Abstract
Acute myeloid leukemia (AML), the most common acute leukemia in adults, is a heterogeneous malignant clonal disorder arising from multipotent hematopoietic progenitor cells characterized by genetic and concerted epigenetic aberrations. Core binding factor-Leukemia (CBFL) is characterized by the recurrent reciprocal translocations t(8;21)(q22;q22) or inv(16)(p13;q22) that, expressing the distinctive RUNX1-RUNX1T1 (also known as Acute myeloid leukemia1-eight twenty-one, AML1-ETO or RUNX1/ETO) or CBFB-MYH11 (also known as CBFβ-SMMHC) translocation product respectively, disrupt the essential hematopoietic function of the CBF. In the past decade, remarkable progress has been achieved in understanding the structure, three-dimensional (3D) chromosomal topology, and disease-inducing genetic and epigenetic abnormalities of the fusion proteins that arise from disruption of the CBF subunit alpha and beta genes. Although CBFLs have a relatively good prognosis compared to other leukemia subtypes, 40–50% of patients still relapse, requiring intensive chemotherapy and allogenic hematopoietic cell transplantation (alloHCT). To provide a rationale for the CBFL-associated altered hematopoietic development, in this review, we summarize the current understanding on the various molecular mechanisms, including dysregulation of Wnt/β-catenin signaling as an early event that triggers the translocations, playing a pivotal role in the pathophysiology of CBFL. Translation of these findings into the clinical setting is just beginning by improvement in risk stratification, MRD assessment, and development of targeted therapies.
Keywords: core binding factor leukemia, AML, RUNX1, RUNX1T1, CBFB, MYH11, miRNA, chromatin remodeling
1. Introduction
The year 2016 coincided with the 25th anniversary of the first cloning of mammalian Runt (Runt domain)-related transcription factor 1 (RUNX1) gene, associated with hematologic disorders [1]. RUNX1 (AML1) is a master transcriptional regulator of adult hematopoiesis also involved in the establishment, maintenance, and functional integrity of hematopoietic stem cells (HSCs) in embryonic and adult blood compartments [2,3,4]. AML1 post-translational modifications help create scaffolds that interact and bind with multiple members recruited to the core binding factor (CBF), promoting or repressing transcription. At about the same time, the gene encoding CBFB (CBFβ) was identified as disrupted by the inv(16) in acute myeloid leukemia [5]. Normally, AML1 and CBFβ form a DNA-binding heterodimer required for binding to the consensus sequence, where it recruits lineage-specifying transcription factors to regulate hematopoietic differentiation. As the Runt-related transcription factor (RUNX) gene family plays important roles in tissue-specific gene expression, it is frequently involved in the malignant transformation of the hematopoietic system. Acute leukemias characterized by the presence of t(8;21) or inv(16) are defined core-binding factor Leukemias (CBFLs), since they both alter the CBF transcription factor complex [6]. Approximately 30% and 13–15% of newly diagnosed pediatric and adult AML patients, respectively, are diagnosed as CBFLs [7]. Although the CBFLs are categorized into a favorable-risk group as compared with other subtypes of AML, approximately 30–40% of the patients still relapse and may require allogeneic hematopoietic cell transplantation (HCT) [8,9]. RUNX1–RUNX1T1 and CBFB–MYH11 translocations may represent acquired initiating events occurring in hematopoietic progenitors. However, little is known about the molecular mechanisms that drive the generation of the t(8;21) or inv(16), after which leukemia clonally evolves through accumulation of secondary mutations. The hypothesis that Wnt signaling promotes genomic proximity between RUNX1 and RUNX1T1 has been recently examined by experiments establishing that Wnt/β-catenin signaling supports RUNX1 and RUNX1T1 expression in hematopoietic precursors and provides spatial information, indicating that transcription of these genes is likely occurring into RNA-polymerase-II nuclear factories (RNAPII-Ser5) [10]. These results suggest a Wnt-mediated model in which an upstream molecular mechanism is capable of favoring and guiding the translocation event [11]. The incremental improvements in understanding the genetic and molecular basis of CBFLs and their association with distinct clinical and biological features provide insights into previously unappreciated cooperating pathways [12,13]. At diagnosis, the disease consists of heterogeneous clusters of cells widely differing from one another in terms of additional genetic lesions, besides sharing the specific chromosomal translocations. Cytogenetic abnormalities that alter the function of the CBF are often associated with specific receptor tyrosine kinase (RTK) mutations, suggesting that additional genetic abnormalities have an essential role in CBFL pathogenesis [14,15]. Despite a common molecular alteration involving a component of the CBF transcription complex, AMLs expressing RUNX1-RUNX1T1 or CBFB-MYH11 alterations display a remarkably different genome-wide spectrum of cooperating mutations [14]. Recent studies clearly indicate that AMLs with t(8;21)(q22;q22) and AMLs with inv(16)(p13q22) show different biological and clinical characteristics, supporting the notion that they represent two distinct diseases [7,16]. A series of concomitant evidence in the CBFL proved the existence of a preleukemic phase confirmed by a prolonged latency observed in experimental models between the occurrence of RUNX1-RUNX1T1 CBF translocation and the development of overt leukemia [17,18], the persistence of CBFL translocations in normal HSC detected from patients in remission [19,20,21], and the maintenance of RUNX1-RUNX1T1 at diagnosis and at relapses. NRAS (neuroblastoma RAS viral oncogene) is the most frequently mutated gene in CBFL, and over 60% of the cases harbor activating mutations in NRAS, KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog), FLT3 (FMS-like tyrosine kinase 3), KRAS (Kirsten rat sarcoma 2 viral oncogene homolog), PTPN11 (protein tyrosine phosphatase non receptor type 11), and/or loss-of-function mutations in NF1 (neurofibromin1) [9,14,15]. Integrated mutational analysis of the genetic and epigenetic changes that are relevant to the pathogenesis of CBFL would be required for a better risk stratification of patients who would benefit from dose-intensified induction chemotherapy or novel targeted therapies. AML1-ETO (eight twenty-one) (RUNX1-RUNX1T1) is the chimeric protein formed as a consequence of the t(8;21) chromosomal rearrangement, which is among the most recurrent cytogenetic rearrangements in de novo AML. The molecular mechanisms through which AML1-ETO fusion protein exerts multiple effects are not fully elucidated, yet all have focused on its strong repressor function. Moreover, several studies documented the multifunctionality of AML1-ETO fusion protein, including impaired differentiation, apoptosis inhibition, and signal activation for cell proliferation. This model might be oversimplified; however, there is convincing evidence supporting the hypothesis that leukemias are induced by cooperation between alterations in protein-coding genes and microRNAs (miRNAs), an entire novel epigenetic targets linked to leukemia development [22]. The consequences of altered expression and epigenetic status of miRNAs in CBF leukemias have been reported by us and other groups, unveiling that microRNAs are extensively integrated into the molecular networks that control leukemic development and progression [23,24,25,26,27,28]. Therefore, in this review, we summarize a synopsis of recent studies on comprehensive molecular profiles in CBF leukemias, providing a rationale for translation of the accumulating molecular evidence into clinical trials for better therapies to CBF leukemia patients.
2. Core Binding Factor Complex: A Critical Role in Hematopoietic Stem Cell Fate
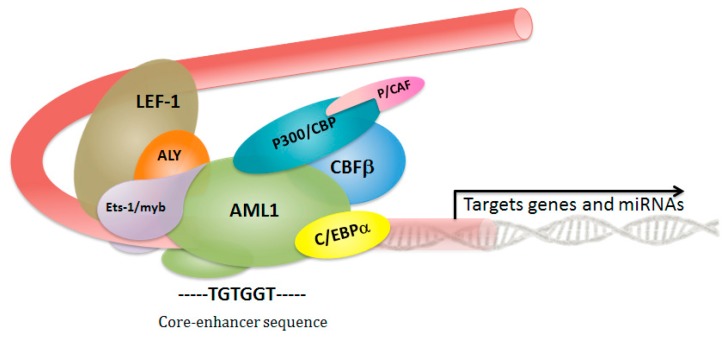
The CBF is a transcription factor complex, which consists of a distinct DNA-binding CBFα subunit (RUNX1, 2, or 3), and its non-DNA-binding heterodimerization partner CBFβ subunit (encoded by the CBFB gene). AML1 is a master regulatory protein expressed throughout all hematopoietic lineages. The RUNX1 and CBFB genes are required for hematopoietic stem cells’ (HSCs) emergence and formation during definitive HSC development through to their terminal differentiation, and are key regulators of hematopoiesis at several steps [29,30]. The loss of definitive hematopoiesis observed in Runx1−/− or Cbfb−/− knockout mice and an expanded HSC compartment in conditional Runx1-deficient mice highlight their complex interplay in orchestrating the accurate maintenance of hematopoietic stem cell differentiation [29,30,31,32,33,34,35]. The heterodimerization with CBFβ leads to the phosphorylation of RUNX1, which in turn induces p300 (encoded by EP300) phosphorylation by homeodomain interacting kinase 2 (HIPK2) in AML1 [36]. By binding to the core-enhancer sequence, AML1/CBFβ complex functions as an organizing element recruiting other DNA-binding proteins, transcription factors, and co-regulators able to activate-or in some cases, repress-the target gene’s transcription. Heterodimerization with core-binding factor-β (CBFβ) confers enhanced DNA binding ability, mediated by the Runt domain in AML1. The presence of CBFβ subunit increases the affinity for DNA and, consistent with predictions, shows a significant enhancement (>40-fold enhancement) of Runt domain DNA binding of full-length AML1 (Figure 1) [37]. RUNX1 and CBFB are frequent targets of gene rearrangements through chromosomal translocations and mutations that are associated with human leukemias. RUNX1 is involved in t(8;21)(q22;q22) and t(12;21)(p13;q22) in acute myeloid and lymphocytic leukemias, and CBFB is rearranged in acute myeloid leukemias by inv(16)(p13;q22), t(16;16), and del(16)(q22). These cytogenetic alterations lead to the expression of fusion proteins that disrupt the heterodimeric CBF complex signaling with a dominant prevalence.
Figure 1.
Schematic representation of the core binding factor transcriptional activation complex. DNA binding and heterodimerization with core binding factor-β (CBFβ) are mediated by the Runt domain in Acute myeloid leukemia1 (AML1 or RUNX1). The interaction with CBFβ leads to the phosphorylation of AML1, which in turn induces p300 phosphorylation, and this is mediated by homeodomain interacting kinase 2 (HIPK2) in AML1. CBP/p300 (CREB binding protein CBP and EP300); C/EBPα (CCAAT/enhancer binding protein alpha); P/CAF (P300/CBP-associated factor); AML1 (acute myeloid leukemia 1 protein); CBFβ (core binding factor subunit beta); LEF1 (lymphoid enhancer-binding factor 1); ALY (Aly/REF export factor); ETS-1 (v-ets erythroblastosis virus E26 oncogene homolog 11).
3. Leukemia Triggered by RUNX1–RUNX1T1
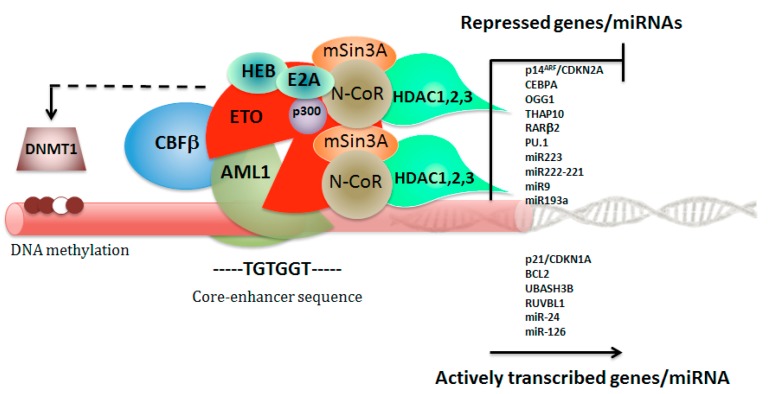
AML1-ETO in frame fusion protein joins the N-terminal 177 amino acids of AML1 (encoded by RUNX1) to nearly all of ETO (encoded by RUNX1T1), and functions as a dominant repressor for the majority of RUNX1-responsive hematopoietic genes and microRNA (miRNA) non-coding genes. Consequently, in RUNX1-RUNX1T1, the DNA-binding Runt domain of RUNX1 is joined to RUNX1T1, creating a fusion transcript lacking the RUNX1 transcription activation domain. The structural insights show four essential interactions for AML1-ETO activity: DNA and CBFβ binding by the Runt domain, oligomerization through the HHR domain, and E-protein binding by the HHR domain [38,39,40,41,42]. The presence of chromosomal rearrangements, such as t(8;21) or inv(16), is associated with a unique DNA methylation patterning that predicts distinct patient outcomes, suggesting for the CBF fusion proteins also a role as epigenotype modifier [43]. The underlying mechanisms involve the AML1-ETO capacity to recruit DNA methyltransferases (DNMTs) to target tumor-suppressor genes by concerted action with certain transcriptional repressors (Figure 2) [44,45,46,47]. AML1-ETO cooperates with HIF1α to transactivate DNMT3a gene and shapes a positive regulatory circuit that contributes to DNA methylation signature, specifically for the non-AML1–ETO targets, leading to a DNA hypermethylation profile in AML [48]. Moreover, Ptasinska and colleagues observed that depletion of AML1-ETO and subsequent cell differentiation involves not only loss of repression, but is also associated with a redistribution of RUNX1-binding activity throughout the genome that restores, on a global scale, the epigenetic alterations mediated by the fusion protein [49]. This effect, therefore, is obtained not only through loss of repression, but involves an increased recruitment of transcription factors to additional sites, depending on AML1 interactions with other transcriptional activators, such as C/EBPα and PU.1, whose activity is altered by AML1-ETO [50,51]. ETO is a member of E-box family of transcription factors, and contains four nervy homology regions (NHR 1–4) that interact with several nuclear repressors (N-CoR, SMRT, mSin3A), including the histone deacetylases (HDACs 1–3), primarily through the NHR2 and NHR4 domains [44,52,53]. AML1-ETO also interacts with the DNA methyltransferase DNMT1 to promote DNA methylation and mediates transcriptional repression (Figure 2) [45]. However, increasing evidences from mammalian cell systems and mouse genetic models suggest that the relationship between AML1-ETO and native AML1 may be more complex, indicating that AML1-ETO depends on some functions of native AML1 to exert its proleukemogenic properties [6,54,55]. Genome-wide ChIP-Seq and RNA-Seq data recently revealed that AML1 is a member of the transcription factor complex containing AML1-ETO, and that relative binding signals on chromatin determine which genes are repressed or activated by AML1/AML1-ETO complex [56]. Thus, these new important findings indicate that the malignant cell phenotype of t(8;21) leukemia is sustained by a delicate balance between AML1-ETO and native AML1. Translocation-prone genes are preferentially recruited into transcription factories with active RNA polymerase II (RNAPII-Ser5) and need to be positioned in close spatial proximity relative to each other prior to translocation. Mechanisms that drive the generation of the RUNX1-RUNX1T1 translocation have been poorly understood, but a recent report established that the Wnt/β-catenin signaling enhances transcription and genomic proximity of RUNX1 and RUNX1T1 genes, which seems to promote the generation of the RUNX1-RUNX1T1 fusion gene [11]. These observations describe the enhanced RUNX1T1 and RUNX1 expression in hematopoietic precursors by Wnt/β-catenin signaling and suggest a nuclear topography of transcription likely occurring into specialized nuclear factories, thereby increasing the potential for a chromosomal translocation event (Figure 3).
Figure 2.
Schematic representation of the AML1–ETO (eight twenty one) repressor complex assembly. Transcriptional repression complex AML1/AML1-ETO recruits corepressors, including NCOR (nuclear receptor corepressor 1), HDACs (histone deacetylases), mSin3A (SIN3 transcription regulator family member A), and also interacts with DNMT1 (DNA methyltransferase 1) to promote DNA methylation and to repress target genes and miRNAs expression.
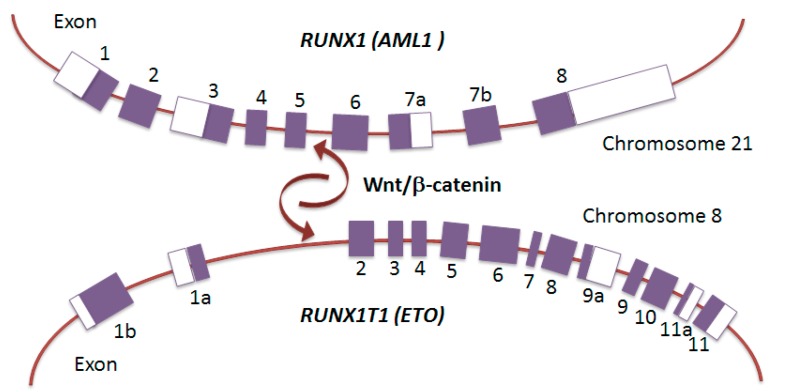
Figure 3.
Wnt/β-catenin promotes spatial proximity and translocation of RUNX1 and RUNX1T1. Genomic structure of RUNX1 on chromosome 21 and RUNX1T1 on chromosome 8. Wnt/β-catenin was shown to induce spatial proximity and translocation of RUNX1 and RUNX1T1, which led to the generation of the RUNX1–RUNX1T1 fusion gene. Exons are depicted as boxes.
4. Leukemia Triggered by CBFβ-MYH11
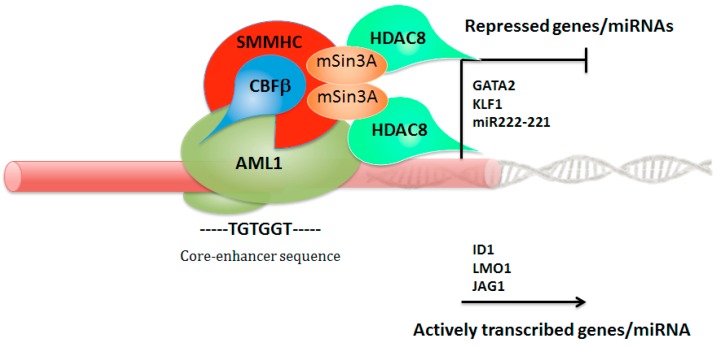
The CBFβ-SMMHC (smooth muscle myosin heavy chain, encoded by MYH11) fusion protein exhibits a higher binding affinity for AML1 than wild-type CBFβ. In addition to the RUNX-binding domain in CBFβ, it contains an additional RUNX-binding domain in the SMMHC portion of the fusion protein. The CBFβ-SMMHC interacts specifically with transcriptional inhibitors and HDACs, such as mSin3A and HDAC8, through an unexpected domain within the SMMHC region, thereby repressing AML1-mediated gene regulation. [57]. Therefore, CBFβ–SMMHC fusion protein acts as a transcriptional repressor, and might do so by sequestering AML1 on actin filaments in the cytoplasm. However, the majority of CBFβ–SMMHC target genes are actively transcribed, including genes such as ID1 (inhibitor of DNA binding 1), LMO1 (LIM domain only 1, rhombotin 1), and JAG1 (Jagged 1) involved in hematopoietic stem cell self-renewal, and repressed upon fusion protein knockdown (Figure 4). An intrinsic inability of CBFβ-SMMHC to provide CBFβ function in vivo has also emerged, based on its failure to complete hematopoietic recovery by Cbfb–/– mouse embryonic stem (ES) cells impaired in their capacity to generate definitive erythroid and myeloid elements [58]. Otherwise, the CBFβ–SMMHC fusion protein is predominantly recruited to promoters engaged by AML1, where it interacts with TAL1 (T-cell acute leukemia 1), FLI1 (Friend leukemia virus integration 1), and TBP-associated factors (TAFs), in synergy with a variety of hematopoietic transcription factors (ERG, GATA2, and PU.1/SPI1), as well as epigenetic coregulators, including EP300 (E1A binding protein p300) and HDAC1 (histone deacetylase 1). Recent results suggest that HDAC1 is an important component of the AML1/CBFβ-SMMHC fusion complex, which functions to activate transcription of specific target genes. The authors found that in vivo treatment with the HDAC1 inhibitor induced differentiation and apoptosis of leukemia cells, indicating HDAC1 as a potential therapeutic target [59]. These findings suggest an important role for CBFβ-SMMHC in regulating the expression of genes essential for emergence of the hematopoietic stem cell [60]. Therefore, AML1 activity is required for CBFB-MYH11-induced leukemogenesis [61], also through the activity of the chromodomain helicase DNA-binding protein-7 (CHD7), which is an ATP-dependent chromatin remodeling factor interacting with AML1/CBFβ–SMMHC complex and altering the expression of its target genes. Chd7 deficiency in Chd7f/fMx1-CreCbfb+/56M mice, which expresses the Cbfb-MYH11 fusion gene, delayed Cbfb–MYH11-induced leukemia in both primary and transplanted mice [62]. Normally, the interacting interface between AML1 and CBFβ, which allows the heterodimerization [63], is retained in CBFβ-SMMHC. Moreover, the fusion between the amino-terminal heterodimerization domain of CBFβ and the C-terminal coiled-coil region of SMMHC creates a novel binding site for AML1, called the AML1 (RUNX1) high-affinity binding domain (HABD) [64]. Therefore, the CBFβ-SMMHC binds AML1 at two sites, resulting in a higher binding affinity for AML1 than wild-type CBFβ [65]. In patients with one allele of the wild-type CBFB and one allele of CBFB-MYH11, AML1 will be preferentially bound to the fusion protein [64,65]. This high-affinity AML1 binding has been proposed to explain the dominant negative role of CBFβ-SMMHC by sequestering AML1 from its targets [64]. However, knock-in mice expressing Cbfb-MYH11 with a HABD deletion unexpectedly potentiated its leukemogenic activity, developed leukemia faster, even though hematopoietic defects associated with Runx1-inhibition were partially rescued, suggesting that AML1-dominant inhibition may not be a critical step for leukemogenesis by CBFβ-SMMHC [55]. CBFβ-SMMHC was also shown to bind and sequester HIPK2, preventing the critical AML1/p300 phosphorylation to mislocalized CBFβ-SMMHC complexes [66]. Martens and colleagues [67] analyzed inv(16) AMLs by multiple transcriptomic and epigenomic profiles with the aim to investigate whether CBFB–MYH11 specifically blocks megakaryocyte/erythrocyte differentiation in the context of human hematopoiesis. Findings revealed that CBFβ-SMMHC seems to be involved in transcription deregulation and occupancy replacement of the transcription factors GATA2 (GATA binding protein 2) and KLF1 (Kruppel-like factor 1), interfering with normal differentiation. These results indicate that the attenuating expression of GATA2/KLF1, induced by CBFβ–SMMHC fusion, inhibits primed megakaryopoiesis [67]. On the other hand, a recent paper [68] showed that Gata2 determines its distinct effects in association with Cbfb-MYH11 in two different stages. Up-regulated Gata2 gene is important in preleukemic Cbfb-MYH11 knock-in mice. Heterozygous knockout of Gata2 in Cbfb-MYH11 mice delayed leukemia onset. However, despite slower development of leukemia, the Cbfb-MYH11 Gata2-deficient mice showed a more aggressive phenotype at the leukemic stage. These findings may reflect the clinical observation of GATA2 recurrent deletions in relapsed CBFL patients [69]. Therefore, GATA2 up-regulation contributes to CBFB-MYH11 leukemogenesis in the early stage, and deficiency could be involved in the relapse/aggressive evolution of CBFL.
Figure 4.
Schematic representation of transcriptional repression mediated by CBFβ-SMMHC. For transcriptional repression, CBFβ-SMMHC retains the RUNX-binding domain in CBFβ and contains an additional RUNX-binding domain in the SMMHC. Fusion protein has the ability to interact specifically with mSin3A and HDAC8 (histone deacetylase 8) through an unexpected repression domain within the SMMHC portion. GATA2 (GATA binding protein 2); KLF1 (Kruppel-like factor 1); ID1 (inhibitor of DNA binding 1); LMO1 (LIM domain only 1); JAG1 (jagged canonical notch ligand 1).
5. MicroRNA Circuitries Contribute to CBF-Mediated Leukemogenesis
In addition to genetic alterations in the chromatin state that affect gene expression and enrichment for mutations in genes encoding proteins essential for mRNA splicing [70], alterations in the expression of long non-coding RNA (lncRNA) [71], as well as microRNAs (miRNAs), are widely suspected to play a critical role in leukemia initiation and outcome prediction. MicroRNAs are a class of small non-coding RNAs elements (≈20–22 nt) implicated in differentiation of mammalian blood cell lineages through the post-transcriptional modulation of gene expression by binding to the 3′ untranslated region of mRNAs and down-regulation of their translation to protein [72]. The role of lineage-specific miRNAs in hematopoiesis was largely determined from profiling studies that revealed distinct miRNA expression patterns at various stages of hematopoietic development [73,74,75,76]. The expression levels of several miRNA genes show abundant natural variation in nearly all physiological processes involving the formation and maintenance of the human blood hierarchy [77,78]. Human granulocytic differentiation is controlled by miR-223, a key member of an integrated regulatory circuit, including C/EBPα and NFI-A [23], and miR-221/miR-222 cluster were found to target the oncogene KIT [73]. The causal mechanisms and molecular player responsible for the widespread dysregulation of miRNA expression in AMLs, which can function as either oncomiRs or tumor suppressors, are only limitedly known. Specific chromosomal and genetic abnormalities in each AML subtype are likely to contribute to the global non-coding transcriptome. The genomic abnormalities, previously described for protein-coding genes such as chromosomal rearrangements, are found to influence the activity of miRNAs through a variety of mechanisms [27], playing a role in nearly all aspects of AML development [79]. The biological phenomenon where several RNA species regulate one another by competing for binding to a limited pool of shared miRNAs has been proposed as competing endogenous RNAs (ceRNAs) [80]. The potential of the aberrantly overexpressed RUNX1T1 3′UTR, acting as ceRNAs and contributing to t(8;21) alterations of transcriptional balance during AML development, has been addressed by a recent report [81]. Results showed a total of 605 RUNX1T1 ceRNAs significantly enriched in gene ontology (GO) categories mainly associated with leukemia, suggesting the hypothesis that RUNX1T1 may also act as a miRNA sponge in t(8;21) AML, and contributing to explain the complex pattern of gene expression alterations observed in CBFL.
5.1. Down-Regulation of miR-222/221 in AML with Deranged Core Binding Factor
One of the most common mechanisms through which miRNA expression dysregulates in AML is yielded by altered transcription factors or oncogenic fusion proteins through epigenetic alterations. Remarkably, the expression of AML1-ETO triggers heterochromatic silencing of genomic regions generating the miR-223 by recruiting chromatin remodeling enzymes at a RUNX1-binding site on the pre-miR-223 gene [82]. The AML1-ETO-associated complex resets the miR-223 gene to a repressed state by changes in chromatin conformation, contributing to the granulocytic differentiation block of the myeloid precursors. Of interest that either ectopic miR-223 expression, down-regulation of AML1-ETO protein levels, or the use of demethylating agents reactivates miR-223 expression and restore myeloid differentiation in t(8;21)-AML blasts [82]. Furthermore, the expression level of both KIT mRNA and proteins is much higher in the CBFL, with either wild-type or mutant KIT, than in leukemia cells negative for CBF rearrangements [83,84]. Gain-of-function KIT mutations, resulting in constitutive tyrosine kinase activity, are significantly enriched in patients with core binding factor leukemia [85,86], and these mutations are associated in t(8;21)-related leukemia with unfavorable outcome [87,88]. Aberrant activation of KIT results in MYC (MYC Proto-Oncogene, BHLH Transcription Factor)-dependent miR-29b down-regulation and an increase in Sp1 expression that results in KIT overexpression by NFkB transactivation, implicating deregulation of the protein–miRNA network Sp1/NFkB/HDAC/miR-29b in KIT-driven leukemia [89]. However, the molecular mechanisms explaining the peculiar association between rearrangements involving the CBF subunits and overexpression of either wild-type or mutant KIT receptor appear much more complex. We reported that CBFL blasts with either t(8;21) or inv(16) rearrangements, characterized by higher expression levels of KIT, display a significantly lower expression levels of miR-222/221 cluster, a negative modulator of KIT, than non-CBFL blasts. Consistently, the t(8;21)-derived fusion protein induces transcriptional repression of the pre-miR-222/221 promoter by binding at the evolutionarily conserved RUNX1-binding sites and thus leading to KIT overexpression [84].
5.2. Epigenetic Silencing of miR-193a Contributes to t(8;21)-Mediated AML
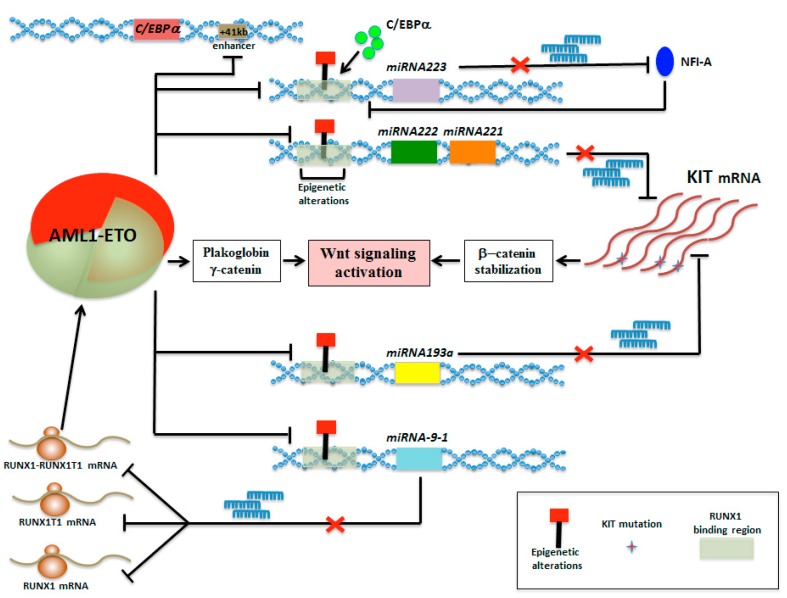
Interestingly, miR-193a represses the expression of multiple target genes, such as RUNX1-RUNX1T1, DNMT3A (DNA methyltransferase 3 alpha), HDAC3 (histone deacetylase3), KIT, CCND1 (B-cell leukemia/lymphoma 1, BCL1), and MDM2 (transformed mouse 3T3 cell double minute 2) directly, and increases PTEN (phosphatase and tensin homolog deleted on chromosome ten) indirectly. AML1-ETO triggers the heterochromatic silencing at the RUNX1-binding sites of miR-193a by recruiting chromatin remodeling enzymes and expanding the oncogenic activity of the fusion protein [90]. These further studies add new insights into understanding the concomitant occurrence of CBF genetic rearrangements and overexpression of wild-type or mutant KIT in CBFL, explaining how CBF fusion proteins may therefore maintain expression of several genes (i.e., KIT, WT1) by repressing the expression patterns of their specific miRNAs (Figure 5) [49,84,90]. Another interesting notion is that KIT-ITD mutant cooperates with canonical Wnt signaling pathway, inactivating GSK3β by hyperphosphorylation, which results in increased β-catenin stability [91]. Hence, activation of Wnt signaling plays an important role in KIT-mediated transformation of myeloid cells. Of note, a step forward suggests that activated Wnt/β-catenin signaling induces RUNX1 transcription mainly through direct β-catenin binding to TBE Site-II, which is located in a highly conserved region within the P1-distal promoter of RUNX1 [92]. Therefore, Wnt/β-catenin signaling rapidly enhances RUNX1 expression in leukemia-derived cell lines and human CD34+ hematopoietic cells, suggesting that transcriptional deregulation of translocation-prone genes occurs prior to translocation [93]. Furthermore, recent results highlight the role of Wnt signaling in AML, describing a new rearrangement leading to WNT10B overexpression, with the exception of clinical contexts with recurrent cytogenetic abnormalities [94], suggesting a different mechanism promoting Wnt signaling activation in CBFL. In line with these findings, it appears interesting the functional relevance of Wnt signaling induced by CBFL fusion proteins via plakoglobin (γ-catenin) induction [95], which provides a possible explanation of the different molecular circuitry involved in Wnt signaling activation as a common feature of several balanced translocations in AML (Figure 5).
Figure 5.
Schematic representation of the AML1-ETO integrated regulatory network comprised micro-RNA minicircuitries promoting Wnt signaling in t(8;21)-AML. AML1-ETO triggers heterochromatic silencing of genomic regions generating the miR-223, miR-222/221 cluster, miR-193a, and miR-9-1 by recruiting chromatin remodeling enzymes at their specific RUNX1-binding sites; in turn, these miRNA repressions generate differentiation impairments and KIT up-modulation that converges with AML1-ETO to activate the Wnt signaling.
5.3. Epigenetic Mini-Circuit AML1-ETO/miR-9-1/miR-383 Contributes to t(8;21) Leukemogenesis
Additionally, new information indicates a feedback minicircuitry through which the expression of miR-9-1 was decreased by AML1-ETO repression activity, leading to increasing level of RUNX1, RUNX1T1, and RUNX1–RUNX1T1, which are all targeted by miR-9-1 (Figure 5) [26]. As thanatos-associated protein 10 (THAP10) is a nuclear protein that inhibits myeloid proliferation and promotes differentiation both in vitro and in vivo, AML1-ETO inhibits expression of the tumor suppressor THAP10 directly via epigenetic suppression of the THAP10 promoter and indirectly through transcriptional activation of miR-383 in t(8;21) AML, unveiling a novel epigenetic mini-circuitry of AML1-ETO/THAP10/miR-383 [28]. Many complexities of miRNA biology have already shed additional light on our understanding of how miRNAs function in AML, substantially improving our understanding of how miRNAs synergize within CBFL cells.
6. The Genomic Landscape of Core-Binding Factor Acute Myeloid Leukemias
Current treatment guidelines for CBFL with t(8;21) do not take into account heterogeneity in these patients, and thus, all CBFL patients generally receive the same induction and consolidation treatments. Many comprehensive genetic analyses recognize that combination of several genetic alterations is associated with the development of CBFL, and is necessary for a better risk stratification in this leukemia. Although the spectrum of mutations for both CBFL subtypes is similar to the reported signature for AML [96], gene expression and mutation profiling of CBFL identified t(8;21) and inv(16) patients as two distinct subgroups [97], reflecting alternative signals activated in each subtype of CBFL [98]. Moreover, 35% of CBFL patients have two or more mutations in tyrosine kinase (TK) genes coding for pathway effectors (especially KIT, FLT3, and RAS genes); these findings highlight the multiclonality of CBFL. NRAS is the most frequently mutated gene in CBFL, more common in CBFB–MYH11 with a different spectrum of mutations, yet its mutations are not associated with outcome. KIT mutations are found in ~40% of CBFL with t(8;21) and ~33% with inv(16); additionally, an enrichment of exon 17 KIT mutations has been documented in RUNX1–RUNX1T1 patients, and are associated with worse outcome [87,99,100,101]. Recent large study created an “International CBF group index for t(8;21)” and validated a new risk scoring system, showing that older age, higher WBC index at diagnosis [102], and KIT D816V/Y mutations were risk factors associated with treatment failure (relapse or death) [103]. These studies strongly support the adverse effect of a KIT mutation in the context of CBFL. In addition, a novel finding indicates that pseudodiploidy was also a risk factor in t(8;21). High-risk score patients may benefit from more intensive approaches in the first complete remission (CR1) [103]. Mutations affecting FLT3–ITD are present in only 3% of inv(16) AML, whereas they occur in 10% of t(8;21) leukemia patients. In addition to mutations in genes involving TK signaling, alterations in MGA (MAX dimerization protein), a negative regulator of MYC signaling, were also recurrently identified in CBFL [104]. Recent results identified CCND2 (cyclin D2) expression as key transmitter of RUNX–RUNX1T1-driven AML, promoting cell cycle progression with the cooperation of the transcription factor Activator protein 1 (AP-1), and suggesting new potentially targetable complexes in CBFL [14,105]. Loss-of-function mutations in genes that regulate chromatin-modifying genes (ASXL1/2, EZH2, KDM6A, BCOR/BCORL1, EED, SETD2, KMT2D, KMT2C, and CREBBP) or in genes implicated in the cohesin complex (RAD21, SMC1A, SMC3, STAG2) were observed almost exclusively in RUNX1–RUNX1T1 AML. Cohesin mutations led to a state of increased chromatin accessibility of binding sites for master hematopoietic transcription factors such as AML1 [106]. These findings suggest links between cohesin-mediated alterations in chromatin structure, or chromatin modifiers mutations, and cooperativity with the AML1–ETO fusion oncoprotein, even if cohesin mutations concerned less than 10% of CBFL [15]. CBFL patients with mutations in the above members of the complex, responsible for sister chromatid cohesion during mitosis and DNA repair, lack evidence of aneuploidy or an increase rate of genetic instability without any effect on the outcome. Recently, mutations in ASXL1 (additional sex combs like 1), ASXL2 (additional sex combs like 2), ZBTB7A (zinc finger and BTB domain conteining 7A), CCND2, and DHX15 (DEAH-box helicase 15) have been frequently identified in RUNX1–RUNX1T1 but not in CBFB–MYH11 AML patients [14,107]. ASXL1 or ASXL2 truncating mutations, which inhibit myeloid differentiation and induce a myelodysplastic syndrome-like disease in mice [108,109], have been described in ~35% of t(8;21) while are absent in inv(16) AML [15,110,111]. Of interest, chromatin modifier ASXL1, as well as cohesin gene mutations, are co-occurring alterations significantly enriched in patients with mutated RUNX1 AML [112,113]. The nature of cooperating mutations associated with t(8;21)-mediated leukemogenesis is evidenced by additional cytogenetic abnormalities such as trisomy 8 and 4, chromosome 9 deletion, and loss of one of the sex chromosomes [114,115,116]. Increased dosage of the mutated KIT (mapped at 4q12) can occur due to trisomy 4, leading to duplication of the mutant KIT allele, and suggesting an additional contribution to leukemogenesis [86]. These observations are supported by a higher dosage of N822K KIT mutated allele linked to an increased segregation of minichromosomes derived from chromosome 4 that preserve the pericentromeric region containing the KIT gene in the t(8;21) positive Kasumi-1 cell line [117]. The most common additional cytogenetic features associated with t(8;21) include loss of either the X or Y chromosome in a disproportionally large number of cases (50–60%), and del(9)(q22) in 15–25% of patients. It has been proposed that haploinsufficiency must be occurring at genes located within shared sequences in the pseudoautosomal regions (PARs) on the X and Y chromosome. A critical event potentially explaining the high incidence of loss of sex chromosomes in t(8;21) may be the loss of CSF2RA (colony stimulating factor 2 receptor alpha subunit) gene, encoding for the α subunit of the heterodimeric receptor CSF2 (colony-stimulating factor 2), which control granulopoiesis [118]. However, given that the whole sex chromosome is typically missing and not only the individual CSF2RA locus, it is likely that additional haploinsufficient factors on the sex chromosome are acting to enhance RUNX1–RUNX1T1-associated leukemogenesis [119]. Sex chromosome loss was reported as a favorable marker for two-year event-free survival (66.9% vs. 43.0%), and in another study showed a modestly favorable, but not significant, effect on disease-free survival (DFS) [103]. Moreover, this last study found that patients with pseudodiploid karyotypes had worse outcome compared with those with hypodiploidy or hyperdiploidy [103]. In contrast, loss of the Y chromosome showed shorter disease-free survival (DFS) for male patients [120].
7. Mouse Models for Core Binding Factor Leukemia
The homozygous disruption of Runx1 or Cbfb in murine knockout models exhibits a similar range of abnormalities associated with developmental defects. These common phenotypes include severe hematopoietic defects such as lack of HSCs and progenitors leading to midgestation embryonic lethality between embryonic days (E) 12.5–13.5. Thus, creating mice deficient in Runx1/Cbfb, lacking the ability to contribute to definitive hematopoiesis, several research groups have shown that the AML1/CBFβ transcription factor complex is essential in the hematopoietic fate process from the hemogenic endothelial cells. [29,121,122,123]. Moreover, all heterozygous Runx1-Runx1t1 knock-in mice die around 12.5 days of embryogenesis and fail to establish definitive hematopoiesis [124,125]. This similarity in phenotypes suggests that AML1-ETO effectively neutralizes the normal biologic activity of the AML1/CBFβ transcriptional factor complex and dominantly blocks AML1 activity during early development. Both mouse and in-vitro models have shown that the expression of Runx1-Runx1t1 or Cbfb-Myh11 contribute to leukemogenesis but require additional “hits”. An interesting observation that emerged from Cre recombinase-mediated conditional AML1-ETO expression transgenic mice is that the non-leukemic AML1–ETO expressing cells were cytokine-dependent, strongly suggesting that one signaling pathway that may collaborate with AML1–ETO is cytokine-mediated proliferation or survival [13,126]. The first step towards CBFL would consist of the acquisition of genetic alterations, like the CBFL translocations, in modulators of differentiation (class 2 mutations) by the preleukemic clone; the second step would be the acquisition of activating mutations in cell cycle and proliferation controllers (class 1 mutations) such as KIT, FLT3, or N-RAS tyrosine kinases [14,86,127,128,129,130,131], and their cooperation with Runx1-Runx1t1 or Cbfb-Myh11 is thought to be crucial for leukemogenesis [132]. CBFβ-SMMHC dominantly represses AML1 function, generates defects in definitive hematopoiesis [133], and predisposes mice to leukemia with cooperating gene mutations [12,134]. Therefore, the fusion protein alone is necessary but not sufficient to cause leukemia, and activating mutations in genes encoding for receptor tyrosine kinases (RTKs) or small GTPase represent the most common genetic events cooperating with CBFL-associated gene fusions [12,126,134]. Although RUNX1-RUNX1T1 and CBFB-MYH11 share a common molecular alteration involving the CBF transcription complex, they up-regulate specific signaling pathways essential for stem cell self-renewal and have remarkably different spectra of cooperating mutations [14,95,96,135,136].
The HSCs from adult chimeric mice generated with CbfbCbfb–MYH11/+ ES cells were found to give rise efficiently to mature erythrocytes, but were unable to differentiate along myeloid and lymphoid lineages [58], suggesting that the differentiation impairment involves the lympho-myeloid primed progenitor (LMPP) as from the revised hematopoietic tree [137]. Knockdown of Runx1 inhibits the growth and survival of Runx1-Runx1t1 leukemia cells [138,139]. Moreover, a variant of the AML1-ETO fusion protein, Runx1-Runx1t19a (ETO9a), which includes an extra exon 9a of the Runx1t1 gene that contains C-terminal truncation, was found to be a much more potent inducer of leukemia than the full-length Runx1-Runx1t1 in mouse retroviral transduction–transplantation model [140]. It has been hypothesized that the deleted region inhibits the leukemogenic potential of AML1-ETO [141]. Interestingly Runx1-Runx1t1tr, a C-terminally truncated protein similar to Runx1-Runx1t19a, lost the ability to inhibit cell cycle progression of myeloid cells, which may contribute to its enhanced leukemogenic potential [142]. Moreover, the ability to regulate the expression of the CD44 gene of both RUNX1-RUNX1T1 and its splice variant RUNX-RUNX1T19a links the t(8;21) translocation to the regulation of a cell adhesion molecule involved in the growth and maintenance of the AML blast/stem cells [141]. Novel integrative data analyses, together with siRNA-mediated depletion of AML1-ETO, strongly support the notion that AML1-ETO binds genes associated with the cell structure and cell-cycle progression affecting transcriptional programs associated with myeloid differentiation, proliferation, and self-renewal, in addition to those promoting DNA synthesis [49]. H3K9Ac at AML1-ETO-binding sites show a significant increase after knockdown, which would be compatible with a repressive role of the fusion protein at these sites.
Critical genes such as CSF2RA (GM-CSF receptor alpha) and IL3RA (IL3 receptor alpha) on human sex chromosomes are localized to syntenic regions on murine 19 chromosome. The receptors for granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin (IL)-3, and IL-5 all share a common β chain (βC) but have a ligand-specific α chain [143]. Matsuura and colleagues [118], in order to explain the selective advantage for sex chromosome loss in t(8;21)-AML, used a transduction/transplantation assay where Runx1-Runx1t1 was expressed from a retroviral vector in βc-knockout bone marrow cells, which were then transplanted into irradiated wild-type recipient animals. Interestingly, loss of βC in association with AML1-ETO significantly accelerated progression to AML. GM-CSF signaling deficiency is favorable for leukemia development driven by Runx1-Runx1t1 in mouse models. Moreover, GM-CSF signaling inhibits RUNX1-RUNX1T1-mediated leukemogenesis by reducing the self-renewal potential of hematopoietic stem and progenitor cells in murine bone marrow and in human t(8;21) Kasumi-1 cells. These findings suggest an unexpected tumor-suppressor role of GM-CSF in t(8;21) leukemias.
8. Molecular Targeted Therapy of CBFL: The Progress and Future Prospect
Individual genomic characterization may be useful to suggest or not targeted interventions, such as the use of RTK inhibitors, for instance dasatinib [144,145] or midostaurin [146], in patients carrying activating KIT and/or FLT3 gene mutations. KIT mutations are common in CBFL and have been associated with worse prognosis (shorter disease-free survival, relapse-free survival, event-free survival, overall survival) [147]. A phase III study of gemtuzumab ozogamicin (Mylotarg®; Pfizer/Wyeth-Ayerst Laboratories) showed that GO abrogated the negative prognostic effect of exon 17 (E17) mutations in treated patients [148]. Moreover, the outcome of patients harboring KIT mutations in the Cancer and Leukemia Group B (CALGB) study, appeared not to be worse than that of patients with KIT wild-type after treatment with dasatinib, implying that a potentially adverse impact of KIT mutations might be abrogated in treated patients [144]. Conversely, within the German-Austrian AML Study Group trial (AMLSG 11- 08 trial), no favorable impact after dasatinib administration was noted for patients with concurrent KIT mutation [145]. Although validation by other studies is needed, the rationale to combine dasatinib with other compounds was supported by data from a mouse model of t(8;21)-positive and KIT-mutated leukemia, where the combination of dasatinib with cytarabine prolonged the survival of the animals compared to the exposure with these drugs as single agents [146,149]. As acquisition of spliceosome gene mutations result to be the initiating events in clonal hematopoiesis, the use of spliceosomal modulation to induce synthetic lethality in splicing factor mutant hematological malignancies, currently being tested in an early phase clinical trial, is quite exciting [150]. CBFL is a still heterogeneous disease entity, and for better characterization of CBFL risk heterogeneity in the spirit of precision medicine, it is necessary to elucidate the combinations of genomic abnormalities and clonal evolutions using refined high-resolution genomic analysis to develop new treatment strategies for CBFL.
9. Conclusions
Genetic definition of CBFL patients using deep sequencing approaches illustrate that we are only beginning to understand how fusion proteins involving the CBF are integrated into the molecular networks of transcriptional and epigenetic regulators. Despite the generation of in vivo models helped us to understand how CBFL originates and propagates, we can imagine a future where the understanding of how expression and CBF fusion proteins activity are modulated during myeloid leukemia transformation and progression will trigger a true progress to translate into the clinic. Such achievements will become extremely useful, in a view of individual treatment on the basis of defined targets. Future studies will be required to identify which CBFL patients could benefit from therapy by each molecular drug combination targeting a specific pathway.
Acknowledgments
The author would like to thank R. Cairoli and R. Brusamolino for helpful discussions.
Funding
This work was supported in part by a grant from Fondazione Regionale per la Ricerca Biomedica (FRRB-2015) and Fondazione Malattie del Sangue Onlus (FMS 2019).
Conflicts of Interest
The author declares no conflicts of interest.
References
- 1.Miyoshi H., Shimizu K., Kozu T., Maseki N., Kaneko Y., Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Link K.A., Chou F.-S., Mulloy J.C. Core binding factor at the crossroads: Determining the fate of the HSC. J. Cell. Physiol. 2010;222:50–56. doi: 10.1002/jcp.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Bruijn M., Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood. 2017;129:2061–2069. doi: 10.1182/blood-2016-12-689109. [DOI] [PubMed] [Google Scholar]
- 4.Menegatti S., de Kruijf M., Garcia-Alegria E., Lacaud G., Kouskoff V. Transcriptional control of blood cell emergence. FEBS Lett. 2019 doi: 10.1002/1873-3468.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P., Tarlé S.A., Hajra A., Claxton D.F., Marlton P., Freedman M., Siciliano M.J., Collins F.S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 6.Goyama S., Mulloy J.C. Molecular pathogenesis of core binding factor leukemia: Current knowledge and future prospects. Int. J. Hematol. 2011;94:126–133. doi: 10.1007/s12185-011-0858-z. [DOI] [PubMed] [Google Scholar]
- 7.Sinha C., Cunningham L.C., Liu P.P. Core Binding Factor Acute Myeloid Leukemia: New Prognostic Categories and Therapeutic Opportunities. Semin. Hematol. 2015;52:215–222. doi: 10.1053/j.seminhematol.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlenk R.F., Benner A., Krauter J., Büchner T., Sauerland C., Ehninger G., Schaich M., Mohr B., Niederwieser D., Krahl R., et al. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: A survey of the German Acute Myeloid Leukemia Intergroup. J. Clin. Oncol. 2004;22:3741–3750. doi: 10.1200/JCO.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Jourdan E., Boissel N., Chevret S., Delabesse E., Renneville A., Cornillet P., Blanchet O., Cayuela J.-M., Recher C., Raffoux E., et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood. 2013;121:2213–2223. doi: 10.1182/blood-2012-10-462879. [DOI] [PubMed] [Google Scholar]
- 10.Ghamari A., van de Corput M.P.C., Thongjuea S., van Cappellen W.A., van Ijcken W., van Haren J., Soler E., Eick D., Lenhard B., Grosveld F.G. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev. 2013;27:767–777. doi: 10.1101/gad.216200.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ugarte G.D., Vargas M.F., Medina M.A., León P., Necuñir D., Elorza A.A., Gutiérrez S.E., Moon R.T., Loyola A., De Ferrari G.V. Wnt signaling induces transcription, spatial proximity, and translocation of fusion gene partners in human hematopoietic cells. Blood. 2015;126:1785–1789. doi: 10.1182/blood-2015-04-638494. [DOI] [PubMed] [Google Scholar]
- 12.Castilla L.H., Garrett L., Adya N., Orlic D., Dutra A., Anderson S., Owens J., Eckhaus M., Bodine D., Liu P.P. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat. Genet. 1999;23:144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Y., Zhou L., Miyamoto T., Iwasaki H., Harakawa N., Hetherington C.J., Burel S.A., Lagasse E., Weissman I.L., Akashi K., et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc. Natl. Acad. Sci. USA. 2001;98:10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faber Z.J., Chen X., Gedman A.L., Boggs K., Cheng J., Ma J., Radtke I., Chao J.-R., Walsh M.P., Song G., et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat. Genet. 2016;48:1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duployez N., Marceau-Renault A., Boissel N., Petit A., Bucci M., Geffroy S., Lapillonne H., Renneville A., Ragu C., Figeac M., et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood. 2016;127:2451–2459. doi: 10.1182/blood-2015-12-688705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solh M., Yohe S., Weisdorf D., Ustun C. Core-binding factor acute myeloid leukemia: Heterogeneity, monitoring, and therapy. Am. J. Hematol. 2014;89:1121–1131. doi: 10.1002/ajh.23821. [DOI] [PubMed] [Google Scholar]
- 17.Corces-Zimmerman M.R., Hong W.-J., Weissman I.L., Medeiros B.C., Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shima T., Miyamoto T., Kikushige Y., Yuda J., Tochigi T., Yoshimoto G., Kato K., Takenaka K., Iwasaki H., Mizuno S., et al. The ordered acquisition of Class II and Class I mutations directs formation of human t(8;21) acute myelogenous leukemia stem cell. Exp. Hematol. 2014;42:955–965. doi: 10.1016/j.exphem.2014.07.267. [DOI] [PubMed] [Google Scholar]
- 19.Nucifora G., Larson R.A., Rowley J.D. Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood. 1993;82:712–715. doi: 10.1182/blood.V82.3.712.712. [DOI] [PubMed] [Google Scholar]
- 20.Jurlander J., Caligiuri M.A., Ruutu T., Baer M.R., Strout M.P., Oberkircher A.R., Hoffmann L., Ball E.D., Frei-Lahr D.A., Christiansen N.P., et al. Persistence of the AML1/ETO fusion transcript in patients treated with allogeneic bone marrow transplantation for t(8;21) leukemia. Blood. 1996;88:2183–2191. doi: 10.1182/blood.V88.6.2183.bloodjournal8862183. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto T., Nagafuji K., Akashi K., Harada M., Kyo T., Akashi T., Takenaka K., Mizuno S., Gondo H., Okamura T., et al. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia. Blood. 1996;87:4789–4796. doi: 10.1182/blood.V87.11.4789.bloodjournal87114789. [DOI] [PubMed] [Google Scholar]
- 22.Croce C.M., Calin G.A. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 23.Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 24.Fischer J., Rossetti S., Datta A., Eng K., Beghini A., Sacchi N., Datta A., Eng K., Beghini A., Sacchi N. miR-17 deregulates a core RUNX1-miRNA mechanism of CBF acute myeloid leukemia. Mol. Cancer. 2015;14 doi: 10.1186/s12943-014-0283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagano F., De Marinis E., Grignani F., Nervi C. Epigenetic role of miRNAs in normal and leukemic hematopoiesis. Epigenomics. 2013;5:539–552. doi: 10.2217/epi.13.55. [DOI] [PubMed] [Google Scholar]
- 26.Fu L., Shi J., Liu A., Zhou L., Jiang M., Fu H., Xu K., Li D., Deng A., Zhang Q., et al. A minicircuitry of microRNA-9-1 and RUNX1-RUNX1T1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Int. J. Cancer. 2017;140:653–661. doi: 10.1002/ijc.30481. [DOI] [PubMed] [Google Scholar]
- 27.Wallace J.A., O’Connell R.M. MicroRNAs and acute myeloid leukemia: Therapeutic implications and emerging concepts. Blood. 2017;130:1290–1301. doi: 10.1182/blood-2016-10-697698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Ning Q., Shi J., Chen Y., Jiang M., Gao L., Huang W., Jing Y., Huang S., Liu A., et al. A novel epigenetic AML1-ETO/THAP10/miR-383 mini-circuitry contributes to t(8;21) leukaemogenesis. EMBO Mol. Med. 2017;9:933–949. doi: 10.15252/emmm.201607180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuda T., van Deursen J., Hiebert S.W., Grosveld G., Downing J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/S0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q., Stacy T., Miller J.D., Lewis A.F., Gu T.L., Huang X., Bushweller J.H., Bories J.C., Alt F.W., Ryan G., et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/S0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 31.Niki M., Okada H., Takano H., Kuno J., Tani K., Hibino H., Asano S., Ito Y., Satake M., Noda T. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl. Acad. Sci. USA. 1997;94:5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichikawa M., Asai T., Saito T., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Ogawa S., Kurokawa M., et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 33.Growney J.D., Shigematsu H., Li Z., Lee B.H., Adelsperger J., Rowan R., Curley D.P., Kutok J.L., Akashi K., Williams I.R., et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putz G., Rosner A., Nuesslein I., Schmitz N., Buchholz F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene. 2006;25:929–939. doi: 10.1038/sj.onc.1209136. [DOI] [PubMed] [Google Scholar]
- 35.Motoda L., Osato M., Yamashita N., Jacob B., Chen L.Q., Yanagida M., Ida H., Wee H.-J., Sun A.X., Taniuchi I., et al. Runx1 protects hematopoietic stem/progenitor cells from oncogenic insult. Stem Cells. 2007;25:2976–2986. doi: 10.1634/stemcells.2007-0061. [DOI] [PubMed] [Google Scholar]
- 36.Aikawa Y., Nguyen L.A., Isono K., Takakura N., Tagata Y., Schmitz M.L., Koseki H., Kitabayashi I. Roles of HIPK1 and HIPK2 in AML1-and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006;25:3955–3965. doi: 10.1038/sj.emboj.7601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu T.L., Goetz T.L., Graves B.J., Speck N.A. Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1) Mol. Cell. Biol. 2000;20:91–103. doi: 10.1128/MCB.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Cheney M.D., Gaudet J.J., Chruszcz M., Lukasik S.M., Sugiyama D., Lary J., Cole J., Dauter Z., Minor W., et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell. 2006;9:249–260. doi: 10.1016/j.ccr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 39.Park S., Speck N.A., Bushweller J.H. The role of CBFbeta in AML1-ETO’s activity. Blood. 2009;114:2849–2850. doi: 10.1182/blood-2009-07-231233. [DOI] [PubMed] [Google Scholar]
- 40.Roudaia L., Cheney M.D., Manuylova E., Chen W., Morrow M., Park S., Lee C.-T., Kaur P., Williams O., Bushweller J.H., et al. CBFbeta is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113:3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corpora T., Roudaia L., Oo Z.M., Chen W., Manuylova E., Cai X., Chen M.J., Cierpicki T., Speck N.A., Bushweller J.H. Structure of the AML1-ETO NHR3-PKA(RIIα) complex and its contribution to AML1-ETO activity. J. Mol. Biol. 2010;402:560–577. doi: 10.1016/j.jmb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X.-J., Wang Z., Wang L., Jiang Y., Kost N., Soong T.D., Chen W.-Y., Tang Z., Nakadai T., Elemento O., et al. A stable transcription factor complex nucleated by oligomeric AML1-ETO controls leukaemogenesis. Nature. 2013;500:93–97. doi: 10.1038/nature12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueroa M.E., Lugthart S., Li Y., Erpelinck-Verschueren C., Deng X., Christos P.J., Schifano E., Booth J., van Putten W., Skrabanek L., et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Hoshino T., Redner R.L., Kajigaya S., Liu J.M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S., Shen T., Huynh L., Klisovic M.I., Rush L.J., Ford J.L., Yu J., Becknell B., Li Y., Liu C., et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res. 2005;65:1277–1284. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 46.Linggi B., Müller-Tidow C., van de Locht L., Hu M., Nip J., Serve H., Berdel W.E., van der Reijden B., Quelle D.E., Rowley J.D., et al. The t(8;21) fusion protein, AML1–ETO, specifically represses the transcription of the p14ARF tumor suppressor in acute myeloid leukemia. Nat. Med. 2002;8:743–750. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- 47.Friedman A.D. C/EBPα in normal and malignant myelopoiesis. Int. J. Hematol. 2015;101:330–341. doi: 10.1007/s12185-015-1764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao X.N., Yan F., Lin J., Gao L., Lu X.L., Wei S.C., Shen N., Pang J.X., Ning Q.Y., Komeno Y., et al. AML1/ETO cooperates with HIF1α to promote leukemogenesis through DNMT3a transactivation. Leukemia. 2015;29:1730–1740. doi: 10.1038/leu.2015.56. [DOI] [PubMed] [Google Scholar]
- 49.Ptasinska A., Assi S.A., Mannari D., James S.R., Williamson D., Dunne J., Hoogenkamp M., Wu M., Care M., McNeill H., et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012;26:1829–1841. doi: 10.1038/leu.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pabst T., Mueller B.U., Harakawa N., Schoch C., Haferlach T., Behre G., Hiddemann W., Zhang D.E., Tenen D.G. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat. Med. 2001;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 51.Vangala R.K., Heiss-Neumann M.S., Rangatia J.S., Singh S.M., Schoch C., Tenen D.G., Hiddemann W., Behre G. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101:270–277. doi: 10.1182/blood-2002-04-1288. [DOI] [PubMed] [Google Scholar]
- 52.Lutterbach B., Westendorf J.J., Linggi B., Patten A., Moniwa M., Davie J.R., Huynh K.D., Bardwell V.J., Lavinsky R.M., Rosenfeld M.G., et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 1998;18:7176–7184. doi: 10.1128/MCB.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J., Hug B.A., Huang E.Y., Chen C.W., Gelmetti V., Maccarana M., Minucci S., Pelicci P.G., Lazar M.A. Oligomerization of ETO is obligatory for corepressor interaction. Mol. Cell. Biol. 2001;21:156–163. doi: 10.1128/MCB.21.1.156-163.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lutterbach B., Westendorf J.J., Linggi B., Isaac S., Seto E., Hiebert S.W. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- 55.Kamikubo Y., Zhao L., Wunderlich M., Corpora T., Hyde R.K., Paul T.A., Kundu M., Garrett L., Compton S., Huang G., et al. Accelerated leukemogenesis by truncated CBFβ-SMMHC defective in high-affinity binding with RUNX1. Cancer Cell. 2010;17:455–468. doi: 10.1016/j.ccr.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y., Wang H., Wang X., Jin W., Tan Y., Fang H., Chen S., Chen Z., Wang K. Genome-wide studies identify a novel interplay between AML1 and AML1/ETO in t(8;21) acute myeloid leukemia. Blood. 2016;127:233–242. doi: 10.1182/blood-2015-03-626671. [DOI] [PubMed] [Google Scholar]
- 57.Durst K.L., Lutterbach B., Kummalue T., Friedman A.D., Hiebert S.W. The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol. Cell. Biol. 2003;23:607–619. doi: 10.1128/MCB.23.2.607-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller J.D., Stacy T., Liu P.P., Speck N.A. Core-binding factor beta (CBFbeta), but not CBFbeta-smooth muscle myosin heavy chain, rescues definitive hematopoiesis in CBFbeta-deficient embryonic stem cells. Blood. 2001;97:2248–2256. doi: 10.1182/blood.V97.8.2248. [DOI] [PubMed] [Google Scholar]
- 59.Richter L.E., Wang Y., Becker M.E., Coburn R.A., Williams J.T., Amador C., Hyde R.K. HDAC1 Is a Required Cofactor of CBFβ-SMMHC and a Potential Therapeutic Target in Inversion 16 Acute Myeloid Leukemia. Mol. Cancer Res. 2019;17:1241–1252. doi: 10.1158/1541-7786.MCR-18-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandoli A., Singh A.A., Jansen P.W.T.C., Wierenga A.T.J., Riahi H., Franci G., Prange K., Saeed S., Vellenga E., Vermeulen M., et al. CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia. 2014;28:770–778. doi: 10.1038/leu.2013.257. [DOI] [PubMed] [Google Scholar]
- 61.Hyde R.K., Zhao L., Alemu L., Liu P.P. Runx1 is required for hematopoietic defects and leukemogenesis in Cbfb-MYH11 knock-in mice. Leukemia. 2015;29:1771–1778. doi: 10.1038/leu.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhen T., Kwon E.M., Zhao L., Hsu J., Hyde R.K., Lu Y., Alemu L., Speck N.A., Liu P.P. Chd7 deficiency delays leukemogenesis in mice induced by Cbfb-MYH11. Blood. 2017;130:2431–2442. doi: 10.1182/blood-2017-04-780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warren A.J., Bravo J., Williams R.L., Rabbitts T.H. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFβ. EMBO J. 2000;19:3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lukasik S.M., Zhang L., Corpora T., Tomanicek S., Li Y., Kundu M., Hartman K., Liu P.P., Laue T.M., Biltonen R.L., et al. Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat. Struct. Biol. 2002;9:674–679. doi: 10.1038/nsb831. [DOI] [PubMed] [Google Scholar]
- 65.Huang G., Shigesada K., Wee H.-J., Liu P.P., Osato M., Ito Y. Molecular basis for a dominant inactivation of RUNX1/AML1 by the leukemogenic inversion 16 chimera. Blood. 2004;103:3200–3207. doi: 10.1182/blood-2003-07-2188. [DOI] [PubMed] [Google Scholar]
- 66.Wee H.-J., Voon D.C.-C., Bae S.-C., Ito Y. PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2: Implications for leukemogenesis. Blood. 2008;112:3777–3787. doi: 10.1182/blood-2008-01-134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi G., Mandoli A., Jussen L., Tijchon E., van Bergen M.G.J.M., Cordonnier G., Hansen M., Kim B., Nguyen L.N., Jansen P.W.T.C., et al. CBFβ-MYH11 interferes with megakaryocyte differentiation via modulating a gene program that includes GATA2 and KLF1. Blood Cancer J. 2019;9:e33. doi: 10.1038/s41408-019-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saida S., Zhen T., Kim E., Yu K., Lopez G., McReynolds L.J., Liu P.P. Gata2 deficiency delays leukemogenesis while contributing to aggressive leukemia phenotype in Cbfb-MYH11 knockin mice. Leukemia. 2019 doi: 10.1038/s41375-019-0605-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sood R., Hansen N.F., Donovan F.X., Carrington B., Bucci D., Maskeri B., Young A., Trivedi N.S., Kohlschmidt J., Stone R.M., et al. Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia. Leukemia. 2016;30:501–504. doi: 10.1038/leu.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Visconte V., Nakashima M.O., Rogers H.J. Mutations in splicing factor genes in myeloid malignancies: Significance and impact on clinical features. Cancers. 2019;11:1844. doi: 10.3390/cancers11121844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 72.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nature. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 73.Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., Liuzzi F., Lulli V., Morsilli O., Santoro S., et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garzon R., Pichiorri F., Palumbo T., Iuliano R., Cimmino A., Aqeilan R., Volinia S., Bhatt D., Alder H., Marcucci G., et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc. Natl. Acad. Sci. USA. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnnidis J.B., Harris M.H., Wheeler R.T., Stehling-Sun S., Lam M.H., Kirak O., Brummelkamp T.R., Fleming M.D., Camargo F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 76.Petriv O.I., Kuchenbauer F., Delaney A.D., Lecault V., White A., Kent D., Marmolejo L., Heuser M., Berg T., Copley M., et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc. Natl. Acad. Sci. USA. 2010;107:15443–15448. doi: 10.1073/pnas.1009320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Georgantas R.W., Hildreth R., Morisot S., Alder J., Liu C., Heimfeld S., Calin G.A., Croce C.M., Civin C.I. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc. Natl. Acad. Sci. USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwarzer A., Emmrich S., Schmidt F., Beck D., Ng M., Reimer C., Adams F.F., Grasedieck S., Witte D., Käbler S., et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nat. Commun. 2017;8:e218. doi: 10.1038/s41467-017-00212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. [(accessed on 1 May 2013)]; doi: 10.1056/NEJMoa1301689. Available online: https://www.nejm.org/doi/full/10.1056/nejmoa1301689. [DOI] [PMC free article] [PubMed]
- 80.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Junge A., Zandi R., Havgaard J.H., Gorodkin J., Cowland J.B. Assessing the miRNA sponge potential of RUNX1T1 in t(8;21) acute myeloid leukemia. Gene. 2017;615:35–40. doi: 10.1016/j.gene.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 82.Fazi F., Racanicchi S., Zardo G., Starnes L.M., Mancini M., Travaglini L., Diverio D., Ammatuna E., Cimino G., Lo-Coco F., et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y.-Y., Zhou G.-B., Yin T., Chen B., Shi J.-Y., Liang W.-X., Jin X.-L., You J.-H., Yang G., Shen Z.-X., et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: Implication in stepwise leukemogenesis and response to Gleevec. Proc. Natl. Acad. Sci. USA. 2005;102:1104–1109. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brioschi M., Fischer J., Cairoli R., Rossetti S., Pezzetti L., Nichelatti M., Turrini M., Corlazzoli F., Scarpati B., Morra E., et al. Down-regulation of microRNAs 222/221 in acute myelogenous leukemia with deranged core-binding factor subunits. Neoplasia. 2010;12:866–876. doi: 10.1593/neo.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beghini A., Cairoli R., Morra E., Lariza L. In vivo differentiation of mast cells from acute myeloid leukemia blasts carrying a novel activating ligand-independent C-kit mutation. Blood Cells Mol. Dis. 1998;24:262–270. doi: 10.1006/bcmd.1998.0191. [DOI] [PubMed] [Google Scholar]
- 86.Beghini A., Peterlongo P., Ripamonti C.B., Larizza L., Cairoli R., Morra E., Mecucci C. C-kit mutations in core binding factor leukemias. Blood. 2000;95:726–727. doi: 10.1182/blood.V95.2.726. [DOI] [PubMed] [Google Scholar]
- 87.Cairoli R., Beghini A., Grillo G., Nadali G., Elice F., Ripamonti C.B., Colapietro P., Nichelatti M., Pezzetti L., Lunghi M., et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: An Italian retrospective study. Blood. 2006;107:3463–3468. doi: 10.1182/blood-2005-09-3640. [DOI] [PubMed] [Google Scholar]
- 88.Paschka P., Marcucci G., Ruppert A.S., Mrózek K., Chen H., Kittles R.A., Vukosavljevic T., Perrotti D., Vardiman J.W., Carroll A.J., et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 89.Liu S., Wu L.-C., Pang J., Santhanam R., Schwind S., Wu Y.-Z., Hickey C.J., Yu J., Becker H., Maharry K., et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y., Gao L., Luo X., Wang L., Gao X., Wang W., Sun J., Dou L., Li J., Xu C., et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121:499–509. doi: 10.1182/blood-2012-07-444729. [DOI] [PubMed] [Google Scholar]
- 91.Tickenbrock L., Hehn S., Sargin B., Evers G., Ng P.R., Choudhary C., Berdel W.E., Müller-Tidow C., Serve H. Activation of Wnt signaling in cKit-ITD mediated transformation and imatinib sensitivity in acute myeloid leukemia. Int. J. Hematol. 2008;88:174–180. doi: 10.1007/s12185-008-0141-0. [DOI] [PubMed] [Google Scholar]
- 92.Medina M.A., Ugarte G.D., Vargas M.F., Avila M.E., Necuñir D., Elorza A.A., Gutiérrez S.E., De Ferrari G.V. Alternative RUNX1 Promoter Regulation by Wnt/β-Catenin Signaling in Leukemia Cells and Human Hematopoietic Progenitors. J. Cell. Physiol. 2016;231:1460–1467. doi: 10.1002/jcp.25258. [DOI] [PubMed] [Google Scholar]
- 93.Mathas S., Kreher S., Meaburn K.J., Jöhrens K., Lamprecht B., Assaf C., Sterry W., Kadin M.E., Daibata M., Joos S., et al. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc. Natl. Acad. Sci. USA. 2009;106:5831–5836. doi: 10.1073/pnas.0900912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lazzaroni F., Del Giacco L., Biasci D., Turrini M., Prosperi L., Brusamolino R., Cairoli R., Beghini A. Intronless WNT10B-short variant underlies new recurrent allele-specific rearrangement in acute myeloid leukaemia. Sci. Rep. 2016;6:e37201. doi: 10.1038/srep37201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Müller-Tidow C., Steffen B., Cauvet T., Tickenbrock L., Ji P., Diederichs S., Sargin B., Köhler G., Stelljes M., Puccetti E., et al. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol. Cell. Biol. 2004;24:2890–2904. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tyner J.W., Tognon C.E., Bottomly D., Wilmot B., Kurtz S.E., Savage S.L., Long N., Schultz A.R., Traer E., Abel M., et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schoch C., Kohlmann A., Schnittger S., Brors B., Dugas M., Mergenthaler S., Kern W., Hiddemann W., Eils R., Haferlach T. Acute myeloid leukemias with reciprocal rearrangements can be distinguished by specific gene expression profiles. Proc. Natl. Acad. Sci. USA. 2002;99:10008–10013. doi: 10.1073/pnas.142103599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hsu C.-H., Nguyen C., Yan C., Ries R.E., Chen Q.-R., Hu Y., Ostronoff F., Stirewalt D.L., Komatsoulis G., Levy S., et al. Transcriptome Profiling of Pediatric Core Binding Factor AML. PLoS ONE. 2015;10:e0138782. doi: 10.1371/journal.pone.0138782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Care R.S., Valk P.J.M., Goodeve A.C., Abu-Duhier F.M., Geertsma-Kleinekoort W.M.C., Wilson G.A., Gari M.A., Peake I.R., Löwenberg B., Reilly J.T. Incidence and prognosis of c-KIT and FLT3 mutations in core binding factor (CBF) acute myeloid leukaemias. Br. J. Haematol. 2003;121:775–777. doi: 10.1046/j.1365-2141.2003.04362.x. [DOI] [PubMed] [Google Scholar]
- 100.Beghini A., Ripamonti C.B., Cairoli R., Cazzaniga G., Colapietro P., Elice F., Nadali G., Grillo G., Haas O.A., Biondi A., et al. KIT activating mutations: Incidence in adult and pediatric acute myeloid leukemia, and identification of an internal tandem duplication. Haematologica. 2004;89:920–925. [PubMed] [Google Scholar]
- 101.Schnittger S., Kohl T.M., Haferlach T., Kern W., Hiddemann W., Spiekermann K., Schoch C. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107:1791–1799. doi: 10.1182/blood-2005-04-1466. [DOI] [PubMed] [Google Scholar]
- 102.Cairoli R., Grillo G., Beghini A., Tedeschi A., Ripamonti C.B., Larizza L., Morra E. C-Kit point mutations in core binding factor leukemias: Correlation with white blood cell count and the white blood cell index. Leukemia. 2003;17:471–472. doi: 10.1038/sj.leu.2402795. [DOI] [PubMed] [Google Scholar]
- 103.Ustun C., Morgan E., Moodie E.E.M., Pullarkat S., Yeung C., Broesby-Olsen S., Ohgami R., Kim Y., Sperr W., Vestergaard H., et al. Core-binding factor acute myeloid leukemia with t(8;21): Risk factors and a novel scoring system (I-CBFit) Cancer Med. 2018;7:4447–4455. doi: 10.1002/cam4.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.. Madan V., Han L., Hattori N., Teoh W.W., Mayakonda A., Sun Q.Y., Ding L.W., Nordin H.B.M., Lim S.L., Shyamsunder P., et al. ASXL2 regulates hematopoiesis in mice and its deficiency promotes myeloid expansion. Haematologica. 2018;103:1980–1990. doi: 10.3324/haematol.2018.189928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Martinez-Soria N., McKenzie L., Draper J., Ptasinska A., Issa H., Poltluri S., Blair H.J., Pickin A., Isa A., Chin P.S., et al. The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation. Cancer Cell. 2018;34:626–642. doi: 10.1016/j.ccell.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mazumdar C., Shen Y., Xavy S., Zhao F., Reinisch A., Li R., Corces M.R., Flynn R.A., Buenrostro J.D., Chan S.M., et al. Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell. 2015;17:675–688. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kawashima N., Akashi A., Nagata Y., Kihara R., Ishikawa Y., Asou N., Ohtake S., Miyawaki S., Sakura T., Ozawa Y., et al. Clinical significance of ASXL2 and ZBTB7A mutations and C-terminally truncated RUNX1-RUNX1T1 expression in AML patients with t(8;21) enrolled in the JALSG AML201 study. Ann. Hematol. 2019;98:83–91. doi: 10.1007/s00277-018-3492-5. [DOI] [PubMed] [Google Scholar]
- 108.Abdel-Wahab O., Gao J., Adli M., Dey A., Trimarchi T., Chung Y.R., Kuscu C., Hricik T., Ndiaye-Lobry D., Lafave L.M., et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Inoue D., Kitaura J., Togami K., Nishimura K., Enomoto Y., Uchida T., Kagiyama Y., Kawabata K.C., Nakahara F., Izawa K., et al. Myelodysplastic syndromes are induced by histone methylation–altering ASXL1 mutations. J. Clin. Investig. 2013;123:4627–4640. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Micol J.-B., Duployez N., Boissel N., Petit A., Geffroy S., Nibourel O., Lacombe C., Lapillonne H., Etancelin P., Figeac M., et al. Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations. Blood. 2014;124:1445–1449. doi: 10.1182/blood-2014-04-571018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jones D., Yao H., Romans A., Dando C., Pierce S., Borthakur G., Hamilton A., Bueso-Ramos C., Ravandi F., Garcia-Manero G., et al. Modeling interactions between leukemia-specific chromosomal changes, somatic mutations, and gene expression patterns during progression of core-binding factor leukemias. Genes Chromosomes Cancer. 2010;49:182–191. doi: 10.1002/gcc.20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chou W.-C., Huang H.-H., Hou H.-A., Chen C.-Y., Tang J.-L., Yao M., Tsay W., Ko B.-S., Wu S.-J., Huang S.-Y., et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood. 2010;116:4086–4094. doi: 10.1182/blood-2010-05-283291. [DOI] [PubMed] [Google Scholar]
- 113.Thota S., Viny A.D., Makishima H., Spitzer B., Radivoyevitch T., Przychodzen B., Sekeres M.A., Levine R.L., Maciejewski J.P. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 2014;124:1790–1798. doi: 10.1182/blood-2014-04-567057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Johansson B., Mertens F., Mitelman F. Secondary chromosomal abnormalities in acute leukemias. Leukemia. 1994;8:953–962. [PubMed] [Google Scholar]
- 115.Nishii K., Usui E., Katayama N., Lorenzo F., Nakase K., Kobayashi T., Miwa H., Mizutani M., Tanaka I., Nasu K., et al. Characteristics of t(8;21) acute myeloid leukemia (AML) with additional chromosomal abnormality: Concomitant trisomy 4 may constitute a distinctive subtype of t(8;21) AML. Leukemia. 2003;17:731–737. doi: 10.1038/sj.leu.2402871. [DOI] [PubMed] [Google Scholar]
- 116.Kuchenbauer F., Schnittger S., Look T., Gilliland G., Tenen D., Haferlach T., Hiddemann W., Buske C., Schoch C. Identification of additional cytogenetic and molecular genetic abnormalities in acute myeloid leukaemia with t(8;21)/AML1-ETO. Br. J. Haematol. 2006;134:616–619. doi: 10.1111/j.1365-2141.2006.06229.x. [DOI] [PubMed] [Google Scholar]
- 117.Beghini A., Magnani I., Ripamonti C.B., Larizza L. Amplification of a novel c-Kit activating mutation Asn(822)-Lys in the Kasumi-1 cell line: A t(8;21)-Kit mutant model for acute myeloid leukemia. Hematol. J. 2002;3:157–163. doi: 10.1038/sj.thj.6200168. [DOI] [PubMed] [Google Scholar]
- 118.Matsuura S., Yan M., Lo M.-C., Ahn E.-Y., Weng S., Dangoor D., Matin M., Higashi T., Feng G.-S., Zhang D.-E. Negative effects of GM-CSF signaling in a murine model of t(8;21)-induced leukemia. Blood. 2012;119:3155–3163. doi: 10.1182/blood-2011-04-350694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Klug C.A. GM-CSFRα: The sex-chromosome link to t(8;21)(+) AML? Blood. 2012;119:2976–2977. doi: 10.1182/blood-2012-01-403691. [DOI] [PubMed] [Google Scholar]
- 120.Krauth M.-T., Eder C., Alpermann T., Bacher U., Nadarajah N., Kern W., Haferlach C., Haferlach T., Schnittger S. High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: Frequency and impact on clinical outcome. Leukemia. 2014;28:1449–1458. doi: 10.1038/leu.2014.4. [DOI] [PubMed] [Google Scholar]
- 121.Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., Speck N.A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A.H., Speck N.A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sasaki K., Yagi H., Bronson R.T., Tominaga K., Matsunashi T., Deguchi K., Tani Y., Kishimoto T., Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc. Natl. Acad. Sci. USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yergeau D.A., Hetherington C.J., Wang Q., Zhang P., Sharpe A.H., Binder M., Marín-Padilla M., Tenen D.G., Speck N.A., Zhang D.E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat. Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 125.Okuda T., Cai Z., Yang S., Lenny N., Lyu C.J., van Deursen J.M., Harada H., Downing J.R. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. doi: 10.1182/blood.V91.9.3134. [DOI] [PubMed] [Google Scholar]
- 126.Castilla L.H., Perrat P., Martinez N.J., Landrette S.F., Keys R., Oikemus S., Flanegan J., Heilman S., Garrett L., Dutra A., et al. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 2004;101:4924–4929. doi: 10.1073/pnas.0400930101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Higuchi M., O’Brien D., Kumaravelu P., Lenny N., Yeoh E.-J., Downing J.R. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/S1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 128.Valk P.J.M., Bowen D.T., Frew M.E., Goodeve A.C., Löwenberg B., Reilly J.T. Second hit mutations in the RTK/RAS signaling pathway in acute myeloid leukemia with inv(16) Haematologica. 2004;89:e106. [PubMed] [Google Scholar]
- 129.Bowen D.T., Frew M.E., Hills R., Gale R.E., Wheatley K., Groves M.J., Langabeer S.E., Kottaridis P.D., Moorman A.V., Burnett A.K., et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106:2113–2119. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 130.Goemans B.F., Zwaan C.M., Miller M., Zimmermann M., Harlow A., Meshinchi S., Loonen A.H., Hählen K., Reinhardt D., Creutzig U., et al. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia. 2005;19:1536–1542. doi: 10.1038/sj.leu.2403870. [DOI] [PubMed] [Google Scholar]
- 131.Boissel N., Leroy H., Brethon B., Philippe N., de Botton S., Auvrignon A., Raffoux E., Leblanc T., Thomas X., Hermine O., et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML) Leukemia. 2006;20:965–970. doi: 10.1038/sj.leu.2404188. [DOI] [PubMed] [Google Scholar]
- 132.Kühn M.W.M., Radtke I., Bullinger L., Goorha S., Cheng J., Edelmann J., Gohlke J., Su X., Paschka P., Pounds S., et al. High-resolution genomic profiling of adult and pediatric core-binding factor acute myeloid leukemia reveals new recurrent genomic alterations. Blood. 2012;119:67–75. doi: 10.1182/blood-2011-09-380444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Speck N.A., Gilliland D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 134.Castilla L.H., Wijmenga C., Wang Q., Stacy T., Speck N.A., Eckhaus M., Marín-Padilla M., Collins F.S., Wynshaw-Boris A., Liu P.P. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/S0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 135.Mulloy J.C., Jankovic V., Wunderlich M., Delwel R., Cammenga J., Krejci O., Zhao H., Valk P.J.M., Lowenberg B., Nimer S.D. AML1-ETO fusion protein up-regulates TRKA mRNA expression in human CD34+ cells, allowing nerve growth factor-induced expansion. Proc. Natl. Acad. Sci. USA. 2005;102:4016–4021. doi: 10.1073/pnas.0404701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sun X., Zhang W., Ramdas L., Stivers D.N., Jones D.M., Kantarjian H.M., Estey E.H., Vadhan-Raj S., Medeiros L.J., Bueso-Ramos C.E. Comparative analysis of genes regulated in acute myelomonocytic leukemia with and without inv(16)(p13q22) using microarray techniques, real-time PCR, immunohistochemistry, and flow cytometry immunophenotyping. Mod. Pathol. 2007;20:811–820. doi: 10.1038/modpathol.3800829. [DOI] [PubMed] [Google Scholar]
- 137.Görgens A., Radtke S., Mollmann M., Durig J., Horn P.A., Giebel B. Revision of the Human Hematopoietic Tree: Granulocyte Subtypes Derive from Distinct Hematopoietic Lineages. Cell Rep. 2013;3:1539–1552. doi: 10.1016/j.celrep.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 138.Ben-Ami O., Friedman D., Leshkowitz D., Goldenberg D., Orlovsky K., Pencovich N., Lotem J., Tanay A., Groner Y. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 2013;4:1131–1143. doi: 10.1016/j.celrep.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 139.Goyama S., Schibler J., Cunningham L., Zhang Y., Rao Y., Nishimoto N., Nakagawa M., Olsson A., Wunderlich M., Link K.A., et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Investig. 2013;123:3876–3888. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yan M., Kanbe E., Peterson L.F., Boyapati A., Miao Y., Wang Y., Chen I.-M., Chen Z., Rowley J.D., Willman C.L., et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 141.Peterson L.F., Boyapati A., Ahn E.-Y., Biggs J.R., Okumura A.J., Lo M.-C., Yan M., Zhang D.-E. Acute myeloid leukemia with the 8q22;21q22 translocation: Secondary mutational events and alternative t(8;21) transcripts. Blood. 2007;110:799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yan M., Burel S.A., Peterson L.F., Kanbe E., Iwasaki H., Boyapati A., Hines R., Akashi K., Zhang D.-E. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc. Natl. Acad. Sci. USA. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ihle J.N. Cytokine receptor signalling. Nature. 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 144.Marcucci G., Geyer S., Zhao W., Caroll A.J., Bucci D., Uy G.L., Blum W., Pardee T., Wetzler M., Stock W., et al. Adding KIT Inhibitor Dasatinib (DAS) to Chemotherapy Overcomes the Negative Impact of KIT Mutation/over-Expression in Core Binding Factor (CBF) Acute Myeloid Leukemia (AML): Results from CALGB 10801 (Alliance) Blood. 2014;124:e8. doi: 10.1182/blood.V124.21.8.8. [DOI] [Google Scholar]
- 145.Paschka P., Schlenk R.F., Weber D., Benner A., Bullinger L., Heuser M., Gaidzik V.I., Thol F., Agrawal M., Teleanu V., et al. Adding dasatinib to intensive treatment in core-binding factor acute myeloid leukemia-results of the AMLSG 11-08 trial. Leukemia. 2018;32:1621–1630. doi: 10.1038/s41375-018-0129-6. [DOI] [PubMed] [Google Scholar]
- 146.Stone R.M., Mandrekar S.J., Sanford B.L., Laumann K., Geyer S., Bloomfield C.D., Thiede C., Prior T.W., Döhner K., Marcucci G., et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017;377:454–464. doi: 10.1056/NEJMoa1614359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.World Health Organization . In: Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Swerdlow S.H., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., Arber D.A., Hasserjian R.P., Le Beau M.M., et al., editors. IARC; Lyon, France: 2017. [Google Scholar]
- 148.Tarlock K., Alonzo T.A., Wang Y.C., Gerbing R.B., Ries R., Loken M.R., Pardo L., Hylkema T., Joaquin J., Sarukkai L., et al. Functional Properties of KIT mitations are associated with differential clinical outcomes and response to targeted therapeutics in CBF acute myeloid leukemia. Clin. Cancer Res. 2019;25:5038–5048. doi: 10.1158/1078-0432.CCR-18-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Y.Y., Zhao L.J., Wu C.F., Liu P., Shi L., Liang Y., Xiong S.M., Mi J.Q., Chen Z., Ren R., et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc. Natl. Acad. Sci. USA. 2011;108:2450–2455. doi: 10.1073/pnas.1019625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Taylor J., Lee S.C. Mutations in spliceosoma genes and therapeutic opportunities in myeloid malignancies. Genes Chromosomes Cancer. 2019;58:889–902. doi: 10.1002/gcc.22784. [DOI] [PMC free article] [PubMed] [Google Scholar]