Abstract
The role of circulating tumor cells (CTCs), tumor microenvironment (TME), and the immune system in the formation of metastasis is evident, yet the details of their interactions remain unknown. This study aimed at exploring the immunotranscriptome of primary tumors associated with the status of CTCs in breast cancer (BCa) patients. The expression of 730 immune-related genes in formalin-fixed paraffin-embedded samples was analyzed using the multigenomic NanoString technology and correlated with the presence and the phenotype of CTCs. Upregulation of 37 genes and downregulation of 1 gene were observed in patients characterized by a mesenchymal phenotype of CTCs when compared to patients with epithelial CTCs. The upregulated genes were involved in NF-kappa B signaling and in the production of type I interferons. The clinical significance of the differentially expressed genes was evaluated using The Cancer Genome Atlas (TCGA) data of a breast invasive carcinoma (BRCA) cohort. Five of the upregulated genes—PSMD7, C2, IFNAR1, CD84, and CYLD—were independent prognostic factors in terms of overall and disease-free survival. To conclude, our data identify a group of genes that are upregulated in BCa patients with mesenchymal CTCs and reveal their prognostic potential, thus indicating that they merit further investigation.
Keywords: breast cancer, circulating tumor cells, epithelial–mesenchymal transition, NF-kappa B signaling, type I interferons, tumor microenvironment, immune-related transcriptome
1. Introduction
Distant metastases account for most of cancer-related deaths. Yet, fundamental questions regarding mechanisms that promote or inhibit the formation of metastasis still remain unanswered. It is evident that in breast cancer (BCa), tumor cell dissemination occurs already at early stages of the disease [1,2], with circulating tumor cells (CTCs) being direct initiators of metastasis [3]. With current analytic methods, CTCs are detectable in the peripheral blood, and their presence is considered an adverse prognostic factor in numerous solid tumors, including BCa [4,5,6,7,8,9,10,11]. Tumor dissemination is known to be facilitated by the process of epithelial–mesenchymal transition (EMT) [12]. CTCs are shown to possess phenotypic plasticity that relates to their ability to display various EMT states in the circulation [13,14,15], with either the mesenchymal [16] or the intermediate [17] phenotype associated with increased tumor-initiating ability. The presence of mesenchymal CTCs is associated with disease progression and worse prognosis for metastatic BCa [18,19,20] and even operable breast cancer patients [21,22,23]. In fact, in metastatic BCa patients, the expression of mesenchymal markers is higher than in early-stage BCa patients [24,25] and correlates with lymph node involvement [26], suggesting that the EMT phenotype is directly related to the metastatic potential of CTCs.
The ability to evade the immune system is one of the hallmarks of cancer [27,28]. Tumor cells use multiple strategies to escape immune surveillance, mainly avoiding immune recognition and instigating an immunosuppressive microenvironment [29]. The interactions between cancer cells and their microenvironment (tumor microenvironment, TME) are involved in tumor dissemination. TME is assumed to modulate the capability of CTCs to evade the innate immune response and CTCs survival [30,31]. We showed that absence of ALDH1-positive stromal cells correlates with the presence of disseminated tumor cells (DTCs) in the bone marrow of BCa patients [32]. Nevertheless, the precise profile of immune-related factors influencing tumor dissemination, in particular the presence and phenotype of CTCs, is scarcely known. Therefore, the current study aimed at exploring the association between the immunotranscriptome of primary breast tumors and the status of CTCs. We hypothesized that the presence and the EMT phenotype of CTCs are connected with a specific immune-related gene signature of primary tumors. To verify this hypothesis, we evaluated the expression of 730 immune-related genes in primary tumors of BCa patients with well-described molecular and clinicopathological features, including CTCs presence and phenotype.
2. Results
2.1. Expression of Immune-Related Genes within Primary Tumours Correlated with the Phenotype of CTCs
To investigate the immune transcriptome associated with each phenotype of CTCs, we applied NanoString multigene expression analysis to samples of primary breast tumors. We observed that the mesenchymal phenotype of CTCs (n = 9) was associated with the upregulation of 37 genes and the downregulation of 1 gene in primary tumors (p-value ≤ 0.05, false discovery rate (FDR) ≤ 0.2) when compared to the epithelial phenotype of CTCs (n = 14) (Table 1, Figure S1; all results in Table S1). Due to the limited number of patients included in our study, we employed the conservative FDR method of multiple testing correction.
Table 1.
Genes up- and downregulated in primary tumors of patients with mesenchymal circulating tumor cells (CTCs) (compared to patients with epithelial CTCs).
| Gene Symbol | Gene Name | FC | p-Value | FDR |
|---|---|---|---|---|
| LAIR2 | Leukocyte-associated immunoglobulin-like receptor 2 | 3.19 | 0.007 | 0.144 |
| FADD | Fas-associated via death domain | 3.14 | 0.003 | 0.082 |
| TLR7 | Toll-like receptor 7 | 2.86 | 0.011 | 0.172 |
| CCRL2 | C–C motif chemokine receptor-like 2 | 2.80 | 0.003 | 0.087 |
| PBK | PDZ-binding kinase | 2.26 | 0.009 | 0.161 |
| CD3EAP | CD3e molecule-associated protein | 2.25 | 0.001 | 0.063 |
| CD84 | CD84 molecule | 2.22 | 0.003 | 0.087 |
| TNFSF13 | TNF superfamily member 13 | 2.21 | 0.007 | 0.144 |
| BIRC5 | Baculoviral IAP repeat-containing 5 | 2.08 | 0.002 | 0.082 |
| KLRC1 | Killer cell lectin-like receptor C1 | 1.98 | 0.001 | 0.063 |
| CD63 | CD63 molecule | 1.93 | 0.003 | 0.082 |
| TNFRSF11A | TNF receptor superfamily member 11a | 1.91 | 0.013 | 0.199 |
| C2 | Complement C2 | 1.85 | 0.001 | 0.069 |
| IL1RAP | Interleukin 1 receptor accessory protein | 1.79 | 0.011 | 0.172 |
| TAPBP | TAP-binding protein | 1.78 | 0.003 | 0.082 |
| NUP107 | nucleoporin 107 | 1.78 | 0.002 | 0.082 |
| PSMD7 | Proteasome 26S subunit, non-ATPase 7 | 1.72 | <0.001 | 0.063 |
| TICAM1 | Toll-like receptor adaptor molecule 1 | 1.67 | 0.013 | 0.199 |
| ICAM3 | Intercellular adhesion molecule 3 | 1.63 | 0.009 | 0.161 |
| IRF3 | Interferon regulatory factor 3 | 1.59 | <0.001 | 0.048 |
| BCL10 | BCL10 immune signaling adaptor | 1.56 | 0.001 | 0.069 |
| IKBKE | Inhibitor of nuclear factor kappa B kinase subunit epsilon | 1.55 | 0.011 | 0.172 |
| ELK1 | ETS transcription factor ELK1 | 1.55 | 0.001 | 0.069 |
| TRAF6 | TNF receptor-associated factor 6 | 1.53 | 0.003 | 0.087 |
| RELA | RELA proto-oncogene, NF-kB subunit | 1.52 | 0.003 | 0.082 |
| IKBKG | Inhibitor of nuclear factor kappa B kinase regulatory subunit gamma | 1.49 | 0.003 | 0.087 |
| TBK1 | TANK-binding kinase 1 | 1.46 | 0.001 | 0.063 |
| STAT6 | Signal transducer and activator of transcription 6 | 1.40 | 0.004 | 0.106 |
| ATG10 | Autophagy-related 10 | 1.40 | 0.007 | 0.144 |
| CD74 | CD74 molecule | 1.38 | 0.009 | 0.161 |
| ICOSLG | Inducible T cell costimulator ligand | 1.38 | 0.002 | 0.082 |
| FCGR2A | Fc fragment of IgG receptor IIa | 1.37 | 0.007 | 0.144 |
| PSMB10 | Proteasome 20S subunit beta 10 | 1.36 | 0.011 | 0.172 |
| CYLD | CYLD lysine 63 deubiquitinase | 1.36 | 0.011 | 0.172 |
| IFNAR1 | Interferon alpha and beta receptor subunit 1 | 1.35 | 0.001 | 0.069 |
| CCND3 | Cyclin D3 | 1.30 | 0.002 | 0.082 |
| MAP2K1 | Mitogen-activated protein kinase kinase 1 | 1.25 | <0.001 | 0.027 |
| MERTK | MER proto-oncogene, tyrosine kinase | 0.68 | 0.003 | 0.082 |
Fold change (FC) based on median normalized counts of the probe in each group; differences in median normalized counts between groups analyzed with Mann–Whitney U test (p-value) with Benjamini–Hochberg correction for multiple comparisons (false discovery rate, FDR); only statistically significant results are presented; gene names according to HUGO Gene Nomenclature.
Our aim was also to explore the association between the expression of immune-related genes within the primary tumors and the overall presence of CTCs. We found no statistically significant differences between the primary tumors’ immunotranscriptomes in relation to patients’ CTC status (positive vs. negative).
2.2. The Mesenchymal Phenotype of CTCs Is Associated with the Upregulation of Genes Involved in NF-kappa B Signalling and Type I Interferons Production in Matched Primary Tumours
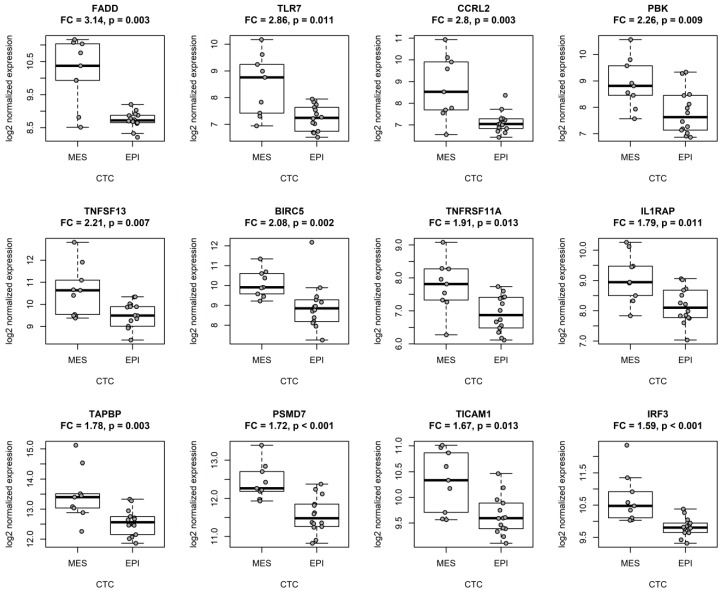
Here, we observed that multiple genes differentially expressed in patients with epithelial and mesenchymal CTC phenotypes (Table 1) play a role in the NF-kappa B signaling pathway. Consequently, we decided to interrogate this link more carefully. We applied the Functional Annotation Tool by DAVID 6.8 [33,34] to associate the selected genes with specific functional annotations. Genes upregulated in tumors with mesenchymal CTCs were generally involved in the activation and regulation of immune response (Table S2). Interestingly, 15 out of 37 upregulated genes (FADD, TLR7, TNFRSF11A, IL1RAP, PSMD7, TICAM1, IRF3, BCL10, IKBKE, TRAF6, RELA, IKBKG, TBK1, PSMB10, and CYLD) were implicated in the regulation of NF-kappa B signaling and activity (GO:0043122, GO:0051092, and GO:0038061). A literature search provided a number of links between other 11 differentially expressed genes (CCRL2, PBK, TNFSF13, BIRC5, TAPBP, ELK1, STAT6, ATG10, IFNAR1, CCND3, and MAP2K1) and the NF-kappa B pathway (top 12 genes depicted in Figure 1).
Figure 1.
Genes implicated in NF-kappa B signaling were upregulated in primary tumors of breast cancer patients with mesenchymal CTCs (MES, n = 9) when compared to patients with epithelial CTCs (EPI, n = 14); the top 12 upregulated genes are presented. Gene expression depicted as number of counts of each probe and normalized to the four most stable reference genes (ABCF1, EDC3, HDAC3, and CNOT4); FC calculated on the basis of the median normalized counts of the probe in each group; differences in median normalized counts between groups analyzed with the Mann–Whitney U test (p); the bars correspond to the interquartile range (IQR), the whiskers cover 1.5 IQR from the median.
Analysis of Gene Ontology also revealed that nine of the upregulated genes regulate the production of type I interferons (GO:0032479; TLR7, TICAM1, IRF3, IKBKE, RELA, TBK1, STAT6, CYLD, and IFNAR1), with a particular role in the stimulation of interferon beta (GO:0032728; TLR7, TICAM1, IRF3, TBK1, and IFNAR1).
2.3. Immune-Related Genes Connected with the Mesenchymal Phenotype of CTCs Are Potent Negative Prognostic Factors in Breast Cancer
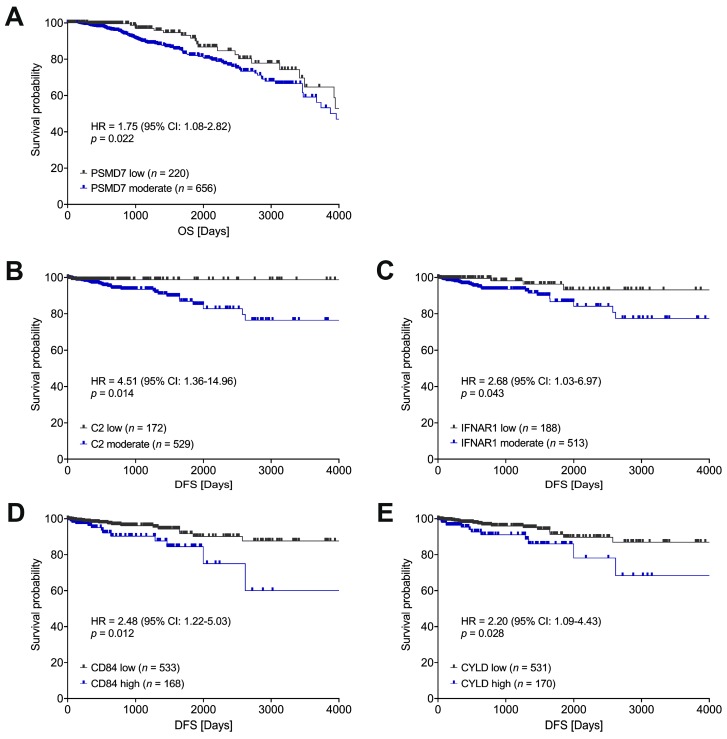
We have previously demonstrated that the mesenchymal phenotype of CTCs correlates with a poor prognosis in breast cancer patients [21]. Consequently, we decided to evaluate the prognostic significance of the immune-related genes that we found significantly up- or downregulated in primary tumors of BCa patients with mesenchymal CTCs. To this end, we turned to The Cancer Genome Atlas (TCGA) database and analyzed the available RNA-seq data on gene expression in a breast invasive carcinoma (BRCA) cohort (n = 877) [35,36]. Five out of 38 genes (PSMD7, C2, IFNAR1, CD84, and CYLD) associated with the mesenchymal phenotype of CTCs demonstrated a negative prognostic impact in the TCGA cohort. Moderate (higher than the first quartile, Q1 in Table S3) expression of PSMD7 correlated with shorter overall survival (OS) in comparison to low expression of PSMD7 in primary tumors (HR = 1.75, 95% CI: 1.08–2.82, p = 0.022; Figure 2A). A higher risk of recurrence was observed for tumors with moderate (higher than the first quartile, Q1 in Table S3) expression of C2 (HR = 4.51, 95% CI: 1.36–14.96, p = 0.014; Figure 2B) and IFNAR1 (HR = 2.68, 95% CI: 1.03–6.97, p = 0.043; Figure 2C) in comparison to tumors with low expression of these genes; high (higher than the third quartile, Q3 in Table S3) expression of CD84 (HR = 2.48, 95% CI: 1.22–5.03, p = 0.012; Figure 2D) and CYLD (HR = 2.20, 95% CI: 1.09–4.43, p = 0.028; Figure 2E) was also linked to shorter disease-free survival (DFS) in comparison to low expression of these genes in the primary tumors. Multivariate analysis including the clinical stage confirmed the significance of the aforementioned genes as independent prognostic factors (Table S3).
Figure 2.
Genes associated with the mesenchymal phenotype of CTCs had a negative impact on survival (A) and recurrence (B–E) in patients in a BRCA cohort of The Cancer Genome Atlas (TCGA). Low/moderate status of gene expression relative to the first quartile (Q1); low/high status of gene expression relative to the third quartile (Q3); hazard ratios (HR) with 95% confidence intervals (95% CI) computed using Cox proportional hazards regression; OS: overall survival, DFS: disease-free survival.
3. Discussion
The knowledge about immune signatures related to tumor dissemination is still limited. Our current study aimed to identify the immunotranscriptomic profiles of primary tumors associated with the presence of CTCs and the CTCs phenotype in non-metastatic BCa patients.
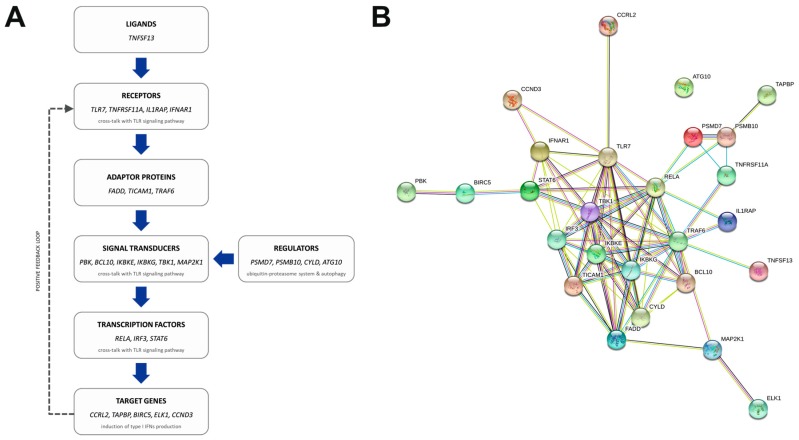
Our data revealed that 38 genes were differentially expressed in the primary tumors of patients with mesenchymal CTCs when compared to patients with epithelial CTCs. Intriguingly, we did not observe any statistically significant difference between primary tumor transcriptomes when comparing patients according to the presence or absence of CTCs in the circulation. The compared groups (CTC-positive and CTC-negative) were not biased in terms of any clinicopathological parameter (Table S4); hence, we believe that the observed difference in the activation of dissemination at BCa tumors may be cell context-dependent and definitely requires a more thorough analysis. On the other hand, our results demonstrate a substantial connection between the mesenchymal phenotype of CTCs and the NF-kappa B pathway. According to the NanoString gene expression assay and a literature search, 26 out of the 37 genes upregulated in mesenchymal-CTC patients in comparison to epithelial-CTC patients are implicated in NF-kappa B signaling at various levels of the transduction pathway (Figure 3A) and demonstrate a complex network of interactions at the protein level (Figure 3B). Importantly, the enrichment of NF-kappa B-related transcripts was consistently observed when we applied a stricter gene inclusion criteria and limited the analysis to a set of 330 genes with the highest expression (log2 mean count of a gene in all samples >9; Table S5).
Figure 3.
Genes upregulated in the primary tumors of patients with mesenchymal CTCs function at various levels of the NF-kappa B signaling pathway (A), within a complex network of interactions (B); image depicting a protein–protein association network, generated using the STRING tool; edge (line) coloring defines the type of interaction: blue—from curated databases, pink—experimentally determined, green—gene neighborhood, red—gene fusions, dark blue—gene co-occurrence, yellow—text mining, black—co-expression, violet—protein homology.
NF-kappa B signaling is a potent regulator of numerous vital physiological processes, including survival, inflammation, and immune responses [37]. The activation of the pathway is mediated by numeros receptors. Our enriched set (Table 1) includes genes encoding both specific ligands (APRIL (TNFSF13)) and receptors (TLR7, TNFRSF11A, IL1RAP, and IFNAR1), as well as universal adaptor proteins (FADD, TICAM1, and TRAF6) that facilitate the transduction of the signal from the receptors in the cell membrane to the effectors in the nucleus. Namely, we observed the upregulation of transducers involved in the canonical cascade (BCL10 and IKBKG) as well as in Toll-like receptor-mediated activation of NF-kappa B signaling (PBK, IKBKE, and TBK1). Moreover, the enhanced expression of MAP2K1 gene points to the possible role of ERK-mediated stimulation of NF-kappa B signaling in tumors with mesenchymal CTCs [38].
On the other hand, the activity of NF-kappa B is known to be regulated by the proteasome and ubiquitin-mediated proteolysis. Here, we report the upregulation of genes that are implicated in the ubiquitin-proteasome system (PSMD7, PSMB10, and CYLD) [39] and the autophagy cascade (ATG10) [40,41,42]. Eventually, we observed an increased expression of one the subunits of the NF-kappa B transcription factor—p65 (RELA), as well as of two other co-operating transcription factors (IRF3 and STAT6). The enhanced signaling resulted in the upregulation of five target genes—CCRL2, TAPBP, BIRC5, ELK1, and CCND3.
The NF-kappa B pathway is a well-known driver of EMT during both embryonic and tumor development [37]. In general, a constant stimulation of this pathway in cancer cells results in abnormal proliferation and differentiation, enhanced metastasis, and treatment resistance [43]. In breast cancer, NF-kappa B directly regulates the transcription of genes encoding EMT-inducing transcription factors [44]. In fact, the increased expression of NF-kappa B is a common feature of breast cancer cell lines and tissues, correlating with intensified activation of both the canonical and the non-canonical pathway [45,46,47]. What is more, several reports point to an interesting association between NF-kappa B and HER2 [47,48,49], with evidence for predominant NF-kappa B activation in ER−/HER2+ breast tumors [45,49].
Our data revealed another interesting pattern of enrichment, with upregulation of several NF-kappa B-related genes that are particularly involved in the positive regulation of type I interferon production (Table S2). The cross-talk between NF-kappa B and Toll-like receptors (TLR)-mediated signaling results in an increased pro-inflammatory response that is additionally enhanced in an autocrine and paracrine manner by a positive feedback loop (Figure 3A) [50,51].
Noteworthy, among the NF-kappa B-unrelated genes, we found markers of platelet activation (CD63 and CD84) [52,53], which is in line with literature reports on the co-operation between platelets and CTCs in the induction of EMT and metastasis formation [54,55]. Of note, tumor dissemination may also be supported by other populations of cells within the intratumoral stroma. The elevated NF-kappa B activity may result from increased release of pro-inflammatory cytokines by macrophages at the tumor site [56,57,58]. In fact, NF-kappa B seems to be involved in the polarization of tumor-associated macrophages [57]. We have previously reported the negative prognostic significance of CTCs of mesenchymal phenotype in BCa patients [21]. Due to the low number of patients in this cohort, in the current study we analyzed the impact of genes linked with mesenchymal CTCs in TCGA BCa cohort. In fact, in TCGA data 5 out of the 38 genes of our interest were associated with worse prognosis (overall survival or risk of recurrence), namely, PSMD7, C2, IFNAR1, CD84, and CYLD. None of the corresponding proteins is currently included in the routine histopathology for breast tumor, thus they need to be validated at the protein level in a large cohort of patients in order to prove their clinical importance and diagnostic applicability.
4. Materials and Methods
4.1. Patients
The study group consisted of 35 breast cancer patients staged I–III, who had undergone surgical treatment at the Medical University Hospital in Gdansk between April 2011 and May 2013. The study was approved by the Ethical Committee of the Medical University of Gdansk (NKBBN 94/2017), and informed consent was collected from all participants. Patients were characterized by different clinicopathological parameters (Table S4), with particular focus on CTC status—negative (n = 12) or positive (n = 23)—and molecular phenotype of CTCs—epithelial (n = 14) or mesenchymal (n = 9)—as described previously [26].
4.2. RNA Extraction
RNA was extracted from formalin-fixed paraffin-embedded (FFPE) primary breast tumor samples (four 10 µm-thick, unstained FFPE sections per patient) using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNA concentration and purity were determined using NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). RNA integrity was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) with Agilent RNA 6000 Pico Kit (Agilent Technologies).
4.3. nCounter Gene Expression Assay
Extracted RNA (4 µl) was pre-amplified using the nCounter Low RNA Input Kit (NanoString Technologies, Seattle, WA, USA) with the dedicated Primer Pool covering the sequences of 730 immune-related genes included in the nCounter PanCancer Immune Profiling Panel (NanoString Technologies). Pre-amplified samples were analyzed using the NanoString nCounter Analysis System (NanoString Technologies) according to the manufacturer’s procedures for hybridization, detection, and scanning.
4.4. Data Analysis
For each tumor sample analyzed with the NanoString technology, the background level was estimated using the mean plus 2 standard deviations of the counts of the negative control probes included in the assay. Data were normalized using the geometric mean of the positive controls included in the assay and 4 most stably expressed housekeeping genes included in the PanCancer Immune Profiling Panel—ABCF1, EDC3, HDAC3, and CNOT4—(expression stability assessed with NormFinder, SD range 173.5–228.4 counts). Background thresholding and normalization were performed using nSolver 4.0 software (NanoString Technologies).
Low-expression genes (log2 mean count of a gene in all samples <6) were excluded, leaving 584 genes for further analysis. Subsequently, the genes differentiating each CTC status were selected on the basis of fold change in comparison to the control; fold change was calculated on basis of the median normalized counts of the probe in each group. The following comparisons were performed: CTC-positive vs. CTC-negative; CTC-epithelial vs. CTC-negative; CTC-mesenchymal vs. CTC-negative; CTC-mesenchymal vs. CTC-epithelial. Genes with FC > 1 were considered upregulated; genes with FC < 1 were considered downregulated.
Data were analyzed using the R statistical computing environment (3.6.1) [59]. Differences in gene expression between groups were analyzed using the Mann–Whitney U test with Benjamini–Hochberg correction for multiple comparisons; p-values ≤ 0.05 and FDR values ≤ 0.2 were considered statistically significant.
For the differing genes, gene ontology was analyzed using the Functional Annotation Tool by DAVID Bioinformatics Resources 6.8 [33,34]. EASE Score, a modified Fisher exact p-value, was used to assess gene enrichment. Multiple testing was corrected using FDR correction.
For the NF-kappa B-related genes, a protein–protein association network was depicted using STRING v11 [60].
4.5. Survival Analysis in TCGA Cohort
RNA-seq (RNASeqV2, RSEM_ normalized) and clinical data of BRCA cohort were obtained from TCGA portal [35,36] (data status of 28 January, 2016). The group was limited to T1-3M0 patients, and records with missing clinical or expression values were excluded, leaving 877 out of 1098 BCa patients for the analysis. OS was defined according to the “days_to_death” variable for survival time and the “vital_status” variable for event; DFS was defined according to the “days_to_last_follow-up” variable for survival time and the “person_neoplasm_cancer_status” variable for event. For the genes of interest, low/moderate status of gene expression was determined according to the 1st quartile (Q1) cut-off, while low/high status of gene expression was determined according to the 3rd quartile (Q3) cut-off. For each gene, the expression status (low vs. moderate; low vs. high) was tested in both univariate and multivariate analyses including the clinical stage. Hazard ratios (HR) with 95% confidence intervals (95% CI) were computed using Cox proportional hazards regression using the R statistical computing environment (3.6.1) [59].
5. Conclusions
To summarize, this study points to the potential link between the expression of immune-related genes in cells within the primary tumor and the EMT state of circulating tumor cells. Increased NF-kappa B signaling-related signatures in the tumor mass might possibly promote EMT in CTCs, thus contributing to their more aggressive phenotype and worse patient prognosis. It merits further investigation whether such effect is due to the action of cancer cells or that of normal cells in the surrounding TME. The potential prognostic relevance of selected genes associated with mesenchymal CTCs is promising and deserves further validation.
Acknowledgments
The authors thank Michał Bieńkowski for the preparation of the pathological material.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/11/12/1961/s1, Table S1: Results of gene expression analysis according to each CTC status or phenotype, Table S2: List of GO terms enriched in the genes upregulated in mesenchymal-CTC patients, Table S3: Results of multivariate survival analysis depending on the expression of genes up- and downregulated in mesenchymal-CTC patients; low/moderate status of gene expression relative to the 1st quartile (Q1); low/high status of gene expression relative to the 3rd quartile (Q3); hazard ratios (HR) with 95% confidence intervals (95% CI) computed using Cox proportional hazards regression; significant results are highlighted in green, Table S4: Clinicopathological characteristics of the patients, Table S5: List of genes upregulated in the primary tumors of patients with mesenchymal CTCs (compared to patients with epithelial CTCs), computed for a set of 330 high-expression genes (log2 mean count of a gene in all samples > 9), Figure S1: Expression of genes up- and downregulated in primary tumors of patients with mesenchymal CTCs (MES, n = 9) and patients with epithelial CTCs (EPI, n = 14); expression depicted as number of counts of each probe and normalized to the 4 most stable reference genes (ABCF1, EDC3, HDAC3, and CNOT4); fold change (FC) based on the median normalized counts of the probe in each group; differences in median normalized counts between groups analyzed with the Mann–Whitney U test (p); bars correspond to IQR, whiskers cover 1.5 IQR from the median.
Author Contributions
Conceptualization, T.S., M.P., A.M., N.B.-K., and A.J.Z.; methodology, T.S., A.J., M.P., M.N., A.M., and N.B.-K.; formal analysis, T.S., M.P., and A.J.Z.; investigation, A.M., N.B.-K., M.P., A.J., and A.B.; resources, J.S., A.K., and L.K.; data curation, M.P.; writing—original draft preparation, M.P., T.S., and A.J.Z.; writing—review and editing, all authors; visualization, T.S. and M.P.; supervision, T.S. and A.J.Z.; project administration, A.J.Z.; funding acquisition, A.J.Z.
Funding
This research was funded by the National Science Centre (Poland), grant number 2016/22/E/NZ4/00664.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Klein C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 2.Marusyk A., Almendro V., Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 3.Aceto N., Toner M., Maheswaran S., Haber D.A. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends Cancer. 2015;1:44–52. doi: 10.1016/j.trecan.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 4.De Bono J.S., Scher H.I., Montgomery R.B., Parker C., Miller M.C., Tissing H., Doyle G.V., Terstappen L.W.W.M., Pienta K.J., Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong A.J., Halabi S., Luo J., Nanus D.M., Giannakakou P., Szmulewitz R.Z., Danila D.C., Healy P., Anand M., Rothwell C.J., et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J. Clin. Oncol. 2019;37:1120–1129. doi: 10.1200/JCO.18.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cristofanilli M., Pierga J.-Y., Reuben J., Rademaker A., Davis A.A., Peeters D.J., Fehm T., Nolé F., Gisbert-Criado R., Mavroudis D., et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit. Rev. Oncol. Hematol. 2019;134:39–45. doi: 10.1016/j.critrevonc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Bidard F.-C., Michiels S., Riethdorf S., Mueller V., Esserman L.J., Lucci A., Naume B., Horiguchi J., Gisbert-Criado R., Sleijfer S., et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. J. Natl. Cancer Inst. 2018;110:560–567. doi: 10.1093/jnci/djy018. [DOI] [PubMed] [Google Scholar]
- 8.Lindsay C.R., Blackhall F.H., Carmel A., Fernandez-Gutierrez F., Gazzaniga P., Groen H.J.M., Hiltermann T.J.N., Krebs M.G., Loges S., López-López R., et al. EPAC-lung: Pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur. J. Cancer. 2019;117:60–68. doi: 10.1016/j.ejca.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Lucci A., Hall C.S., Lodhi A.K., Bhattacharyya A., Anderson A.E., Xiao L., Bedrosian I., Kuerer H.M., Krishnamurthy S. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012;13:688–695. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 10.Cobain E.F., Paoletti C., Smerage J.B., Hayes D.F. Clinical Applications of Circulating Tumor Cells in Breast Cancer. Recent Results Cancer Res. 2020;215:147–160. doi: 10.1007/978-3-030-26439-0_8. [DOI] [PubMed] [Google Scholar]
- 11.Todenhöfer T., Pantel K., Stenzl A., Werner S. Pathophysiology of Tumor Cell Release into the Circulation and Characterization of CTC. Recent Results Cancer Res. 2020;215:3–24. doi: 10.1007/978-3-030-26439-0_1. [DOI] [PubMed] [Google Scholar]
- 12.Pearson G.W. Control of Invasion by Epithelial-to-Mesenchymal Transition Programs during Metastasis. J. Clin. Med. 2019;8:646. doi: 10.3390/jcm8050646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McInnes L.M., Jacobson N., Redfern A., Dowling A., Thompson E.W., Saunders C.M. Clinical implications of circulating tumor cells of breast cancer patients: Role of epithelial-mesenchymal plasticity. Front. Oncol. 2015;5:42. doi: 10.3389/fonc.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulfoni M., Gerratana L., Del Ben F., Marzinotto S., Sorrentino M., Turetta M., Scoles G., Toffoletto B., Isola M., Beltrami C.A., et al. In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal transition is associated with a poor prognosis. Breast Cancer Res. 2016;18:30. doi: 10.1186/s13058-016-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markiewicz A., Żaczek A.J. The Landscape of Circulating Tumor Cell Research in the Context of Epithelial-Mesenchymal Transition. Pathobiology. 2017;84:264–283. doi: 10.1159/000477812. [DOI] [PubMed] [Google Scholar]
- 16.Pattabiraman D.R., Bierie B., Kober K.I., Thiru P., Krall J.A., Zill C., Reinhardt F., Tam W.L., Weinberg R.A. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 2016;351:aad3680. doi: 10.1126/science.aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolly M.K., Somarelli J.A., Sheth M., Biddle A., Tripathi S.C., Armstrong A.J., Hanash S.M., Bapat S.A., Rangarajan A., Levine H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 2019;194:161–184. doi: 10.1016/j.pharmthera.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Yu M., Bardia A., Wittner B.S., Stott S.L., Smas M.E., Ting D.T., Isakoff S.J., Ciciliano J.C., Wells M.N., Shah A.M., et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satelli A., Brownlee Z., Mitra A., Meng Q.H., Li S. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule- and cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin. Chem. 2015;61:259–266. doi: 10.1373/clinchem.2014.228122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan X., Ma F., Li C., Wu S., Hu S., Huang J., Sun X., Wang J., Luo Y., Cai R., et al. The prognostic and therapeutic implications of circulating tumor cell phenotype detection based on epithelial-mesenchymal transition markers in the first-line chemotherapy of HER2-negative metastatic breast cancer. Cancer Commun. 2019;39:1. doi: 10.1186/s40880-018-0346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markiewicz A., Nagel A., Szade J., Majewska H., Skokowski J., Seroczynska B., Stokowy T., Welnicka-Jaskiewicz M., Zaczek A.J. Aggressive Phenotype of Cells Disseminated via Hematogenous and Lymphatic Route in Breast Cancer Patients. Transl. Oncol. 2018;11:722–731. doi: 10.1016/j.tranon.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markiewicz A., Topa J., Nagel A., Skokowski J., Seroczynska B., Stokowy T., Welnicka-Jaskiewicz M., Zaczek A.J. Spectrum of Epithelial-Mesenchymal Transition Phenotypes in Circulating Tumour Cells from Early Breast Cancer Patients. Cancers. 2019;11:59. doi: 10.3390/cancers11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mego M., Karaba M., Minarik G., Benca J., Silvia J., Sedlackova T., Manasova D., Kalavska K., Pindak D., Cristofanilli M., et al. Circulating Tumor Cells with Epithelial-to-mesenchymal Transition Phenotypes Associated With Inferior Outcomes in Primary Breast Cancer. Anticancer Res. 2019;39:1829–1837. doi: 10.21873/anticanres.13290. [DOI] [PubMed] [Google Scholar]
- 24.Kallergi G., Papadaki M.A., Politaki E., Mavroudis D., Georgoulias V., Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadaki M.A., Kallergi G., Zafeiriou Z., Manouras L., Theodoropoulos P.A., Mavroudis D., Georgoulias V., Agelaki S. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14:651. doi: 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markiewicz A., Książkiewicz M., Wełnicka-Jaśkiewicz M., Seroczyńska B., Skokowski J., Szade J., Żaczek A.J. Mesenchymal phenotype of CTC-enriched blood fraction and lymph node metastasis formation potential. PLoS ONE. 2014;9:e93901. doi: 10.1371/journal.pone.0093901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingangi V., Minopoli M., Ragone C., Motti M.L., Carriero M.V. Role of Microenvironment on the Fate of Disseminating Cancer Stem Cells. Front. Oncol. 2019;9:82. doi: 10.3389/fonc.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohme M., Riethdorf S., Pantel K. Circulating and disseminated tumour cells—Mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 2017;14:155–167. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 32.Bednarz-Knoll N., Nastały P., Żaczek A., Stoupiec M.G., Riethdorf S., Wikman H., Müller V., Skokowski J., Szade J., Sejda A., et al. Stromal expression of ALDH1 in human breast carcinomas indicates reduced tumor progression. Oncotarget. 2015;6:26789–26803. doi: 10.18632/oncotarget.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciriello G., Gatza M.L., Beck A.H., Wilkerson M.D., Rhie S.K., Pastore A., Zhang H., McLellan M., Yau C., Kandoth C., et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pires B.R.B., Silva R.C.M.C., Ferreira G.M., Abdelhay E. NF-kappaB: Two Sides of the Same Coin. Genes. 2018;9:24. doi: 10.3390/genes9010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi J.E., Matthews A.J., Michel G., Vuong B.Q. AID Phosphorylation Regulates Mismatch Repair-Dependent Class Switch Recombination and Affinity Maturation. J. Immunol. 2019 doi: 10.4049/jimmunol.1900809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kravtsova-Ivantsiv Y., Ciechanover A. The ubiquitin-proteasome system and activation of NF-κB: Involvement of the ubiquitin ligase KPC1 in p105 processing and tumor suppression. Mol. Cell Oncol. 2015;2:e1054552. doi: 10.1080/23723556.2015.1054552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song C., Mitter S.K., Qi X., Beli E., Rao H.V., Ding J., Ip C.S., Gu H., Akin D., Dunn W.A., et al. Oxidative stress-mediated NFκB phosphorylation upregulates p62/SQSTM1 and promotes retinal pigmented epithelial cell survival through increased autophagy. PLoS ONE. 2017;12:e0171940. doi: 10.1371/journal.pone.0171940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duran A., Linares J.F., Galvez A.S., Wikenheiser K., Flores J.M., Diaz-Meco M.T., Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Trocoli A., Djavaheri-Mergny M. The complex interplay between autophagy and NF-κB signaling pathways in cancer cells. Am. J. Cancer Res. 2011;1:629–649. [PMC free article] [PubMed] [Google Scholar]
- 43.Xia L., Tan S., Zhou Y., Lin J., Wang H., Oyang L., Tian Y., Liu L., Su M., Wang H., et al. Role of the NFκB-signaling pathway in cancer. Onco Targets Ther. 2018;11:2063. doi: 10.2147/OTT.S161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pires B.R.B., Mencalha A.L., Ferreira G.M., de Souza W.F., Morgado-Díaz J.A., Maia A.M., Corrêa S., Abdelhay E.S.F.W. NF-kappaB Is Involved in the Regulation of EMT Genes in Breast Cancer Cells. PLoS ONE. 2017;12:e0169622. doi: 10.1371/journal.pone.0169622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakshatri H., Bhat-Nakshatri P., Martin D.A., Goulet R.J., Sledge G.W. Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Mol. Cell. Biol. 1997;17:3629–3639. doi: 10.1128/MCB.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cogswell P.C., Guttridge D.C., Funkhouser W.K., Baldwin A.S. Selective activation of NF-kappa B subunits in human breast cancer: Potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene. 2000;19:1123–1131. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y., Eppenberger-Castori S., Eppenberger U., Benz C.C. The NFkappaB pathway and endocrine-resistant breast cancer. Endocr. Relat. Cancer. 2005;12(Suppl. S1):S37–S46. doi: 10.1677/erc.1.00977. [DOI] [PubMed] [Google Scholar]
- 48.Hou Y., Liang H., Rao E., Zheng W., Huang X., Deng L., Zhang Y., Yu X., Xu M., Mauceri H., et al. Non-canonical NF-κB Antagonizes STING Sensor-Mediated DNA Sensing in Radiotherapy. Immunity. 2018;49:490–503.e4. doi: 10.1016/j.immuni.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biswas D.K., Shi Q., Baily S., Strickland I., Ghosh S., Pardee A.B., Iglehart J.D. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc. Natl. Acad. Sci. USA. 2004;101:10137–10142. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hervas-Stubbs S., Perez-Gracia J.L., Rouzaut A., Sanmamed M.F., Le Bon A., Melero I. Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 2011;17:2619–2627. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]
- 51.Jin J., Hu H., Li H.S., Yu J., Xiao Y., Brittain G.C., Zou Q., Cheng X., Mallette F.A., Watowich S.S., et al. Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity. Immunity. 2014;40:342–354. doi: 10.1016/j.immuni.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuenca M., Sintes J., Lányi Á., Engel P. CD84 cell surface signaling molecule: An emerging biomarker and target for cancer and autoimmune disorders. Clin. Immunol. 2019;204:43–49. doi: 10.1016/j.clim.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 53.Pols M.S., Klumperman J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009;315:1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 54.Labelle M., Begum S., Hynes R.O. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labelle M., Begum S., Hynes R.O. Platelets guide the formation of early metastatic niches. Proc. Natl. Acad. Sci. USA. 2014;111:E3053–E3061. doi: 10.1073/pnas.1411082111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoesel B., Schmid J.A. The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ben-Neriah Y., Karin M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 58.Xia Y., Shen S., Verma I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 60.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.