Abstract
Components of the pre-messenger RNA splicing machinery are frequently mutated in myeloid malignancies. Mutations in LUC7L2, PRPF8, SF3B1, SRSF2, U2AF1, and ZRSR2 genes occur at various frequencies ranging between 40% and 85% in different subtypes of myelodysplastic syndrome (MDS) and 5% and 10% of acute myeloid leukemia (AML) and myeloproliferative neoplasms (MPNs). In some instances, splicing factor (SF) mutations have provided diagnostic utility and information on clinical outcomes as exemplified by SF3B1 mutations associated with increased ring sideroblasts (RS) in MDS-RS or MDS/MPN-RS with thrombocytosis. SF3B1 mutations are associated with better survival outcomes, while SRSF2 mutations are associated with a shorter survival time and increased AML progression, and U2AF1 mutations with a lower remission rate and shorter survival time. Beside the presence of mutations, transcriptomics technologies have shown that one third of genes in AML patients are differentially expressed, leading to altered transcript stability, interruption of protein function, and improper translation compared to those of healthy individuals. The detection of SF mutations demonstrates the importance of splicing abnormalities in the hematopoiesis of MDS and AML patients given the fact that abnormal splicing regulates the function of several transcriptional factors (PU.1, RUNX1, etc.) crucial in hematopoietic function. This review provides a summary of the significance of the most frequently mutated SF genes in myeloid malignancies and an update on novel targeted therapies in experimental and clinical trial stages.
Keywords: splicing factor genes, mutations, AML, MDS, therapies
1. Genetics of Myeloid Malignancies
Myelodysplastic syndromes (MDS) are a genetically and clinically diverse group of clonal stem cell malignancies characterized by inefficient hematopoiesis, peripheral blood cytopenias, and an increased risk of transformation to acute myeloid leukemia (AML). Genetic alterations occurring at the level of a multipotent stem cell are believed to accelerate clonal evolution from MDS, sometimes resulting in transformation to acute leukemia, although no specific disease-initiating defect has been identified to date. In the last decade, several gene mutations acting as driver events have been identified with various frequencies across the spectrum of MDS and AML. Disease evolution is driven by positive selection of driver mutations. So far, more than 30 driver genes have been associated with the pathogenesis of leukemia [1,2]. Gene functional analysis has clustered these genes into pathways including DNA methylation, chromatin remodeling, RNA-splicing, cohesion complex, RAS family signaling, gene transcription, and DNA repair. One of the major classes of driver mutations is mutations in splicing factors (SF) (SF3B1, U2AF1, SRSF2, ZRSR2, PRPF8, LUC7L2) (Figure 1). SF genes encode for components of the spliceosomes. Spliceosomes are nuclear structures composed of five small nuclear RNAs (snRNA) and approximately 150 proteins, which catalyze the splicing reaction. The complex removes non-coding sequences (introns) from precursor messenger RNA and ligates coding sequences (exons) in order to form mature mRNA transcripts through early and late steps. Early steps involve recognition of the 5′ and 3′ exon/intron junctions and late steps recall all the spliceosome together. The information to define the splicing regions are included in short and conserved sequences at the 5′ splice site (SS), the 3′SS, and the branch site (BS). In higher eukaryotes, the BS is located approximately 18–40 nucleotides upstream of the 3′SS and is followed by a polypyrimidine tract. Many genes in humans are spliced into two or more transcripts with altered sequences through a process called alternative splicing. There are two types of pre-mRNA introns: U2-dependent (major spliceosome), which accounts for almost all the human introns, and U12-dependent (minor spliceosome), which accounts for less than a thousand introns [3,4].
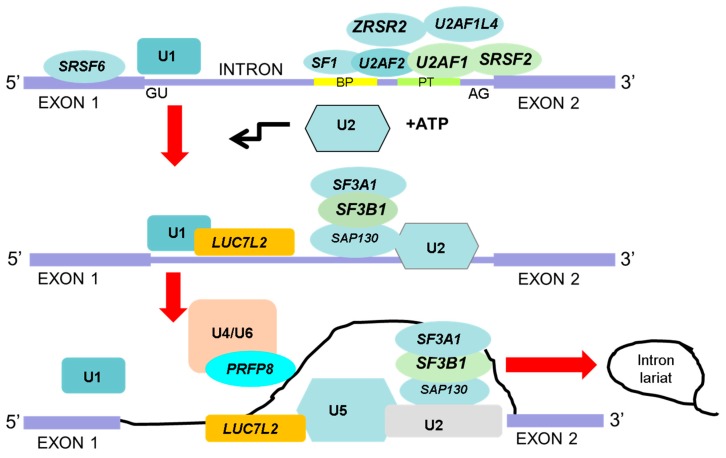
Figure 1.
Spliceosome complex. Pre-mRNA splicing initiates with the recruitment of U1 snRNP to the 5′ splice site (SS). Serine-arginine rich (SR) proteins such as SRSF6 are localized nearby the 5′SS regulating alternative splicing. The SF1 protein and the larger subunit of the U2 auxiliary factor (U2AF), U2AF2, both gather to bind the branch point (BP) sequence and the polypyrimidine tract (PT). The smaller subunit of U2AF (U2AF1) binds to the AG dinucleotide of the 3′SS, interacting with both U2AF2 and the splicing factor SRSF2. ZRSR2 and U2AF1L4 (other two RNA-binding proteins) are recruited to interact with U2AF complex (U2AF1 and U2AF2) in order to recognize the 3′SS. After the recognition of the 3′SS, the U2 snRNP, together with SF3A1, SF3B1, and SAP130*, is recruited to the 3′SS. In the meantime, the U1 and LUC7L2 are recruited to the 5′SS and the U4/U6 and U5 complex, together with PRFP8, localize on the BP and PT regions to catalyze the release of the intron lariat and ligation of exons. *SAP130: Official gene name is SF3B3.
As a whole, SF mutations are detected in 45–85% of different MDS subtypes and in 5–10% of primary AML [5,6]. Mutations in the SF genes occur in the same spots in MDS and AML. In MDS, SF mutations are associated with mutations in epigenetic genes, e.g., SF3B1 with DNMT3A mutations, SRSF2 with RUNX1, IDH1/2 and ASXL1 mutations and U2AF1 with ASXL1 and DNMT3A mutations. In AML, SF mutations are associated mainly with RUNX1, ASXL1, IDH2 and TET2 [6].
The mutations often occur in genes controlling 3′SS selection. Missense mutations are a hallmark of SF3B1, SRSF2, and U2AF1 and inactivating mutations (nonsense or frameshift) are often detected in the ZRSR2 gene. These SF mutations are the most commonly observed mutations in MDS and AML, and are found in approximately two-thirds of SF-mutated cases. Except for ZRSR2, the other SF are part of the U2-type spliceosome. The heterozygous configuration of the mutations at selective sites and the absence of nonsense or truncating mutations in major SF (SF3B1, SRSF2, U2AF1) suggest that SF are likely gain-of-function oncogenes [7,8]. The consequences of SF mutations and the production of alternative transcripts are presented in Figure 2.
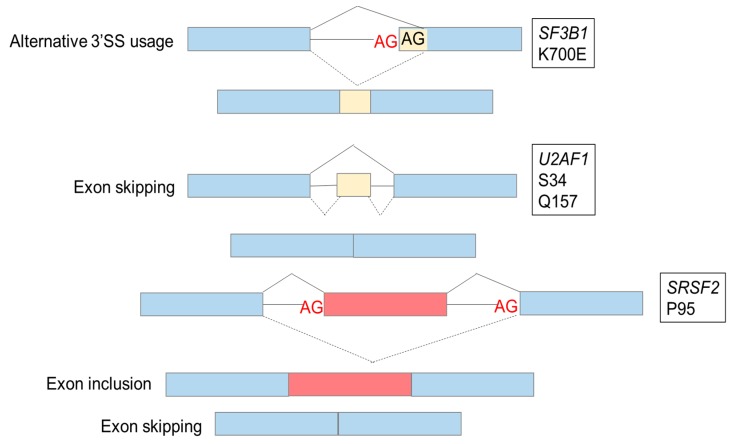
Figure 2.
Consequences of mutations in splicing factors. Examples of consequences of splicing factors mutations are alternative 3′ splicing site (SS) usage caused by SF3B1 mutations, increased exon skipping caused by U2AF1 mutations, exon skipping and exon inclusion caused by SRSF2 mutations.
Mutations in SF3B1, SRSF2, and U2AF1 have been associated with specific disease subtypes with SF3B1 being mostly mutated in MDS-RS, SRSF2 occurring mostly in chronic myelomonocytic leukemia (CMML), and U2AF1 in secondary AML [9]. Representative cases of SF mutant patients and morphologic features are presented in Figure 3.
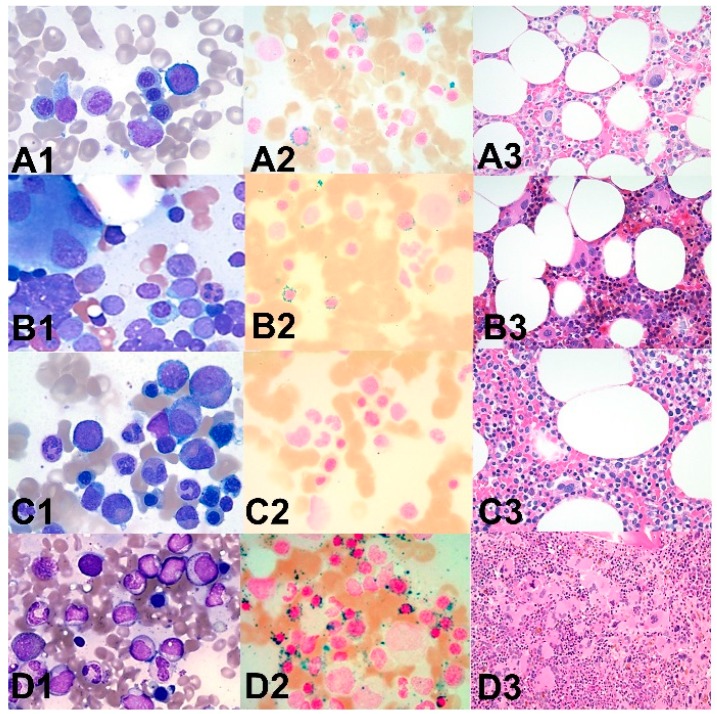
Figure 3.
Morphologic features of representative patients with mutations in splicing factors. Bone marrow aspiration smears (1: Wright–Giemsa stain, ×500) and iron stains (2, ×400) and core biopsies (3: H&E stain, ×200) in representative cases with myeloid malignancies and mutation in splicing factor genes. A1–3: A patient with myelodysplastic syndrome with ring sideroblasts and multilineage dysplasia with SF3B1 and ZRSR2 mutation (detected by next generation sequencing) showing mild dyserythropoiesis (A1), increased ring sideroblasts (A2) and dysmegakaryopoiesis (A3). B1–3: A patient with acute myeloid leukemia with SF3B1 mutation showing increased myeloblasts and dysmegakaryopoiesis (B1,B3) and increased ring sideroblasts (B2). C1–3: A patient with myelodysplastic syndrome with U2AF1 mutation showing minimal dyserythropoiesis and no increased blasts (C1), no increased ring sideroblasts (C2) and mild dysmegakaryopoiesis (C3). D1–3: A patient with myelodysplastic/myeloproliferative neoplasm with thrombocytosis and ring sideroblasts with SF3B1 mutation showing no significant dysplasia in granulocytes and erythroid cells (D1), increased ring sideroblasts (D2) and dysmegakaryopoiesis (D3).
2. Overview of Splicing Factor Mutations in Myeloid Malignancies
SF3B1. The SF3B1 gene (chromosome 2q33.1) encodes a core component of the U2 nuclear ribonucleoprotein, which recognizes the 3′SS at intron-exon junctions. Mutations are located preferentially in four consecutive HEAT (Huntington elongation factor 3 protein phosphatase 2A, and the yeast PI3-kinase TOR1) domains of the C-terminal region, with the lysine to glutamic acid substitution at codon 700 (K700E) accounting for more than 50% of all mutant cases. Other common hotspot mutations involve the conserved amino acids 622, 625, 662, and 666 [10]. As such, all mutations occur quite distant to the region of the protein involved in the 3′ branch site recognition suggesting that these alterations do not influence the RNA-binding properties of the protein. Studies in murine models have shown that homozygous mutations of the SF3B1 gene are incompatible with life [11,12]. Clonal analysis and in vitro experiments of human leukemia cells have shown that SF3B1 mutations are initiating events which occur in rare lympho-myeloid hematopoietic stem cells (HSCs) (Lin− CD34+ CD38− CD90+ CD45RA−) [13,14]. The cells seem to provide a marked clonal advantage to the MDS-RS HSCs. Indeed, the percentage of RS in the bone marrow correlates with the SF3B1 variant allele frequency [15]. Xenotransplantation studies showed that clones with SF3B1 mutations are often not suppressed by other clones with different mutations (e.g., DNMT3A, JAK2) [14]. SF3B1 mutations are significantly associated with older age [16]. More recently, SF3B1 mutations have also been found in a proportion of individuals with clonal hematopoiesis of indeterminate potential (CHIP) at risk for developing malignancies including MDS [17]. Mutual exclusivity in SF mutations suggest that co-occurring mutations are incompatible with life. SF mutations induce a variety of splicing changes which are unique to specific SF. RNA-sequencing analysis of many tumor types have found that splicing abnormalities in SF3B1 mutant cells are exclusively related to the utilization of a cryptic splice acceptor site located upstream of the canonical 3′SS [18,19]. These splicing events often produce framing errors. Among the several target genes of SF, most have been studied for effects of mutations in the SF3B1 gene. SF3B1 mutant cells have a defect in the splicing of ABCB7 (ATP Binding Cassette Subfamily B, Member 7) due to the usage of an alternative 3′SS which causes an early termination of the protein [20,21]. The change in splicing seems to be the major determinant in decreasing ABCB7 mRNA levels and consequent increase in iron deposition (a feature of MDS-RS). Moreover, alternative 3′SS were also observed in PPOX (1q23.3) and TMEM14C (6p24.2) genes, particularly during erythroid maturation, using in vitro cellular systems [22]. Other events of aberrant splicing with resultant low mRNA production were also found in tumor suppressor genes including NF1, DICER1, PML, PDS5A, MAP3K7, and PPP2R5A [23]. Mice conditionally expressing K700E mutation (Mx1-Cre Sf3b1+/K700E) develop progressive macrocytic anemia, inefficient cellular differentiation, and failure in bone marrow reconstitution capacity [12].
SF3B1 mutations often co-occur with TET2 mutations [24]. Double mutant mice generated by the breeding of Sf3b1+/K700E and Tet2 knock-out mice manifested a more severe anemia compared to mice with a sole alteration in Sf3b1 or Tet2 [12]. RNA-sequencing analysis of human and Sf3b1+/K700E mice have shown that about 68% of splicing defects are due to alternative 3′SS usage occurring at −15 and −24 nucleotides upstream the canonical 3′ [12]. In vitro experiments expressing mutations often observed in human MDS in the yeast orthologue of SF3B1 (Hsh155) have shown that those mutations alter the interaction of SF3B1 and Prp5 a protein helicase involved in the first ATP-dependent catalysis of the splicing cascade [25].
U2AF1. The U2-complex auxiliary factor 1 gene (U2AF1, 21q22.3) encodes a 35-kDa protein (alias U2AF35) of the U2-spliceosome responsible for recognition of the terminal 3′ AG dinucleotide in pre-messenger RNA introns. The protein has four major domains including two zinc finger regions, a serine-arginine (SR) domain, and an U2AF-homology domain. The U2AF1 unit forms a complex by heterodimerization with the 65-Kda protein called U2AF2 in order to bind the polypyrimidine tract upstream the 3′SS and recognize the branch point and the AG dinucleotide of the 3′SS. Mutations in U2AF1 at codon S34 and Q157 are found in about 11% of patients with MDS and in 4% of patients with AML. Mutations are associated with a worse survival and are associated with an increased risk of AML transformation. U2AF1 mutations result in the production of neomorphic phenotypes by chancing splicing patterns for many RNA downstream genes. These changes are lineage-specific and seem to influence the division of erythroid progenitors and subsequently change the differentiation trajectory of granulocytes and monocytes [26]. Analysis of the transcriptome of U2AF1 mutant cells has shown changes in the splicing patterns (cassette exons) of genes present in the granulocytic compartment (H2AFY, STRAP) [27]. Primary cells expressing U2AF1 mutations showed missplicing in mitotic (ATR, CEP164, EHMT1, WAC) and RNA processing (PABPC4, PPWD1, PTBP1, STRAP, UPF3B) genes [28]. Studies have also shown that cells expressing U2AF1S34F have a decrease in ATG7 protein levels due to aberrant splicing resulting in a decrease in autophagy (as ATG7 is one of the key genes in autophagy). U2AF1S34F cells with low levels of ATG7 and reduced autophagy had also increased reactive oxygen species and increased chromosomal instability [29]. Presence of U2AF1 mutations was also correlated with increased IRAK4 isoforms which activate the NF-KB signaling pathway [30]. Genetically engineered murine models expressing U2AF1S34F have been generated using the Cre recombinase tool and by activating Cre specifically in hematopoietic tissues. Upon Cre activation, mice expressing the S34 mutation developed features recapitulating MDS (dysplasia, cytopenias) as well as abnormal splicing profiles similar to the splicing patterns observed in human MDS [31]. Transgenic U2AF1 mice also are sensitive to pharmacologic treatment with the splicing modulator, sudemycin [26].
SRSF2. The SRSF2 gene is located on chromosome 17q25.2 and encodes a member of the serine/arginine (SR)-rich family of pre-mRNA splicing components. It contains an RNA recognition motif (RRM) for binding RNA and an arginine and serine domain for binding other proteins. The arginine and serine domain is enriched in SR residues facilitating the interaction between SR and SF. SR proteins have versatile functions such as regulating pre-mRNA splicing, RNA stability, and translation. SRSF2 mutations occur almost exclusively at proline 95 and alter binding affinity of the RRM motif. SRSF2 mutations have been found in 28–47% of patients with CMML and about 14% of patients with MDS [32] and have been associated with increased age, higher levels of hemoglobin, and normal cytogenetics. SRSF2 mutations are almost never sole mutations. In CMML, SRSF2 mutations are often found with TET2 mutations, while in AML are typically associated with RUNX1, IDH2, and ASXL1 mutations. Pooled meta-analysis studies of MDS patients have shown that patients with SRSF2 mutations predict for a worse survival and an increased risk for AML transformation [32,33] and have no prognostic effects in CMML [34].
In AML, SRSF2 mutations have been found in about 25% of patients and associated with older age [35]. Studies have shown that proline 95 in normal SRSF2 forms a strong interaction with the second cytosine in the UCCAGU site and the second guanine in the UGGAGU site of the DNA. When mutations alter proline to histidine, the change in amino acid creates an alteration in the hydrogen bond leading to a change in the structural conformation. SRSF2 mutations seem to induce differential splicing in EZH2, a known gene implicated in the pathogenesis of MDS and AML, and in several HNRNP proteins, including HNRNPA2B1, HNRNPH1, HNRNPM, and HNRNPH3 [36,37]. The diverse biological effects of SRSF2 mutations are also demonstrated by the fact that other pathways are affected by the mutations, e.g., perturbation of double-strand DNA breaks, increase in p53 phosphorylation, and cell cycle arrest. In inducible Mx1-Cre Srsf2+/P95H knock-in mice, sole alterations in SRSF2 produce a phenotype similar to MDS. These features are also recapitulated by mice carrying homozygous Srsf2 [36,38]
3. Other Splicing Factor Mutations
Mutations in other SF have been found in myeloid malignancies at lower frequencies and with patterns different from those of SF3B1, SRSF2, and U2AF1.
ZRSR2 is a gene located on chromosome Xp22.2, which is mutated in about 5% of patients with MDS, predominantly males. The protein is another member of the SR-rich family of SF, and the gene encodes a component of the U2 auxiliary factor heterodimer which is responsible for the recognition of the 3′ splice acceptor site. The nature of the mutations resembles loss-of-function mutations [39]. Out-of-frame insertions and deletions, nonsense, missense, and splice site mutations have all been detected. No mutation hotspots have been observed and the mutations scatter across the entire coding region. In terms of splicing abnormalities, ZRSR2 mutations cause abnormal splicing via intron retention of U12-depedent introns [39].
PRPF8 is a gene located on chromosome 17p13.3 and has been found to be affected by somatic mutations or hemizygous deletions. A majority of patients (50%) with PRPF8 mutations and del(17p) were found to be AML patients with poor prognosis. PRPF8 alterations were found to correlate with increased RS and myeloblasts [40]. A subsequent analysis of a large cohort showed that PRPF8 mutations were common and found in 4% (65/1700) of patients with MDS and AML [41]. Mutations were mainly missense, nonsense, frameshift, and splice site mutations, and showed a strong association with dismal prognosis. Based on studies of the yeast Prp8 protein, PRPF8 protein appears to be involved in spliceosome assembly, however a clear function in myeloid malignancies has not yet been demonstrated [42]. Cryo-electron microscopy technology has helped to resolve the structure of many components of the spliceosome and added more information on the function of PRPF8, specifically the N domain of PRPF8. PRPF8 interacts with U5 snRNA to stabilize the pre-mRNA in the spliceosome active site and it potentiates the U2 and U6 snRNA interactions with the intron [43].
LUC7L2 is located on chromosome 7q34, and thus is a common target of gene deletions given the frequency of deletion of the long arm of chromosome 7 [del(7q)] and monosomy 7 (−7) in myeloid malignancies. The gene encodes a protein with a C2H2-type zinc finger, a coiled-coil region, and an SR domain. To date the function of LUC7L2 is not well characterized and the majority of what is known about the protein is based on observations of its ortholog splicing factor LUC7 which is involved in recruitment and interaction of SF. LUC7L2 mutations can be hemizygous, heterozygous, and homozygous. LUC7L2 mutations have been associated with shorter survival in patients with -7/del7q compared to patients with normal LUC7L2 expression [44,45]. LUCL7L2 has been found aberrantly spliced in MDS cases harboring SRSF2 small deletions [46]. Very rare mutations (<1%) have been found in other SF genes including SF3A1, SF1, PRPF40B, and U2AF2 [5,47].
4. Therapeutic Intervention for Splicing Factor Mutations
As we have described, there are multiple ways by which pathologically altered splicing can promote the initiation and maintenance of cancer. In the last eight years, interest has grown in targeting splicing catalysis, splicing regulatory proteins, and specific altered splicing events. Splicing requires protein–protein and protein–RNA interaction, and is directed by a number of proteins, which are also subjected to regulation via post-translational modifications and protein–RNA interactions. This variety of interactions provides a range of possibilities to manipulate splicing for pharmacologic purposes. Pan-splicing modulators have been developed and are being tested in myeloid malignancies associated with SF mutations. Because splicing is ubiquitous and SF interact with many different proteins, it is possible that splicing modulators might also have effects against leukemic cells that do not harbor SF mutations [48]. Bacterially-derived products and their derivatives have been shown to bind the SF3B component to disrupt the early stages of the spliceosome cascade. These compounds have different stabilities in in vitro systems, and include low stability-agents such as FR901463, FR901464, FR901465, herboxidienes, and pladienolides, and high stability-agents such as E7107 (an analog of pladienolide B), spliceostatin A (SSA; from FR901464), and sudemycins, which block early spliceosome assembly [49]. While several compounds have only been shown to biologically alter splicing in vitro, a few compounds have been tested in both in vitro and in vivo. For instance, E7107 seems to induce marked splicing inhibition in several cellular and animal models [50]. The susceptibility of the spliceosomal mutant leukemia to splicing modulation via E7107 has also been validated in patient-derived xenografts obtained from primary AML cells with SF mutations [48].
Recently, H3B-8800 (a semi-derivative of pladienolides), a selective and orally bioavailable modulator of normal and mutant SF3b complex, has shown dose-dependent modulation of splicing in pre-clinical studies. Oral administration of H3B-8800 demonstrated preferential induction of cell growth arrest in several pre-clinical xenograft models of MDS/AML carrying SF mutations. H3B-8800 entered an open-label, multicenter phase 1 trial to evaluate pharmacokinetics/pharmacodynamics in AML, CMML, and MDS with SF mutations [51]. Several studies are ongoing using agents with non-spliceosome functions. More recently, several hematopoietic cells have been found more sensitive to the treatment with aryl sulfonamides (e.g., indisulam) compared to other cancer cell lines. Drug-sensitivity has been correlated with increased DCAF15 expression levels and copy number variation. The mechanism of action of indisulam seems to be shared with other sulfonamides such as tasisulam and chloroquinoxaline sulfonamide. Indeed, indisulam seems to induce the degradation of RBM39 (RNA binding motif protein 39) which in turn causes abnormal mRNA splicing (intron retention, exon skipping). RBM39 is a nuclear protein which associates with the E3 ubiquitin ligase complex (CUL4-DCAF15) in the presence of indisulam [52]. Inhibition of protein arginine methyltransferases (PRMTs) has also been correlated with increased anti-proliferative activity of SF mutant cell lines. The possibility of using protein arginine methyltransferases (PRMTs) inhibitors was demonstrated after subjecting murine AML cells driven by MLL-AF9 fusion with and without Srsf2P95H mutation to 45 compounds. Srsf2 mutant cells were more sensitive than wild type cells to two PRMTs inhibitors, GSK591 (PRMT5 inhibitor) and MS023 (a pan type I PRMTs inhibitor) [53]. Very recently, SF3B1 mutations have been shown to provoke interruption of the open reading frame of BRD9 with resultant decrease in the half-life of the protein. Cells derived from patients harboring SF3B1 mutations have a reduced expression of BRD9 compared to cells of patients with wild type SF3B1 [54].
Recently, luspatercept (ACE-536), a fusion protein containing a modified extracellular domain of the human activin receptor type IIB combined with a human IgG1 Fc domain and influencing the TGF-β and SMAD family proteins, is a phase II clinical trial for MDS patients and seems to induce a better clinical response in patients with SF3B1 mutations [55]. In addition, SF3B1 mutations represent a positive factor for erythroid response in a randomized phase II clinical trial of azacitidine and epoietin-β [56].
5. Conclusions
Mutations in SF genes are common in myeloid malignancies and induce different disease phenotypes according to cellular context. Each SF is also associated with a distinct pattern of mutations in other genes commonly detected in myeloid malignancies, as in the case of SRSF2 mutations with TET2 mutations in CMML and with RUNX1, IDH2, and ASXL1 mutations in AML. Targeting SF mutations represents a novel avenue of drug discovery and development. In vitro studies have shown several different ways to impact splicing and restore splicing defects. Future experimental trials will shed light on the safety of pharmacologic agents targeting SF mutations.
Abbreviations
| Gene Symbol | Official Gene Name |
| ABCB7 | ATP Binding Cassette Subfamily B Member 7 |
| ASXL1 | Additional Sex Combs Like 1 |
| ATR | ATR Serine/Threonine Kinase |
| BRD9 | Bromodomain Containing 9 |
| CEP164 | Centrosomal Protein 164 |
| DCAF15 | DDB1 And CUL4 Associated Factor 15 |
| DICER1 | Dicer 1, Ribonuclease III |
| DNMT3A | DNA Methyltransferase 3 Alpha |
| EHMT1 | Euchromatic Histone Lysine Methyltransferase 1 |
| EZH2 | Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit |
| HNRNPA2B1 | Heterogeneous Nuclear Ribonucleoprotein A2/B1 |
| HNRNPH1 | Heterogeneous Nuclear Ribonucleoprotein H1 |
| HNRNPH3 | Heterogeneous Nuclear Ribonucleoprotein H3 |
| HNRNPM | Heterogeneous Nuclear Ribonucleoprotein M |
| IDH1 | Isocitrate Dehydrogenase (NADP(+)) 1 |
| IDH2 | Isocitrate Dehydrogenase (NADP(+)) 2 |
| IRAK4 | Interleukin 1 Receptor Associated Kinase 4 |
| JAK2 | Janus Kinase 2 |
| LUC7L2 | LUC7 Like 2, Pre-MRNA Splicing Factor |
| ¥MACROH2A1 | MacroH2A.1 Histone |
| MAP3K7 | Mitogen-Activated Protein Kinase Kinase Kinase 7 |
| NF1 | Neurofibromin 1 |
| PABPC4 | Poly(A) Binding Protein Cytoplasmic 4 |
| PDS5A | PDS5 Cohesin Associated Factor A |
| PML | Promyelocytic Leukemia |
| PPOX | Protoporphyrinogen Oxidase |
| PPP2R5A | Protein Phosphatase 2 Regulatory Subunit B’Alpha |
| PPWD1 | Peptidylprolyl Isomerase Domain And WD Repeat Containing 1 |
| PRMT | Protein Arginine Methyltransferase |
| PRPF40B | Pre-MRNA Processing Factor 40 Homolog B |
| PRPF8 | Pre-MRNA Processing Factor 8 |
| PTBP1 | Polypyrimidine Tract Binding Protein 1 |
| RBM39 | RNA Binding Motif Protein 39 |
| RUNX1 | RUNX Family Transcription Factor 1 |
| SF1 | Splicing Factor 1 |
| SF3A1 | Splicing Factor 3a Subunit 1 |
| SF3B1 | Splicing Factor 3b Subunit 1 |
| ≠SF3B3 | Splicing Factor 3b Subunit 3 |
| SRSF2 | Serine And Arginine Rich Splicing Factor 2 |
| SRSF6 | Serine And Arginine Rich Splicing Factor 6 |
| STRAP | Serine/Threonine Kinase Receptor Associated Protein |
| TET2 | Tet Methylcytosine Dioxygenase 2 |
| TMEM14C | Transmembrane Protein 14C |
| U2AF1 | U2 Small Nuclear RNA Auxiliary Factor 1 |
| U2AF1L4 | U2 Small Nuclear RNA Auxiliary Factor 1 Like 4 |
| U2AF2 | U2 Small Nuclear RNA Auxiliary Factor 2 |
| UPF3B | UPF3B Regulator Of Nonsense Mediated MRNA Decay |
| WAC | WW Domain Containing Adaptor With Coiled-Coil |
| ZRSR2 | Zinc Finger CCCH-Type, RNA Binding Motif And Serine/Arginine Rich 2 |
Gene symbol and official names follow the nomenclature of the GeneCards [57]; ≠SF3B3 alias SAP130 ¥MACROH2A1 alias H2AFY.
Funding
V.V. thank the Vera and Joseph Dresner Foundation grant for MDS.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Spaulding T.P., Stockton S.S., Savona M.R. The evolving role of next generation sequencing in myelodysplastic syndromes. Br. J. Haematol. 2019 doi: 10.1111/bjh.16212. [DOI] [PubMed] [Google Scholar]
- 2.Hosono N. Genetic abnormalities and pathophysiology of MDS. Int. J. Clin. Oncol. 2019;24:885–892. doi: 10.1007/s10147-019-01462-6. [DOI] [PubMed] [Google Scholar]
- 3.Maciejewski J.P., Padgett R.A. Defects in spliceosomal machinery: A new pathway of leukaemogenesis. Br. J. Haematol. 2012;158:165–173. doi: 10.1111/j.1365-2141.2012.09158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahl M.C., Will C.L., Luhrmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa S. Splicing factor mutations in AML. Blood. 2014;123:3216–3217. doi: 10.1182/blood-2014-04-566752. [DOI] [PubMed] [Google Scholar]
- 6.Hou H.A., Liu C.Y., Kuo Y.Y., Chou W.C., Tsai C.H., Lin C.C., Lin L.I., Tseng M.H., Chiang Y.C., Liu M.C., et al. Splicing factor mutations predict poor prognosis in patients with de novo acute myeloid leukemia. Oncotarget. 2016;7:9084–9101. doi: 10.18632/oncotarget.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pellagatti A., Boultwood J. Splicing factor mutant myelodysplastic syndromes: Recent advances. Adv. Biol. Regul. 2019:100655. doi: 10.1016/j.jbior.2019.100655. [DOI] [PubMed] [Google Scholar]
- 8.Visconte V., Makishima H., Maciejewski J.P., Tiu R.V. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia. 2012;26:2447–2454. doi: 10.1038/leu.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obeng E.A., Ebert B.L. Charting the “Splice” Routes to MDS. Cancer Cell. 2015;27:607–609. doi: 10.1016/j.ccell.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 11.Isono K., Mizutani-Koseki Y., Komori T., Schmidt-Zachmann M.S., Koseki H. Mammalian polycomb-mediated repression of Hox genes requires the essential spliceosomal protein Sf3b1. Genes Dev. 2005;19:536–541. doi: 10.1101/gad.1284605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obeng E.A., Chappell R.J., Seiler M., Chen M.C., Campagna D.R., Schmidt P.J., Schneider R.K., Lord A.M., Wang L., Gambe R.G., et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell. 2016;30:404–417. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortera-Blanco T., Dimitriou M., Woll P.S., Karimi M., Elvarsdottir E., Conte S., Tobiasson M., Jansson M., Douagi I., Moarii M., et al. SF3B1-initiating mutations in MDS-RSs target lymphomyeloid hematopoietic stem cells. Blood. 2017;130:881–890. doi: 10.1182/blood-2017-03-776070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mian S.A., Rouault-Pierre K., Smith A.E., Seidl T., Pizzitola I., Kizilors A., Kulasekararaj A.G., Bonnet D., Mufti G.J. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat. Commun. 2015;6:10004. doi: 10.1038/ncomms10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D., Pellagatti A., Wainscoat J.S., Hellstrom-Lindberg E., Gambacorti-Passerini C., et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C.C., Hou H.A., Chou W.C., Kuo Y.Y., Wu S.J., Liu C.Y., Chen C.Y., Tseng M.H., Huang C.F., Lee F.Y., et al. SF3B1 mutations in patients with myelodysplastic syndromes: The mutation is stable during disease evolution. Am. J. Hematol. 2014;89:E109–E115. doi: 10.1002/ajh.23734. [DOI] [PubMed] [Google Scholar]
- 17.Dorsheimer L., Assmus B., Rasper T., Ortmann C.A., Ecke A., Abou-El-Ardat K., Schmid T., Brüne B., Wagner S., Serve H., et al. Association of Mutations Contributing to Clonal Hematopoiesis with Prognosis in Chronic Ischemic Heart Failure. JAMA Cardiol. 2019;4:25–33. doi: 10.1001/jamacardio.2018.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darman R.B., Seiler M., Agrawal A.A., Lim K.H., Peng S., Aird D., Bailey S.L., Bhavsar E.B., Chan B., Colla S., et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 19.DeBoever C., Ghia E.M., Shepard P.J., Rassenti L., Barrett C.L., Jepsen K., Jamieson C.H., Carson D., Kipps T.J., Frazer K.A. Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput. Biol. 2015;11:e1004105. doi: 10.1371/journal.pcbi.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolatshad H., Pellagatti A., Liberante F.G., Llorian M., Repapi E., Steeples V., Roy S., Scifo L., Armstrong R.N., Shaw J., et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia. 2016;30:2322–2331. doi: 10.1038/leu.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikpour M., Scharenberg C., Liu A., Conte S., Karimi M., Mortera-Blanco T., Giai V., Fernandez-Mercado M., Papaemmanuil E., Högstrand K., et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia. 2013;27:889–896. doi: 10.1038/leu.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolatshad H., Pellagatti A., Fernandez-Mercado M., Yip B.H., Malcovati L., Attwood M., Przychodzen B., Sahgal N., Kanapin A.A., Lockstone H., et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29:1092–1103. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiozawa Y., Malcovati L., Galli A., Sato-Otsubo A., Kataoka K., Sato Y., Watatani Y., Suzuki H., Yoshizato T., Yoshida K., et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat. Commun. 2018;9:3649. doi: 10.1038/s41467-018-06063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J., Moscinski L., Zhang H., Zhang X., Hussaini M. Does SF3B1/TET2 Double Mutation Portend Better or Worse Prognosis Than Isolated SF3B1 or TET2 Mutation? Cancer Genom. Proteom. 2019;16:91–98. doi: 10.21873/cgp.20115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Q., Rodriguez-Santiago S., Wang J., Pu J., Yuste A., Gupta V., Moldón A., Xu Y.Z., Query C.C. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev. 2016;30:2710–2723. doi: 10.1101/gad.291872.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirai C.L., Ley J.N., White B.S., Kim S., Tibbitts J., Shao J., Ndonwi M., Wadugu B., Duncavage E.J., Okeyo-Owuor T., et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer Cell. 2015;27:631–643. doi: 10.1016/j.ccell.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yip B.H., Steeples V., Repapi E., Armstrong R.N., Llorian M., Roy S., Shaw J., Dolatshad H., Taylor S., Verma A., et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J. Clin. Invest. 2017;127:3557. doi: 10.1172/JCI96202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Przychodzen B., Jerez A., Guinta K., Sekeres M.A., Padgett R., Maciejewski J.P., Makishima H. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013;122:999–1006. doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S.M., Ou J., Chamberlain L., Simone T.M., Yang H., Virbasius C.M., Ali A.M., Zhu L.J., Mukherjee S., Raza A., et al. U2AF35(S34F) Promotes Transformation by Directing Aberrant ATG7 Pre-mRNA 3′ End Formation. Mol. Cell. 2016;62:479–490. doi: 10.1016/j.molcel.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith M.A., Choudhary G.S., Pellagatti A., Choi K., Bolanos L.C., Bhagat T.D., Gordon-Mitchell S., Ahrens D.V., Pradhan K., Steeples V., et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 2019;21:640–650. doi: 10.1038/s41556-019-0314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fei D.L., Zhen T., Durham B., Ferrarone J., Zhang T., Garrett L., Yoshimi A., Abdel-Wahab O., Bradley R.K., Liu P., et al. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor gene U2af1. Proc. Natl. Acad. Sci. USA. 2018;115:E10437–E10446. doi: 10.1073/pnas.1812669115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou H.A., Tsai C.H., Lin C.C., Chou W.C., Kuo Y.Y., Liu C.Y., Tseng M.H., Peng Y.L., Liu M.C., Liu C.W., et al. Incorporation of mutations in five genes in the revised International Prognostic Scoring System can improve risk stratification in the patients with myelodysplastic syndrome. Blood Cancer J. 2018;8:39. doi: 10.1038/s41408-018-0074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X., Zhan Z., Naren D., Li J., Yan T., Gong Y. Prognostic value of SRSF2 mutations in patients with de novo myelodysplastic syndromes: A meta-analysis. PLoS ONE. 2017;12:e0185053. doi: 10.1371/journal.pone.0185053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arbab Jafari P., Ayatollahi H., Sadeghi R., Sheikhi M., Asghari A. Prognostic significance of SRSF2 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: A meta-analysis. Hematology. 2018;23:778–784. doi: 10.1080/10245332.2018.1471794. [DOI] [PubMed] [Google Scholar]
- 35.Prassek V.V., Rothenberg-Thurley M., Sauerland M.C., Herold T., Janke H., Ksienzyk B., Konstandin N.P., Goerlich D., Krug U., Faldum A., et al. Genetics of acute myeloid leukemia in the elderly: Mutation spectrum and clinical impact in intensively treated patients aged 75 years or older. Haematologica. 2018;103:1853–1861. doi: 10.3324/haematol.2018.191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim E., Ilagan J.O., Liang Y., Daubner G.M., Lee S.C., Ramakrishnan A., Li Y., Chung Y.R., Micol J.B., Murphy M.E., et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang Y., Tebaldi T., Rejeski K., Joshi P., Stefani G., Taylor A., Song Y., Vasic R., Maziarz J., Balasubramanian K., et al. SRSF2 mutations drive oncogenesis by activating a global program of aberrant alternative splicing in hematopoietic cells. Leukemia. 2018;32:2659–2671. doi: 10.1038/s41375-018-0152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kon A., Yamazaki S., Nannya Y., Kataoka K., Ota Y., Nakagawa M.M., Yoshida K., Shiozawa Y., Morita M., Yoshizato T., et al. Physiological Srsf2 P95H expression causes impaired hematopoietic stem cell functions and aberrant RNA splicing in mice. Blood. 2018;131:621–635. doi: 10.1182/blood-2017-01-762393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madan V., Kanojia D., Li J., Okamoto R., Sato-Otsubo A., Kohlmann A., Sanada M., Grossmann V., Sundaresan J., Shiraishi Y., et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 2015;6:6042. doi: 10.1038/ncomms7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurtovic-Kozaric A., Przychodzen B., Singh J., Konarska M.M., Clemente M.J., Otrock Z.K., Nakashima M., His E.D., Yoshida K., Shiraishi Y., et al. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia. 2015;29:126–136. doi: 10.1038/leu.2014.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adema V., Hirsch C., Przychodzen B.P., Nagata Y., Lemelle E., Nazha A., Carraway H.E., Sekeres M.A., Visconte V., Maciejewski J.P., et al. Somatic PRPF8 Mutations in Myeloid Neoplasia. Blood. 2017;130:584. [Google Scholar]
- 42.Keightley M.C., Crowhurst M.O., Layton J.E., Beilharz T., Markmiller S., Varma S., Hogan B.M., de Jong-Curtain T.A., Heath J.K., Lieschke G.J. In vivo mutation of pre-mRNA processing factor 8 (Prpf8) affects transcript splicing, cell survival and myeloid differentiation. FEBS Lett. 2013;587:2150–2157. doi: 10.1016/j.febslet.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacRae A.J., Mayerle M., Hrabeta-Robinson E., Chalkley R.J., Guthrie C., Burlingame A.L., Jurica M.S. Prp8 positioning of U5 snRNA is linked to 5′ splice site recognition. Rna. 2018;24:769–777. doi: 10.1261/rna.065458.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hershberger C.E., Hosono N., Singh J., Dietrich R.C., Gu X., Makishima H., Saunthararajah Y., Maciejewski J.P., Padgett R.A. The Role of LUC7L2 in Splicing and MDS. Blood. 2016;128:5504. doi: 10.1182/blood.V128.22.5504.5504. [DOI] [Google Scholar]
- 45.Sperling A.S., Gibson C.J., Ebert B.L. The genetics of myelodysplastic syndrome: From clonal haematopoiesis to secondary leukaemia. Nat. Rev. Cancer. 2017;17:5–19. doi: 10.1038/nrc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madan V., Li J., Zhou S., Teoh W.W., Han L., Meggendorfer M., Malcovati L., Cazzola M., Ogawa S., Haferlach T., et al. Distinct and convergent consequences of splice factor mutations in myelodysplastic syndromes. Am. J. Hematol. 2019 doi: 10.1002/ajh.25673. [DOI] [PubMed] [Google Scholar]
- 47.Larsson C.A., Cote G., Quintas-Cardama A. The changing mutational landscape of acute myeloid leukemia and myelodysplastic syndrome. Mol. Cancer Res. 2013;11:815–827. doi: 10.1158/1541-7786.MCR-12-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee S.C., Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat. Med. 2016;22:976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webb T.R., Joyner A.S., Potter P.M. The development and application of small molecule modulators of SF3b as therapeutic agents for cancer. Drug Discov. Today. 2013;18:43–49. doi: 10.1016/j.drudis.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Folco E.G., Coil K.E., Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seiler M., Yoshimi A., Darman R., Chan B., Keaney G., Thomas M., Agrawal A.A., Caleb B., Csibi A., Sean E., et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 2018;24:497–504. doi: 10.1038/nm.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han T., Goralski M., Gaskill N., Capota E., Kim J., Ting T.C., Xie Y., Williams N.S., Nijhawan D. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356:eaan7977. doi: 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- 53.Fong J.Y., Pignata L., Goy P.A., Kawabata K.C., Lee S.C., Koh C.M., Musiani D., Massignani D., Kotini A.G., Penson A., et al. Therapeutic Targeting of RNA Splicing Catalysis through Inhibition of Protein Arginine Methylation. Cancer Cell. 2019;36:194–209. doi: 10.1016/j.ccell.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue D., Chew G.L., Liu B., Michel B.C., Pangallo J., D’Avino A.R., Hitchman T., North K., Lee S., Bitner L., et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature. 2019 doi: 10.1038/s41586-019-1646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Platzbecker U., Germing U., Gotze K.S., Kiewe P., Mayer K., Chromik J., Radsak M., Wolff T., Zhang X., Laadem A., et al. Luspatercept for the treatment of anaemia in patients with lower-risk myelodysplastic syndromes (PACE-MDS): A multicentre, open-label phase 2 dose-finding study with long-term extension study. Lancet Oncol. 2017;18:1338–1347. doi: 10.1016/S1470-2045(17)30615-0. [DOI] [PubMed] [Google Scholar]
- 56.Thepot S., Ben Abdelali R., Chevret S., Renneville A., Beyne-Rauzy O., Prebet T., Park S., Stamatoullas A., Guerci-Bresler A., Cheze S., et al. A randomized phase II trial of azacitidine +/− epoetin-beta in lower-risk myelodysplastic syndromes resistant to erythropoietic stimulating agents. Haematologica. 2016;101:918–925. doi: 10.3324/haematol.2015.140988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.GeneCards®: The Human Gene Database Version 4.12 (v5.0 preview) [(accessed on 5 November 2019)]; Available online: https://www.genecards.org.