. Appearance of hair follicle-inducible mesenchymal cells in the rat embryo. Dev Growth Differ. 2000;42:19–27. doi: 10.1046/j.1440-169x.2000.00485.x. [DOI] [PubMed] [Google Scholar]
- 11.Ouji Y, Yoshikawa M, Moriya K, Ishizaka S. Effects of Wnt-10b on hair shaft growth in hair follicle cultures. Biochem Biophys Res Commun. 2007;359:516–522. doi: 10.1016/j.bbrc.2007.05.135. [DOI] [PubMed] [Google Scholar]
- 12.Jahoda CA, Horne KA, Oliver RF. Induction of hair growth by implantation of cultured dermal papilla cells. Nature. 1984;311:560–562. doi: 10.1038/311560a0. [DOI] [PubMed] [Google Scholar]
- 13.Jeong KH, Joo HJ, Kim JE, Park YM, Kang H. Effect of mycophenolic acid on proliferation of dermal papilla cells and induction of anagen hair follicles. Clin Exp Dermatol. 2015;40:894–902. doi: 10.1111/ced.12650. [DOI] [PubMed] [Google Scholar]
- 14.Park S, Shin WS, Ho J. Fructus panax ginseng extract promotes hair regeneration in C57BL/6 mice. J Ethnopharmacol. 2011;138:340–344. doi: 10.1016/j.jep.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Rendl M, Polak L, Fuchs E. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 2008;22:543–557. doi: 10.1101/gad.1614408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohyama M, Kobayashi T, Sasaki T, Shimizu A, Amagai M. Restoration of the intrinsic properties of human dermal papilla in vitro. J Cell Sci. 2012;125:4114–4125. doi: 10.1242/jcs.105700. [DOI] [PubMed] [Google Scholar]
- 17.Galluzzi L, Spranger S, Fuchs E, Lopez-Soto A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. 2019;29:44–65. doi: 10.1016/j.tcb.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 20.Ouji Y, Ishizaka S, Yoshikawa M. Dermal papilla cells serially cultured with Wnt-10b sustain their hair follicle induction activity after transplantation into nude mice. Cell Transplant. 2012;21:2313–2324. doi: 10.3727/096368912X636867. [DOI] [PubMed] [Google Scholar]
- 21.Soma T, Fujiwara S, Shirakata Y, Hashimoto K, Kishimoto J. Hair-inducing ability of human dermal papilla cells cultured under Wnt/β-catenin signalling activation. Exp Dermatol. 2012;21:307–309. doi: 10.1111/j.1600-0625.2012.01458.x. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa T, Matsuda K, Inui S, Takenaka H, Katoh N, Itami S, Kishimoto S, Kawata M. Keratinocyte growth inhibition through the modification of Wnt signaling by androgen in balding dermal papilla cells. J Clin Endocrinol Metab. 2009;94:1288–1294. doi: 10.1210/jc.2008-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin H, Kwack MH, Shin SH, Oh JW, Kang BM, Kim AA, Kim J, Kim MK, Kim JC, Sung YK. Identification of transcriptional targets of Wnt/beta-catenin signaling in dermal papilla cells of human scalp hair follicles: EP2 is a novel transcriptional target of Wnt3a. J Dermatol Sci. 2010;58:91–96. doi: 10.1016/j.jdermsci.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Ouji Y, Nakamura-Uchiyama F, Yoshikawa M. Canonical Wnts, specifically Wnt-10b, show ability to maintain dermal papilla cells. Biochem Biophys Res Commun. 2013;438:493–499. doi: 10.1016/j.bbrc.2013.07.108. [DOI] [PubMed] [Google Scholar]
- 25.Li YH, Zhang K, Ye JX, Lian XH, Yang T. Wnt10b promotes growth of hair follicles via a canonical Wnt signalling pathway. Clin Exp Dermatol. 2011;36:534–540. doi: 10.1111/j.1365-2230.2011.04019.x. [DOI] [PubMed] [Google Scholar]
- 26.Lei M, Guo H, Qiu W, Lai X, Yang T, Widelitz RB, Chuong CM, Lian X, Yang L. Modulating hair follicle size with Wnt10b/DKK1 during hair regeneration. Exp Dermatol. 2014;23:407–413. doi: 10.1111/exd.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J, Yang T, Guo H, Tang Y, Deng F, Li Y, Xing Y, Yang L, Yang K. Wnt10b promotes differentiation of mouse hair follicle melanocytes. Int J Med Sci. 2013;10:691–698. doi: 10.7150/ijms.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li YH, Zhang K, Yang K, Ye JX, Xing YZ, Guo HY, Deng F, Lian XH, Yang T. Adenovirus-mediated Wnt10b overexpression induces hair follicle regeneration. J Invest Dermatol. 2013;133:42–48. doi: 10.1038/jid.2012.235. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Xing Y, Guo H, Ma X, Li Y. Immunohistochemical study of hair follicle stem cells in regenerated hair follicles induced by Wnt10b. Int J Med Sci. 2016;13:765–771. doi: 10.7150/ijms.16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai X, Lei M, Shi J, Yu Y, Qiu W, Lai X, Liu Y, Yang T, Yang L, Widelitz RB, et al. Roles of gasdermina3 in catagen-telogen transition during hair cycling. J Invest Dermatol. 2015;135:2162–2172. doi: 10.1038/jid.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll LS, Capecchi MR. Hoxc8 initiates an ectopic mammary program by regulating Fgf10 and Tbx3 expression and Wnt/β-catenin signaling. Development. 2015;142:4056–4067. doi: 10.1242/dev.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang T, Liu L, Fan J, Tian J, Gan C, Yang Z, Jiao H, Han B, Liu Z. Low-level laser treatment stimulates hair growth via upregulating Wnt10b and β-catenin expression in C3H/HeJ mice. Lasers Med Sci. 2017;32:1189–1195. doi: 10.1007/s10103-017-2224-8. [DOI] [PubMed] [Google Scholar]
- 33.Enshell-Seijffers D, Lindon C, Wu E, Taketo MM, Morgan BA. Beta-catenin activity in the dermal papilla of the hair follicle regulates pigment-type switching. Proc Natl Acad Sci USA. 2010;107:21564–21569. doi: 10.1073/pnas.1007326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy S, Andl T, Bagasra A, Lu MM, Epstein DJ, Morrisey EE, Millar SE. Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of sonic hedgehog in hair follicle morphogenesis. Mech Dev. 2001;107:69–82. doi: 10.1016/S0925-4773(01)00452-X. [DOI] [PubMed] [Google Scholar]
- 35.Matheson J, Bühnemann C, Carter EJ, Barnes D, Hoppe HJ, Hughes J, Cobbold S, Harper J, Morreau H, Surakhy M, Hassan AB. Epithelial-mesenchymal transition and nuclear β-catenin induced by conditional intestinal disruption of Cdh1 with Apc is E-cadherin EC1 domain dependent. Oncotarget. 2016;7:69883–69902. doi: 10.18632/oncotarget.11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheon SS, Cheah AY, Turley S, Nadesan P, Poon R, Clevers H, Alman BA. beta-Catenin stabilization dysregulates mesenchymal cell proliferation, motility, and invasiveness and causes aggressive fibromatosis and hyperplastic cutaneous wounds. Proc Natl Acad Sci USA. 2002;99:6973–6978. doi: 10.1073/pnas.102657399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin GL, Hankenson KD. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J Cell Biochem. 2011;112:3491–3501. doi: 10.1002/jcb.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouji Y, Yoshikawa M, Moriya K, Nishiofuku M, Matsuda R, Ishizaka S. Wnt-10b, uniquely among Wnts, promotes epithelial differentiation and shaft growth. Biochem Biophys Res Commun. 2008;367:299–304. doi: 10.1016/j.bbrc.2007.12.091. [DOI] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Nilforoushzadeh MA, Zare M, Zarrintaj P, Alizadeh E, Taghiabadi E, Heidari-Kharaji M, Amirkhani MA, Saeb MR, Mozafari M. Engineering the niche for hair regeneration - A critical review. Nanomedicine. 2019;15:70–85. doi: 10.1016/j.nano.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 41.El Agha E, Kramann R, Schneider RK, Li X, Seeger W, Humphreys BD, Bellusci S. Mesenchymal stem cells in fibrotic disease. Cell Stem Cell. 2017;21:166–177. doi: 10.1016/j.stem.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Madaan A, Verma R, Singh AT, Jaggi M. Review of hair follicle dermal papilla cells as in vitro screening model for hair growth. Int J Cosmet Sci. 2018;40:429–450. doi: 10.1111/ics.12489. [DOI] [PubMed] [Google Scholar]
- 43.Kim OY, Cha HJ, Ahn KJ, An IS, An S, Bae S. Identification of microRNAs involved in growth arrest and cell death in hydrogen peroxide-treated human dermal papilla cells. Mol Med Rep. 2014;10:145–154. doi: 10.3892/mmr.2014.2158. [DOI] [PubMed] [Google Scholar]
- 44.Kim J, Shin JY, Choi YH, Jang M, Nam YJ, Lee SY, Jeon J, Jin MH, Lee S. Hair growth promoting effect of hottuynia cordata extract in cultured human hair follicle dermal papilla cells. Biol Pharm Bull. 2019;42:1665–1673. doi: 10.1248/bpb.b19-00254. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Xiao S, Liu B, Miao Y, Hu Z. Use of extracellular matrix hydrogel from human placenta to restore hair-inductive potential of dermal papilla cells. Regen Med. 2019 doi: 10.2217/rme-2018-0112. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 46.Shim JH, Lee TR, Shin DW. Novel in vitro culture condition improves the stemness of human dermal stem/progenitor cells. Mol Cells. 2013;36:556–563. doi: 10.1007/s10059-013-0260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jansson L, Kim GS, Cheng AG. Making sense of Wnt signaling-linking hair cell regeneration to development. Front Cell Neurosci. 2015;9:66. doi: 10.3389/fncel.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myung PS, Takeo M, Ito M, Atit RP. Epithelial Wnt ligand secretion is required for adult hair follicle growth and regeneration. J Invest Dermatol. 2013;133:31–41. doi: 10.1038/jid.2012.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buszczak M, Signer RA, Morrison SJ. Cellular differences in protein synthesis regulate tissue homeostasis. Cell. 2014;159:242–251. doi: 10.1016/j.cell.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peltier J, O'Neill A, Schaffer DV. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol. 2007;67:1348–1361. doi: 10.1002/dneu.20506. [DOI] [PubMed] [Google Scholar]
- 52.Du KT, Deng JQ, He XG, Liu ZP, Peng C, Zhang MS. MiR-214 regulates the human hair follicle stem cell proliferation and differentiation by targeting EZH2 and Wnt/β-catenin signaling way in vitro. Tissue Eng Regen Med. 2018;15:341–350. doi: 10.1007/s13770-018-0118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gentile P, Scioli MG, Bielli A, De Angelis B, De Sio C, De Fazio D, Ceccarelli G, Trivisonno A, Orlandi A, Cervelli V, Garcovich S. Platelet-rich plasma and micrografts enriched with autologous human follicle mesenchymal stem cells improve hair re-growth in androgenetic alopecia. Biomolecular pathway analysis and clinical evaluation. Biomedicines. 2019;7:E27. doi: 10.3390/biomedicines7020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woo H, Lee S, Kim S, Park D, Jung E. Effect of sinapic acid on hair growth promoting in human hair follicle dermal papilla cells via Akt activation. Arch Dermatol Res. 2017;309:381–388. doi: 10.1007/s00403-017-1732-5. [DOI] [PubMed] [Google Scholar]
- 55.Feutz AC, Barrandon Y, Monard D. Control of thrombin signaling through PI3K is a mechanism underlying plasticity between hair follicle dermal sheath and papilla cells. J Cell Sci. 2008;121:1435–1443. doi: 10.1242/jcs.018689. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Nan W, Wang S, Zhang T, Si H, Yang F, Li G. Epidermal growth factor promotes proliferation and migration of follicular outer root sheath cells via wnt/β-catenin signaling. Cell Physiol Biochem. 2016;39:360–370. doi: 10.1159/000445630. [DOI] [PubMed] [Google Scholar]
- 57.Yin G, Liang Y, Wang Y, Yang Y, Yang M, Cen XM, Xie QB. mTOR complex 1 signalling regulates the balance between lipid synthesis and oxidation in hypoxia lymphocytes. Biosci Rep. 2017;37:BSR20160479. doi: 10.1042/BSR20160479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen JJ, Zhang S. Heme-regulated eIF2alpha kinase in erythropoiesis and hemoglobinopthies. Blood. 2019:2019001915. doi: 10.1182/blood.2019001915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, Yu Y, Liu X, He L, Cavaliere PM, et al. Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell. 2017;171:1545–1558. e1518. doi: 10.1016/j.cell.2017.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar D, Nitzan E, Kalcheim C. YAP promotes neural crest emigration through interactions with BMP and Wnt activities. Cell Commun Signal. 2019;17:69. doi: 10.1186/s12964-019-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massey J, Liu Y, Alvarenga O, Saez T, Schmerer M, Warmflash A. Synergy with TGFβ ligands switches WNT pathway dynamics from transient to sustained during human pluripotent cell differentiation. Proc Natl Acad Sci USA. 2019;116:4989–4998. doi: 10.1073/pnas.1815363116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kahn M. Wnt signaling in stem cells and cancer stem cells: A tale of two coactivators. Prog Mol Biol Transl Sci. 2018;153:209–244. doi: 10.1016/bs.pmbts.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 63.Wend P, Wend K, Krum SA, Miranda-Carboni GA. The role of WNT10B in physiology and disease. Acta Physiol (Oxf) 2012;204:34–51. doi: 10.1111/j.1748-1716.2011.02296.x. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, Lehmann R. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell. 2016;18:276–290. doi: 10.1016/j.stem.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.