Abstract
Purpose
This article reports on FLVCR-AS1 effects on osteosarcoma (OS) growth.
Methods
Tumor tissue and adjacent normal tissue of 48 OS patients were collected. HOS and 143B cells were transfected. Gene expression was examined with qRT-PCR and Western blot. CCK8 assays and cell cloning was performed to measure cell proliferation. Cell cycle and apoptosis were assessed. Luciferase-reporter gene assays and RNA pull-down tests were used to detect targeting relationships between genes.
Results
Prominently higher FLVCR-AS1 expression was found in OS tissue and cells, and was associated with poor prognosis (P<0.05, P<0.01, or P<0.001). Compared with the siCtrl group, 143B and HOS cells of the siFLVCR-AS1 group had significantly lower OD450 values and clone numbers and obviously higher percentages of cells in the G1 phase and apoptosis (P<0.01 or P<0.001). miR381-3p expression was directly inhibited by FLVCR-AS1, and CCND1 expression was directly suppressed by miR381-3p. Compared with the FLVCR-AS1 group, 143B cells of the FLVCR-AS1+ miR381-3p mimic group and FLVCR-AS1+ siCCND1 group showed remarkably lower OD450 values and clone numbers obviously higher apoptosis and percentage of cells in the G1 phase (P<0.05, P<0.01, or P<0.001).
Conclusion
FLVCR-AS1 promoted OS growth by upregulating CCND1 expression via downregulation of miR381-3p.
Keywords: OS, FLVCR-AS1, miR381-3p, CCND1, proliferation, apoptosis
Introduction
Osteosarcoma (OS) is a malignant tumor that originates in mesenchymal tissue. It occurrs in children and adolescents, and has characteristics of high malignancy, strong invasiveness, easy recurrence, and being prone to early distant metastasis.1 Complete radical resection is a common method for the treatment of OS, and postoperative chemotherapy is an important strategy to improve the prognosis of patients. However, approximately 40% of OS patients have been found to have metastasized at the time of initial diagnosis, which poses a serious challenge to patient prognosis and long-term survival.2 Therefore, early diagnosis and treatment of OS is critical to improving patient outcomes.
In the past few decades, medical researchers have made breakthroughs in research on molecular targeted therapy for human tumors. The discovery of effective diagnostic and therapeutic targets provided important clues for improving the therapeutic effects of human malignancies, including OS. Abnormal expression of some important coding genes (such as DDX46 and SOX3) and signaling pathways (including PTEN–Akt and PI3K–Akt) have been found to be involved in the occurrence and progression of OS.3–6 As an important class of noncoding genes, microRNAs have also been discovered to be abnormally expressed in OS. Some microRNAs act as tumor suppressors in OS, such as miR144, miR451, and miR598,7–9 while some microRNAs are oncogenes in OS, such as miR664, miR92a, and miR504.10–12 The discovery of numerous molecular targets has provided important biomarkers for the early diagnosis of OS and important therapeutic targets for the treatment of OS, which have important clinical significance for improving the prognosis of OS patients.
In recent years, lncRNAs, another kind of important ncRNA in the human body, was proved to be abnormally expressed in human diseases (especially malignant tumors) and to play an important regulatory role in the development of tumors. A variety of lncRNAs have also been confirmed to participate in the occurrence and progression of OS. FLVCR-AS1 is an important member of lncRNA family, yet its expression in OS and impact on the growth of OS have not been previously explored. The lncRNA–miRNA–mRNA axis have been found to be one of the important ways for lncRNAs to regulate tumor development. We have noticed that FLVCR-AS1 has a binding site for miR381-3p, and thus whether FLVCR-AS1 regulates OS progression by affecting miR381-3p expression was explored in this study. In addition, CCND1, located on the 11q13 chromosome, has been reported to be involved in regulation of the cell cycle and tumor proliferation.13 Cyclin D1, an important protein involved in tumor development by regulating the cell cycle, is encoded by CCND1 and has also been discovered to be directly regulated by some miRNAs.14,15 This article reports for the first time the effect of FLVCR-AS1 expression on the growth of OS via exploring the miR381-3p–CCND1 axis. This study might provide a novel target for early diagnosis and targeted therapy of OS.
Methods
Patients and tissue
A total of 48 pairs of OS tissue and matched adjacent normal tissue samples were obtained during surgery. All tissue was immediately frozen in liquid nitrogen after being obtained. All these patients, enrolled from May 2012.5 to September 2014, had been diagnosed with OS for the first time and did not have a history of treatment for cancer-related diseases. All patients volunteered to join the study and were followed for 5 years. Written informed consent was obtained from each patient, and this research was approved by the Ethics Committee of the Second Affiliated Hospital of Soochow University. The study was conducted on the basis of the guidelines of Declaration of Helsinki.
Cell lines
Human osteoblasts (hFOB1.19) weres purchased from Shanghai Bioye Biological Technology. OS cells (143B, HOS, MG63, and SaOS2) were provided by the American Type Culture Collection (Manassas, VA, USA). Each cell line was cultured in DMEM with 10% FBS at 37°C, 5% CO2.
Cell transfection
At approximately 80% confluence, 143B and HOS cells were harvested and resuspended in serum-free DMEM at a density of 2×105 cells/mL. Sufficient six-well plates were prepared and 1 mL cell suspension added to each well. Lentivector-mediated FLVCR-AS1 shRNAs (shFLVCR-AS1-1, shFLVCR-AS1-2, and shFLVCR-AS1-3) and corresponding FLVCR-AS1 shRNA–negative control (NC) were provided by RiboBio (Guangzhou, China). With Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA), these shRNAs were transfected into 143B and HOS cells and grouped: siFLVCR-AS1-1 group, siFLVCR-AS1-2 group, siFLVCR-AS1-3 group, and si-control (Ctrl) group. At the same time, pcDNA3.1 vectors containing miR381-3p mimics and miR381-3p mimics as NCs (GeneCopoeia, Guangzhou, China) were also used to transfect 143B and HOS cells (set as miR381-3p-mimic group and NC-mimic group, respectively) using Lipofectamine 2000. In addition, 143B cells were subjected to transfection with FLVCR-AS1 vector (FLVCR-AS1 group) and cotransfection with FLVCR-AS1 vector and CCND1 siRNA (FLVCR-AS1 + siCCND1 group) or FLVCR-AS1 vector and miR381-3p mimics (FLVCR-AS1 + miR381-3p-mimic group). Those 143B cells without any treatment served as the Ctrl group. Cells of each group were maintained at 37°C, 5% CO2 for 6 hours, and cells successfully transfected were harvested for continued culture with DMEM (10% FBS).
qRT-PCR
Trizol regent purchased from Thermo Fisher Scientific was used to extract total RNA in tissue and cells. Synthesis of cDNA was carried out using PrimeScript RT reagent (Takara, Tokyo, Japan). qRT-PCR was conducted with the ABI 7500 fast real-time PCR system (Thermo Fisher Scientific) according to reaction procedures of 95°C for 5 minutes and 39 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. The expression level of genes to be tested was obtained by the 2–ΔΔCt method with U6 as internal Ctrl.
CCK8 assays
A total of 5×103 cells suspended in 200 μL DMEM were seeded in 96-well plates. After 12, 24, 48, and 72 hours of incubation at 37°C, 5% CO2, CCK8 (Solarbio, Beijing, China) with a volume of 10 μL was added to each well and cells continuously incubated for 2 hours at 37°C. At a wavelength of 450 nm, the optical density (OD) of each well was measured using a microplate reader (BioTek).
Cell clones
Cells were seeded in six-well plates with 500 cells in each well. All plates were placed at 37°C, 5% CO2 for 2 weeks and fresh DMEM containing 10% FBS was changed every 2 days. After being fixed with paraformaldehyde for 15 minutes and stained with crystal violet for 15 minutes, cells were placed under a microscope and number of clones of >50 cells counted.
Cell cycle
A total of 106 cells were fixed overnight using 75% ethanol in an ice bath. After being washed with PBS and centrifuged, cells were resuspended with 100 μL RNaseA and incubated for 30 minuts at 37 °C. Next, propidium iodide (PI) 400 μL was added to cells for 30 minutes' incubation at 4°C. Cell cycle analysis was performed using flow cytometry (Becton Dickinson, Heidelberg, Germany).
Apoptosis
Apoptosis was detected with an Annexin V-Fluos kit (Roche). Approximately 106 cells were resuspended in 100 μL solutions containing Annexin V-Fluos and PI. After being incubated for 20 minutes at room temperature, a total of 500 μL incubation buffer was then added to cells. Apoptosis was subsequently analyzed by flow cytometry and cell-apoptosis percentage calculated.
Luciferase-reporter gene assay
Mutant (mut) and wild-type (wt) fragments of FLVCR-AS1 containing miR381-3p, miR513c-5p, and miR877-5p binding sites were designed by GenePharma (Shanghai, China) and cloned into pmirGLO reporter vectors. CCND1-wt and CCND1-mut fragments were also designed and cloned into pmirGLO reporter vectors. These four types of reporter vector were used to transfect 143B cells of the miR381-3p-mimic, miR513c-5p-mimic, miR877-5p-mimic, and corresponding NC-mimic groups, respectively, with Lipofectamine 2000. Cells were incubated at 37°C, 5% CO2 for 48 hours after transfection, and then the relative luciferase activity of these cells was determined with adual-luciferase reporter assay system (Promega, Madison, WI, USA).
RNA pull-down test
RNA pull-down tests were used to determine targeting relationships between miRNAs (miR513c-5p, miR381-3p, and miR877-5p) and FLVCR-AS1. FLVCR-AS1-sense as well as FLVCR-AS1-antisense was transcribed from the pGEM-T-FLVCR-AS1 vector in vitro. Then, 143B cells were cotransfected with biotin-labeled FLVCR-AS1 plasmids and miRNAs (miR513c-5p, miR381-3p, and miR877-5p) mimics. Cells were collected after 48 hours' incubation at 37°C, 5% CO2 and washed with PBS three times. After treatment with formaldehyde (0.37%), cells were further incubated with ice-cold lysis buffer. Agarose beads (Pierce, Rockford, IL, USA) were used to preclean the cell lysate for 1 hour at 4°C. Subsequently, the cell lysate was rotated in streptavidin beads (Biotage) for 3 hours at 4°C. Lastly, the obtained streptavidin beads were washed with elution buffer and heated for 45 minutes at 70°C. Expression of the three miRNAs was determined by qRT-PCR.
Western blot
CCND1 protein expression in cells was detected by Western blot. In short, total protein concentration was determined with a BCA kit (Pierce) after total protein in cells being obtained by RIPA lysis buffer. Equal amounts of each protein sample were subjected to SDS-PAGE. Next, these proteins were transferred onto PVDF membranes for 1 hour's blocking in 5% skimmed milk, followed by being incubated with primary antibody (rabbit antihuman CCND1, 1:1,000; Biocare Medical) for 12 hours at 4°C and secondary antibody (1:2,000; Boaosen Biotechnology, Beijing, China) for 2 hours at room temperature. ECL regent was used to incubate PVDF membranes, and with β-actin as internal Ctrl, the density of target bands was analyzed using ImageJ.
Statistical analysis
All data in this paper were processed with SPSS 19.0 and are presented in the form of means ± SD. Data were obtained from three independent experiments. Differences between two groups were compared by Student’s t-test. Pearson correlation was performed to research the correlation between FLVCR-AS1 and miR381-3p, as well as between miR381-3p and CCND1. Patient 5-year survival was tested by the KaplanMeier method. P<0.05 was considered statistically significant.
Results
FLVCR-AS1 expression was increased in OS
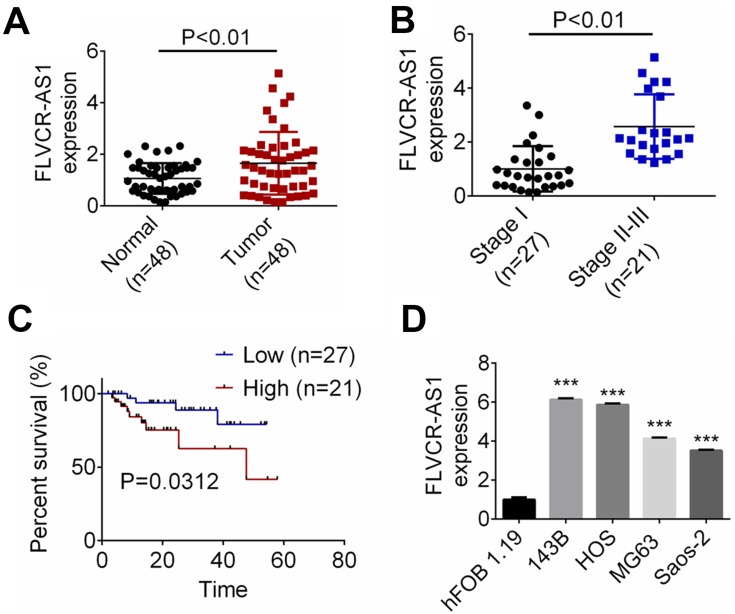
FLVCR-AS1 expression in OS and paracancerous tissue was determined by qRT-PCR, and the results are shown in Figure 1A. Notably, compared with normal tissue, FLVCR-AS1 expression in tumor tissue was prominently increased (P<0.01). The relationship between FLVCR-AS1 expression level and clinicopathological characteristics of OS patients was evaluated, and we noticed that high FLVCR-AS1 expression level was closely related to large tumors, high Enneking stage, and positive distant metastasis (P<0.05, Table 1). For patients with stage II and III, FLVCR-AS1 expression in their OS tissue was markedly higher than patients with stage I (P<0.01, Figure 1B). All patients participating in this study were followed up for 5 years. As a result, compared with patients with low FLVCR-AS1 expression, those with high FLVCR-AS1 expression exhibited much lower 5-year survival (P<0.05, Figure 1C). We also noted that compared with hFOB1.19 cells, significantly increased FLVCR-AS1 expression was found in 143B, HOS, MG63, and SaOS2 cells (P<0.001, Figure 1D).
Figure 1.
FLVCR-AS1 expression was increased in OS.
Notes: (A) Compared with normal tissue, FLVCR-AS1 expression in tumor tissue was prominently increased. (B) For patients with stage II and III, FLVCR-AS1 expression in OS tissue was markedly higher than patients with stage I. (C) Compared with patients with low FLVCR-AS1 expression, those with high FLVCR-AS1 expression exhibited much lower 5-year survival. (D) Compared with hFOB1.19 cells, significantly increased FLVCR-AS1 expression was found in 143B, HOS, MG63, and SaOS2 cells. ***P<0.001.
Table 1.
Relationship of FLVCR-AS1 expression with clinicopathologic characteristics of OS patients
| n | FLVCR-AS1 expression | P-value | ||
|---|---|---|---|---|
| Low (n=27) | High (n=21) | |||
| Sex | ||||
| Male | 24 | 15 | 9 | 0.548 |
| Female | 24 | 12 | 12 | |
| Age, years | ||||
| <18 | 34 | 19 | 15 | 0.426 |
| ≥18 | 14 | 8 | 6 | |
| Size of tumor | ||||
| <5 cm | 25 | 18 | 7 | 0.015 |
| ≥5 cm | 23 | 9 | 14 | |
| Enneking stage | ||||
| Low | 19 | 13 | 6 | 0.034 |
| High | 29 | 14 | 15 | |
| Distant metastasis | ||||
| Yes | 20 | 7 | 13 | 0.011 |
| No | 28 | 20 | 8 | |
Downregulation of FLVCR-AS1 inhibited OS-cell proliferation and promoted OS-cell apoptosis
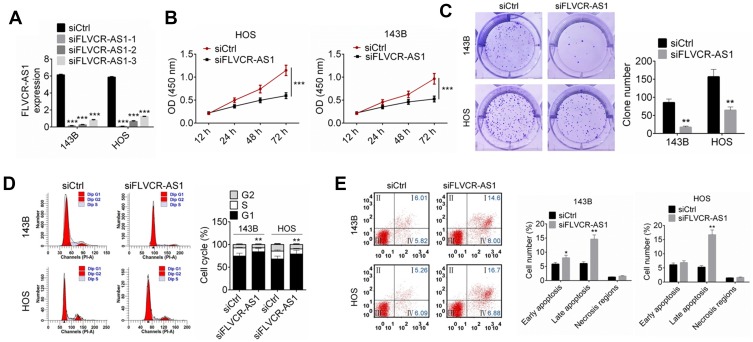
Compared with the siCtrl group, FLVCR-AS1 expression in 143B and HOS cells of the siFLVCR-AS1-1 group, siFLVCR-AS1-2 group, and siFLVCR-AS1-3 group was all effectively silenced (P<0.001, Figure 2A). It should be explained here that cells of the siFLVCR-AS1-1 group had the lowest FLVCR-AS1 expression, after which cells of the siFLVCR-AS1-1 group (renamed siFLVCR-AS1 group in the following) were used as the object in subsequent studies. Test results related to cell proliferation indicated that compared with the siCtrl group, 143B and HOS cells of the siFLVCR-AS1 group showed obviously decreased OD450 values at 72 hours (P<0.001, Figure 2B) and much-declined clone numbers (P<0.01, Figure 2C). Moreover, 143B and HOS cells of the siFLVCR-AS1 group showed obviously higher percentages in the G1 phase and much lower percentages in S and G2 phrases compared to the siCtrl group (P<0.01, Figure 2D). Apoptosis-detection results further indicated that compared with the siCtrl group, prominently elevated apoptosis (including early apoptosis and late apoptosis) percentages in 143B and HOS cells were discovered in the siFLVCR-AS1 group (P<0.05 or P<0.01, Figure 2E).
Figure 2.
Downregulation of FLVCR-AS1 inhibited OS-cell proliferation and promoted OS-cell apoptosis.
Notes: (A) Compared with the siCtrl group, the expression of FLVCR-AS1 in 143B and HOS cells of the siFLVCR-AS1, siFLVCR-AS1-2, and siFLVCR-AS1-3 groups was effectively silenced. (B, C) Compared with the siCtrl group, 143B and HOS cells of the siFLVCR-AS1 group showed obviously decreased OD450 at 72 hours and much-declined clone numbers. (D) HOS and 143B cells of the siFLVCR-AS1 group had obviously higher percentages in the G1 phase and much lower percentages in the S and G2 phases compared to the siCtrl group. (E) Compared with the siCtrl group, prominently elevated apoptotic 143B- and HOS-cell percentages were discovered in the siFLVCR-AS1 group. The lower-left quadrant shows viable cells negative for both AV and PI. The lower-right quadrant shows early apoptotic cells positive for AV and negative for PI. The upper-right quadrant shows late apoptotic cells positive for both AV and PI. The upper-left quadrant shows cells damaged during the procedure. *P<0.05; **P<0.01; ***P<0.001.
miR381-3p expression was unidirectionally inhibited by FLVCR-AS1 through a ceRNA mechanism
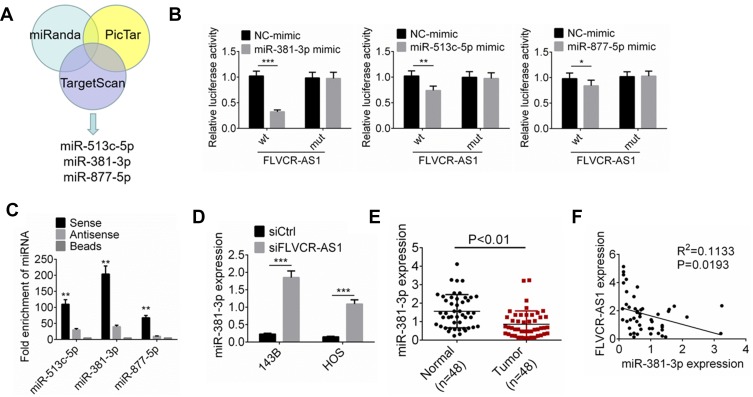
According to online bioinformatic analysis (miRanda, PicTar, and TargetScan), we noticed that three miRNAs (miR513c-5p, miR381-3p, and miR877-5p) had potential binding sites for FLVCR-AS1 (Figure 3A). Results from luciferase-reporter gene assays indicated that after the wt-FLVCR-AS1 sequence had been inserted, the relative luciferase activity of the miR381-3p-mimic (P<0.001), miR513c-5p (P<0.01), and miR877-5p groups (P<0.05) was dramatically lower than their corresponding NC-mimic group (Figure 3B). RNA pull-down tests also indicated that compared with the beads group and antisense groups, the sense group exhibited much higher enrichment of miR513c-5p, miR381-3p, and miR877-5p (P<0.01). More importantly, the enrichment of miR381-3p was obviously higher than miR513c-5p and miR877-5p. Therefore, miR381-3p was identified to interact with FLVCR-AS1 (Figure 3C). In addition, compared with the siCtrl group, the expression of miR381-3p expression in 143B and HOS cells of the siFLVCR-AS1 group was significantly increased (P<0.001, Figure 3D). We also detected miR381-3p expression in OS patients and discovered significantly reduced miR381-3p expression in OS tissue compared to corresponding normal tissue (P<0.01, Figure 3E). A negative correlation was found in expression levels between FLVCR-AS1 and miR381-3p in OS tissue of patients (Figure 3F). All these data confirmed that miR381-3p was directly and unidirectionally inhibited by FLVCR-AS1.
Figure 3.
FLVCR-AS1 downregulated miR381-3p expression.
Notes: (A) According to online bioinformatics analysis (miRanda, PicTar, and TargetScan), miR513c-5p, miR381-3p, and miR877-5p had potential binding sites for FLVCR-AS1. (B) Luciferase-reporter gene assay. (C) RNA pull-down tests indicated that miR381-3p interacted with FLVCR-AS1. (D) Compared with the siCtrl group, miR381-3p expression of 143B and HOS cells of the siFLVCR-AS1 group was significantly increased. (E) Significantly reduced miR381-3p expression was discovered in OS tissue compared to corresponding normal tissue. (F) A negative correlation was found in expression levels between FLVCR-AS1 and miR381-3p. *P<0.05; **P<0.01; ***P<0.001.
Expression of CCND1 was directly suppressed by miR381-3p
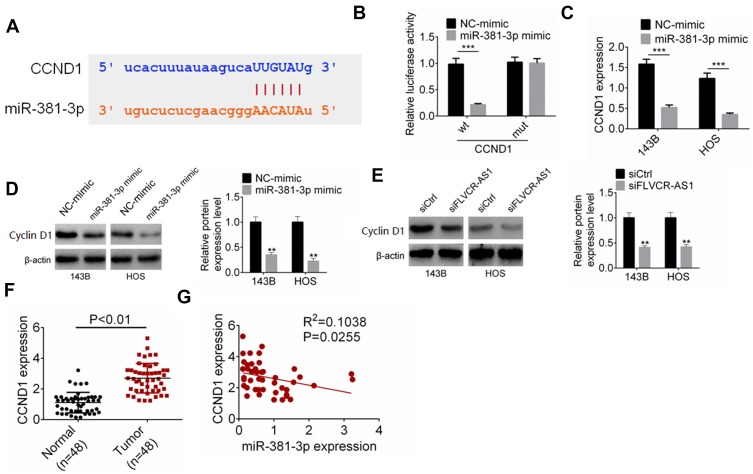
CCND1 is a well-known gene that regulates tumor development by accelerating cell-cycle progression. The protein encoded by CCND1 is cyclin D1, which promotes the cell cycle from the G1 phase to the S phase, thereby promoting tumor-cell proliferation.14,16 In this research, TargetScan and StarBase were employed to predict the target gene of miR381-3p. We noticed that CCND1 possessed binding sites for miR381-3p in the 3ʹUTR (Figure 4A). Based on the data from luciferase-reporter gene assays, CCND1 was a target gene of miR381-3p, whose expression was directly inhibited by miR381-3p (Figure 4B). Compared with the NC-mimic group, 143B and HOS cells of the miR381-3p mimic group had much lower CCND1 mRNA and protein expression (P<0.001 or P<0.01, Figure 4CD). In addition, obviously lower CCND1 protein expression occurred in 143B and HOS cells of the siFLVCR-AS1 group than the siCtrl group (P<0.01, Figure 4E). In OS patients, CCND1 expression in OS tissue was significantly higher than in normal tissue (P<0.01, Figure 4F), and in OS tissue a negative correlation was found in between CCND1 and miR381-3p expression levels (P<0.05, Figure 4G). Therefore, CCND1 was directly inhibited by miR381-3p.
Figure 4.
miR381-3p suppressed the expression of CCND1.
Notes: (A) CCND1 possessed binding sites for miR381-3p. (B) Luciferase reporter gene assay. (C, D) Compared with the NC-mimic group, 143B and HOS cells of the miR381-3p–mimic group had much lower CCND1 mRNA and protein expression. (E) Obviously lower CCND1 protein expression occurred in 143B and HOS cells of the siFLVCR-AS1 group than the siCtrl group. (F) CCND1 expression in OS tissue was significantly higher than in normal tissue. (G) In OS tissue, a negative correlation was found in expression levels between CCND1 and miR381-3p. **P<0.01; ***P<0.001.
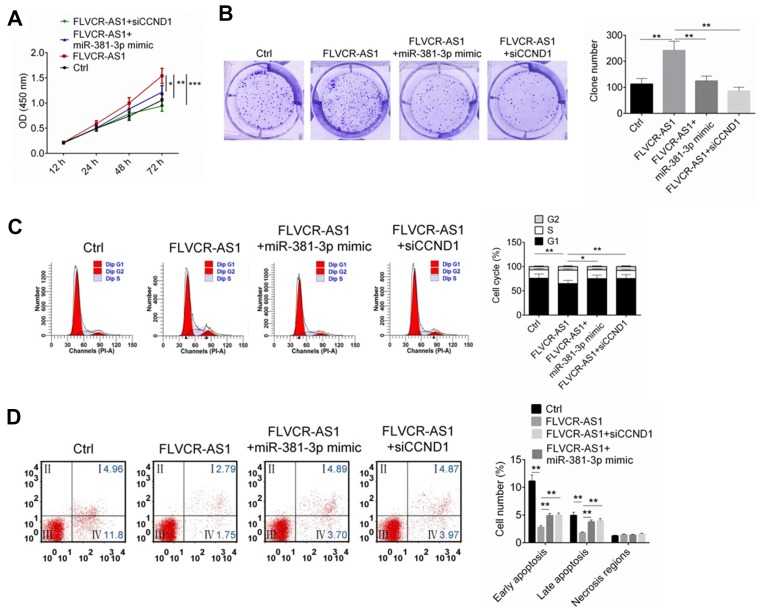
FLVCR-AS1 promoted OS growth by targeting miR381-3p/CCND1
As shown in Figure 5, A and B, compared with the Ctrl group, 143B cells of the FLVCR-AS1 group had much higher OD450 values at 72 hours and obviously higher clone numbers (P<0.01). However, when compared with the FLVCR-AS1 group, prominently lower OD450 values at 72 hours and markedly lower clone numbers were found in 143B cells of the FLVCR-AS1 + miR381-3p-mimic group and FLVCR-AS1 + siCCND1 (P<0.05, P<0.01, or P<0.001). Further testing of the cell cycle indicated that 143B cells of FLVCR-AS1 group had much fewer cells in the G1 phase and more cells in the S and G2 phases than the Ctrl group (P<0.01). However, when compared with the FLVCR-AS1 group, he FLVCR-AS1 + miR381-3p-mimic group and FLVCR-AS1 + siCCND1 showed obviously more 143B cells in the G1 phase and fewer cells in the S and G2 phases (P<0.05 or P<0.01, Figure 5C). In the FLVCR-AS1 group, 143B cells exhibited much lower apoptosis percentages (including early apoptosis and late apoptosis) than the Ctrl group (P<0.01). Meanwhile, compared to the FLVCR-AS1 group, remarkably higher apoptosis percentage(including early apoptosis and late apoptosis) was discovered in 143B cells of the FLVCR-AS1 + miR381-3p-mimic group and FLVCR-AS1 + siCCND1 (P<0.01, Figure 5D).
Figure 5.
FLVCR-AS1 promoted OS development by targeting miR381-3p/CCND1.
Notes: (A, B) Much higher OD450 at 72 hours and obviously higher clone numbers were found in 143B cells of the FLVCR-AS1 group thanthe other three groups. (C) Compared to the other three groups, 143B cells of the FLVCR-AS1 group showed markedly fewer cells in the G1 phase and more cells in the S and G2 phases. (D) Compared to the other three groups, 143B cells of the FLVCR-AS1 group had obviously lower apoptosis. The lower-left quadrant shows viable cells negative for both AV and PI. The lower-right quadrant shows early apoptotic cells positive for AV and negative for PI. The upper-right quadrant shows late apoptotic cells positive for both AV and PI. The upper-left quadrant shows cells damaged during the procedure. *P<0.05; **P<0.01; ***P<0.001.
Discussion
ncRNAs were a class of RNAs that do not possess the ability to encode proteins. There are two main types: small ncRNAs (microRNAs), about 22 nucleotides in length, and lncRNAs >200 bp in length.17 lncRNAs can participate in epigenetic regulation, transcriptional regulation, and posttranscriptional regulation through different mechanisms.18 lncRNAs have been confirmed to participate in the development of numerous tumors, including OS, by regulating the proliferation, differentiation, invasion, and apoptosis of tumor cells. According to the existing literature, GAS5, MEG3, RAB11B-AS1, and WWOX‑AS1 are considered tumor suppressors in OS, and can inhibit OS development by suppressing OS-cell proliferation and epithelial–mesenchymal transition.19–22 On the contrary, some other lncRNAs, such as DANCR, C2dat1, XIST, SATB2-AS1, ITGB2-AS1, MALAT1, HULC, CCAL, CBR3-AS1, ZEB1-AS1, and FGFR3-AS1, can promote OS progression by enhancing OS-cells proliferation, migration, and invasion and weakening OS-cells apoptosi. Therefore, these above lncRNAs are regarded as oncogenes in OS.23–33 To date, no studies have documented the expression of FLVCR-AS1 in human tumors. In this study, we noticed for the first time that FLVCR-AS1 expression level was remarkably increased in OS patients and high FLVCR-AS1 expression closely related to poor outcome and low 5-year survival rate of OS patients. Data also indicated weakened proliferative capacity and enhanced apoptosis ability of OS cells after FLVCR-AS1 had been knocked out. Therefore, we propose FLVCR-AS1 as an oncogene in OS that should be applied in early diagnosis and targeted therapy of OS.
Cumulative studies have demonstrated that expression levels of several microRNAs in various tumor tissue are up- or downregulated to different degrees, including in OS.34,35 miR381-3p is an important member of the microRNAs, and has been shown to be involved in the regulation of a variety of human diseases, such as intestinal ischemia–reperfusion injury and chondrogenesis and cartilage degradation.36,37 However, there are few records related to the study of miR381-3p in human tumors. Yang et al38 revealed low expression of miR381-3p in oral squamous-cell carcinoma. miR381-3p was a promising tumor suppressor in oral squamous-cell carcinoma. Following miR381-3p overexpression, oral squamous cell carcinoma–cell apoptosis was enhanced, whereas these cells' proliferation ability was attenuated. Our data also demonstrate that miR381-3p expression was reduced in OS tissue and that FLVCR-AS1 can directly inhibit the expression of miR381-3p. miR381-3p might be a potent tumor suppressor in OS. The main function of microRNAs is to bind specifically to the 3ʹUTR of the target mRNA and inhibit protein translation or degradation of mRNA to silence gene expression.39 In this research, we discovered that miR381-3p was able to inhibit the expression of CCND1 in OS cells by directly targeting the 3ʹUTR of CCND1.
CCND1 is ubiquitous in eukaryotic cells, and its main function is to regulate the transition of cells from the G1 to S phase.40 The translation product of CCND1 (also known as cyclin D1 protein) is a kind of cell cycle–associated protein whose abnormal expression has been confirmed to interfere with the cell cycle.41 Many researchers have pointed out that CCND1 is abnormally overexpressed in several human tumor cell types, such as gastric carcinoma, endometrial carcinoma, breast cancer, and hepatocellular carcinoma.42–45 In OS, Wu et al46 suggested that of 30 OS tissue samples, 22 had CCND1-positive expression. However, among 30 benign bone tumor–tissue samples, only one exhibited positive CCND1 expression. Results from this article also illustrated that CCND1 was obviously overexpressed in tumor tissue of OS patients and that the upregulation of CCND1 expression might be due to the promotion of FLVCR-AS1 via downregulation of miR381-3p. Knockdown of CCND1 was able to significantly increase the proportion of 143B cells in the G1 phase and decrease the proportion of 143B cells in the S phase. Therefore, knockdown of CCND1 might promote 143B-cell apoptosis through preventing the transition of 143B cells from the G1 phase to the S phase.
In conclusion, we explored FLVCR-AS1 expression in OS and noticed that FLVCR-AS1 might promote OS growth by upregulating CCND1 expression via downregulating miR381-3p. FLVCR-AS1 might be a novel biomarker for early diagnosis of OS and a potential target for targeted therapy of OS. In future, we would be committed to more in-depth research on the impact of FLVCR-AS1 on OS.
Acknowledgment
This study was supported by the Changzhou Municipal Application Foundation Project (grant CJ20159043).
Highlights
FLVCR-AS1 expression was increased in OS patients.
FLVCR-AS1 downregulation inhibited proliferation and promoted apoptosis of OS cells.
FLVCR-AS1 promoted OS growth by targeting miR381-3p/CCND1.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13(8):480. doi: 10.1038/nrendo.2017.16 [DOI] [PubMed] [Google Scholar]
- 2.Chang J, Li Y, Wang X, et al. Polyphyllin I suppresses human osteosarcoma growth by inactivation of Wnt/β-catenin pathway in vitro and in vivo. Sci Rep. 2017;7(1):7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li T, Xiao Y, Huang T. HIF-1α-induced upregulation of lncRNA UCA1 promotes cell growth in osteosarcoma by inactivating the PTEN/AKT signaling pathway. Oncol Rep. 2018;39(3):1072–1080. doi: 10.3892/or.2018.6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu B, Cheng D, Hou L, Zhou S, Ying T, Yang Q. SLC3A2 is upregulated in human osteosarcoma and promotes tumor growth through the PI3K/Akt signaling pathway. Oncol Rep. 2017;37(5):2575–2582. doi: 10.3892/or.2017.5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang F, Zhang D, Li G, Wang X. Knockdown of DDX46 inhibits the invasion and tumorigenesis in osteosarcoma cells. Oncol Res. 2017;25(3):417–425. doi: 10.3727/096504016X14747253292210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Yin J, Tang M, Yu X. Downregulation of SOX3 leads to the inhibition of the proliferation, migration and invasion of osteosarcoma cells. Int J Oncol. 2018. doi: 10.3892/ijo [DOI] [PubMed] [Google Scholar]
- 7.Ren YF, Zhang TH, Zhong S, Zhao YT, Lv YN. miR144 suppresses proliferation and induces apoptosis of osteosarcoma cells via direct regulation of mTOR expression. Oncol Lett. 2018;15(1):1163–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Liu S-Y, He Y-B, et al. MiR-451 suppresses proliferation, migration and promotes apoptosis of the human osteosarcoma by targeting macrophage migration inhibitory factor. Biomed Pharmacother. 2017;87:621–627. doi: 10.1016/j.biopha.2016.12.121 [DOI] [PubMed] [Google Scholar]
- 9.Liu K, Sun X, Zhang Y, Liu L, Yuan Q. MiR-598: A tumor suppressor with biomarker significance in osteosarcoma. Life Sci. 2017;188:141–148. doi: 10.1016/j.lfs.2017.09.003 [DOI] [PubMed] [Google Scholar]
- 10.Bao Y, Chen B, Wu Q, et al. Overexpression of miR-664 is associated with enhanced osteosarcoma cell migration and invasion ability via targeting SOX7. Clin Exp Med. 2017;17(1):51–58. doi: 10.1007/s10238-015-0398-6 [DOI] [PubMed] [Google Scholar]
- 11.Xiao J, Yu W, Hu K, Li M, Chen J, Li Z. miR-92a promotes tumor growth of osteosarcoma by targeting PTEN/AKT signaling pathway. Oncol Rep. 2017;37(4):2513–2521. doi: 10.3892/or.2017.5484 [DOI] [PubMed] [Google Scholar]
- 12.Cai Q, Zeng S, Dai X, Wu J, Ma W. miR-504 promotes tumour growth and metastasis in human osteosarcoma by targeting TP53INP1. Oncol Rep. 2017;38(5):2993. doi: 10.3892/or.2017.5851 [DOI] [PubMed] [Google Scholar]
- 13.Zhang YJ, Chen SY, Chen CJ, Santella RM. Polymorphisms in cyclin D1 gene and hepatocellular carcinoma. Mol Carcinog. 2002;33(2):125–129. [PubMed] [Google Scholar]
- 14.Ramos-Garcã-A P, Gil-Montoya JA, Scully C, et al. An update on the implications of cyclin D1 in oral carcinogenesis. Oral Dis. 2017;23(7):897. doi: 10.1111/odi.12620 [DOI] [PubMed] [Google Scholar]
- 15.Li J, Wei J, Mei Z, et al. Suppressing role of miR-520a-3p in breast cancer through CCND1 and CD44. Am J Transl Res. 2017;9(1):146. [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Li X, Li C, et al. Transcription factor OCT4 promotes cell cycle progression by regulating CCND1 expression in esophageal carcinoma. Cancer Lett. 2014;354(1):77–86. doi: 10.1016/j.canlet.2014.07.049 [DOI] [PubMed] [Google Scholar]
- 17.Zhang R, Hardin H, Chen J, Guo Z, Lloyd RV. Non-coding RNAs in thyroid cancer. Endocr Pathol. 2016;27(1):12–20. doi: 10.1007/s12022-016-9417-8 [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Sun Y, Cai R, Wang G, Shu X, Pang W. Long noncoding RNA: multiple players in gene expression. BMB Rep. 2018;51(6):280–289. doi: 10.5483/bmbrep.2018.51.6.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.v118.12 [DOI] [PubMed] [Google Scholar]
- 20.Zhang SZ, Cai L, Li B. MEG3 long non-coding RNA prevents cell growth and metastasis of osteosarcoma. Bratisl Lek Listy. 2018;118(10):632. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, Liu Z, Yang Y, et al. Long non-coding RNA RAB11B-AS1 prevents osteosarcoma development and progression via its natural antisense transcript RAB11B. Oncotarget. 2018;9(42):26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu G, Ma Z, Tong W, Yang J. LncRNA WWOX‑AS1 inhibits the proliferation, migration and invasion of osteosarcoma cells. Mol Med Rep. 2018;18(1):779–788. doi: 10.3892/mmr.2018.9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009 [DOI] [PubMed] [Google Scholar]
- 24.Jia D, Niu Y, Li D, Liu Z. LncRNA C2dat1 promotes cell proliferation, migration, and invasion by targeting miR-34a-5p in osteosarcoma cells. Oncol Res. 2017;26(5):753–764. doi: 10.3727/096504017X15024946480113 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Lv GY, Miao J, Zhang XL. Long non-coding RNA XIST promotes osteosarcoma progression by targeting ras-related protein RAP2B via miR-320b. Oncol Res. 2017;26(6):837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu SH, Zhu J-W, Xu -H-H, et al. A novel antisense long non-coding RNA SATB2-AS1 overexpresses in osteosarcoma and increases cell proliferation and growth. Mol Cell Biochem. 2017;430(1–2):1–10. doi: 10.1007/s11010-017-2953-9 [DOI] [PubMed] [Google Scholar]
- 27.Dai J, Xu LJ, Han GD, et al. Down-regulation of long non-coding RNA ITGB2- AS1 inhibits osteosarcoma proliferation and metastasis by repressing Wnt/β-catenin signalling and predicts favourable prognosis. Artif Cells Nanomed Biotechnol. 2018;46(sup3):S783–S790. [DOI] [PubMed] [Google Scholar]
- 28.Gao KT, Lian D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20(17):3561–3565. [PubMed] [Google Scholar]
- 29.Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8(3):2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou DK, Yang X-W, Li H, Yang Y, Zhu Z-J, Wu N. Up-regulation of long noncoding RNA CCAL predicts poor patient prognosis and promotes tumor metastasis in osteosarcoma. Int J Biol Markers. 2017;32(1):108–112. doi: 10.5301/jbm.5000240 [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Meng W, Cui H. LncRNA CBR3-AS1 predicts unfavorable prognosis and promotes tumorigenesis in osteosarcoma. Biomed Pharmacother. 2018;102:169. doi: 10.1016/j.biopha.2018.02.081 [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res. 2016;8(10):4095–4105. [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Wang X, Fu C, et al. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol Biol Rep. 2016;43(5):427–436. doi: 10.1007/s11033-016-3975-1 [DOI] [PubMed] [Google Scholar]
- 34.Liu YJ, Li W, Chang F, Liu JN, Lin JX, Chen DX. MicroRNA-505 is downregulated in human osteosarcoma and regulates cell proliferation, migration and invasion. Oncol Rep. 2018;39(2):491–500. doi: 10.3892/or.2017.6142 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Xiao Y, Zhao Q, Du B, Chen H-Y, Zhou D-Z. MicroRNA-187 inhibits growth and metastasis of osteosarcoma by downregulating S100A4. Cancer Invest. 2018;36:1–9. doi: 10.1080/07357907.2017.1415348 [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Yao J, Li Z, et al. miR-381-3p knockdown improves intestinal epithelial proliferation and barrier function after intestinal ischemia/reperfusion injury by targeting nurr1. Cell Death Dis. 2018;9(3):411. doi: 10.1038/s41419-018-0450-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou C, Zhang Z, Yang Z, Wu P, Gu M, Liao W. miR-381-3p participates in chondrogenesis and cartilage degradation by alleviating collagen 2 expression and enhancing mmp13 expression. Osteoarthr Cartilage. 2014;22(4):S320–S321. doi: 10.1016/j.joca.2014.02.593 [DOI] [Google Scholar]
- 38.Yang X, Ruan H, Hu X, Cao A, Song L. miR-381-3p suppresses the proliferation of oral squamous cell carcinoma cells by directly targeting FGFR2. Am J Cancer Res. 2017;7(4):913–922. [PMC free article] [PubMed] [Google Scholar]
- 39.Haneklaus M. Analysis of post-transcriptional gene regulation of nod-like receptors via the 3′UTR. Methods Mol Biol. 2016;1390:197–211. doi: 10.1007/978-1-4939-3335-8_13 [DOI] [PubMed] [Google Scholar]
- 40.Zheng M, Wan L, He X, Qi X, Liu F, Zhang DH. Effect of the CCND1 A870G polymorphism on prostate cancer risk: a meta-analysis of 3,820 cases and 3,825 controls. World J Surg Oncol. 2015;13(1):1–7. doi: 10.1186/1477-7819-13-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fusté NP, Castelblanco E, Felip I, et al. Characterization of cytoplasmic cyclin D1 as a marker of invasiveness in cancer. Oncotarget. 2016;7(19):26979–26991. doi: 10.18632/oncotarget.8876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumari S, Prasad SB, Yadav SS, et al. Cyclin D1 and cyclin E2 are differentially expressed in gastric cancer. Med Oncol. 2016;33(5):40. doi: 10.1007/s12032-016-0754-8 [DOI] [PubMed] [Google Scholar]
- 43.Khabaz MN, Abdelrahman AS, Butt NS, Al-Maghrabi B, Al-Maghrabi J. Cyclin D1 is significantly associated with stage of tumor and predicts poor survival in endometrial carcinoma patients. Ann Diagn Pathol. 2017;30:47–51. doi: 10.1016/j.anndiagpath.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 44.Pestell TG, Jiao X, Kumar M, et al. Stromal cyclin D1 promotes heterotypic immune signaling and breast cancer growth. Oncotarget. 2017;8(47):81754–81775. doi: 10.18632/oncotarget.19953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou JJ, Cheng D, He X-Y, Meng Z, Li W-Z, Chen R-F. Knockdown of Hotair suppresses proliferation and cell cycle progression in hepatocellular carcinoma cell by downregulating CCND1 expression. Mol Med Rep. 2017;16(4):4980. doi: 10.3892/mmr.2017.7162 [DOI] [PubMed] [Google Scholar]
- 46.Wu J, Cui -L-L, Yuan J, Wang Y, Song S. Clinical significance of the phosphorylation of MAPK and protein expression of cyclin D1 in human osteosarcoma tissues. Mol Med Rep. 2017;15(4):2303. doi: 10.3892/mmr.2017.6224 [DOI] [PubMed] [Google Scholar]