Abstract
In a case–control study of patients with Clostridium difficile infection, we found no statistically significant association between the presence of trehalose utilization variants in infecting C. difficile strains and development of severe infection outcome. These results do not support trehalose utilization conferring enhanced virulence in the context of human C. difficile infections.
Keywords: case–control study, Clostridium difficile, comparative genomics, trehalose
Clostridium difficile infection (CDI) is a health care–associated infection that can result in a range of patient outcomes. Of greatest concern is the development of severe disease outcome, defined as intensive care unit admission, intra-abdominal surgery (such as colectomy), and/or death attributable to the infection [1]. Specific patient factors such as age, antibiotic use, and female gender have been associated with severe infection outcome. The genetic background of the infecting C. difficile isolate may also influence clinical outcome. Prior studies have reported an increased risk of severe infection outcome for ribotypes RT027 and RT078 [2, 3]. One recently proposed mechanism for increased virulence of specific C. difficile lineages is the differential capacity to utilize the dietary disaccharide trehalose [4]. These authors observed enhanced trehalose utilization in RT027 strains, identified an additional trehalose operon in RT078, and demonstrated increased virulence of an RT027 strain in a mouse model of infection when physiologically relevant quantities of trehalose were given. This report also noted the coincidence between the introduction of trehalose as a food additive in 2000 and the global emergence of RT027 and RT078 shortly thereafter.
Subsequently, Eyre et al. examined clinical C. difficile isolates for the ability to use trehalose, noting that variants conferring improved trehalose utilization were not confined to successful epidemic lineages. Moreover, Eyre et al. found no evidence of association between a trehalose utilization variant and 30-day all-cause mortality in RT015, a ribotype in which enhanced trehalose utilization is variably present [5]. Here, we set out to more comprehensively evaluate the potential contribution of trehalose utilization to clinical outcomes by quantifying the independent contribution of trehalose utilization variants to infection severity across ribotypes, when controlling for all clinical factors independently associated with risk for severe infection outcome.
METHODS
Study Population
All subjects signed written consent to participate in this study. This study was approved by the University of Michigan Institutional Review Board (Study HUM33286). We considered a cohort of 1144 cases of CDI from hospitalized adults diagnosed with CDI between October 2010 and January 2013 at the University of Michigan Health System [2]. The following predictors of severe outcome were noted in the window 24–48 hours post–CDI diagnosis: age, gender, metastatic cancer, concurrent antibiotic use, systolic blood pressure, creatinine, bilirubin, and white blood cell count. Of the 981 unique patients, 898 had complete clinical information. CDI was classified as severe outcome if any of the following outcomes attributable to CDI occurred within 30 days of diagnosis: admission to an intensive care unit, intra-abdominal surgery, or death [1]. Ribotyping was performed [6], and 137 ribotypes were identified (Supplementary Table 1). Cases were diagnosed as health care–associated CDI if symptoms developed >48 hours after admission [7].
Data Analysis
Summary statistics, matching, modeling, and imputation were conducted in R, version 3.5.0 [8]. The R code is available at: github.com/katiesaund/clinical_cdifficile_trehalose_variants.
Severe Outcome Risk Score Matching
Unique subjects with complete clinical information (n = 898) were assigned a severe outcome risk score based on a logistic regression model with severe CDI outcome as the response variable and the following predictors: age (years), female gender, metastatic cancer, concurrent antibiotic use, systolic blood pressure (mm Hg), creatinine (>1.5 mg/dL), bilirubin (>1.2 mg/dL), and white blood cell count (cells/µL) [7]. Where possible, isolates were sorted into strata, each with exactly 1 case (severe CDI outcome) and at least 1 and up to 4 controls (nonsevere CDI outcome), with control scores within ±0.10 of the case score. The matching process identified 323 CDI case isolates, of which only 247 C. difficile isolates were successfully grown, isolated, and sequenced. Due to growth or sequencing failures, only 235 isolates were from 59 complete strata (strata with at least 1 control and 1 case). Each stratum contained 1 case (n = 59, severe CDI outcome) and up to 4 controls (n = 176, nonsevere CDI outcome; total n = 235). The median number of controls per case (range) was 3 (1–4). There were 61 RT027 isolates (19/59 cases; 46/176 controls), 5 RT078 isolates (0/59 cases; 5/176 controls), and only 1 RT015 (1/59 cases; 0/176 controls) in this matched data set.
Conditional Logistic Regression Model for Matched Samples
Logistic regressions were modeled with severe CDI outcome as the response variable, conditioned on strata, with a trehalose variant as the predictor. Bonferroni-corrected P values were reported.
Genomic Analysis
Genomic DNA extracted with the MoBio PowerMag Microbial DNA Isolation Kit (Qiagen) from C. difficile isolates (n = 247) was prepared for sequencing using the Illumina Nextera DNA Flex Library Preparation Kit. Sequencing was performed on either an Illumina HiSeq 4000 System at the University of Michigan Advanced Genomics Core or on an Illumina MiSeq System at the University of Michigan Microbial Systems Molecular Biology Laboratories. Quality of reads was assessed with FastQC [9]. Adapter sequences and low-quality bases were removed with Trimmomatic [10]. Details on sequenced strains are available in Supplementary Table 1. Sequence data are available under Bioproject PRJNA561087. For variant calling details, see the Supplementary Methods. Pangenome analysis was performed with roary [11], and the insertion putatively conferring enhanced trehalose utilization was inferred based on the presence of 4 genes: treA2, ptsT, treX, and treR2 (Supplementary Data).
RESULTS
Given the limited clinical data regarding the potential contribution of trehalose utilization variants to CDI severity, we set out to test for an association between trehalose utilization across all strains of C. difficile–causing infection while comprehensively controlling for patient factors associated with severe outcome. Patient factors associated with CDI severity in our clinical cohort (n = 1144) were age, female gender, metastatic cancer, concurrent antibiotic use, systolic blood pressure, creatinine, bilirubin, and white blood cell count [2]. To quantify the independent contribution of trehalose utilization variants to severe patient outcomes, we implemented a retrospective, matched case–control study to control for these patient factors. Each C. difficile episode was assigned a severe outcome risk score utilizing variables available around the time of diagnosis, which is the patient’s predicted probability of having severe CDI outcome, based on a logistic regression model of CDI built from the 8 patient factors. Unique patients were grouped into strata based on their risk score.
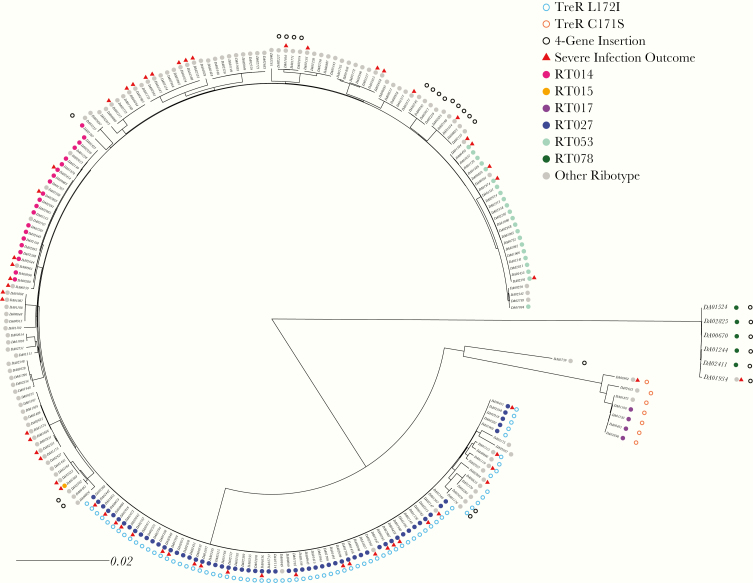
We identified the presence of reported trehalose utilization variants: 2 amino acid substitutions in the transcriptional repressor, treR (TreR C171S and TreR L172I), and a set of 4 accessory genes, called the 4-gene trehalose insertion, that contain an additional phosphotrehalase enzyme and transcriptional regulator [4, 12]. Consistent with previous reports, we found TreR C171S in all 4 RT017 isolates and 3 closely related ribotypes (Figure 1). Similarly, we found TreR L172I in all 61 RT027 isolates and in 7 closely related ribotypes. However, in contrast to Collins et al., who found the 4-gene trehalose insertion only in RT078, we identified the insertion in 25 isolates that were broadly distributed across the phylogeny [4, 5].
Figure 1.
Comparative analysis of trehalose genetic variants in Clostridium difficile. A maximum-likelihood phylogeny was constructed (n = 235) using variants identified in the genome (scale in mutations per site in the whole genome). Trehalose variants are either point mutations in treR (C171S empty orange circle, L172I empty blue circle) or the presence of the 4-gene trehalose insertion (empty black circle). A severe infection outcome is indicated by a red triangle. Ribotype is indicated by the filled circles.
To investigate if trehalose utilization variants were individually associated with severe CDI outcome, we performed unadjusted logistic regressions conditioned on severe outcome risk strata (Supplementary Table 2). The presence of the TreR L172I trehalose utilization variant was associated with an increased, but not statistically significant, odds of severe CDI outcome (OR, 1.60; 95% CI, 0.84–3.05; P = .52). The presence of the 4-gene trehalose insertion was associated with decreased, but not statistically significant, odds of a severe CDI outcome (OR, 0.35; 95% CI, 0.09–1.32; P = .52). None of the other 4 amino acid substitutions observed in TreR were significantly associated with severe CDI outcome (Supplementary Table 2). Due to low prevalence (n = 7), we did not test for an association between the presence of the TreR C171S trehalose utilization variant and severe CDI outcome.
To evaluate the overall effect of the 3 previously described trehalose utilization variants on CDI outcome, we combined the 3 variants into a single predictor of severe CDI outcome and performed a logistic regression conditioned on severe outcome risk strata. When combined, the presence of any of the 3 trehalose utilization variants does not significantly affect the odds of severe CDI outcome (OR, 1.10; 95% CI, 0.61–1.99; P = .86). To evaluate whether these results translated to the original unmatched cohort, we also tested for associations between trehalose utilization variants and severe infection outcome using severe outcome risk score as a covariate rather than a matching criterion. To infer trehalose utilization genotypes in the larger cohort for which genomic data were unavailable, we performed an imputation where the presence of trehalose utilization variants (TreR C171S, TreR L172I, and the 4-gene trehalose insertion) was assumed to have the same distribution across ribotypes as was observed in the matched cohort (Supplementary Methods). We then performed a logistic regression modeling severe outcome with presence of any of the 3 trehalose utilization variants as a predictor and severe outcome risk score as a covariate. When combined, the presence of any of the 3 trehalose utilization variants did not significantly affect the odds of severe CDI outcome (median OR [range], 0.99 [0.87–0.99]; median P [range] = .97 [.64–.99]; n = 586). Note that using this logistic regression framework we were powered to recapitulate the original finding in Rao et al., that RT027 is significantly associated with severe outcome when not conditioning on risk score (OR, 2.22; 95% CI, 1.20–4.04; P = .01).
Discussion
The ability to predict the clinical outcome of C. difficile infection based on patient and pathogen characteristics could help guide therapy for this important nosocomial infection. An enhanced ability to utilize trehalose was shown to be associated with increased virulence in mice, prompting us to evaluate this relationship using a case–control study performed with a large clinical data set. Our controlled analysis, in patients from a diverse set of ribotypes, failed to detect a significant association between any of the previously described trehalose utilization variants and severe CDI outcome. Although it is possible that an effect may be observed in a larger cohort, this is not supported by the odds ratio being close to 1 both in our matched analysis and when imputing to the larger cohort. We also note that while the TreR L172I variant alone has an odds ratio >1, the tight association between this variant and RT027 leave unclear its independent contribution to worse outcomes in patients with RT027 infection. Our results are consistent with Eyre et al.’s study of patients infected with RT015, which observed a lack of association between the 4-gene trehalose insertion and 30-day all-cause mortality. We also confirmed Eyre et al.’s observation that the 4-gene trehalose insertion is not exclusive to RT078, but rather is found throughout the C. difficile phylogeny [5].
The lack of association between trehalose utilization variants and severe CDI outcome in hospitalized patients emphasizes the need to incorporate clinical results earlier into the genetic variant discovery pipeline. Indeed, a more relevant hypothesis-generating process could begin by identifying genetic loci of interest first through comparative genomic analysis of clinical isolates, followed by validation in lab strains with molecular and animal studies. A critical component of this discovery work is to control for both patient factors and strain background, as both may confound analysis, which we did above in separate analyses using 2 distinct modeling strategies. Given the association between ribotype, recent acquisition, and CDI outcome, controlling for patient factors is critical for identifying the genetic variants associated with severe CDI outcome [13].
Although these clinical data cannot rule out a potential role of trehalose utilization variants in the success of hypervirulent C. difficile lineages, they do emphasize the difference between human infection and murine models. More broadly, generating hypotheses in controlled laboratory systems and having them generalize to genetically and clinically heterogeneous human populations will limit insights to only the most penetrant phenotypes. With increasing availability of electronic health record and microbial genomic data, it is now becoming feasible to flip the script and generate hypotheses through analysis of human data, which can subsequently be tested in appropriate animal and in vitro systems with a priori knowledge of relevance to human disease.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Ali Pirani for bioinformatics support. We thank Arianna Miles-Jay for her helpful suggestions.
Financial support. This work was supported by the National Institutes of Health (1U01Al124255). K.S. was supported by the National Institutes of Health T32GM007544.
Potential conflicts of interest. K.S. reports grants from National Institutes of Health during the conduct of the study; K.R. reports personal fees from Bio-K+, Inc., outside the submitted work; V.B.Y. reports grants from National Institutes of Health, during the conduct of the study; non-financial support from Vedanta Biosciences, non-financial support from Bio-K+ International, non-financial support from Pantheryx, outside the submitted work; E.S.S. reports grants from National Institutes of Health, during the conduct of the study.
References
- 1. McDonald LC, Coignard B, Dubberke E, Song X, Horan T, Kutty PK. Recommendations for surveillance of Clostridium difficile–associated disease. Infect Control Hosp Epidemiol 2007; 28(2):140–145. [DOI] [PubMed] [Google Scholar]
- 2. Rao K, Micic D, Natarajan M, et al. Clostridium difficile ribotype 027: relationship to age, detectability of toxins A or B in stool with rapid testing, severe infection, and mortality. Clin Infect Dis 2015; 61:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walker AS, Eyre DW, Wyllie DH, et al. ; Infections in Oxfordshire Research Database Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis 2013; 56:1589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collins J, Robinson C, Danhof H, et al. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 2018; 553:291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eyre DW, Didelot X, Buckley AM, et al. Clostridium difficile trehalose metabolism variants are common and not associated with adverse patient outcomes when variably present in the same lineage. EBioMedicine 2019; 43:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martinson JNV, Broadaway S, Lohman E, et al. Evaluation of portability and cost of a fluorescent PCR ribotyping protocol for Clostridium difficile epidemiology. J Clin Microbiol 2015; 53:1192–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rao K, Micic D, Natarajan M, et al. Clostridium difficile ribotype 027: relationship to age, detectability of toxins A or B in stool with rapid testing, severe infection, and mortality. Clin Infect Dis 2015; 61:233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. R Core Team. R: a language and environment for statistical computing. 2018; Available at: https://www.r-project.org/. Accessed 13 September 2019. [Google Scholar]
- 9. Si A. FastQC: a quality control tool for high throughput sequence data. 2010. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 13 September 2019. [Google Scholar]
- 10. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015; 31:3691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins J, Danhof H, Britton RA. The role of trehalose in the global spread of epidemic Clostridium difficile. Gut Microbes 2019; 10:204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin JSH, Eyre DW, Fawley WN, et al. Patient and strain characteristics associated with Clostridium difficile transmission and adverse outcomes. Clin Infect Dis 2018; 67:1379–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.