Abstract
Osteoporosis (OP) is an age-related bone disease occurring worldwide. Osteoporotic fracture is one of the leading causes of disability and death in elderly patients. MicroRNAs (miRNAs/miRs) are key molecular regulatory factors in bone remodeling processes. The present study investigated the expression and mechanism of miR-135b-5p in patients with osteoporosis. The present results suggested that miR-135b-5p was upregulated in bone tissue fragments of patients with osteoporosis compared with the control patients. MC3T3-E1 cells were used to perform osteogenic differentiation induction. Reverse transcription-quantitative PCR and western blot assay were used to detect the mRNA and protein expression levels of the osteogenic markers osteocalcin (OC), Osterix and alkaline phosphatase (ALP). A specific kit was used for detecting ALP activity. The present results indicated that the mRNA expression levels of OC, Osterix and ALP significantly increased on the 7 and 14th day after osteogenic differentiation induction compared with the control group. Protein expression levels of OC, Osterix and ALP also increased on the 7 and 14th day after induction. ALP assay showed that ALP activity was significantly increased on the 7 and 14th day after induction. In addition, the present study found that miR-135b-5p was downregulated in MC3T3-E1 cells 7 and 14 days after osteogenic differentiation induction. The results of TargetScan analysis and dual luciferase reporter gene assay indicated that runt-related transcription factor 2 (RUNX2) was a direct target gene of miR-135b-5p. RUNX2 was upregulated in MC3T3-E1 cells on the 7 and 14th day after induction. Moreover, the present study found that compared with the osteogenic differentiation induction group, miR-135b-5p mimic significantly decreased OC, Osterix and ALP expression, and reduced ALP activity in MC3T3-E1 cells. However, these reductions were reversed following overexpression of RUNX2. The present results showed that miR-135b-5p mimic significantly reduced cell viability in MC3T3-E1 cells and induced cell apoptosis, and these effects were significantly reversed following RUNX2 overexpression. In summary, the present results suggested that miR-135-5p participated in the occurrence and development of osteoporosis via inhibition of osteogenic differentiation and osteoblast growth by targeting RUNX2. The present study suggested a novel potential target that may faciliate the treatment of osteoporosis, and further study is required to examine this possibility.
Keywords: osteoporosis, osteogenic differentiation, microRNA-135b-5p, runt-related transcription factor 2
Introduction
Osteoporosis (OP) is the most common type of bone disorder (1). OP is a public health issue with gradually increasing incidence (2). In China, the incidence of OP was 14.94% in 2008, and reached 27.96% between 2012 and 2015 (3,4). In Europe, ~27.5 million people were affected by OP in 2010 (5). As a bone disease, the main characteristics of OP include low bone density, easy bone damage, decreasing bone mass and increased risk of fracture (6). Hip fractures are frequently observed in patients with OP (7). Diagnosis of OP is very difficult until a fracture occurs (8). With an increasing number of elderly people in China, it is important to investigate methods for the prevention and treatment of osteoporosis. Elderly patients can suffer from motor dysfunction and impaired body functions, which can have adverse effects on mental health. Such mental health issues can lead to complications with the treatment and care of patients with OP. Therefore, elderly patients with OP require nursing interventions to aid their physical and mental health in order to promote quick recovery and improve their life quality (9). There is an urgent need to understand the pathogenesis and molecular mechanisms of OP to identify potential biomarkers and therapeutic targets.
MicroRNAs (miRNAs) are small non-coding regulatory RNAs (~23 nucleotides in length) that play a crucial role in the post-transcriptional regulation of gene expression by suppressing target mRNA translation by binding to its 3′untranslated region (UTR) region (10–12). Therefore, miRNAs can regulate multiple mRNAs to create a complex regulatory network of signaling pathways including oncogene pathways and tumor suppressor pathways, such as the Wnt and PI3K pathways (13–16). Altering the expression of these miRNAs alters the function of their target genes, some of which regulate crucial cellular mechanisms such as growth, proliferation, migration, invasion and metastasis (17–19). Previous studies have reported that changes in circulating miRNAs are associated with osteoporotic fractures. Seeliger et al (20) reported that five miRNAs were upregulated both in serum and bone tissue of patients with OP. Long non-coding RNA maternally expressed 3 (MEG3) inhibited osteogenic differentiation by decreasing miR-133a-3p expression, and the expression of MEG3 was found to be upregulated in bone marrow stem cells in ovariectomized mice and in patients with OP (21). An increasing number of miRNAs have been shown to play an important role in osteoblastogenesis and osteoporosis (22–24). However, the function of miR-135b-5p in osteoporosis remains unclear.
Therefore, the aims of the present study were to determine the expression of miR-135b-5p in bone tissue fragments of patients with OP, to investigate the role of miR-135b-5p in osteogenic differentiation and osteoblast growth, and to examine the underlying mechanism of miR-135b-5p.
Materials and methods
Cell culture and tissue samples
The mouse preosteoblast cell line MC3T3-E1 was purchased from American Type Culture Collection (cat. no. CRL-2594). The cells were cultured for 24 h in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5% CO2.
For osteogenic differentiation, MC3T3-E1 cells were cultured at 37°C in osteogenic induction medium (25) containing 10% FBS, 100 nM dexamethasone, 5 mM L-glycerophosphate and 50 mg/ml ascorbic acid (Sigma-Aldrich; Merck KGaA) for 14 days.
Bone fragments (bone tissues, 100 mg) were collected from 30 patients with OP [female, 15; male, 15; age, 52–73 years; body mass index (BMI), 23.6±5.4 kg/m2; lumbar spine bone mineral density, 0.79±0.10 g/cm2; femoral neck bone mineral density, 0.62±0.10 g/cm2; total hip bone mineral density, 0.65±0.07 g/cm2] and 30 patients with osteoarthritis (control; female, 15; male, 15; 51–72 years; BMI, 25.1±3.7 kg/m2; lumbar spine bone mineral density, 1.00±0.15 g/cm2; femoral neck bone mineral density, 0.85±0.13 g/cm2; total hip bone mineral density, 0.87±0.14 g/cm2) at the Third Affiliated Hospital of Sun Yat-sen University, between May 2015 and May 2017. Bone fragments extracted from the transcervical region of the femoral neck, were dissected into smaller fragments, washed three times in PBS and stored at −80°C until further use. The present study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University and every patient provided written informed consent.
Cell transfection
MC3T3-E1 cells were seeded in 6-well plates at a density of 1×106 cells/well and cultured at 37°C for 24 h. Then, 100 nM mimic control (5′-UAUAUCGUGUUAUUAGCGUUCCU-3′; Shanghai GenePharma Co., Ltd.), 100 nM miR-135b-5p mimic (5′-UAUGGCUUUUCAUUCCUAUGUGA-3′; Shanghai GenePharma Co., Ltd.), 2 µl runt-related transcription factor 2 (RUNX2)-plasmid (cat. no. sc-400183-ACT; Santa Cruz Biotechnology, Inc.), 2 µl control-plasmid (cat. no. sc-108083; Santa Cruz Biotechnology, Inc.) or 100 nM miR-135b-5p mimic + 2 µl RUNX2-plasmid were transfected into MC3T3-E1 cells using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) as per the manufacturer's protocol. Cells without any treatment were used the control group, and 48 h after cell transfection, transfection efficiency was detected using reverse transcription-quantitative PCR (RT-qPCR).
Alkaline phosphatase assay
MC3T3-E1 cells were seeded in 96-well plates and then transfected with mimic control, miR-135b-5p mimic or miR-135b-5p mimic+RUNX2-plasmid for 48 h. Then, the ALP assay kit (cat. no. P0321; Beyotime Institute of Biotechnology) was used to determine the extracellular ALP activity in MC3T3-E1 cells following the manufacturer's protocol.
RT-qPCR
Total RNA was extracted with the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) from tissue samples or cells according to the manufacturer's instructions. Then, complementary DNA was synthesized from total RNA by using the TaqMan MicroRNA Reverse Transcription kit (Invitrogen; Thermo Fisher Scientific, Inc.). The temperature protocol for the reverse transcription reaction consisted of primer annealing at 25°C for 5 min, cDNA synthesis at 42°C for 60 min and termination at 80°C for 2 min. RT-qPCR assay was performed using SYBR Premix Ex TaqTM II (TliRNaseH Plus) kit (cat. no. RR820a; Takara Bio, Inc.). The reaction volume was 20 µl. The thermocycling conditions were as follows: Initial denaturation at 94°C for 30 sec; 40 cycles of denaturation at 94°C for 5 sec, annealing at 60°C for 20 sec; and a final extension at 72°C for 30 sec. GAPDH or U6 were used as an internal reference. Primer sequences were as following: U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; GAPDH forward, 5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse, 5′-GTAGAGGCAGGGATGATGTTCT-3′; miR-135b-5p forward, 5′-GGTATGGCTTTTCATTCCT-3′ and reverse, 5′-CAGTGCGTGTCGTGGAGT3′; RUNX2 forward, 5′-GATGATGACACTGCCACCTCT-3′ and reverse, 5′-AGGGCCCAGTTCTGAAGC-3′; ALP forward, 5′-CCGAATTCATGTTGGCCTGTTCAACT-3′ and reverse, 5′-ATGTCGACTTAGTTATTTTCATAATACCAAATTCC-3′; OC forward, 5′-CAGACCTAGCAGACACCATGA-3′ and reverse, 5′-CTGCCAGAGTTTGGCTTTAGG-3′; Osterix forward, 5′-AGAGATCTGAGCTGGGTAG-3′ and reverse, 5′-AAGAGAGCCTGGCAAGAGG-3′. The relative expression levels of the genes were analyzed using 2−ΔΔCq method (26). All experiments were performed in triplicate.
Western blot assay
The proteins were extracted from cells or tissues samples using RIPA lysis buffer (cat. no. P0013E; Beyotime Institute of Biotechnology). The protein concentration was detected using the bicinchoninic acid assay kit (cat. no. BCA1-1KT; Sigma-Aldrich; Merck KGaA). Equal amount of proteins (30 µg per lane) was separated by 12% SDS-PAGE for 40 min. The protein was transferred to a PVDF membrane (EMD Millipore). The membrane was blocked with 5% skimmed milk for 1 h at room temperature. The membrane was washed three times with 1X PBS-0.1% Tween-20 (PBST). The membranes were then incubated with primary antibodies: Runx2 (1:1,000; cat. no. 12556; Cell Signaling Technology, Inc.), ALP (1:1,000; cat. no. sc-365765; Santa Cruz Biotechnology, Inc.), OC (1:1,000; cat. no. ab93876; Abcam), Osterix (1:1,000; cat. no. sc-393060; Santa Cruz Biotechnology, Inc.), and β-actin (1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.) overnight at 4°C. Subsequently, the protein was incubated with a horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibody (1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.) overnight at 4°C. Finally, protein blots were visualized and analyzed using a chemiluminescence system (Beyotime Institute of Biotechnology). β-actin was used as an internal control.
MTT assay
Cell viability was evaluated using an MTT assay. For the MTT detection, MC3T3-E1 cells were seeded in 96-well plates and then transfected with mimic control, miR-135b-5p mimic or miR-135b-5p mimic+RUNX2-plasmid for 48 h. Subsequently, 20 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA) was added into each well and cultured at 37°C for 4 h. To assess cell viability, absorbance was measured at 490 nm using a microplate spectrophotometer (Bio-Rad Laboratories, Inc.).
Flow cytometry analysis
Cells were centrifuged (500 × g, 5 min, 4°C), and re-suspended in 100 µl of FITC-binding buffer. Then, ~5 µl of ready-to-use Annexin V-FITC and 5 µl propidium iodide (PI) (cat. no. 70-AP101-100; MultiSciences Biotech Co., Ltd.) were added to the mixture. Cells were incubated for 30 min in the dark at room temperature. Annexin V-FITC and PI fluorescence were analyzed using a BD FACSCalibur flow cytometer (Becton, Dickinson and Company), Apoptosis in cultured MC3T3-E1 cells was determined by flow cytometry according to the manufacturer's instructions. Data were analyzed by Cell Quest software (version 5.1; BD Biosciences).
Luciferase reporter gene assay
TargetScan 7.2 (http://www.targetscan.org/vert_72/) was used to predict the potential targets of miR-135b-5p, and the binding sites between RUNX2 and miR-135b-5p were observed. To confirm the binding sites between RUNX2 and miR-135b-5p, luciferase reporter gene assays were performed. Luciferase reporter plasmids (psi-CHECK2) containing the wild-type 3′UTRs of RUNX2 (WT-RUNX2), as well as mutant 3′UTRs of RUNX2 (MUT-RUNX2) were manufactured by TsingKe Biotech Co., Ltd. Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to cotransfect MC3T3-E1 cells with the wild-type or mutant 3′UTR luciferase reporter plasmids, and the miR-135b-5p mimic or mimic control respectively. Cells were harvested after transfection for 48 h, and the luciferase activities were measured using the Dual-Luciferase® Reporter Assay Kit (Promega Corporation) following the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Data are presented as mean ± SD. SPSS 22.0 software (IBM Corp.) was used for statistical analysis. Differences between groups were determined using a one-way ANOVA with Tukey's post hoc test or Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-135b-5p in bone tissue fragments of patients with OP
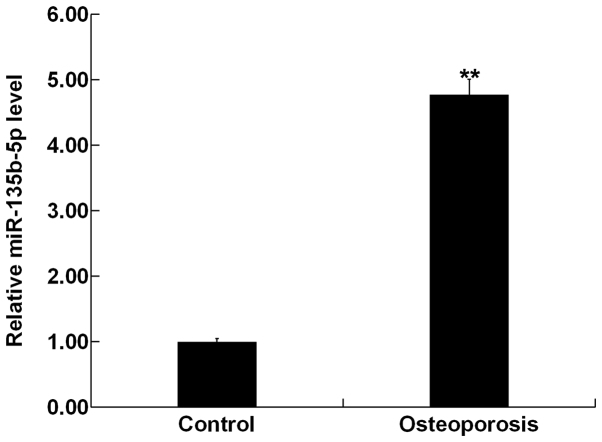
The present study used RT-qPCR to investigate the expression level of miR-135b-5p in bone tissue fragments from 30 patients with OP and 30 patients with osteoarthritis (control group). The present results showed that compared with the control group, the expression level of miR-135b-5p was significantly increased in bone tissue fragments of patients with OP (Fig. 1).
Figure 1.
miR-135b-5p is upregulated in bone tissue fragments of patients with osteoporosis. Reverse transcription-quantitative PCR was used to detect the relative mRNA expression levels of miR-135b-5p in bone tissue fragments from 30 patients with osteoarthritis (control) and 30 patients with osteoporosis. **P<0.01 vs. the control group. miR-135b-5p, microRNA-135b-5p.
Expression of miR-135b-5p during osteogenic differentiation
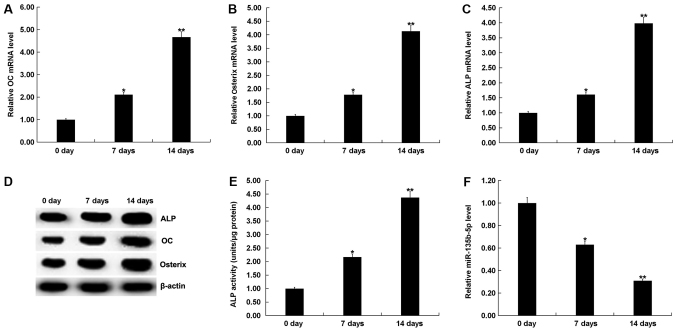
To determine whether osteogenesis could be induced in MC3T3-E1 cells, RT-qPCR and western blot assay were performed to detect the mRNA and protein expression levels of osteogenic markers such as OC, Osterix and ALP. A specific kit was used to examine the activity of ALP. RT-qPCR results showed that mRNA expression levels of OC, Osterix and ALP in MC3T3-E1 cells were significantly increased the 7 and 14th day after induction, compared with the control group (Fig. 2A-C). Protein expression levels of OC, Osterix and ALP in MC3T3-E1 cells were also increased on the 7 and 14th day after induction, compared with the control group (Fig. 2D). The ALP assay indicated that ALP activity was significantly increased (Fig. 2E). To investigate the role of miR-135b-5p in osteogenic differentiation, the present study examined the expression of miR-135b-5p by RT-qPCR. The present results indicated that, compared with the control group, the expression level of miR-135b-5p was significantly decreased in in MC3T3-E1 cells on days 7 and 14 after induction (Fig. 2F). The present results suggested that miR-135b-5p may be involved in the development and progression of OP by affecting osteogenic differentiation.
Figure 2.
Expression of miR-135b-5p is gradually decreased with the prolongation of osteogenic differentiation induction time. (A) RT-qPCR analysis of OC expression level on 0, 7 and 14 days after osteogenic differentiation induction of MC3T3-E1 cells. (B) RT-qPCR analysis of Osterix expression level on 0, 7 and 14 days after induction of MC3T3-E1 cells. (C) RT-qPCR analysis of ALP on 0, 7 and 14 days after induction of MC3T3-E1 cells. (D) Western blot analysis of OC, Osterix and ALP protein expression levels on 0, 7 and 14 days after induction of MC3T3-E1 cells. (E) ALP assay was used to detect ALP activity on 0, 7 and 14 days after induction of MC3T3-E1 cells. (F) RT-qPCR analysis of miR-135b-5p on 0, 7 and 14 days after induction of MC3T3-E1 cells. *P<0.05, **P<0.01 vs. 0 day. miR-135b-5p, microRNA-135b-5p; RT-qPCR, reverse transcription-quantitative PCR; OC, osteocalcin; ALP, alkaline phosphatase.
RUNX2 is a direct target of miR-135b-5p
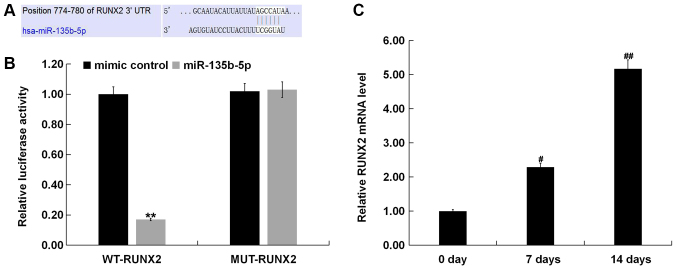
To reveal the molecular mechanism of the role of miR-135-5p in OP, TargetScan was used to predict potential targets of miR-135b-5p, and RUNX2 was identified as a potential target of miR-135b-5p (Fig. 3A). To further examine the bioinformatic prediction, a dual-luciferase reporter assay was performed to detect the relationship between RUNX2 and miR-135b-5p. The present results showed that miR-135b-5p mimic significantly suppressed luciferase activity when MC3T3-E1 cells were cotransfected with a reporter plasmid containing the WT 3′-UTR and miR-135b-5p mimic (Fig. 3B). However, the luciferase activity of the MUT 3′-UTR did not change. RT-qPCR was used to detect the expression of RUNX2, and the results showed that the expression of RUNX2 was significantly increased in MC3T3-E1 cells 7 and 14 days after induction compared with the control group (Fig. 3C).
Figure 3.
RUNX2 is a direct target gene of miR-135b-5p. (A) TargetScan was used to predict miR-135b-5p binding sites in the 3′UTR of RUNX2. (B) Luciferase reporter gene assay was used to identify the binding sites between miR-135b-5p and RUNX2. Firefly luciferase activity was normalized to Renilla luciferase activity. (C) Reverse transcription-quantitative PCR assay measured the relative mRNA expression of RUNX2 on 0, 7 and 14 days after induction of MC3T3-E1 cells. **P<0.01 vs. mimic control. #P<0.05, ##P<0.01 vs. 0 day. miR-135b-5p, microRNA-135b-5p; RUNX2, runt-related transcription factor 2; WT, wild-type; MUT, mutant; 3′UTR, 3′untranslated region.
Role of miR-135b-5p in osteogenic differentiation
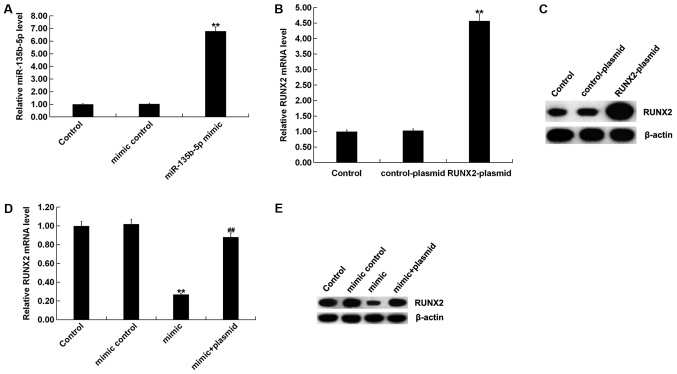
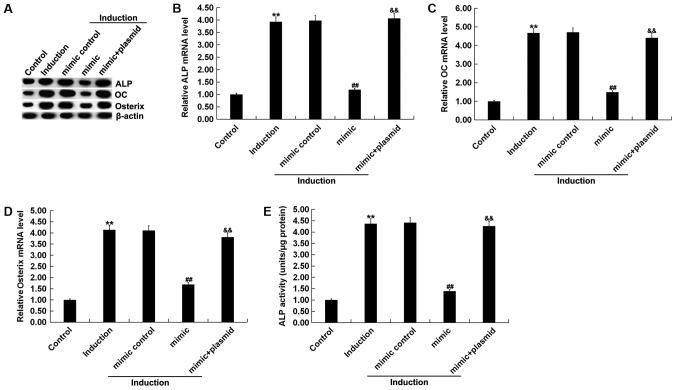
To investigate the role of miR-135b-5p in osteogenic differentiation, MC3T3-E1 cells were transfected with mimic control, miR-135b-5p mimic, RUNX2-plasmid, control-plasmid or miR-135b-5p mimic+RUNX2-plasmid for 48 h. RT-qPCR assay and western blot assay were performed to detect transfection efficiency. The miR-135b-5p mimic significantly increased the expression level of miR-135b-5p in MC3T3-E1 cells compared with the control group (Fig. 4A). RUNX2-plasmid significantly increased the expression level of RUNX2 in MC3T3-E1 cells (Fig. 4B and C). The miR-135b-5p mimic significantly reduced the expression level of RUNX2 in MC3T3-E1 cells, which was reversed by RUNX2 overexpression, compared with the control group (Fig. 4D and E). Osteogenic differentiation was induced in MC3T3-E1 cells after transfection. The levels of mRNA and protein expression of the osteogenic markers OC, Osterix and ALP were detected by RT-qPCR and western blot assay. In addition, levels of ALP activity were analyzed using a commercially available kit. RT-qPCR assay and western blot assay results showed that compared with the control group, the mRNA and protein expression levels of OC, Osterix and ALP in the induction group were markedly increased (Fig. 5A-D), and ALP activity was significantly increased (Fig. 5E). Compared with the induction group, miR-135b-5p mimic significantly reduced the mRNA and protein expression levels of OC, Osterix and ALP, and decreased ALP activity; these reductions were significantly reversed by RUNX2 overexpression. The present results suggested that miR-135b-5p may be involved in the development and progression of OP by affecting osteogenic differentiation.
Figure 4.
miR-135b-5p negatively regulates RUNX2 expression level in MC3T3-E1 cells. (A) RT-qPCR detected the relative expression level of miR-135b-5p in MC3T3-E1 cells transfected with mimic control or miR-135b-5p mimic for 48 h. (B) RT-qPCR assay and (C) western blotting identified the relative mRNA expression level and protein expression level of RUNX2 in MC3T3-E1 cells transfected with control-plasmid or RUNX2 plasmid for 48 h. (D) RT-qPCR and (E) western blotting results of the relative mRNA expression level and protein expression level of RUNX2 in MC3T3-E1 cells transfected with mimic control, miR-135b-5p mimic or miR-135b-5p mimic + RUNX2-plasmid. **P<0.01 vs. control group. ##P<0.01 vs. mimic. miR-135b-5p, microRNA-135b-5p; RT-qPCR, reverse transcription-quantitative PCR; RUNX2, runt-related transcription factor 2.
Figure 5.
Role of miR-135b-5p in osteogenic differentiation. (A) Western blot assay results of the protein expression level of ALP, OC and Osterix. Reverse transcription-quantitative PCR detected the mRNA expression levels of (B) ALP, (C) OC and (D) Osterix. (E) ALP assay was used to measure ALP activity. Control, MC3T3-E1 cells without any treatment. Induction, MC3T3-E1 cells subjected to osteogenic differentiation induction. Mimic control, MC3T3-E1 cells were transfected with mimic control for 48 h, then subjected to osteogenic differentiation induction. Mimic, MC3T3-E1 cells were transfected with miR-135b-5p mimic for 48 h, then subjected to osteogenic differentiation induction. Mimic + plasmid, MC3T3-E1 cells were transfected with miR-135b-5p mimic + RUNX2-plasmid for 48 h, then subjected to osteogenic differentiation induction. **P<0.01 vs. control group. ##P<0.01 vs. induction group. &&P<0.01 vs. mimic group. miR-135b-5p, microRNA-135b-5p; OC, osteocalcin; ALP, alkaline phosphatase; RUNX2, runt-related transcription factor 2.
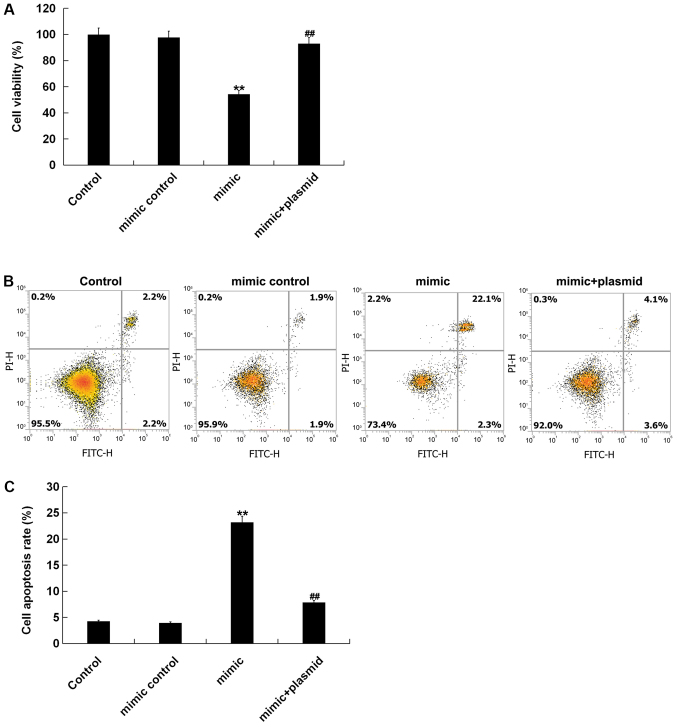
Effect of miR-135b-5p on cell viability and apoptosis of MC3T3-E1 cells
The present study investigated whether miR-135-5p had an effect on the viability and apoptosis of MC3T3-E1 cells. MC3T3-E1 cells were transfected with mimic control, miR-135b-5p mimic or miR-135b-5p mimic+RUNX2-plasmid for 48 h. Then, cell viability was detected by MTT. In addition, cell apoptosis was measured by flow cytometry. The present results showed that compared with the control group, miR-135b-5p mimic significantly reduced cell viability of MC3T3-E1 cells and induced cell apoptosis. These effects, however, were significantly reversed by RUNX2 overexpression (Fig. 6).
Figure 6.
miR-135b-5p mimic suppresses MC3T3-E1 cell viability and induces apoptosis. (A) MTT assay was performed to detect cell viability in MC3T3-E1 cells transfected with mimic control, miR-135b-5p mimic or miR-135b-5p mimic + RUNX2-plasmid. (B) Flow cytometry was used to detect cell apoptosis and (C) the results were quantified. **P<0.01 vs. control group. ##P<0.01 vs. mimic group. miR-135b-5p, microRNA-135b-5p; PI, propidium iodide.
Discussion
The present study investigated the expression of miR-135b-5p in the bone tissues of patients with OP and explored its mechanism of action. The present results suggested that miR-135b-5p was highly expressed in the bone tissues of patients with OP compared with patients with osteoarthritis. Osteoblasts are responsible for the formation and mineralization of the skeleton (27), and play important roles in OP; therefore, research on osteoblasts has become an important topic for OP in in vitro research (28,29). The present study focused on the effects of miR-135b-5p on osteogenic differentiation and osteoblast proliferation. Therefore, the mouse pre-osteoblast cell line MC3T3-E1, which is widely used in osteogenic differentiation investigation (30–32), was selected for this study. Osteogenic differentiation was induced in MC3T3-E1 cells. The present results showed that on 7 and 14th day after induction, the mRNA expression levels of OC, Osterix and ALP were significantly increased. Protein expression levels of OC, Osterix and ALP were also increased in MC3T3-E1 cells 7 and 14 days after osteogenic differentiation induction. ALP activity was increased and the expression of miR-135b-5p was significantly decreased in MC3T3-E1 cells on 7 and 14th day after osteogenic induction. Therefore, the present results suggested that miR-135b-5p may be involved in the occurrence and development of osteoporosis by affecting osteogenic differentiation. In addition, bioinformatics analysis predicted that RUNX2 was a potential target of miR-135b-5p. Luciferase reporter gene assay was performed to investigate this interaction. The present results suggested that the expression level of RUNX2 was increased in MC3T3-E1 cells on 7 and 14th day after osteogenic differentiation induction. The present results indicated that miR-135b-5p mimic significantly reduced cell viability and induced cell apoptosis, and that these effects were significantly reversed by RUNX2-plasmid.
OP is a common and complex disease that has a higher rate of incidence in the aged population (33,34). OP is a multifactorial bone disease with microstructural deterioration and impaired bone strength, making bones susceptible to fragility and fracture (35). OP is the most frequent geriatric disease, especially in postmenopausal women (36). OP occurs due to an imbalance between osteoclastic bone resorption and osteoblastic bone formation (37). Restoring and maintaining the balance between bone formation and bone resorption is an effective way to treat OP (38,39). Physical exercise is an important stimulus for prevention and treatment of osteoporosis (40,41). Due to the high prevalence and severity of OP, it is important to understand its pathogenicity and molecular mechanisms for drug development and treatment.
Previous studies have reported that miRNAs play an important role in regulating osteoblast differentiation and bone formation (42,43). miR-135b-5p is a specific miRNA that has been well studied in cancer and has been identified as an oncogene or tumor suppressor (44,45). Li et al (44) showed that the expression of miR-135b-5p was downregulated in human osteoblastoma tissues. miR-135b-5p inhibited lipopolysaccharide-induced tumor necrosis factor-α secretion by suppressing reactive oxygen species production and NF-κB activation (46). miR-135b-5p was previously found to be highly expressed in pancreatic cancer (45). However, to the best of our knowledge, there is little research on the effect and mechanism of miR-135b-5p in OP. The present results suggested that the expression level of miR-135b-5p was upregulated in bone tissue fragments of patients with OP. However, the present study did not analyzed the correlation of miR-135b-5p expression and the characteristics of patients with OP (age, sex, BMI and Bone Mineral Density). This is limitation of the present study, and will need to be investigated in future studies.
To further examine the specific mechanism in which miR-135b-5p may regulate MC3T3-E1 cells, the bioinformatics software TargetScan was used to predict the target gene of miR-135b-5p. Dual luciferase reporter gene assay results indicated that RUNX2 was a direct target of miR-135b-5p. A previous study indicated that miR-135b inhibited osteoblastoma cells by mediating the expression of protein phosphatase Mg2+/Mn2+ dependent 1E (47). RUNX2 is a key regulator of bone osteogenic differentiation (48–50). RUNX2 promotes bone formation and inhibits bone resorption by regulating specific extracellular matrix protein expression levels and the cell cycle of osteoblasts (50). Previous studies have reported that miR-338-3p can suppress tumor cell proliferation and metastasis in osteosarcoma cells by targeting RUNX2 (51). Li et al (52) demonstrated that miR-129-1-3p regulated circulating stretch-induced differentiation of endothelial progenitor cells by targeting RUNX2. The present results suggested that osteogenic differentiation inhibition caused by miR-135b-5p was suppressed by RUNX2 overexpression. In addition, the present results suggested that miR-135b-5p mimic significantly reduced cell viability and induced cell apoptosis in MC3T3-E1 cells, and that all these effects were significantly reversed by RUNX2 overexpression.
In conclusion, the present results suggested that miR-135-5p may be involved in the occurrence and development of OP via the inhibition of osteogenic differentiation and osteoblast proliferation by targeting RUNX2. However, the present study was a preliminary study of the role of miR-135b-5p in OP. Therefore, further research is required to understand the role of miR-135b-5p, such as the analysis of the relationship between miR-135b-5p expression and the characteristics of patients with OP. The mechanism of miR-135b-5p in human osteoblast should also be investigated. In addition, in vivo studies should be performed, and will be addressed in future experiments.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BC and WY contributed to study design, data collection, statistical analysis, data interpretation and manuscript preparation. HZ and KL contributed to data collection, statistical analysis and manuscript preparation. AD, GZ and KP contributed to data collection and data interpretation.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University and every patient provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cosman F, de Beur SJ, LeBoff MS, Lewiecki EM, Tanner B, Randall S, Lindsay R. Erratum to: Clinician's guide to prevention and treatment of osteoporosis. Osteoporos Int. 2015;26:2045–2047. doi: 10.1007/s00198-015-3037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sotorník I. Osteoporosis-epidemiology and pathogenesis. Vnitr Lek. 2016;62(Suppl 6):S84–S87. (In Czech) [PubMed] [Google Scholar]
- 3.Body JJ, Bergmann P, Boonen S, Boutsen Y, Devogelaer JP, Goemaere S, Kaufman JM, Rozenberg S, Reginster JY. Evidence-based guidelines for the pharmacological treatment of postmenopausal osteoporosis: A consensus document by the Belgian Bone Club. Osteoporos Int. 2010;21:1657–1680. doi: 10.1007/s00198-010-1223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen P, Li Z, Hu Y. Prevalence of osteoporosis in China: A meta-analysis and systematic review. BMC Public Health. 2016;16:1039. doi: 10.1186/s12889-016-3712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svedbom A, Hernlund E, Ivergård M, Compston J, Cooper C, Stenmark J, McCloskey EV, Jönsson B, Kanis JA, EU Review Panel of IOF Osteoporosis in the European Union: A compendium of country-specifc reports. Arch Osteoporos. 2013;8:137. doi: 10.1007/s11657-013-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge DW, Wang WW, Chen HT, Yang L, Cao XJ. Functions of microRNAs in osteoporosis. Eur Rev Med Pharmacol Sci. 2017;21:4784–4789. [PubMed] [Google Scholar]
- 7.Dadra A, Aggarwal S, Kumar P, Kumar V, Dibar DP, Bhadada SK. High prevalence of vitamin D deficiency and osteoporosis in patients with fragility fractures of hip: A pilot study. J Clin Orthop Trauma. 2019;10:1097–1100. doi: 10.1016/j.jcot.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leutner M, Matzhold C, Bellach L, Deischinger C, Harreiter J, Thurner S, Klimek P, Kautzky-Willer A. Diagnosis of osteoporosis in statin-treated patients is dose-dependent. Ann Rheum Dis. 2019;78:1706–1711. doi: 10.1136/annrheumdis-2019-215714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loh FE, Stuart B, Sturpe D, Davidoff A, Onukwugha E, Hochberg M. Osteoporosis medication use: A comparison of elderly females living in long-term care facilities versus community Dwellers. Sr Care Pharm. 2019;34:109–126. doi: 10.4140/TCP.n.2019.109. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Darnet S, Moreira FC, Hamoy IG, Burbano R, Khayat A, Cruz A, Magalhães L, Silva A, Santos S, Demachki S, et al. High-throughput sequencing of miRNAs reveals a tissue signature in gastric cancer and suggests novel potential biomarkers. Bioinform Biol Insights. 2015;9(Suppl 1):S1–S8. doi: 10.4137/BBI.S23773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tseng CW, Lin CC, Chen CN, Huang HC, Juan HF. Integrative network analysis reveals active microRNAs and their functions in gastric cancer. BMC Syst Biol. 2011;5:99. doi: 10.1186/1752-0509-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui H, Wang L, Gong P, Zhao C, Zhang S, Zhang K, Zhou R, Zhao Z, Fan H. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PLoS One. 2015;10:e0123926. doi: 10.1371/journal.pone.0123926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Wang J, Wang J, Yung VY, Hsu E, Li A, Kang Q, Ma J, Han Q, Jin P, et al. MicroRNA-135b regulates apoptosis and chemoresistance in colorectal cancer by targeting large tumor suppressor kinase 2. Am J Cancer Res. 2015;5:1382–1395. [PMC free article] [PubMed] [Google Scholar]
- 15.Imam JS, Plyler JR, Bansal H, Prajapati S, Bansal S, Rebeles J, Chen HI, Chang YF, Panneerdoss S, Zoghi B, et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One. 2012;7:e52397. doi: 10.1371/journal.pone.0052397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao F, Ding J, Liang L, Wang Q, Liu L, et al. MicroRNA-135b, a HSF1 target, promotes tumor invasion and metastasis by regulating RECK and EVI5 in hepatocellular carcinoma. Oncotarget. 2015;6:2421–2433. doi: 10.18632/oncotarget.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, Hu J, Sun W, Li S, Deng S, Li M. MiR-29c inhibits cell growth, invasion, and migration of pancreatic cancer by targeting ITGB1. Onco Targets Ther. 2016;9:99–109. doi: 10.2147/OTT.S92758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang L, Zhang H, Chen X, Yang Y, Liu G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin β1 and matrix metalloproteinase2 (MMP2) PLoS One. 2013;8:e70192. doi: 10.1371/journal.pone.0070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeliger C, Karpinski K, Haug AT, Vester H, Schmitt A, Bauer JS, van Griensven M. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res. 2014;29:1718–1728. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Li Y, Zhang Y, Ma L, Lin L, Meng J, Jiang L, Wang L, Zhou P, Zhang Y. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother. 2017;89:1178–1186. doi: 10.1016/j.biopha.2017.02.090. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Xie H, Liu W, Hu R, Huang B, Tan YF, Xu K, Sheng ZF, Zhou HD, Wu XP, Luo XH. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;19:3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125:1509–1522. doi: 10.1172/JCI77716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 25.Zheng F, Wang F, Xu Z. MicroRNA-98-5p prevents bone regeneration by targeting high mobility group AT-Hook 2. Exp Ther Med. 2019;18:2660–2666. doi: 10.3892/etm.2019.7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Zou W, Greenblatt MB, Brady N, Lotinun S, Zhai B, de Rivera H, Singh A, Sun J, Gygi SP, Baron R, et al. The microtubule-associated protein DCAMKL1 regulates osteoblast function via repression of Runx2. J Exp Med. 2013;210:1793–1806. doi: 10.1084/jem.20111790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu B, Xue F, Zhang C, Li G. Ginkgolide B promotes osteoblast differentiation via activation of canonical Wnt signalling and alleviates osteoporosis through a bone anabolic way. J Cell Mol Med. 2019;23:5782–5793. doi: 10.1111/jcmm.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao W, Zhang WL, Yang B, Sun J, Yang MW. NIPA2 regulates osteoblast function via its effect on apoptosis pathways in type 2 diabetes osteoporosis. Biochem Biophys Res Commun. 2019;513:883–890. doi: 10.1016/j.bbrc.2019.04.030. [DOI] [PubMed] [Google Scholar]
- 30.Yuan M, Wang Y, Qin YX. SPIO-Au core-shell nanoparticles for promoting osteogenic differentiation of MC3T3-E1 cells: Concentration-dependence study. J Biomed Mater Res A. 2017;105:3350–3359. doi: 10.1002/jbm.a.36200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai F, Song N, Ma J, Gong W, Tian H, Li X, Jiang C, Wang H. FGF18 inhibits MC3T3-E1 cell osteogenic differentiation via the ERK signaling pathway. Mol Med Rep. 2017;16:4127–4132. doi: 10.3892/mmr.2017.7088. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Xu F, Pei HX, Zhu X, Lin X, Song CY, Liang QH, Liao EY, Yuan LQ. Vaspin regulates the osteogenic differentiation of MC3T3-E1 through the PI3K-Akt/miR-34c loop. Sci Rep. 2016;6:25578. doi: 10.1038/srep25578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuiling KD, Robinia K, Nye R. Osteoporosis update. J Midwifery Womens Health. 2011;56:615–627. doi: 10.1111/j.1542-2011.2011.00135.x. [DOI] [PubMed] [Google Scholar]
- 34.Tucker KL. Dietary intake and bone status with aging. Curr Pharm Des. 2003;9:2687–2704. doi: 10.2174/1381612033453613. [DOI] [PubMed] [Google Scholar]
- 35.Caillet P. Consensus development conference: Diagnosis, prophylaxis, and treatment of osteoporosis. Osteoporosis International. 1987;295:914–915. doi: 10.1136/bmj.295.6603.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade SW, Strader C, Fitzpatrick LA, Anthony MS, O'Malley CD. Estimating prevalence of osteoporosis: Examples from industrialized countries. Arch Osteoporos. 2014;9:182. doi: 10.1007/s11657-014-0182-3. [DOI] [PubMed] [Google Scholar]
- 37.Raisz LG. Pathogenesis of osteoporosis: Concepts, conflicts, and prospects. J Clin Invest. 2005;115:3318–3325. doi: 10.1172/JCI27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marie PJ, Kassem M. Osteoblasts in osteoporosis: Past, emerging, and future anabolic targets. Eur J Endocrinol. 2011;165:1–10. doi: 10.1530/EJE-11-0132. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Gaspà S, Blanch-Rubió J, Ciria-Recasens M, Monfort J, Tío L, Garcia-Giralt N, Nogués X, Monllau JC, Carbonell-Abelló J, Pérez-Edo L. Reduced proliferation and osteocalcin expression in osteoblasts of male idiopathic osteoporosis. Calcif Tissue Int. 2010;86:220–226. doi: 10.1007/s00223-010-9336-5. [DOI] [PubMed] [Google Scholar]
- 40.Kerschan-Schindl K. Prevention and rehabilitation of osteoporosis. Wien Med Wochenschr. 2016;166:22–27. doi: 10.1007/s10354-015-0417-y. [DOI] [PubMed] [Google Scholar]
- 41.Moreira LD, Oliveira ML, Lirani-Galvão AP, Marin-Mio RV, Santos RN, Lazaretti-Castro M. Physical exercise and osteoporosis: Effects of different types of exercises on bone and physical function of postmenopausal women. Arq Bras Endocrinol Metabol. 2014;58:514–522. doi: 10.1590/0004-2730000003374. [DOI] [PubMed] [Google Scholar]
- 42.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valenti MT, Dalle Carbonare L, Mottes M. Role of microRNAs in progenitor cell commitment and osteogenic differentiation in health and disease (Review) Int J Mol Med. 2018;41:2441–2449. doi: 10.3892/ijmm.2018.3452. [DOI] [PubMed] [Google Scholar]
- 44.Li ZW, Zhu YR, Zhou XZ, Zhuo BB, Wang XD. microRNA-135b expression silences Ppm1e to provoke AMPK activation and inhibit osteoblastoma cell proliferation. Oncotarget. 2017;8:26424–26433. doi: 10.18632/oncotarget.15477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P, Fan JB, Gao Y, Zhang M, Zhang L, Yang N, Zhao X. miR-135b-5p inhibits LPS-induced TNFα production via silencing AMPK phosphatase Ppm1e. Oncotarget. 2016;7:77978–77986. doi: 10.18632/oncotarget.12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han X, Saiyin H, Zhao J, Fang Y, Rong Y, Shi C, Lou W, Kuang T. Overexpression of miR-135b-5p promotes unfavorable clinical characteristics and poor prognosis via the repression of SFRP4 in pancreatic cancer. Oncotarget. 2017;8:62195–62207. doi: 10.18632/oncotarget.19150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, Pockwinse SM. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23:4315–4329. doi: 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- 48.Xiao ZS, Hjelmeland AB, Quarles LD. Selective deficiency of the ‘bone-related’ Runx2-II unexpectedly preserves osteoblast-mediated skeletogenesis. J Biol Chem. 2004;279:20307–20313. doi: 10.1074/jbc.M401109200. [DOI] [PubMed] [Google Scholar]
- 49.Yang S, Wei D, Wang D, Phimphilai M, Krebsbach PH, Franceschi RT. In vitro and in vivo synergistic interactions between the Runx2/Cbfa1 transcription factor and bone morphogenetic protein-2 in stimulating osteoblast differentiation. J Bone Miner Res. 2003;18:705–715. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komori T. Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol. 2010;658:43–9. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- 51.Jia F, Zhang Z, Zhang X. MicroRNA-338-3p inhibits tumor growth and metastasis in osteosarcoma cells by targeting RUNX2/CDK4 and inhibition of MAPK pathway. J Cell Biochem. 2019;120:6420–6430. doi: 10.1002/jcb.27929. [DOI] [PubMed] [Google Scholar]
- 52.Li N, Wang WB, Bao H, Shi Q, Jiang ZL, Qi YX, Han Y. MicroRNA-129-1-3p regulates cyclic stretch-induced endothelial progenitor cell differentiation by targeting Runx2. J Cell Biochem. 2018;120:5256–5267. doi: 10.1002/jcb.27800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.