Abstract
Acute myocardial infarction (AMI), as the first manifestation of ischemic heart disease, is the most common cause of death in developed countries. A recent study showed that metastasis associated lung adenocarcinoma transcript 1 (MALAT1), a prognostic marker for lung cancer metastasis, could promote myocardial ischemia-reperfusion injury by regulating the levels of microRNA (miR)-145. In order to elucidate the biological function of MALAT1 in the pathogenesis of AMI and to explore the mechanisms underlying its action, an AMI rat model was established by ligation of the left anterior descending coronary artery. Downregulation of MALAT1 by siRNA transfection attenuated heart damage in an AMI model rat. The mouse cardiomyocyte cell line HL-1 was used to show that downregulation of nucleotide binding and oligomerization domain-like receptor C5 (NLRC5) and upregulation of miR-125b-5p were the results of MALAT1 silencing. TargetScan and a dual-luciferase reporter assay indicated that NLRC5 is a direct target of miR-125b-5p. Overexpression of miR-125b-5p significantly reduced hypoxia/reperfusion-induced apoptosis of HL-1 cells, an effect that could be blocked by NLCR5 overexpression. Taken together, these results suggest that MALAT1 reduced the protective effect of miR-125b-5p on injured cells through upregulation of NLCR5. This study highlights the role of MALAT1 in the pathogenesis of AMI and may guide future genetic therapeutic strategies for AMI treatment.
Keywords: acute myocardial infarction, cardiomyocyte, apoptosis, long non-coding RNA, metastasis associated lung adenocarcinoma transcript 1, microRNA-125b-5p, nucleotide binding and oligomerization domain-like receptor C5
Introduction
Acute myocardial infarction (AMI) is an ischemic heart disease, which is characterized by myocardial necrosis due to persistent and severe myocardial ischemia (1). AMI is the cause of more than a third of deaths in developed nations annually, and is responsible for a large number of annual deaths in the USA and more than 4 million annual deaths in Europe and Northern Asia (2,3). In recent decades the incidence of AMI has begun to decline due to the development of new therapies and changing lifestyles. However, approximately 7 million individuals still suffer from AMI worldwide each year (4).
If reperfusion is not performed shortly after ischemic injury following AMI, it results in irreversible necrosis of cardiomyocytes, which leads to myocardial fibrosis and scar tissue formation. Progression of this fibrosis affects contraction of the ventricle, leading to ventricular remodeling, and ultimately contributes to the occurrence of heart failure (5).
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is a long non-coding RNA (lncRNA) also known as nuclear-enriched abundant transcript 2, and was originally identified as a prognostic marker for lung cancer metastasis (6). Two modes of action have been proposed for MALAT1: i) Alternative splicing of a subset of pre-micro RNAs is regulated by MALAT1's recruitment of phosphorylated SR splicing factors (7); and ii) MALAT1 is associated with transcriptional control of the gene expression of cell cycle genes (8). Though the regulatory mechanisms of MALAT1 action in other conditions remains to be clarified, it has been reported that MALAT1 is expressed at high levels in tumor tissue and its expression has been linked to tumor cell proliferation, apoptosis, migration, invasion and metastatic spread (9). A study showed that the development of bladder cancer is controlled by MALAT1 through regulation of NAD-dependent protease sirtuin-7 and microRNA (miR)-125b (10). Additional research indicated that MALAT1 also promotes the proliferation, migration and invasion of esophageal squamous cell carcinoma (11). Studies have also shown that MALAT1 contributes to endothelial cell function, vessel growth and inflammation (12,13). In 2017, Zhao et al (14) found that MALAT1 functions as a mediator of the cardioprotective effect of fentanyl during myocardial ischemia-reperfusion injury. However, the detailed molecular mechanisms involved in MALAT1-mediated modulation of AMI need further elucidation.
Nucleotide binding and oligomerization domain-like receptors (NLRs) are a group of pattern-recognition receptors, which have various functions in innate immunity (15). Nucleotide binding and oligomerization domain-like receptor C5 (NLRC5), the largest member of the NLR protein family, has been well characterized as a regulator of antigen presentation via modulation of major histocompatibility complex class I gene expression (16). NLRC5 has also been linked to suppression of type I interferon production through the IκB kinase complex or retinoic-acid inducible protein I/melanoma-differentiation-associated gene 5 (17). It has been confirmed that NLRC5 participates in the transformation and invasion of malignant cancer cells (18,19) and an increasing number of studies have found that NLRC5 is associated with non-immunological diseases (20). MiR-125b-5p is a known regulator of apoptosis in several types of cells (21–23). Recently, NLRC5 was reported to have a protective effect against heart disease (24). This led to the hypothesis that the effect of MALAT1 on AMI-induced heart damage may be associated with NLRC5 and miR-125b-5p.
In the present study, downregulation of MALAT1 was found to prevent AMI-induced tissue damage in rats. In vitro, expression of NLRC5 and miR-125b-5p were both influenced by MALAT1. TargetScan and dual-luciferase reporter assays provided further confirmation that NLRC5 is the direct target gene of miR-125b-5p. Furthermore, NLRC5 overexpression inhibited miR-125b-5p-associated cytoprotection in ischemia/reperfusion-treated HL-1 cells. These results suggest that MALAT1 abrogates the protective effect of miR-125b-5p on cardiomyocytes through upregulation of NLCR5 expression. These findings suggest an emerging role for MALAT1 in the pathogenesis of AMI and will guide future development of genetic therapeutic strategies to treat AMI.
Materials and methods
Animals
A total of 32 adult male Sprague-Dawley (SD) rats (14 weeks) were purchased from the experimental animal center of the Dezhou People's Hospital. All procedures were approved by the Dezhou People's Hospital Ethics Review Committee for Animal Experimentation (experiment number 2017-TX-007). Rats were divided into four groups (sham NC-siRNA, sham MALAT1-siRNA, AMI NC-siRNA and AMI MALAT-siRNA, 8 rats per each group) and housed at room temperature (22–26°C), humidity 50–70% on a 12-h light/dark cycle. Food and water were provided ad libitum. The body weight of each rat was recorded weekly. The SD rats (220–250 g) were anesthetized using 10% chloral hydrate (350 mg/kg; Sinopharm Chemical Reagent Co., Ltd.) intraperitoneally and were mechanically ventilated by a TOPO Dual Mode Ventilator (Kent Scientific Corporation). Peritonitis was not observed following the administration of 10% chloral hydrate.
A rat model of AMI was developed by ligation of the left anterior descending coronary artery. In brief, the anesthetized rats were placed in a supine position and the trachea carefully exposed. An endotracheal tube was introduced into the trachea (polyethylene size 90). Mechanical ventilation was achieved by connecting the endotracheal tube to a TOPO Dual Mode Ventilator (Kent Scientific Corporation) cycling at 80 breaths per minute with a tidal volume of 1.2 ml per 100 g body weight. Once steady breathing was established, the left ventricle was exposed by opening the chest at the 4th left intercostal space. Ischemia was induced by extruding the heart and quickly and accurately ligating the left anterior descending coronary artery with a 6-0 suture. Reperfusion was established by cutting the knot in the ligature, 30 min after ligation. Before closing the chest, rats were monitored for 5 min to ensure there was no bleeding due to the surgery. The chest was closed using 6-0 prolene sutures with a continuous suture pattern. Post-surgery, the vital signs of rats were measured until sample collection. In sham control rats, the procedure was identical, but there was no transient artery ligation. For the MALAT1 knockdown experiment, the rats were intravenously injected through the tail vein with small interfering (si)RNA against MALAT1 and siRNA-non-coding (NC) at the final concentration of 10 mg/kg for 24 h before surgery. When pulsatile blood was observed in the needle hub, the siRNA was injected with a continuous and slow movement. The plasmids of siRNA against MALAT1 and NC were generated using the pcDNA3.1 vector, and purchased from Shanghai GenePharma Co., Ltd. siMALAT1, 5′-AAGAAAAAUAAAAGCUUUCCU-3′ and siNC, 5′-ACGUCACACGUUCGGAGAATT-3′.
Determination of myocardial infarction
The left ventricle was cut into 2-mm-thick sections perpendicular to the long axis of the heart. A section from each heart was wrapped with gauze, followed by incubation of the heart in 1% 2,3,5-triphenyl-2H-tetrazolium chloride (TTC; Beijing Solarbio Science & Technology Co., Ltd.) for 60 min at 37°C. An additional section was incubated with phosphate-buffered saline (PBS) as a negative control for the same length of time. To avoid sample oxidation, each section was sealed with the PBS in a closed freezer bag and gently shaken in a hot bath to obtain uniform staining throughout the incubation at 37°C for 2 h. The sizes of the TTC-stained area (red staining, ischemic but viable tissue) and unstained area [infarct myocardium (INF)] were evaluated using Image Pro Plus 6.0 (Media Cybernetics Inc.). The myocardial INF size was expressed as a percentage of INF over total area at risk.
Analysis of cardiac function
Cardiac function was evaluated by motion-mode echocardiography using the VEVO 770 high-resolution in vivo imaging system (FUJIFILM VisualSonics Inc.). Left ventricular fractional shortening (LVFS) was calculated by computerized algorithms. In brief, the left ventricular end-systolic diameter (LVESD) and the left ventricular end-diastolic diameter (LVEDD) were measured by M-mode of echocardiography. These parameters refer to the size of the ventricle at the end of systole and diastole. Based on the formula: (LVEDD-LVESD/LVEDD) ×100, the percentage of size differences of the left ventricle as a parameter of how well the left ventricle is contracting itself (25). All measurements represented the mean of 5 consecutive cardiac cycles. Left ventricular internal dimension (LVIDs) were measured at end-systole (s) through four-chamber view. By using this, the endocardial margin of the lateral wall and the septum were analyzed through Vevo 770 (V2.2.0; VisualSonics, Toronto, Canada).
Cell culture
The mouse cardiomyocyte cell line HL-1 (cat. no. SCC065) was purchased from Sigma-Aldrich (Merck KGaA). HL-1 cells were cultured in Claycomb medium (Sigma-Aldrich; Merck KGaA) supplemented with 10% heat-inactivated fetal bovine serum (MP Biomedicals LLC.), 0.1 mM norepinephrine and 1% penicillin-streptomycin (Nacalai Tesque Inc.) in a 5% CO2 humidified incubator at 37°C. To establish a hypoxia/reoxygenation (H/R) model the cells were cultured in a Napco 8000WJ hypoxia (1% O2−5% CO2−94% N2) incubator (Thermo Fisher Scientific, Inc.). After 10 h of hypoxia, the HL-1 cells were plated at 5×105 cells/ml and incubated under normoxic conditions in a CO2 incubator for an additional 4 h. The HL-1 cells passage time was less than 20.
Plasmid construction
The MALAT1 siRNA were designed and obtained from Invitrogen (4455877; Thermo Fisher Scientific, Inc.). The sequence of MALAT1 siRNA was 5′-GCAGAGGCAUUUCAUCCUU-3′. The sequence of the negative control siRNA was 5′-ACGUCACACGUUCGGAGAATT-3′ and it was provided by Thermo Fisher Scientific, Inc. The human NLRC5 (NCBI reference sequence, NP_115582) was obtained by nested PCR from a human cDNA library (Marathon-ready cDNA; Clontech Laboratories, Inc.) using the following primers: Forward, 5′-CGTGGGGACCCTAGAGCACCTATCA-3′ and reverse, 5′-GCATCACTTGGCTGGATTCCAAAGG-3′. The PCR products were cloned using a Takara one step PCR kit (Takara Biotechnology Co., Ltd.) and initial denaturation at 98°C for 30 sec, 30 cycles of denaturation at 98°C for 5 sec, annealing at 60°C for 10 sec, and extension at 72°C for 10 sec, finally, extension at 72°C for 2 min. For MALAT1 overexpression, the human MALAT1-expressing vector (ORF023250, NR_002819.3) was obtained from Applied Biological Materials Inc (Richmond, BC, Canada).
Transfection
The function of MALTA1 was inhibited using siRNA. HL-1 cells were plated on 24-well plates at a density of 1×105 cell per well and cultured overnight. The following day, the cells were transfected with MALAT1 siRNA (500 ng/well) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Overexpression of NLRC5 or empty vector in HL-1 cells was also carried out using the Lipofectamine 2000 system (500 ng/well of plasmid). For the overexpression of miR-125b-5p, cells were transfected with miR-125b-5p mimic or negative control mimic (NC mimic) using a NanoFectin transfection reagent (Shanghai Excell Biology, Inc.) according to the manufacturer's protocol. The miR-125b-5p mimic (5′-AUUUGCGGCCGCUCAGCUGAGAUGUUCUGAGGU-3′) or NC-mimic (5′-UUCUCCGAACGUGUCACGUTT-3′) was used at a concentration of 100 nM (Shanghai GenePharma Co., Ltd.).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells or tissue using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNase-free DNase (Takara Biotechnology Co., Ltd.) was then added to the RNA following a manufacturer's protocol. A total of 2 µl of RNA was reverse-transcribed to cDNA using the SuperScript IV First-Strand Synthesis System (Invitrogen; Thermo Fisher Scientific, Inc.) following the instructions provided by the manufacturer. The qPCR amplification conditions consisted of pre-denaturation for 3 min at 94°C, followed by a total of 30 cycles of denaturation for 30 sec at 94°C, annealing at 58°C for 30 sec and extension for 60 sec at 72°C. The expression of genes was measured by SYBR-green master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the primer sets described below using an ABIViiA7 Real-Time PCR System. U6 was used as an internal control for miR-125b-5p. Expression of β-actin was used as internal control for the analysis of other genes while the 2−ΔΔCq method was used for data analysis (26). The primer sequences were (5′→3′) as follows: Malat1 forward, TCTTAGAGGGTGGGCTTTTGTT and reverse, CTGCATCTAGGCCATCATACTG; miR-125b-5p forward, TGCGCTCCCTGAGACCCTAAC and reverse, CCAGTGCAGGGTCCGAGGTATT; nlrc5 forward, CTCAGCCAGGAGCACGTAG and reverse, CCAGCAGGTTTGCTGAGAGA; β-actin forward, CCTCTATGCCAACACAGTGC and reverse, CATCGTACTCCTGCTTGCTG; and U6 forward, CTCGCTTCGGCAGCACA and reverse, AACGCTTCACGAATTTGCG.
Western blotting
The samples were lysed and prepared using Laemmli sample buffer (cat. no. 161-0747, Bio-Rad Laboratories, Inc.). The concentration of protein in each sample was determined using a BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). A total of 15 µg of protein were subjected to 10% SDS-PAGE and separated proteins were transferred to polyvinylidene difluoride membrane (Immobilon-P; EMD Millipore). The membrane was blocked with 5% skimmed milk in TBS-T for 1 h at room temperature and then incubated with the primary antibody against NLRC5 (1:1,000, cat. no. sc-515668; Santa Cruz Biotechnology, Inc.) overnight at 4°C. After washing three times in TBS-T, the membrane was incubated with secondary antibody (1:5,000, anti-mouse; AP31535BT-N, OriGene Technologies Inc.) for 1 h at room temperature. The expression of β-actin was measured by using an anti-β-actin antibody for 1 h at room temperature (1:1,000; cat. no. 3700, CST Biological Reagents Co., Ltd.), before incubation with the same secondary antibody following the aforementioned conditions. Protein expression was detected using Luminata Forte Western HRP Substrate (EMD Millipore) with a Bio-Rad ChemiDoc XRS + imaging system (Bio-Rad Laboratories, Inc.).
Luciferase activity
The TargetScan 7.2 (targetscan.org) was used for prediction of the direct target gene of miR-125b-5p, and the confirmation was further performed by dual-luciferase reporter assay. To construct a luciferase reporter containing the wild type NLRC5 3′untranslated region (3′UTR) with a predicted miR-125b-5p binding site and mutant NLRC5 3′UTR, the region of NLRC5 and its mutant were amplified using genomic DNA as a template. PCR products were inserted into the pMIR-report plasmid (Ambion; Thermo Fisher Scientific, Inc.). In brief, HL-1 cells were cultured in 24-well plates at the density of 2.5×104 cells/well and transfected with 0.3 µg of firefly luciferase reporter plasmid (Ambion, Thermo Fisher Scientific, Inc.) using Lipofectamine® 2000. An equal amount of β-galactosidase expression vector (Ambion; Thermo Fisher Scientific, Inc.) was transfected as a control. After a 48-h transfection, the relative activity of luciferase was determined using the Dual-Luciferase Reporter Assay kit (Promega Corporation) according to the manufacturer's instructions. The miR-125b-5p mimic or negative control mimic (Guangzhou RiboBio Co., Ltd.) was co-transfected with the wild-type or mutant reporter vectors. The luciferase activities were then determined using Beckman Coulter DTX880 Multimode Detector (Beckman Coulter, Inc.) at 560 nm and the data were normalized against Renilla luciferase activity.
Measurement of L-lactate dehydrogenase (LDH) and malondialdehyde (MDA)
The LDH level in the supernatant was quantified using an LDH assay kit (ab102526, Abcam) according to the manufacturer's instructions. In brief, the HL-1 cell culture supernatants were incubated for 1 h at room temperature in 96-well plates coated with an anti-LDH antibody. After washing, a biotin-conjugated capture antibody was added and incubated for 1 h at room temperature, followed by addition of an avidin-horseradish peroxidase conjugate to bind to any captured antigen. Absorbance was measured at 450 nm. The values were expressed as the percentage of LDH released relative to the value obtained following permeabilization of the cells with digitonin. The MDA level was analyzed using a Cellular Malondialdehyde Test kit (A003-2, Nanjing Jiancheng Bioengineering Institute). The standards, of known concentrations, were incubated with N-methyl-2-phenylindole for 1 h at 45°C The resulting chromophore was analyzed with a spectrophotometer at 586 nm. The MDA concentration in the samples was interpolated from the standard curve.
Fluorescence activated cell sorting (FACs)
After 48 h of transfection, the HL-1 cells were recovered, washed with PBS and resuspended. The culture supernatants were discarded by centrifugation at 100 × g at 4°C for 10 min. Cells were then resuspended in 150 µl of 10 mM HEPES (pH 7.4), 140 mM NaCl and 2.5 mM CaCl2, and placed on ice for 5 min. A further 50 µl of buffer containing 5 µl propidium iodide and 10 µl Annexin V-FITC (FITC Annexin V Apoptosis Detection Kit I; cat. no. 556547; BD Biosciences) was added. After mixing, the cells were incubated at room temperature in the dark for 15 min. A FACS Calibur cell analyzer (BD Biosciences) and FlowJo 10.6.1 (FlowJo LLC) were used to analyze cell apoptosis and calculate the apoptosis rate. The apoptotic cells including apoptotic cells in early stage (Annexin V+/PI− cells) and late stage (Annexin V+/PI+ cells), which were used to calculation of apoptotic cells/viable cells.
Statistical analysis
All data are presented as the mean ± standard deviation. To determine statistical significance, GraphPad Prism 7 (GraphPad Software, Inc.) was used. One-way ANOVA with Tukey's post hoc test was used for multiple comparisons. Student's t-test was used when comparing between 2 conditions. P<0.05 indicated statistical significance.
Results
Downregulation of MALAT1 attenuates heart damage in a rat model of AMI
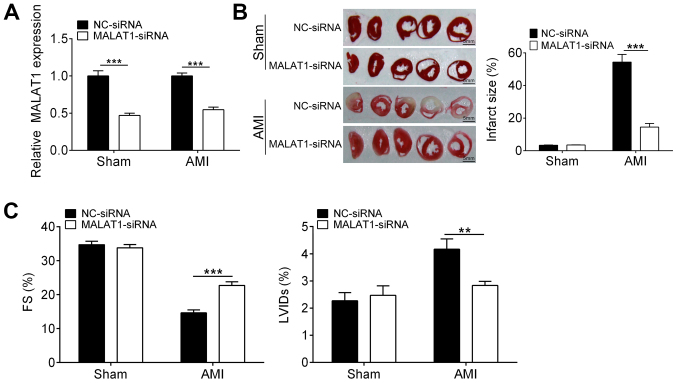
To investigate the exact role of MALAT1 in myocardial infarction, a rat model of AMI was established and MALAT1 was knocked down by siRNA transfection. As shown in Fig. 1A, the mRNA levels of MALAT1 in both sham and AMI rats were significantly downregulated through siRNA. Infarction size of heart from sham rats were not affected by downregulation of MALAT1. Interestingly, knockdown of MALAT1 strongly decreased the myocardial infarction area in AMI rats as compared with the NC-siRNA group (Fig. 1B). Although AMI caused a reduction in the LVFS and increased the LVIDs (Fig. 1C), downregulation of MALAT1 increased the LVFS and reduced LVIDs in AMI rats when compared with the control (Fig. 1C). These findings suggested that MALAT1 knockdown ameliorated AMI-induced heart damage.
Figure 1.
Downregulation of MALAT1 attenuates heart damage in an AMI model rat. (A) The efficiency of MALAT1 knockdown was confirmed by reverse transcription-quantitative PCR. (B) The infarcted area of heart was evaluated by 2,3,5-triphenyl-2H-tetrazolium chloride staining. (C) The cardiac function of rats was evaluated. The data are represented as the mean ± standard deviation (n=10). **P<0.01, ***P<0.001. AMI, acute myocardial infarction; LVFS, left ventricular fractional shortening; LVID, left ventricular internal dimension; MALAT1, metastasis associated lung adenocarcinoma transcript 1; NC, negative control; siRNA, short interfering RNA.
MALAT1 negatively regulates miR-125b-5p expression and positively regulates the expression of NLRC5
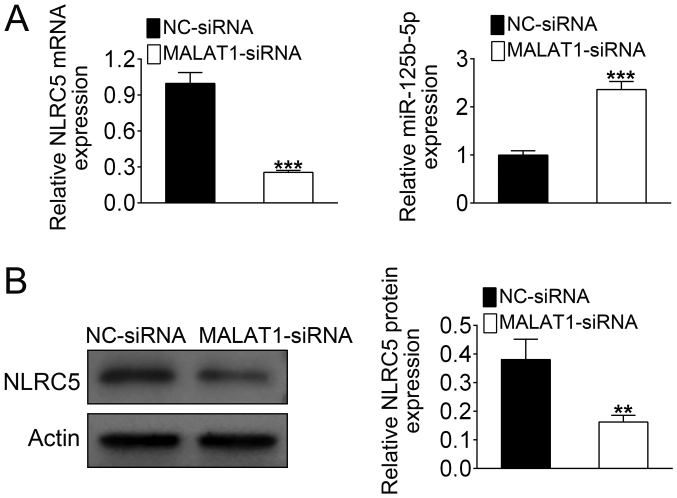
MALAT1 siRNA was transfected into the mouse cardiomyocyte cell line HL-1. The mRNA level of NLRC5 was reduced by more than 60% with MALAT1 knockdown compared with the control group (Fig. 2A). By contrast, MALAT1 siRNA treatment caused a significant increase in the expression of miR-125b-5p (Fig. 2A). Western blotting confirmed that downregulation of MALAT1 significantly reduced the expression of NLRC5 (Fig. 2B). These data indicated that MALAT1 could regulate the expression levels of NLRC5 and miR-125b-5p.
Figure 2.
MALAT1 negatively regulates miR-125b-5p expression and positively regulates the expression of NLRC5. (A) The mRNA expression levels of NLRC5 and miR-125b-5p were analyzed by reverse transcription-quantitative PCR. (B) The protein level of NLRC5 was confirmed by western blotting. The data are presented as the mean ± standard deviation (n=3). **P<0.01, ***P<0.001. MALAT1, metastasis associated lung adenocarcinoma transcript 1; miR, microRNA; NC, negative control; NLRC5, nucleotide binding and oligomerization domain-like receptor C5; siRNA, small interfering RNA.
NLRC5 is a target of miR-125b-5p
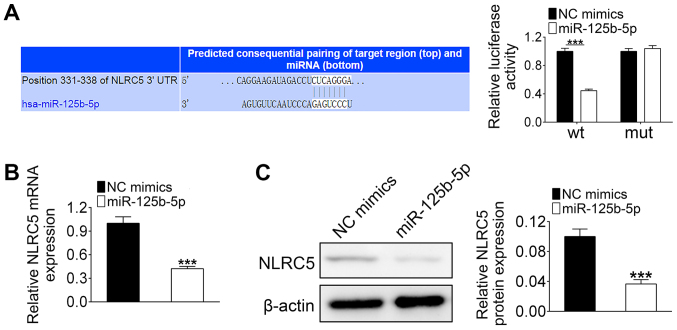
Since MALAT1 directly regulates the expression of miR-125b-5p, it was hypothesized that MALAT1 could regulate NLRC5 expression via miR-125b-5p. The binding sites between miR-125b-5p and NLRC5 are shown in Fig. 3A. Luciferase activity was significantly decreased in HL-1 cells that were co-transfected with miR-125b-5p mimics and wild-type 3′UTR of NLRC5 compared with that co-transfected with the miR-125b-5p mimics and the mutant 3′UTR of NLRC5 (Fig. 3A). Additionally, both mRNA and protein levels of NLRC5 in HL-1 cells were significantly decreased by miR-125b-5p overexpression (Fig. 3B and C). These results indicated that NLRC5 is a target of miR-125b-5p.
Figure 3.
NLRC5 is a target of miR-125b-5p. (A) Complementary sequences between wt or mut NLRC5 3′UTR and miR-125b-5p. (B) HL-1 cells were transfected with miR-125b-5p mimics or NC mimics. (C) The expression of NLRC5 in miR-125b-5p-transfected HL-1 cells was measured by western blotting. The data are presented as the mean ± standard deviation (n=3). ***P<0.001. miR, microRNA; mut, mutant; NC, negative control; NLRC5, nucleotide binding and oligomerization domain-like receptor C5; UTR, untranslated region; wt, wild-type.
MiR-125b-5p exerts an inhibitory effect on H/R-induced apoptosis of a cardiomyocyte cell line by suppressing NLRC5 expression
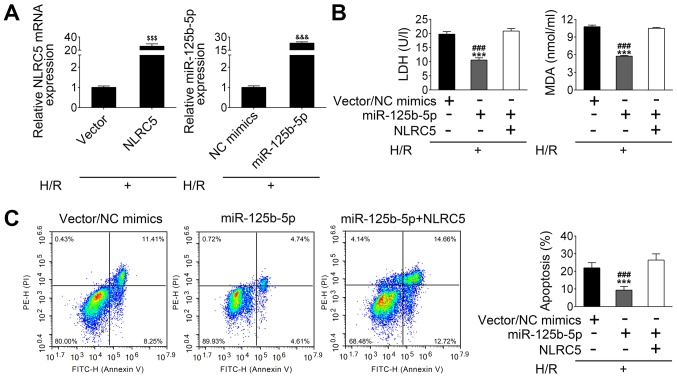
HL-1 cells were treated with H/R to establish an in vitro ischemia/reperfusion I/R model. By using these cells, NLRC5 and miR-125b-5p mRNA expression was increased following the transfection of H/R-treated HL-1 cells with the NLRC5 vector or the miR-125b-5p mimics, respectively (Fig. 4A). As shown in Fig. 4B, the concentration of lactate dehydrogenase (LDH), a marker of cell pyroptosis, was decreased by a miR-125b-5p mimic in H/R-treated HL-1 cells compared with control vector/mimic transfected cells. However, NLRC5 overexpression significantly reversed these results (Fig. 4B). The level of MDA, an index of reactive nitrogen species formation, was also decreased in miR-125b-5p-overexpressing HL-1 cells under H/R conditions. Similarly, the decreased MDA level in miR-125b-5p-overexpressing H/R-treated HL-1 cells was blocked by co-transfection with miR-125b-5p mimics and NLRC5 overexpression plasmid (Fig. 4B). FACs analysis showed that H/R treatment causes HL-1 cells apoptosis including 8.25% at early stage and 11.41% at late stage, and reduction of cell death (~10% reduction), including 4.61% at early stage and 4.74% at late stage, was observed when the miR-125b-5p expression was raised in HL-1 cells, as compared with control. Meanwhile, overexpression of NLRC5 in raised miR-125b-5p expressing HL-1 cells suppressed the cytoprotective effect of miR-125b-5p, as observed by the increased apoptotic cell number (Fig. 4C). These data suggest that miR-125b-5p overexpression may exert an inhibitory effect on H/R-induced apoptosis through negatively regulating NLRC5.
Figure 4.
miR-125b-5p exerts inhibitory effects on hypoxia/reoxygenation-induced apoptosis of cardiomyocytes by suppressing NLRC5 expression. (A) The expression of NLRC5 and miR-125b-5p in hypoxia/reoxygenation-treated HL-1 cells were determined by reverse transcription-quantitative PCR after transfection of the cells with the vectors or mimics. (B) Levels of LDH and MDA in the culture supernatant were measured by assay kits. (C) The fluorescence-activated cell sorting analysis of cardiomyocyte apoptosis following transfection with NLRC5 and miR-125b-5p. The data are presented as the mean ± standard deviation (n=3). $$$P<0.001 vs. Vector; &&&P<0.001 vs. NC mimics; ###P<0.001 vs. miR-125b-5p + NLRC5; ***P<0.001 vs. Vector/NC mimics. LDH, L-lactate dehydrogenase; MDA, malondialdehyde; miR, microRNA; NLRC5, nucleotide binding and oligomerization domain-like receptor C5.
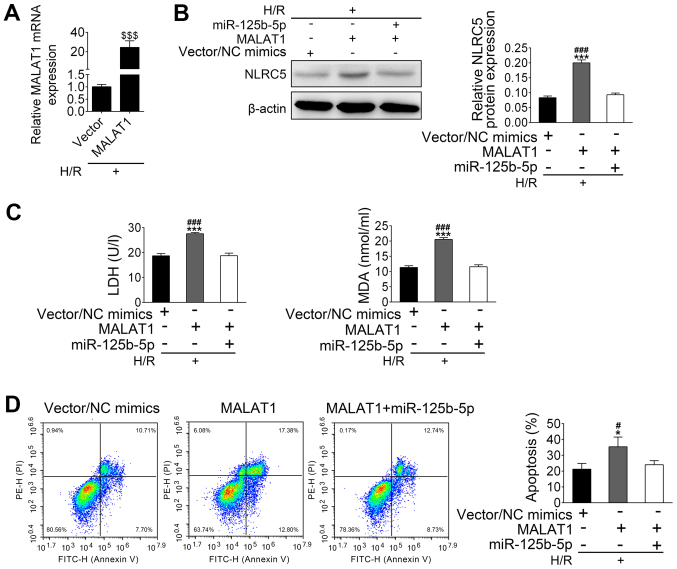
MALAT1 alleviates the protective effect of miR-125b-5p on H/R-induced apoptosis by upregulation of NLRC5
After establishing that transfected HL-1 cells overexpressed MALAT-1 (Fig. 5A), further experiments aimed to investigate whether MALAT1 could inhibit the protective effect of miR-125b-5p on H/R-induced apoptosis. H/R treated HL-1 cells were co-transfected with MALAT1 vector and miR-125b-5p. In H/R-treated HL-1 cells, the expression of NLRC5 was increased by MALAT1 overexpression and suppressed by transfection of miR-125b-5p and MALAT1 (Fig. 5B). Higher levels of LDH and MDA were seen in the cultured supernatants of MALAT1-overexpressing H/R-treated HL-1 cells compared with the control group. This increased LDH release and MDA production induced by MALAT1 overexpression was reduced after overexpression of miR-125b-5p in H/R-treated HL-1 cells (Fig. 5C). MALAT1 overexpression also induced apoptosis in H/R-treated HL-1 cells, which was inhibited by upregulation of miR-125b-5p (Fig. 5D). Taken together, these data suggest that MALAT1 may induce HL-1 apoptosis through downregulation of miR-125b-5p, which leads to upregulation of NLRC5.
Figure 5.
MALAT1 alleviates the protective effect of miR-125b-5p on hypoxia/reoxygenation-induced apoptosis of cardiomyocytes by upregulation of NLRC5. (A) mRNA levels of MALAT-1 in transfected and control cells were confirmed. (B) The expression of NLRC5 in transfected cell was confirmed by western blotting. (C) Levels of LDH and MDA in the culture supernatant were quantified using assay kits. (D) The fluorescence-activated cell sorting analysis of cardiomyocyte apoptosis in transfected HL-1 cells. The data are presented as the mean ± standard deviation (n=3). $$$P<0.001 vs. Vector, #P<0.05 and ###P<0.001 vs. miR-125b-5p + NLRC5, *P<0.05 and ***P<0.001 vs. Vector/NC mimics. LDH, L-lactate dehydrogenase; MDA, malondialdehyde; MALAT1, metastasis associated lung adenocarcinoma transcript 1; miR, microRNA; NLRC5, nucleotide binding and oligomerization domain-like receptor C5.
Discussion
Although lncRNA MALAT1 has been investigated in multiple human cancers (6,26), little is known about its involvement and regu3lation in the pathogenesis of heart disease. A recent study revealed that MALAT1 acts as an inducer of myocardial injury through negatively regulating miR-204 expression to increase cardiomyocyte autophagy and I/R-induced myocardial injury (27). Huang et al (28) showed that MALAT1 mediates cardiac fibrosis and reduces cardiac function in a MI mouse model. Furthermore, MALAT1 has been linked with inflammation in myocardial I/R injury (29). It has also been shown to block the cardioprotective effects of fentanyl by negatively regulating the miR-145/Bcl2 interacting protein 3 pathway (14). By contrast, Zhao et al (30) reported that MAPK1 upregulated the expression of MALAT1 to promote the proliferation of cardiomyocytes. MALAT1 has also been found to promote cardiac stem cell proliferation and migration (29). In the present study, downregulation of MALAT1 significantly reduced infarction area and LVIDs, and enhanced LVFS. However, the reason for the differential expression of MALAT1 in the myocardium is unclear and requires further investigation in the future.
The regulatory mechanisms underlying MALAT1 activity in cardiovascular biology and disease are complex (31). It has been reported that lncRNA can modify mRNA expression by acting as an miRNA sponge and blunting the regulatory effects of relevant miRNAs (32). Oral squamous cell carcinoma development is known to be promoted by MALAT1 via the miR-125b/STAT3 axis (33). Han et al (10) reported that miR-125b suppressed bladder cancer development via inhibition of MALAT1. Both of these publications suggest that miR-125b is a direct target of MALAT1. In the present study the expression of miR-125b-5p was negatively regulated by MALAT1 in cardiomyocyte cell line HL-1. In H/R-treated HL-1 cells, cell apoptosis was significantly reduced by miR-125b-5p overexpression, as were LDH release and MDA production compared with controls. However, these protective effects of miR-125b-5p on H/R-treated HL-1 cells were reversed by MALAT1 overexpression. These data suggested that miR-125b-5p is a potential apoptosis regulator, which was downregulated by MALAT1.
NLR proteins act as pattern-recognition receptors and are critical components of the innate immune response (34). NLRC5, the largest member of the NLR protein family, is widely expressed in numerous tissues (35). NLRC5 has been linked with major histocompatibility complex class I gene expression (36). The involvement of NLRC5 in major histocompatibility complex class I mediated CD8+ T cell activation, proliferation and cytotoxicity is critical for host defense against intracellular bacterial infections (37,38). The biological function of NLRC5 in MI remains poorly understood. In 2017, Zhou et al (24) reported that silencing of NLRC5 ameliorates cardiac fibrosis. Consistent with these data, the present study showed that downregulation of NLRC5 by miR-125b-5p mimic transfection decreased apoptosis and intracellular oxidative stress, evidenced by measurement of LDH and MDA levels as well as FACs analysis.
The expression of NLRC5 was downregulated by treatment of HL-1 cells with MALAT1 siRNA and it was also observed that its expression was upregulated by MALAT1 overexpression. Together, these results indicated that MALAT1 modulated the expression of NLRC5 in AMI. Previous studies demonstrated that MALAT1 directly regulates the expression of miR-125b-5p (10,34). Bioinformatics analysis as well as miR-125b-5p overexpression experiments showed that NLRC5 is the target of miR-125b-5p. Indeed, MALAT1 abrogated the miR-125b-5p-mediated cytoprotection in H/R-induced apoptosis through regulation of NLRC5 expression levels. Ma et al (39) found that knockout of NLRC5 led to myocardial damage in mice on a high fat diet. These different results may be due to differences in experimental model, aim or conditions.
Taken together, to the best of our knowledge, the results of the present study provide the first evidence for a crosstalk between MALAT1, miR-125b and NLRC5 in AMI. It was demonstrated that MALAT1 may function as a negative regulator of miR-125b-5p to abrogate the protective effects of miR-125b-5p on myocardial cells in AMI through modulating NLRC5 expression. The MALAT1/NCLR5/miR-125b-5p axis could provide an effective therapeutic target in AMI treatment.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- AMI
acute myocardial infarction
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- TTC
2,3,5-triphenyl-2H-tetrazolium chloride
- MDA
malondialdehyde
- SD
Sprague Dawley
Funding
This work was supported by the Medical and Health Science and Technology development project in Shandong province (grant no. 2017WS766).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZL and JL performed the experiments and wrote the manuscript. YW, JX and ZW collected and analyzed the experimental data. PW and HS analyzed the experimental data and revised the manuscript. ZS and QL designed the experiments and approved the final version manuscript.
Ethics approval and consent to participate
The Dezhou People's Hospital Ethics Review Committee for Animal Experimentation (experiment number 2017-TX-007) approved this experiment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA, et al. Fourth universal definition of myocardial infarction (2018) Eur Heart J. 2018 doi: 10.1093/eurheartj/ehy655. [DOI] [PubMed] [Google Scholar]
- 2.Nichols M, Townsend N, Scarborough P, Rayner M. Cardiovascular disease in Europe 2014: Epidemiological update. Eur Heart J. 2014;35:2950–2959. doi: 10.1093/eurheartj/ehu299. [DOI] [PubMed] [Google Scholar]
- 3.Yeh RW, Sidney S, Chandra M, Sorel M, Selby JV, Go AS. Population trends in the incidence and outcomes of acute myocardial infarction. N Engl J Med. 2010;362:2155–2165. doi: 10.1056/NEJMoa0908610. [DOI] [PubMed] [Google Scholar]
- 4.Writing Group Members. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després JP, et al. Heart disease and stroke statistics-2016 update: A report from the American Heart Association. Circulation. 2016;133:e38–e360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 5.Talman V, Ruskoaho H. Cardiac fibrosis in myocardial infarction-from repair and remodeling to regeneration. Cell Tissue Res. 2016;365:563–581. doi: 10.1007/s00441-016-2431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 7.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutschner T, Hammerle M, Diederichs S. MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 10.Han Y, Liu Y, Zhang H, Wang T, Diao R, Jiang Z, Gui Y, Cai Z. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long non-coding RNA MALAT1. FEBS Lett. 2013;587:3875–3882. doi: 10.1016/j.febslet.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 13.Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19:1418–1425. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao ZH, Hao W, Meng QT, Du XB, Lei SQ, Xia ZY. Long non-coding RNA MALAT1 functions as a mediator in cardioprotective effects of fentanyl in myocardial ischemia-reperfusion injury. Cell Biol Int. 2017;41:62–70. doi: 10.1002/cbin.10701. [DOI] [PubMed] [Google Scholar]
- 15.Lupfer C, Kanneganti TD. Unsolved mysteries in NLR biology. Front Immunol. 2013;4:285. doi: 10.3389/fimmu.2013.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downs I, Vijayan S, Sidiq T, Kobayashi KS. CITA/NLRC5: A critical transcriptional regulator of MHC class I gene expression. Biofactors. 2016;42:349–357. doi: 10.1002/biof.1285. [DOI] [PubMed] [Google Scholar]
- 17.Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, Jung S, Grötzinger J, Fickenscher H, Schreiber S, Rosenstiel P. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- 18.Neerincx A, Jakobshagen K, Utermöhlen O, Buning H, Steimle V, Kufer TA. The N-terminal domain of NLRC5 confers transcriptional activity for MHC class I and II gene expression. J Immunol. 2014;193:3090–3100. doi: 10.4049/jimmunol.1401065. [DOI] [PubMed] [Google Scholar]
- 19.Yoshihama S, Roszik J, Downs I, Meissner TB, Vijayan S, Chapuy B, Sidiq T, Shipp MA, Lizee GA, Kobayashi KS. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc Natl Acad Sci USA. 2016;113:5999–6004. doi: 10.1073/pnas.1602069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chelbi ST, Guarda G. NLRC5, a promising new entry in tumor immunology. J Immunother Cancer. 2016;4:39. doi: 10.1186/s40425-016-0143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolha L, Ravnik-Glavač M, Glavač D. Long noncoding RNAs as biomarkers in cancer. Dis Markers. 2017;2017:7243968. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsang FH, Au V, Lu WJ, Shek FH, Liu AM, Luk JM, Fan ST, Poon RT, Lee NP. Prognostic marker microRNA-125b inhibits tumorigenic properties of hepatocellular carcinoma cells via suppressing tumorigenic molecule eIF5A2. Dig Dis Sci. 2014;59:2477–2487. doi: 10.1007/s10620-014-3184-5. [DOI] [PubMed] [Google Scholar]
- 23.Bayoumi AS, Park K-m, Wang Y, Teoh JP, Aonuma T, Tang Y, Su H, Weintraub NL, Kim IM. A carvedilol-responsive microRNA, miR-125b-5p protects the heart from acute myocardial infarction by repressing pro-apoptotic bak1 and klf13 in cardiomyocytes. J Mol Cell Cardiol. 2018;114:72–82. doi: 10.1016/j.yjmcc.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H, Yu X, Zhou G. NLRC5 silencing ameliorates cardiac fibrosis by inhibiting the TGFbeta1/Smad3 signaling pathway. Mol Med Rep. 2017;16:3551–3556. doi: 10.3892/mmr.2017.6990. [DOI] [PubMed] [Google Scholar]
- 25.Pasha Z, Haider HKh, Ashraf M. Efficient non-viral reprogramming of myoblasts to stemness with a single small molecule to generate cardiac progenitor cells. PLoS One. 2011;6:e23667. doi: 10.1371/journal.pone.0023667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Yu W, Luo X, Chen J, Deng F. MALAT1/miR-204/LC3-II: A potential regulated axis of autophagy in myocardial ischemia-reperfusion injury. Int J Cardiol. 2019;277:222. doi: 10.1016/j.ijcard.2018.08.075. [DOI] [PubMed] [Google Scholar]
- 28.Huang S, Zhang L, Song J, Wang Z, Huang X, Guo Z, Chen F, Zhao X. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J Cell Physiol. 2019;234:2997–3006. doi: 10.1002/jcp.27117. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Lu G, Chen Z. MALAT1 promoted cell proliferation and migration via MALAT1/miR-155/MEF2A pathway in hypoxia of cardiac stem cells. J Cell Biochem. 2019;120:6384–6394. doi: 10.1002/jcb.27925. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, Li L, Peng L. MAPK1 up-regulates the expression of MALAT1 to promote the proliferation of cardiomyocytes through PI3K/AKT signaling pathway. Int J Clin Exp Pathol. 2015;8:15947–15953. [PMC free article] [PubMed] [Google Scholar]
- 31.Hou J, Zhou C, Long H, Zheng S, Guo T, Wu Q, Wu H, Zhong T, Wang T. Long noncoding RNAs: Novel molecules in cardiovascular biology, disease and regeneration. Exp Mol Pathol. 2016;100:493–501. doi: 10.1016/j.yexmp.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 33.Chang SM, Hu WW. Long non-coding RNA MALAT1 promotes oral squamous cell carcinoma development via microRNA-125b/STAT3 axis. J Cell Physiol. 2018;233:3384–3396. doi: 10.1002/jcp.26185. [DOI] [PubMed] [Google Scholar]
- 34.Motta V, Soares F, Sun T, Philpott DJ. NOD-like receptors: Versatile cytosolic sentinels. Physiol Rev. 2015;95:149–178. doi: 10.1152/physrev.00009.2014. [DOI] [PubMed] [Google Scholar]
- 35.Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185:1681–1691. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi KS, van den Elsen PJ. NLRC5: A key regulator of MHC class I-dependent immune responses. Nat Rev Immunol. 2012;12:813–820. doi: 10.1038/nri3339. [DOI] [PubMed] [Google Scholar]
- 37.Biswas A, Meissner TB, Kawai T, Kobayashi KS. Cutting edge: Impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J Immunol. 2012;189:516–520. doi: 10.4049/jimmunol.1200064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Y, Wang Y, Chen F, Huang Y, Zhu S, Leng Q, Wang H, Shi Y, Qian Y. NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. 2012;22:836–847. doi: 10.1038/cr.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma SR, Xie XW. NLRC5 deficiency promotes myocardial damage induced by high fat diet in mice through activating TLR4/NF-κB. Biomed Pharmacother. 2017;91:755–766. doi: 10.1016/j.biopha.2017.03.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.