Abstract
Expression and clinical value of long non-coding RNA (lncRNA) MALAT1 and lncRNA ANRIL in glaucoma patients were investigated. Altogether 86 glaucoma patients who were diagnosed (study group) and 86 people who underwent physical examinations and were confirmed to be healthy (control group) in the Hospital of Chengdu University of Traditional Chinese Medicine from January 2016 to June 2018 were enrolled. Expression of the serum lncRNA MALAT1, lncRNA ANRIL, pigment epithelium-derived factor (PEDF), homocysteine (Hcy), and inflammatory cytokines [interleukin-12 (IL-12), interleukin-4 (IL-4) and interferon-γ (IFN-γ)] was detected. The clinical significance of lncRNA MALAT1 and lncRNA ANRIL was analyzed. Compared with those in the control group, patients in the study group had significantly lower expression of serum lncRNA MALAT1 and lncRNA ANRIL (P<0.05), significantly lower expression of serum PEDF and IL-12 (P<0.05), and significantly higher expression of serum Hcy and IL-4 (P<0.05), without significant difference in the expression of serum IFN-γ between the two groups (P>0.05). Serum lncRNA MALAT1 and lncRNA ANRIL were positively correlated with PEDF and IL-12 (P<0.05), but negatively correlated with Hcy and IL-4 (P<0.05). The diagnostic value of the combined detection of lncRNA MALAT1 and lncRNA ANRIL was higher than that of lncRNA MALAT1 alone and lncRNA ANRIL alone. The expression of lncRNA MALAT1 and lncRNA ANRIL was significantly related to the pathological staging of the patients (P<0.05), not the sex, age, body mass index (BMI), types, and presence or absence of myopia (P>0.05). lncRNA MALAT1 and lncRNA ANRIL are poorly expressed in the serum of glaucoma patients and related to the patients' conditions. Their combined detection has high diagnostic value for the disease. Therefore, they may be used as new molecular targets for the diagnosis and severity evaluation of glaucoma patients.
Keywords: lncRNA MALAT1, lncRNA ANRIL, glaucoma, expression, clinical value
Introduction
Glaucoma is an eye disease characterized by optic atrophy and visual field defect. Primary glaucoma is divided into open-angle and angle-closure glaucoma. The incidence of glaucoma has been increasing with the development of science and technology and the overuse of eyes in recent years (1,2). The disease has a very covert onset, so its pathogenesis remains unclear. Studies have shown that the onset is related to mechanical and vascular factors (3), so early diagnosis and timely treatment are crucial to prevent glaucoma patients from severe visual impairment (4).
Currently, glaucoma is mainly diagnosed by optic nerve examination and ultrasonic biomicroscopy. However, the two methods are expensive and susceptible to the subjective judgment of operators (5), which is not conducive to the diagnosis and the severity evaluation of the disease. Long non-coding RNA (lncRNA) is a long-chain non-coding RNA and its role in various diseases has been widely valued in recent years (6). lncRNA MALAT1, a typical and multifunctional non-coding RNA, has been reported to promote tumor metastasis by inducing epithelial-mesenchymal transformation (7).
According to a previous study, lncRNA MALAT1 plays a pivotal role in proliferative vitreoretinopathy, and affects apoptosis of retinal ganglion cells in glaucoma rats by regulating PI3K/Akt signaling pathway (8). As an lncRNA that has been widely studied, lncRNA ANRIL has been found to protect human trabecular meshwork cells of glaucoma mice by conditioning miR-7 (9). Although these studies have shown that lncRNA MALAT1 and lncRNA ANRIL play an important role in the development of glaucoma, their clinical value in glaucoma patients has been rarely analyzed.
Therefore, the expression of serum lncRNA MALAT1 and lncRNA ANRIL in glaucoma patients was detected to analyze their clinical significance in the patients, to provide more possibilities for the diagnosis and treatment of the disease.
Patients and methods
General information
Altogether 86 glaucoma patients diagnosed in the Hospital of Chengdu University of Traditional Chinese Medicine from January 2016 to June 2018 were enrolled (the study group), including 49 males and 37 females, with an average age of 51.19±1.33 years. A total of 86 people who underwent physical examinations and were confirmed to be healthy in the hospital during the same period were also enrolled (the control group). Inclusion criteria were as follows: patients diagnosed with glaucoma by optic nerve examination, ultrasound biomicroscopy, and visual field examination. Exclusion criteria were as follows: Patients complicated with other eye diseases; pregnant or lactating patients; patients complicated with other malignant tumor diseases; patients with severe diseases of the immune system; patients with severe hepatic and renal dysfunction. All patients and their families agreed to participate in the study, which was approved by the Hospital Ethics Committee.
Detection of indices
Fasting venous blood (5 ml) was drawn from the patients in the morning of the next day after admission, anticoagulated with heparin, and centrifuged at 3,000 rpm for 5 min. The serum was taken out for the detection of the indices.
qRT-PCR detection of lncRNA MALAT1 and lncRNA ANRIL
TRIzol reagent was used to extract total RNA from the serum, and an ultraviolet spectrophotometer was used to detect its purity and concentration. SYBR-Green Real-time PCR Master mix was used to reverse transcribe the total RNA of lncRNA MALAT1 and lncRNA ANRIL, with the steps carried out in strict accordance with the manufacturer's kit. Then, PCR amplification was carried out. The reaction conditions were: pre-degeneration at 95°C for 10 min, degeneration at 95°C for 15 sec, annealing at 60°C for 60 sec, and finally extension at 72°C for 30 sec, for 40 cycles. The primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. GAPDH was used as an internal reference and 2−ΔΔCt was used to calculate the relative expression (Table I).
Table I.
Related primers.
| Factors | Upstream primers | Downstream primers |
|---|---|---|
| MALAT1 | 5′-CAGTGGGGAACTCTGACTCG-3′ | 5′-GTGCCTGGTGCTCTCTTACC-3′ |
| ANRIL | 5′-TGCTCTATCCGCCAATCAGG-3′ | 5′-GGGCCTCAGTGGCACATACC-3′ |
| GAPDH | 5′-ACAGTCAGCCGCATCTTCTT-3′ | 5′-GACAAGCTTCCCGTTCTCAG-3′ |
Detection of other relevant indices
The expression of serum pigment epithelium-derived factor (PEDF), homocysteine (Hcy), and inflammatory cytokines [interleukin-12 (IL-12), interleukin-4 (IL-4) and interferon-γ (IFN-γ)] was detected. Enzymatic cycling assay was used to determine the expression of serum Hcy. A fully automatic biochemical analyzer was used to analyze the expression of serum PEDF. Enzyme-linked immunosorbent assay (ELISA) was used to detect the expression of serum IL-12, IL-4 and IFN-γ.
Statistical methods
In this study, SPSS 18.0 was used to statistically analyze the experimental data. A Chi-square test was used for count data. Measurement data were expressed as mean ± standard deviation. A t-test was used for comparison between two groups. Pearson was used for correlation analysis. Receiver operating characteristic (ROC) curves were plotted to analyze the diagnostic value of lncRNA MALAT1 alone, lncRNA ANRIL alone, and their combined detection for glaucoma. Multivariate Logistic regression was used for the multivariate analysis of risk factors for the disease. P<0.05 indicates a statistically significant difference.
Results
Comparison of general information
There were no significant differences in sex, age, body mass index (BMI), and other information between the study and control groups (P>0.05) (Table II).
Table II.
General information.
| Factors | Study group (n=86) | Control group (n=86) | t/χ2 value | P-value |
|---|---|---|---|---|
| Sex | 0.024 | 0.878 | ||
| Male | 47 (54.65) | 48 (55.81) | ||
| Female | 39 (45.35) | 38 (44.19) | ||
| Age (years) | 0.023 | 0.879 | ||
| ≤51 | 41 (47.67) | 42 (48.84) | ||
| >51 | 45 (52.33) | 44 (51.16) | ||
| BMI (kg/m2) | 0.094 | 0.759 | ||
| ≤23 | 40 (45.51) | 38 (44.19) | ||
| >23 | 46 (54.49) | 48 (55.81) | ||
| History of smoking | 0.223 | 0.637 | ||
| Yes | 31 (36.05) | 34 (39.53) | ||
| No | 55 (63.95) | 52 (60.47) | ||
| Educational background | 0.097 | 0.755 | ||
| Below junior high school | 33 (38.37) | 35 (40.70) | ||
| Junior high school or above | 53 (61.63) | 51 (59.30) | ||
| Place of residence | 0.216 | 0.642 | ||
| Countryside | 49 (56.98) | 52 (60.47) | ||
| City | 37 (43.02) | 34 (39.53) | ||
| Types | 0.024 | 0.878 | ||
| Open-angle | 37 (43.02) | 38 (44.19) | ||
| Angle-closure | 49 (56.98) | 48 (55.81) | ||
| Creatinine (µmol/l) | 62.77±4.24 | 63.05±4.31 | 0.668 | 0.430 |
| Blood urea nitrogen (mmol/l) | 6.08±1.08 | 6.11±1.10 |
BMI, body mass index.
Comparison of expression of lncRNA MALAT1 and lncRNA ANRIL
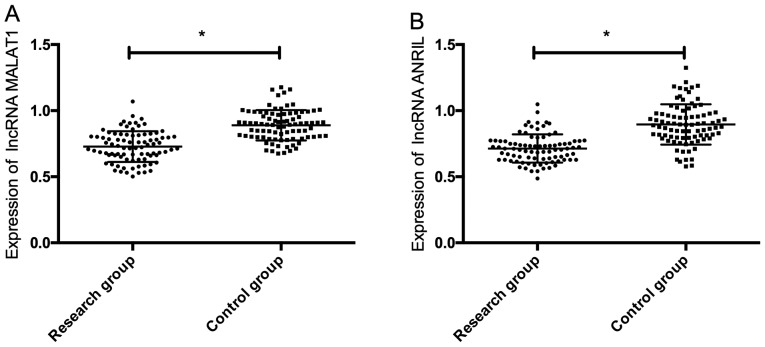
The expression of serum lncRNA MALAT1 and lncRNA ANRIL in the study group was significantly lower than that in the control group (P<0.05) (Fig. 1).
Figure 1.
Comparison of expression of lncRNA MALAT1 and lncRNA ANRIL. (A) Comparison of the expression of lncRNA MALAT1. (B) Comparison of the expression of lncRNA ANRIL. *P<0.05. lncRNA, long non-coding RNA.
Comparison of other related indices
The expression of serum PEDF and IL-12 in the study group was significantly lower than that in the control group, while the expression of serum Hcy and IL-4 was significantly higher than that in the control group (P<0.05), without significant difference in the expression of serum IFN-γ between the two groups (P>0.05) (Table III).
Table III.
Detection of other related indices.
| Indices | Study group (n=86) | Control group (n=86) | t value | P-value |
|---|---|---|---|---|
| PEDF (pg/ml) | 9.57±1.24 | 16.39±2.41 | 23.34 | <0.001 |
| Hcy (mmol/l) | 18.52±2.96 | 5.79±1.36 | 36.24 | <0.001 |
| IL-12 (pg/ml) | 93.24±12.83 | 138.55±18.28 | 18.81 | <0.001 |
| IL-4 (pg/ml) | 258.41±31.58 | 182.49±24.96 | 17.49 | <0.001 |
| IFN-γ (pg/ml) | 110.75±26.93 | 104.54±35.93 | 1.283 | 0.201 |
PEDF, pigment epithelium-derived factor; Hcy, homocysteine; IL, interleukin; IFN-γ, interferon-γ.
Correlation of serum lncRNA MALAT1 and lncRNA ANRIL with PEDF, Hcy, IL-12 and IL-4
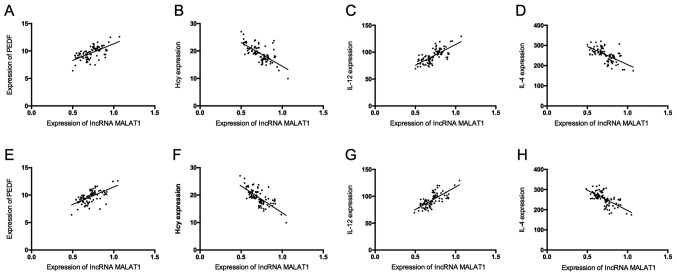
Serum lncRNA MALAT1 and lncRNA ANRIL were positively correlated with PEDF and IL-12 (P<0.05), but negatively correlated with Hcy and IL-4 (P<0.05) (Table IV and Fig. 2).
Table IV.
Correlation analysis.
| lncRNA MALAT1 | lncRNA ANRIL | |||
|---|---|---|---|---|
| Factors | r value | P-value | r value | P-value |
| PEDF | 0.632 | P<0.001 | 0.606 | P<0.001 |
| Hcy | −0.686 | P<0.001 | −0.911 | P<0.001 |
| IL-12 | 0.720 | P<0.001 | 0.717 | P<0.001 |
| IL-4 | −0.633 | P<0.001 | −0.650 | P<0.001 |
lncRNA, long non-coding RNA; PEDF, pigment epithelium-derived factor; Hcy, homocysteine; IL, interleukin.
Figure 2.
Correlation analysis. (A) Correlation analysis of serum MALAT1 with PEDF. (B) Correlation analysis of serum MALAT1 with Hcy. (C) Correlation analysis of serum MALAT1 with IL-12. (D) The correlation analysis of serum MALAT1 with IL-4. (E) The correlation analysis of serum ANRIL with PEDF. (F) Correlation analysis of serum ANRIL with Hcy. (G) Correlation analysis of serum ANRIL with IL-12. (H) Correlation analysis of serum ANRIL with IL-4. lncRNA, long non-coding RNA.
Diagnostic value of lncRNA MALAT1 alone, lncRNA ANRIL alone, and their combined detection
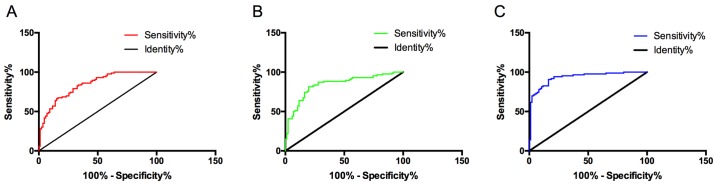
The sensitivity, specificity, and area under the curve (AUC) of lncRNA MALAT1 for diagnosing glaucoma were 83.72, 66.28 and 0.836%, respectively. Those of lncRNA ANRIL were 81.40, 76.74 and 0.842%, respectively. With lncRNA MALAT1 and lncRNA ANRIL considered as independent variables, binary Logistic regression analysis was conducted, and a Logistic regression model: Logit (P)=0.884+1.673 lncRNA MALAT1+-2.960 lncRNA ANRIL was obtained. The sensitivity, specificity, and AUC of the model were 93.02, 79.07 and 0.932, respectively. The diagnostic value of the combined detection was higher than that of lncRNA MALAT1 alone and lncRNA ANRIL alone (Fig. 3).
Figure 3.
Diagnostic value of lncRNA MALAT1 alone, lncRNA ANRIL alone, and their combined detection. (A) The diagnostic value of lncRNA MALAT1 for diagnosing glaucoma. (B) The diagnostic value of lncRNA ANRIL for diagnosing glaucoma. (C) The diagnostic value of the combined detection for diagnosing glaucoma. lncRNA, long non-coding RNA.
Correlation of lncRNA MALAT1 and lncRNA ANRIL with clinicopathological features
The expression of lncRNA MALAT1 and lncRNA ANRIL was significantly related to the pathological staging of the patients (P<0.05), but not to sex, age, BMI, types, and presence or absence of myopia (P>0.05) (Table V).
Table V.
Correlation of lncRNA MALAT1 and lncRNA ANRIL with clinicopathological features.
| Factors | Relative expression of lncRNA MALAT1 | t value | P-value | Relative expression of lncRNA ANRIL | t value | P-value |
|---|---|---|---|---|---|---|
| Sex | 0.674 | 0.464 | 0.644 | |||
| Male (n=47) | 0.62±0.10 | 0.422 | 0.63±0.09 | |||
| Female (n=39) | 0.61±0.12 | 0.64±0.11 | ||||
| Age | 0.886 | 0.879 | 0.382 | |||
| <57 years (n=41) | 0.65±0.09 | 0.388 | 0.67±0.12 | |||
| ≥57 years (n=45) | 0.63±0.12 | 0.65±0.09 | ||||
| BMI | 1.316 | 0.192 | 0.422 | 0.675 | ||
| ≤23 (n=40) | 0.67±0.10 | 0.65±0.12 | ||||
| >23 (n=46) | 0.64±0.11 | 0.66±0.10 | ||||
| Types | 0.130 | 0.124 | ||||
| Angle-closure (n=37) | 0.63±0.09 | 1.530 | 0.61±0.10 | 1.555 | ||
| Open-angle (n=49) | 0.66±0.09 | 0.65±0.13 | ||||
| Myopia | 1.463 | 0.147 | 1.414 | 0.161 | ||
| Yes (n=52) | 0.65±0.08 | 0.64±0.10 | ||||
| No (n=34) | 0.62±0.11 | 0.67±0.09 | ||||
| Stages | <0.001 | |||||
| I and II (n=47) | 0.75±0.07 | 18.28 | <0.001 | 0.74±0.08 | 17.63 | |
| III and IV (n=39) | 0.49±0.06 | 0.48±0.05 |
lncRNA, long non-coding RNA; BMI, body mass index.
Discussion
Glaucoma is an eye disease that can lead to blindness. Intraocular hypertension causes damage to retinal ganglion cells, and then to vision and visual field, eventually resulting in glaucoma (10,11). There are many theories about the pathogenesis of the disease, such as mechanical theory, vascular theory, and nervous activity theory. These theories believe that intraocular hypertension leads to apoptosis of retinal ganglion cells through different pathways (12,13). LncRNA is a widely studied biomolecule. Its expression changes are closely related to the development and progression of many diseases, so it can be used as a molecular target for diagnosis or treatment (14).
lncRNA ANRIL regulates adjacent tumor suppressor gene CDKN2A/CDKN2B through an epigenetic mechanism and then regulates the proliferation and apoptosis of cells (15). According to a previous study exploring the mechanism of action of lncRNA ANRIL in glaucoma, lncRNA ANRIL reduces oxidative stress responses of human trabecular meshwork cells, thus inhibiting the pathogenesis of the disease (15). Previous studies have found that lncRNA MALAT1 plays a regulatory role in angiogenesis of endothelial cells (16), and inhibits apoptosis of neuronal cells by upregulating the expression of Bcl-2 (17). The clinical significance of lncRNA ANRIL and lncRNA MALAT1 in glaucoma was evaluated in our study. The two genes were poorly expressed in the serum of glaucoma patients. A previous study has shown that the downregulation of lncRNA MALAT1 expression leads to the decline of visual function and apoptosis of retinal cells, which also confirms the downregulation of lncRNA MALAT1 expression in patients with eye diseases (18), consistent with our findings.
Subsequently, the expression of serum Hcy, PEDF, IL-12, IL-4 and IFN-γ in the patients was detected to analyze the correlation of lncRNA ANRIL and lncRNA MALAT1 expression with glaucoma. The results showed that the expression of serum PEDF and IL-12 in the study group was significantly lower than that in the control group, while the expression of serum Hcy and IL-4 was significantly higher than that in the control group, without significant difference in the expression of serum IFN-γ between the two groups. Hcy, a sulfur-containing amino acid that is highly expressed in glaucoma patients, can damage vascular endothelial cells of eyes through multiple pathways, and induce optic atrophy (19). PEDF can be produced in many parts of the eyes inhibiting angiogenesis, so the reduction in its expression significantly increases the number of new vessels in the eyes, thus promoting the progression of glaucoma (20). IL-12 inhibits apoptosis of ganglion cells and then protects the optic nerve. IL-4 plays an important role during the process of optic nerve injury, and its high expression represents serious optic nerve injury (21,22). These studies are consistent with our conclusions. The correlation of lncRNA ANRIL and lncRNA MALAT1 with Hcy, PEDF, IL-12 and IL-4 were analyzed. The results showed that serum lncRNA MALAT1 and lncRNA ANRIL were positively correlated with PEDF and IL-12, but negatively correlated with Hcy and IL-4, which further indicates that lncRNA MALAT1 and lncRNA ANRIL may be closely related to the progression of glaucoma.
The diagnostic value of lncRNA MALAT1 alone, lncRNA ANRIL alone, and their combined detection for glaucoma, and the correlation of the two genes with the clinicopathological features were analyzed. The results showed that the AUCs of lncRNA MALAT1 alone and lncRNA ANRIL alone were >0.8, but the AUC of the combined detection was 0.93, which suggests that the latter has higher diagnostic value for glaucoma. According to the analysis of the clinicopathological features, the expression of lncRNA MALAT1 and lncRNA ANRIL was correlated with the pathological staging of glaucoma, which indicates that the low expression of the two genes represents serious glaucoma. This demonstrates that lncRNA MALAT1 and lncRNA ANRIL may be used as important markers for the severity evaluation of the disease. There are currently only few studies on this aspect, so more research is needed to confirm our conclusions.
In conclusion, lncRNA MALAT1 and lncRNA ANRIL are poorly expressed in the serum of glaucoma patients, significantly correlated with serum Hcy, PEDF, IL-12 and IL-4, and related to the pathological staging of the disease. Their combined detection has high diagnostic value for the disease. Therefore, lncRNA MALAT1 and lncRNA ANRIL may be used as new molecular targets for the diagnosis and severity evaluation of glaucoma patients. However, whether the two genes have a common mechanism of action in glaucoma is still unclear. Our conclusions should be further verified due to the few relevant studies and the small sample size in this study.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MZ conceived the study and drafted the manuscript. YZ, MG and HM detected indices. XZ and YL collected subjects and compared their information. FW and HH performed qRT-PCR. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the ethics committee of the Hospital of Chengdu University of Traditional Chinese Medicine. Patients who participated in this research, signed an informed consent and had complete clinical data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bashir H, Sridhar U, Mazumdar S, Tripathy K. Panscleritis masquerading as an attack of primary acute angle closure glaucoma. GMS Ophthalmol Cases. 2019;9:Doc31. doi: 10.3205/oc000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youngblood H, Hauser MA, Liu Y. Update on the genetics of primary open-angle glaucoma. Exp Eye Res. 2019;188:107795. doi: 10.1016/j.exer.2019.107795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinreb RN, Khaw PT. Primary open-angle glaucoma. Lancet. 2004;363:1711–1720. doi: 10.1016/S0140-6736(04)16257-0. [DOI] [PubMed] [Google Scholar]
- 4.Ribeiro A, Veiga F, Santos D, Torres-Labandeira JJ, Concheiro A, Alvarez-Lorenzo C. Hydrophilic acrylic hydrogels with built-in or pendant cyclodextrins for delivery of anti-glaucoma drugs. Carbohydr Polym. 2012;88:977–985. doi: 10.1016/j.carbpol.2012.01.053. [DOI] [Google Scholar]
- 5.Garcia-Medina JJ, Garcia-Medina M, Garrido-Fernandez P, Galvan-Espinosa J, Garcia-Maturana C, Zanon-Moreno V, Pinazo-Duran MD. A two-year follow-up of oral antioxidant supplementation in primary open-angle glaucoma: An open-label, randomized, controlled trial. Acta Ophthalmol. 2015;93:546–554. doi: 10.1111/aos.12629. [DOI] [PubMed] [Google Scholar]
- 6.Islam MN, Moriam S, Umer M, Phan HP, Salomon C, Kline R, Nguyen NT, Shiddiky MJ. Naked-eye and electrochemical detection of isothermally amplified HOTAIR long non-coding RNA. Analyst (Lond) 2018;143:3021–3028. doi: 10.1039/C7AN02109G. [DOI] [PubMed] [Google Scholar]
- 7.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 8.Li HB, You QS, Xu LX, Sun LX, Abdul Majid AS, Xia XB, Ji D. Long non-coding RNA-MALAT1 mediates retinal ganglion cell apoptosis through the PI3K/Akt signaling pathway in rats with glaucoma. Cell Physiol Biochem. 2017;43:2117–2132. doi: 10.1159/000484231. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Sun H, Zhang JM, Wang M, Du XJ, Zhang JL. Long non-coding RNA ANRIL downregulates microRNA-7 to protect human trabecular meshwork cells in an experimental model for glaucoma. Eur Rev Med Pharmacol Sci. 2019;23:3173–3182. doi: 10.26355/eurrev_201904_17675. [DOI] [PubMed] [Google Scholar]
- 10.Francis BA, Varma R, Vigen C, Lai MY, Winarko J, Nguyen B, Azen S, Los Angeles Latino Eye Study Group Population and high-risk group screening for glaucoma: The Los Angeles Latino Eye Study. Invest Ophthalmol Vis Sci. 2011;52:6257–6264. doi: 10.1167/iovs.09-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CA. The Glenn A. Fry Award Lecture. Early losses of visual function in glaucoma. Optom Vis Sci. 1995;72:359–370. doi: 10.1097/00006324-199506000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Ashok A, Kang MH, Wise AS, Pattabiraman P, Johnson WM, Lonigro M, Ravikumar R, Rhee DJ, Singh N. Prion protein modulates endothelial to mesenchyme-like transition in trabecular meshwork cells: Implications for primary open angle glaucoma. Sci Rep. 2019;9:13090. doi: 10.1038/s41598-019-49482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swamy R, Francis BA, Akil H, Yelenskiy A, Francis BA, Chopra V, Huang A. Clinical results of ab interno trabeculotomy using the trabectome in patients with uveitic glaucoma. Clin Exp Ophthalmol. 2019 Sep 10; doi: 10.1111/ceo.13639. (Epub ahead of print). doi: 10.1111/ceo.13639. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Zhang F, Pan Z, Luo H, Liu K, Duan X. LncRNA NR_003923 promotes cell proliferation, migration, fibrosis, and autophagy via the miR-760/miR-215-3p/IL22RA1 axis in human Tenon's capsule fibroblasts. Cell Death Dis. 2019;10:594. doi: 10.1038/s41419-019-1829-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Congrains A, Kamide K, Ohishi M, Rakugi H. ANRIL: Molecular mechanisms and implications in human health. Int J Mol Sci. 2013;14:1278–1292. doi: 10.3390/ijms14011278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou RM, Wang XQ, Yao J, Shen Y, Chen SN, Yang H, Jiang Q, Yan B. Identification and characterization of proliferative retinopathy-related long noncoding RNAs. Biochem Biophys Res Commun. 2015;465:324–330. doi: 10.1016/j.bbrc.2015.07.120. [DOI] [PubMed] [Google Scholar]
- 17.Yao J, Wang XQ, Li YJ, Shan K, Yang H, Wang YN, Yao MD, Liu C, Li XM, Shen Y, et al. Long non-coding RNA MALAT1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol Med. 2016;8:346–362. doi: 10.15252/emmm.201606749. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Nakazawa T, Shimura M, Tomita H, Akiyama H, Yoshioka Y, Kudou H, Tamai M. Intrinsic activation of PI3K/Akt signaling pathway and its neuroprotective effect against retinal injury. Curr Eye Res. 2003;26:55–63. doi: 10.1076/ceyr.26.1.55.14254. [DOI] [PubMed] [Google Scholar]
- 19.Lee YJ, Ke CY, Tien N, Lin PK. Hyperhomocysteinemia causes chorioretinal angiogenesis with placental growth factor upregulation. Sci Rep. 2018;8:15755. doi: 10.1038/s41598-018-34187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Midena E, Bini S, Frizziero L, Pilotto E, Esposito G, Micera A. Aqueous humour concentrations of PEDF and Erythropoietin are not influenced by subthreshold micropulse laser treatment of diabetic macular edema. Biosci Rep. 2019;39:39. doi: 10.1042/BSR20190328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benitez-Del-Castillo J, Cantu-Dibildox J, Sanz-González SM, Zanón-Moreno V, Pinazo-Duran MD. Cytokine expression in tears of patients with glaucoma or dry eye disease: A prospective, observational cohort study. Eur J Ophthalmol. 2019;29:437–443. doi: 10.1177/1120672118795399. [DOI] [PubMed] [Google Scholar]
- 22.Aketa N, Yamaguchi T, Suzuki T, Higa K, Yagi-Yaguchi Y, Satake Y, Tsubota K, Shimazaki J. Iris damage is associated with elevated cytokine levels in aqueous humor. Invest Ophthalmol Vis Sci. 2017;58:BIO42–BIO51. doi: 10.1167/iovs.17-21421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.