Abstract
Intellectual disability (ID) is a neurodevelopmental condition affecting 1–3% of the world’s population. Genetic factors play a key role causing the congenital limitations in intellectual functioning and adaptive behavior. The heterogeneity of ID makes it more challenging for genetic and clinical diagnosis, but the advent of large-scale genome sequencing projects in a trio approach has proven very effective. However, many variants are still difficult to interpret. A combined approach of next-generation sequencing and functional, electrophysiological, and bioinformatics analysis has identified new ways to understand the causes of ID and help to interpret novel ID-causing genes. This approach offers new targets for ID therapy and increases the efficiency of ID diagnosis. The most recent functional advancements and new gene editing techniques involving the use of CRISPR–Cas9 allow for targeted editing of DNA in in vitro and more effective mammalian and human tissue-derived disease models. The expansion of genomic analysis of ID patients in diverse and ancient populations can reveal rare novel disease-causing genes.
Keywords: Intellectual Disability, Neurological Disorders, Gene Editing, Mental Retrdation, NGS, WES, CRISPR/Cas9.
Introduction
Intellectual disability (ID) occurs in the developmental period before the age of 18 years. ID is a heterogeneous group of disorders characterized by significantly impaired intellectual functioning and deficits in adaptive behaviors 1. It affects 1–3% of the world population, is the most common developmental disorder, and represents an important socio-economic problem in healthcare. However, owing to the heterogeneity of ID, its frequency ratio changes worldwide 2. Diagnosis of ID is also performed with the identification of clinical phenotype symptoms such as delayed speech, hypotonia, and seizures 3. In the past decade, the genetic background of ID was believed to be mostly autosomal dominant ( de novo mutations) in the outbred countries such as the USA and those in Western Europe, while in the middle-east countries where inbreeding is common, autosomal recessive ID has some preponderance 4, 5.
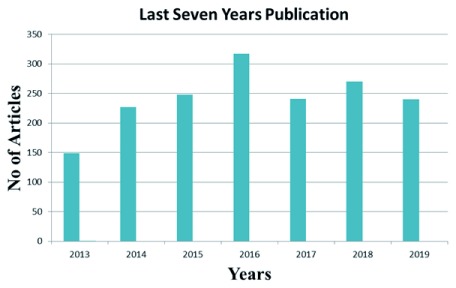
Next-generation sequencing (NGS) provided tremendous power to sequence personal genomes and detect a large number of genetic variants. The discovery of disease-causing variants by whole exome or genome sequencing in patients has dramatically changed our perspective on precision medicine 6. Through NGS methods, now it is possible to find pathogenic mutations, including novel mutations, associated with ID 7. A combined NGS and bioinformatics approach is used to identify novel ID genes and screen candidate ID genes as well. NGS has efficiently expedited ID research and provided new strategies at the clinical level in the last few years, as shown in Figure 1. Genomics England’s PanelApp database ( https://panelapp.genomicsengland.co.uk/panels/285/) shows that around 1,396 genes cause ID (see Table 1, Extended data).
Figure 1. The number of articles on intellectual disability published in the last seven years identified using the PubMed search terms “ID”, “mental retardation”, “next-generation sequencing”, and “exome sequencing”.
Etiological classification of intellectual disability
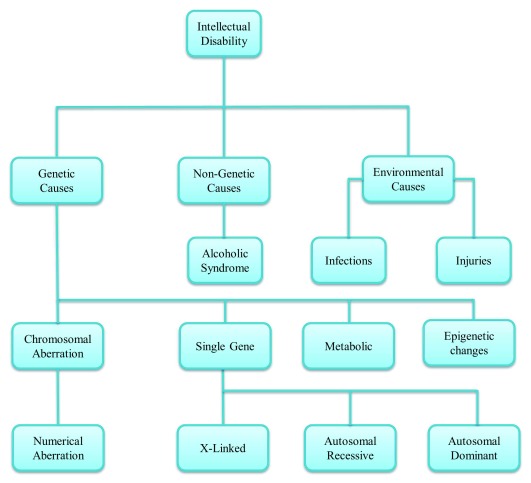
Multiple factors are involved in causing ID. Genetic factors include genetic variations such as aneuploidies, copy number variations (CNVs), and tandem repeats in specific genes 8. DNA is liable to mutation, mediating genetic plasticity. The expansion of tandem repeats can cause a range of disorders associated with ID such as X-linked ID (XLID) 9, various ataxias, motor neuron disease, and epilepsy. Emerging data suggest that tandem repeat polymorphisms (TRPs) can contribute to the missing heritability of polygenic disorders 10– 13. Furthermore, various metabolic factors, repeat expansions of nucleotides, and mitochondrial DNA variants can also contribute to ID 7. Environmental factors such as hazardous chemical exposures, infections during pregnancy, and UV radiation are also reported to cause ID. In addition to these, lack of nutrition, cultural deprivation, childhood diseases such as measles, meningitis, and severe head injury can cause malfunction of the nervous system, leading to ID 14. Even though the etiological factors of ID are very broad, as shown in Figure 2, in 50% of individuals the cause of ID is still unknown, but the most prominent causes are genetic 15.
Figure 2. Intellectual disability classification. Multiple factors are involved in intellectual disability including genetic inheritance and environmental conditions.
Cytogenetic abnormalities
The identification of genetic factors causing ID has advanced in terms of number and type with recent developments in cytogenetic techniques. The first genetic test used in the investigation of ID was karyotyping 16 to identify aneuploidies, such as Down’s syndrome (trisomy 21) and Edwards syndrome (trisomy 18), and large structural re-arrangements such as insertions, deletions, and duplications 17, 18. Karyotyping detects only large deletions or insertions owing to its low resolution. Fluorescence in situ hybridization (FISH) is used to detect structural abnormalities and numerical changes on chromosomes. Specific probes are used in FISH for the analysis of chromosomal aberration 19. A number of studies on ID describe the growing benefits of using NGS in clinics for diagnostic purposes 20.
Chromosomal aberrations
An extra copy of chromosome 21 as a result of an error in cell division can cause a trisomy known as Down’s syndrome 21. It is the most frequent form of ID. The clinical features of Down’s syndrome include dysmorphic features, seizures, psychomotor slowing, and congenital malformation. These conditions may not be present in each affected individual 22. Edwards syndrome is a trisomy of chromosome 18 characterized by psychomotor and cognitive brain impairment, malformation, and growth deficiency in infants 23. The occurrence of Edwards syndrome is 1 out of 8,000 live births. The mortality rate is very high with this condition; only 5–10% of affected children survive after the first year of life 24. Patau syndrome is a trisomy of chromosome 13, characterized by malformations of the central nervous system (CNS). The occurrence of this syndrome is 1 in 12,000 in the general population 23. The survival rate is very low in infants with trisomy 13, but it depends on the severity of the condition in infants, i.e. whether or not there are cerebral, cardiac, or other congenital malformations 25.
Copy number variation
CNVs are small segments of DNA that vary in number. Usually each individual carries two copies: one that comes from the maternal side and one from the paternal side. Variations in copy number occur through duplications and deletions of the small DNA segments. But not all copy numbers, that are either a deletion or duplication, are pathogenic to humans 26, 27. The largest database for CNVs (DGV) is available online ( www.dgv.tcag.ca) 28. Decipher is another database used by clinicians to compare clinical and genetic information for the identification and interpretation of pathogenic variants (sequence variants and copy number variants) in patients with ID 29. CNVs causing de novo and inherited mutations have been associated with ID. A study on a large cohort reported 118 rare de novo CNVs 30. Analysis of these reported CNVs and candidate genes pinpointed 10 genes with loss of function associated with ID 31. CNVs cannot be visibly detected with a light microscope. Array comparative genomic hybridization (CGH) can perform rapid genome-wide analysis at a high resolution and detect CNVs, gain or loss, at the chromosomal level 32. Array CGH analysis of unresolved ID cases increased the identification of pathogenic CNVs up to 13% in recent years. Phenotypes associated with these 13% of cases are congenital defects, primary microcephaly, and short stature 33. A disorder (Online Mendelian Inheritance in Man [OMIM] #612001) with microdeletion on 15q13.3 shows an ID phenotype with a complex variety of seizures, autism, and psychiatric conditions 34, 35. SNP array is also a new technique as compared to array CGH which can detect CNVs 36. A novel genetic disorder related to ID with a 3q29 microdeletion was reported using microarray techniques 37.
Whole exome and genome sequencing
Whole exome sequencing (WES) is an efficient technology that can increase the diagnostic yield when searching for alleles causing rare Mendelian disorders. Exome analysis examines the protein’s encoding region, where an estimated 85% of disease-causing mutations are believed to occur 38. It has been an invaluable tool in gene discovery for ID. WES performed on three members of a family with autosomal dominant ID (MRD44; 617061) showed a heterozygous mutation, a 1 bp deletion (c.4466delA, NM_007118) in exon 30 of the TRIO gene. This mutation resulted in a framshift and premature termination (Gln1489ArgfsTer11) in the GEFD1 domain. A 9-year-old girl with autosomal dominant ID was also found to carry a de novo heterozygous c.3239A-T transversion (c.3239A-T, NM_007118) in exon 19 of the TRIO gene, which causes changes in the spectrin repeat domain 39. Whole genome sequencing (WGS) allows examination of single-nucleotide variants (SNVs), indels, structural variants (SVs), and CNVs in both the ~1% part of the genome that encodes protein sequences and the ~99% of remaining non-coding sequences. Therefore, WGS has more reliable sequence coverage with more uniformity. It is likely to reveal many novel variants and genes and derive new scientific and clinical findings for ID. With the rapid drop in sequencing cost and the ability of WGS to rapidly produce large volumes of data, it is becoming a powerful tool for genomic research.
Inherited mutations causing intellectual disability
Single gene disorders are grouped into different types on the basis of inheritance pattern 40.
Autosomal recessive intellectual disability
Autosomal recessive ID is a genetically heterogeneous group of disorders 41. Autosomal recessive ID occurs in syndromic and non-syndromic forms. The syndromic type of ID is characterized by intellectual problems occurring with a group of other phenotypic features 42. Non-syndromic ID is characterized by a lack of associated pathology. Genes linked with non-syndromic ID are being studied to understand the normal variation in intelligence 43. Distinction in intelligence quotient (IQ) is linked with those genes that can also cause large variations in intellectual ability when mutated. Homozygosity mapping is performed to check the autosomal recessive causes of ID in consanguineous families with affected siblings 44. OMIM and SysID ( http://sysid.cmbi.umcn.nl/) search results show that 399 genes can cause autosomal recessive ID (see Table 2, Extended data).
Autosomal dominant intellectual disability
The inheritance pattern of autosomal dominant ID is when an individual carries one copy of a mutant allele and one normal allele on a gene. Autosomal dominant ID is caused by heterozygous mutations in different reported genes and CNVs. Tuberous sclerosis, neurofibromatosis, and myotonic dystrophy are autosomal dominant disorders linked with ID 45. SNP microarray analysis of the methyl binding domain gene on chromosome 2q23.1 showed that a 200 kb deletion in exon 6 of a female patient was associated with autosomal dominant ID 46. It is difficult to find the estimated frequency of mutations in autosomal dominant ID genes. ARID1B, SYNGAP1, DYRK1A, MED13L, KCNQ2, CTNNB1, STXB1, KMT2A, PACS1, FOXP1, and SMARCA2 are the most commonly mutated autosomal dominant ID genes 47, 48. Some of these genes are essential for neuronal differentiation in the developing brain and play important roles in synaptic formation and transmission 49. The outcomes of our OMIM search show that in total around 180 genes or loci, as reported in the literature, are involved in autosomal dominant ID (see Table 3, Extended data).
X-linked intellectual disability
The human X-chromosome comprises 5% of the human genome, but an increasing number of genetic diseases are associated with the X chromosome; approximately 10–12% of X-chromosome genes have been linked with ID 50. X-linked recessive fragile X syndrome occurs on chromosome Xq27.3 in the FMR1 gene at the 5ʹ untranslated region (UTR) owing to the expansion of CGG trinucleotide repeats 51. Fragile X syndrome is clinically characterized as ID; phenotypically, patients show dysmorphic facial features and protruded ears. FMR1 encodes for a protein that provides RNA stability and plays a key role in brain development and neuronal plasticity. Deficiency of the FMRP protein causes suppression or excitation of GABA that results in low synaptic connections, leading to syndromic features in patients 52. The number of new X-linked ID genes identified has increased rapidly over the last few years with the use of NGS. More than 140 known X-linked genes have been reported 53, 54.
Identification of candidate and novel intellectual disability genes
Different techniques have been used to identify novel ID-causing genes over the past few decades. For the identification of novel or candidate genes in affected families, it is necessary to reconstruct the family pedigree and perform some clinical investigation before reaching any conclusion 55, 56. The introduction of robust microarray technologies in research has increased the power of identification; SNP arrays can detect small deletions and micro duplications in the probands 57. NGS has accelerated the speed of identification of novel ID-causing genes. NGS technology has become very popular over the past 5 years, with a considerable number of new ID genes and candidate genes reported. NGS can detect SNVs and small insertions/deletions in the whole genome.
In vitro and in vivo study of intellectual disability
Biological assays can be used to study any undefined variant and its role or pathogenicity. These assays can be used in mutated cells and also directly in patient-derived cells 58. To identify the pathogenicity of candidate ID genes, electrophysiological studies of SH-SY5Y neuronal cell lines are a very powerful approach 59 to capture early cortical development with high fidelity, thus helping to study genes that are relevant in early cortical development and associated with ID 60. In vivo studies of candidate ID genes provide additional information regarding pathogenicity. Different animal models can be used to study ID genes. CRBN knockout (CrbnKO) mice have been used to study learning and memory tasks. Loss of CRBN results in memory problems, learning problems when AMPK activity is accelerated, blocking of mTORC1 signaling, and a decreased level of glutamatergic synaptic proteins. These findings show that the CrbnKO mouse is an ideal animal model to understand the molecular mechanisms of learning and memory problems in ID patients 61. Zebrafish can also be used as a model organism to study the function of genes associated with ID. Zebrafish have unique features including the rapid development of embryos and easy visualization of the nervous system during developmental stages, which make them an ideal organism to study the function of genes. Overexpression comparison can be performed between the wild-type PK1A gene and the zebrafish ortholog of PK1A. In this case, zebrafish mutants show severe phenotypes as compared to the wild-type, and a mutant PK1A gene has been reported in patients with epilepsy. These results signify that the mutation changes the in vivo function of PK1A 62. In addition, the TAF1 gene is associated with ID. Functional study of this gene was performed by using a zebrafish knockout model. Severe phenotypes were observed during embryogenesis and neurodevelopment in the zebrafish TAF1 knockout model 63. The heterogeneity of ID makes it very difficult to validate candidate genes as causative ID genes, but in vitro and in vivo studies provide grounds for conclusively identifying certain genes. Single-cell transcriptomics and quantitative proteomics can also be used to improve our understanding of the global changes in the central nervous system when the genome-edited organism is available.
CRISPR–Cas9 gene editing tool
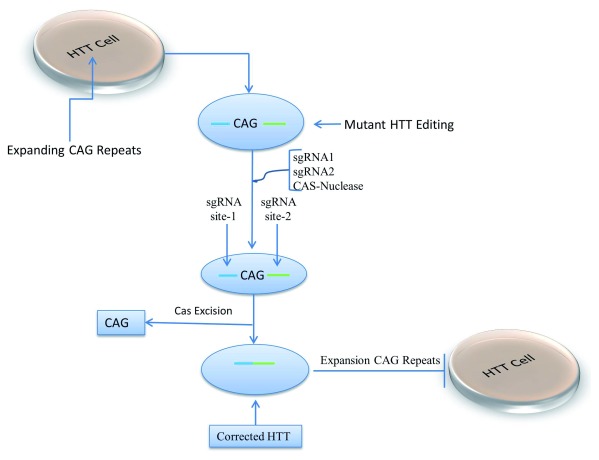
The identification of causal genes and their functional analysis requires further understanding of disease mechanisms, especially for potential therapies, even if we do not have a full understanding of the biological functions. Acknowledging this, clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 is an RNA control nuclease system that has become essential for gene editing and correcting mutated genes. It provides potential treatment options for genetic disorders that cause ID ( Figure 3). This system has been applied recently to mammalian genomes to stop the expression of the disease gene or to edit mutated genes, thereby correcting the mutation. CRISPR–Cas9 technology will possibly cure diseases that have no treatment option available, such as trinucleotide repeat expansion diseases causing neurological disorders 64. CRISPR–Cas9 systems have been used on fragile X syndrome caused by the extension of trinucleotide repeats, which results in deteriorated levels of FMR1 protein and Huntington disease (HD) models. No treatment is available for these conditions 65. Induced pluripotent stem cells (iPSCs) of patients with fragile X syndrome in the FRX gene upstream of the CGH repeat were targeted with expressed CRISPR–Cas9 nuclease along with single guide RNA (sgRNA) and resulted in the reactivation of the FMR1 protein 66. Further studies show that two sgRNA-guided methods that flanked the trinucleotide CGG repeat produced two double-stranded breaks and the recombination of the breaks resulted in the deletion of repeats. The deletion of repeats resulted in an increased FMR1 protein level 67. HD is caused by trinucleotide repeat extensions in the coding region of the huntingtin gene ( HTT) 68. The CRISPR–Cas9 system has been used recently in the treatment of HD. iPSCs derived from a HD patient were corrected by using CRISPR–Cas9 that selectively inactivated the mutant HTT gene without altering the normal allele of the same gene 69, 70. HD patient fibroblast cell lines were used to show that CAG repeats can be precisely excised using CRISPR–Cas9 nickase from the HTT gene 71. Strict guidelines must be adopted to avoid the exploitation of the safety and security weakness in genome editing techniques as well as to reduce the risk of off-site editing of genome and epigenetic changes with the help of further research. Adopting appropriate biosafety levels for genome editing to stop contamination is important but so is applying rules to cover the biosecurity of gene editing 72. Further studies are needed before the application of gene therapy to the treatment of these diseases.
Figure 3. The CRISPR–Cas9 system used to correct extension repeats in the huntingtin gene ( HTT) by using single-guide RNA (sgRNA) on both sides of the repeats and Cas9 nuclease, creating nicks and removing the CAG repeats, blocking further extension.
Conclusions and the future direction of intellectual disability genetics
The prevalence of ID varies from country to country, and it is especially low in developed countries as compared to less-developed countries 73, 74. The identification of genes causing ID rapidly increased over the past 3 to 5 years owing to the use of sophisticated sequencing techniques. The diagnosis of ID patients became easy with new massively parallel sequencing methods and the help of different human variant databases. The heterogeneity of ID makes it difficult for etiological diagnosis; however, WES is likely to be used as the first-line test for ID probands. NGS improves our understanding of the genetic origin of ID. Not all ID-causing genes have been identified yet, but a combined approach of sequencing techniques, functional analysis, and bioinformatics will help to identify new ID-causing genes. This approach will potentially provide a new way of treating ID. Although ID genes can be therapeutically targeted using the CRISPR–Cas-9 system, this method and its application in mammals is still emerging, and many regulatory, methodological, and off-target effects still need to be understood.
The genomic understanding of ID has primarily come from developed countries, and our knowledge in developing countries is very limited. This is important, as many of these countries have a young population and culturally prefer large families, indicating that the burden of ID and other childhood disorders will increase before technology is developed enough to treat these conditions. Therefore, early diagnosis, education, and genetic counseling will be important for families suffering with these conditions. In addition, many of the genes in developing countries are likely to have founder effects that may be amenable to diagnosis in cultural groups and demographic areas, accelerating diagnosis and revealing carrier status to help family planning.
Abbreviations
CGH, comparative genomic hybridization; CNV, copy number variation; CRISPR, clustered regularly interspaced short palindromic repeats; FISH, fluorescence in situ hybridization; HD, Huntington's disease; ID, intellectual disability; iPSC, induced pluripotent stem cell; NGS, next-generation sequencing; OMIM, Online Mendelian Inheritance in Man; sgRNA, single guide RNA; SNV, single nucleotide variation; WES, whole exome sequencing.
Data availability
Extended data
Harvard Dataverse: Replication Data for: The Genetics of Intellectual Disability Review Tables, https://doi.org/10.7910/DVN/MMUNLR 75
This project contains the following extended data:
Table 1: Estimated total number of genes involved in Intellectual Disability Genes from Literature search and from genome England Database
Harvard Dataverse: The Genetics of Intellectual Disability, https://doi.org/10.7910/DVN/AEKQOL 76
This project contains the following extended data:
Table 2: List of Genes causing Autosomal Recessive Intellectual Disability
Harvard Dataverse: Replication Data for: The Genetics of Intellectual Disability, https://doi.org/10.7910/DVN/9BJDI6 77
This project contains the following extended data:
Table 3: List of Autosomal Dominant Genes in Omim and PubMed DataBase
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Acknowledgements
The authors would like to thank the participants and their families for their essential help with this work. They also thank the National Institute for Health Research Biomedical Research Centre at UCLH/UCL.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Jennifer Winter, Institute of Human Genetics, University Medical Center of the Johannes Gutenberg University, Mainz, Germany
Hao Hu, Guangzhou Women and Children’s Medical Center, Guangzhou, China; Max-Planck Institute for Molecular Genetics, Berlin, Germany
Funding Statement
The authors’ work is funded by the MRC (MR/S01165X/1, MR/S005021/1, G0601943), The National Institute for Health Research University College London Hospitals Biomedical Research Centre, Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK (MDUK), Muscular Dystrophy Association (MDA USA), and Higher Education Commission of Pakistan (Project 7028_NRPU).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
References
- 1. Bertelli MO, Munir K, Harris J, et al. : “Intellectual developmental disorders”: reflections on the international consensus document for redefining “mental retardation-intellectual disability” in ICD-11. Adv Ment Health Intellect Disabil. 2016;10(1):36–58. 10.1108/AMHID-10-2015-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harripaul R, Vasli N, Mikhailov A, et al. : Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol Psychiatry. 2018;23(4):973–984. 10.1038/mp.2017.60 [DOI] [PubMed] [Google Scholar]
- 3. Redin C, Gérard B, Lauer J, et al. : Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet. 2014;51(11):724–736. 10.1136/jmedgenet-2014-102554 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Kahrizi K, Hu H, Hosseini M, et al. : Effect of inbreeding on intellectual disability revisited by trio sequencing. Clin Genet. 2019;95(1):151–159. 10.1111/cge.13463 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Hu H, Kahrizi K, Musante L, et al. : Genetics of intellectual disability in consanguineous families. Mol Psychiatry. 2019;24(7):1027–1039. 10.1038/s41380-017-0012-2 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Teng S: NGS for Sequence Variants. In Translational Biomedical Informatics.Springer. Adv Exp Med Biol. 2016;939:1–20. 10.1007/978-981-10-1503-8_1 [DOI] [PubMed] [Google Scholar]
- 7. Harripaul R, Noor A, Ayub M, et al. : The Use of Next-Generation Sequencing for Research and Diagnostics for Intellectual Disability. Cold Spring Harb Perspect Med. 2017;7(3): pii: a026864. 10.1101/cshperspect.a026864 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Palmer E, Speirs H, Taylor PJ, et al. : Changing interpretation of chromosomal microarray over time in a community cohort with intellectual disability. Am J Med Genet A. 2014;164A(2):377–385. 10.1002/ajmg.a.36279 [DOI] [PubMed] [Google Scholar]
- 9. Zablotskaya A, Van Esch H, Verstrepen KJ, et al. : Mapping the landscape of tandem repeat variability by targeted long read single molecule sequencing in familial X-linked intellectual disability. BMC Med Genomics. 2018;11(1):123. 10.1186/s12920-018-0446-7 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Hannan AJ: Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet. 2018;19(5):286–298. 10.1038/nrg.2017.115 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Cortese A, Simone R, Sullivan R, et al. : Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat Genet. 2019;51(4):649–658. 10.1038/s41588-019-0372-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishiura H, Doi K, Mitsui J, et al. : Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet. 2018;50(4):581–590. 10.1038/s41588-018-0067-2 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Kim K, Bang S, Yoo D, et al. : De novo emergence and potential function of human-specific tandem repeats in brain-related loci. Hum Genet. 2019;138(6):661–672. 10.1007/s00439-019-02017-5 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Winnepenninckx B, Rooms L, Kooy RF: Mental retardation: a review of the genetic causes. Br J Dev Disabil. 2003;49(96):29–44. 10.1179/096979503799104138 [DOI] [Google Scholar]
- 15. Milani D, Ronzoni L, Esposito S: Genetic Advances in Intellectual Disability. J Pediatr Genet. 2015;4(3):125–127. 10.1055/s-0035-1564438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tjio JH, Levan A: The chromosome number of man. Hereditas. 1956;42(1–2):1–6. 10.1111/j.1601-5223.1956.tb03010.x [DOI] [Google Scholar]
- 17. Raymond FL, Tarpey P: The genetics of mental retardation. Hum Mol Genet. 2006;15 Spec No 2:R110–R116. 10.1093/hmg/ddl189 [DOI] [PubMed] [Google Scholar]
- 18. Curry CJ, Stevenson RE, Aughton D, et al. : Evaluation of mental retardation: recommendations of a Consensus Conference: American College of Medical Genetics. Am J Med Genet. 1997;72(4):468–477. [DOI] [PubMed] [Google Scholar]
- 19. Fauzdar A, Chowdhry M, Makroo RN, et al. : Rapid-prenatal diagnosis through fluorescence in situ hybridization for preventing aneuploidy related birth defects. Indian J Hum Genet. 2013;19(1):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Ligt J, Willemsen MH, van Bon BW, et al. : Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367(20):1921–1929. 10.1056/NEJMoa1206524 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Asim A, Kumar A, Muthuswamy S, et al. : "Down syndrome: an insight of the disease". J Biomed Sci. 2015;22(1):41. 10.1186/s12929-015-0138-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ostermaier KK: Down syndrome: Clinical features and diagnosis. UpToDate. Welleseley, MA: UpToDate,2010. Reference Source [Google Scholar]
- 23. Smith DW, Patau K, Therman E, et al. : A new autosomal trisomy syndrome: multiple congenital anomalies caused by an extra chromosome. J Pediatr. 1960;57(3):338–345. 10.1016/s0022-3476(60)80241-7 [DOI] [PubMed] [Google Scholar]
- 24. Cereda A, Carey JC: The trisomy 18 syndrome. Orphanet J Rare Dis. 2012;7(1):81. 10.1186/1750-1172-7-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Plaiasu V, Ochiana D, Motei G, et al. : Clinical relevance of cytogenetics to pediatric practice. Postnatal findings of Patau syndrome - Review of 5 cases. Maedica (Buchar). 2010;5(3):178–85. [PMC free article] [PubMed] [Google Scholar]
- 26. Sebat J, Lakshmi B, Troge J, et al. : Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. 10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Canales CP, Walz K: Copy number variation and susceptibility to complex traits. EMBO Mol Med. 2011;3(1):1–4. 10.1002/emmm.201000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. MacDonald JR, Ziman R, Yuen RK, et al. : The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–D992. 10.1093/nar/gkt958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bragin E, Chatzimichali EA, Wright CF, et al. : DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic Acids Res. 2014;42(Database issue):D993–D1000. 10.1093/nar/gkt937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hehir-Kwa JY, Rodríguez-Santiago B, Vissers LE, et al. : De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J Med Genet. 2011;48(11):776–778. 10.1136/jmedgenet-2011-100147 [DOI] [PubMed] [Google Scholar]
- 31. Coe BP, Witherspoon K, Rosenfeld JA, et al. : Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46(10):1063–71. 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitra A, Liu G, Song J: A genome-wide analysis of array-based comparative genomic hybridization (CGH) data to detect intra-species variations and evolutionary relationships. PLoS One. 2009;4(11):e7978. 10.1371/journal.pone.0007978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shoukier M, Klein N, Auber B, et al. : Array CGH in patients with developmental delay or intellectual disability: are there phenotypic clues to pathogenic copy number variants? Clin Genet. 2013;83(1):53–65. 10.1111/j.1399-0004.2012.01850.x [DOI] [PubMed] [Google Scholar]
- 34. , , Sharp AJ Mefford HC Li K et al. : A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40(3):322–8. 10.1038/ng.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ben-Shachar S, Lanpher B, German JR, et al. : Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46(6):382–388. 10.1136/jmg.2008.064378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernardini L, Alesi V, Loddo S, et al. : High-resolution SNP arrays in mental retardation diagnostics: how much do we gain? Eur J Hum Genet. 2010;18(2):178–85. 10.1038/ejhg.2009.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Digilio MC, Bernardini L, Mingarelli R, et al. : 3q29 Microdeletion: a mental retardation disorder unassociated with a recognizable phenotype in two mother-daughter pairs. Am J Med Genet A. 2009;149(8):1777–1781. 10.1002/ajmg.a.32965 [DOI] [PubMed] [Google Scholar]
- 38. Choi M, Scholl UI, Ji W, et al. : Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–19101. 10.1073/pnas.0910672106 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Pengelly RJ, Greville-Heygate S, Schmidt S, et al. : Mutations specific to the Rac-GEF domain of TRIO cause intellectual disability and microcephaly. J Med Genet. 2016;53(11):735–742. 10.1136/jmedgenet-2016-103942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Naviaux RK, Nguyen KV: POLG mutations associated with Alpers' syndrome and mitochondrial DNA depletion. Ann Neurol. 2004;55(5):706–712. 10.1002/ana.20079 [DOI] [PubMed] [Google Scholar]
- 41. Marangi G, Leuzzi V, Manti F, et al. : TRAPPC9-related autosomal recessive intellectual disability: report of a new mutation and clinical phenotype. Eur J Hum Genet. 2013;21(2):229–32. 10.1038/ejhg.2012.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. You J, Sobreira NL, Gable DL, et al. : A Syndromic Intellectual Disability Disorder Caused by Variants in TELO2, a Gene Encoding a Component of the TTT Complex. Am J Hum Genet. 2016;98(5):909–918. 10.1016/j.ajhg.2016.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hill WD, Davies G, Liewald DC, et al. : Examining non-syndromic autosomal recessive intellectual disability (NS-ARID) genes for an enriched association with intelligence differences. Intelligence. 2016;54:80–89. 10.1016/j.intell.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Basel-Vanagaite L, Attia R, Yahav M, et al. : The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet. 2006;43(3):203–210. 10.1136/jmg.2005.035709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nelson DL: Mental retardation and intellectual disability. In Vogel and Motulsky's Human Genetics Springer.2010;663–680. 10.1007/978-3-540-37654-5_27 [DOI] [Google Scholar]
- 46. Wagenstaller J, Spranger S, Lorenz-Depiereux B, et al. : Copy-number variations measured by single-nucleotide-polymorphism oligonucleotide arrays in patients with mental retardation. Am J Hum Genet. 2007;81(4):768–79. 10.1086/521274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deciphering Developmental Disorders Study: Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519(7542):223–8. 10.1038/nature14135 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Wieczorek D: Autosomal dominant intellectual disability. Med Genet. 2018;30(3):318–322. 10.1007/s11825-018-0206-2 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Ka M, Chopra DA, Dravid SM, et al. : Essential Roles for ARID1B in Dendritic Arborization and Spine Morphology of Developing Pyramidal Neurons. J Neurosci. 2016;36(9):2723–2742. 10.1523/JNEUROSCI.2321-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ropers HH, Hamel BC: X-linked mental retardation. Nat Rev Genet. 2005;6(1):46–57. 10.1038/nrg1501 [DOI] [PubMed] [Google Scholar]
- 51. Bagni C, Tassone F, Neri G, et al. : Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J Clin Invest. 2012;122(12):4314–22. 10.1172/JCI63141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saldarriaga W, Tassone F, González-Teshima LY, et al. : Fragile X syndrome. Colomb Med (Cali). 2014;45(4):190–8. [PMC free article] [PubMed] [Google Scholar]
- 53. Schirwani S, Wakeling E, Smith K, et al. : Expanding the molecular basis and phenotypic spectrum of ZDHHC9-associated X-linked intellectual disability. Am J Med Genet A. 2018;176(5):1238–1244. 10.1002/ajmg.a.38683 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Neri G, Schwartz CE, Lubs HA, et al. : X-linked intellectual disability update 2017. Am J Med Genet A. 2018;176(6):1375–1388. 10.1002/ajmg.a.38710 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Ropers HH: Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet. 2010;11:161–87. 10.1146/annurev-genom-082509-141640 [DOI] [PubMed] [Google Scholar]
- 56. van Karnebeek CD, Stockler S: Treatable inborn errors of metabolism causing intellectual disability: a systematic literature review. Mol Genet Metab. 2012;105(3):368–381. 10.1016/j.ymgme.2011.11.191 [DOI] [PubMed] [Google Scholar]
- 57. Collin RW, van den Born LI, Klevering BJ, et al. : High-resolution homozygosity mapping is a powerful tool to detect novel mutations causative of autosomal recessive RP in the Dutch population. Invest Ophthalmol Vis Sci. 2011;52(5):2227–39. 10.1167/iovs.10-6185 [DOI] [PubMed] [Google Scholar]
- 58. Fattahi Z, Sheikh TI, Musante L, et al. : Biallelic missense variants in ZBTB11 can cause intellectual disability in human. Hum Mol Genet. 2018;27(18):3177–3188. 10.1093/hmg/ddy220 [DOI] [PubMed] [Google Scholar]
- 59. Vlachos A, Korkotian E, Schonfeld E, et al. : Synaptopodin regulates plasticity of dendritic spines in hippocampal neurons. J Neurosci. 2009;29(4):1017–33. 10.1523/JNEUROSCI.5528-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chiocchetti A, Haslinger D, Stein JL, et al. : Transcriptomic signatures of neuronal differentiation and their association with risk genes for autism spectrum and related neuropsychiatric disorders. Transl Psychiatry. 2016;6(8):e864. 10.1038/tp.2016.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bavley CC, Rice RC, Fischer DK, et al. : Rescue of Learning and Memory Deficits in the Human Nonsyndromic Intellectual Disability Cereblon Knock-Out Mouse Model by Targeting the AMP-Activated Protein Kinase-mTORC1 Translational Pathway. J Neurosci. 2018;38(11):2780–2795. 10.1523/JNEUROSCI.0599-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Bassuk AG, Wallace RH, Buhr A, et al. : A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy-ataxia syndrome. Am J Hum Genet. 2008;83(5):572–581. 10.1016/j.ajhg.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gudmundsson S, Wilbe M, Filipek-Górniok B, et al. : TAF1, associated with intellectual disability in humans, is essential for embryogenesis and regulates neurodevelopmental processes in zebrafish. Sci Rep. 2019;9(1):10730. 10.1038/s41598-019-46632-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Cong L, Ran FA, Cox D, et al. : Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Santoro MR, Bray SM, Warren ST: Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 2012;7:219–45. 10.1146/annurev-pathol-011811-132457 [DOI] [PubMed] [Google Scholar]
- 66. Park CY, Halevy T, Lee DR, et al. : Reversion of FMR1 Methylation and Silencing by Editing the Triplet Repeats in Fragile X iPSC-Derived Neurons. Cell Rep. 2015;13(2):234–41. 10.1016/j.celrep.2015.08.084 [DOI] [PubMed] [Google Scholar]
- 67. Xie N, Gong H, Suhl JA, et al. : Reactivation of FMR1 by CRISPR/Cas9-Mediated Deletion of the Expanded CGG-Repeat of the Fragile X Chromosome. PLoS One. 2016;11(10):e0165499. 10.1371/journal.pone.0165499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bates GP, Dorsey R, Gusella JF, et al. : Huntington disease. Nat Rev Dis Primers. 2015;1:15005. 10.1038/nrdp.2015.5 [DOI] [PubMed] [Google Scholar]
- 69. Shin JW, Kim KH, Chao MJ, et al. : Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. 2016;25(20):4566–4576. 10.1093/hmg/ddw286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mattioli F, Schaefer E, Magee A, et al. : Mutations in Histone Acetylase Modifier BRPF1 Cause an Autosomal-Dominant Form of Intellectual Disability with Associated Ptosis. Am J Hum Genet. 2017;100(1):105–116. 10.1016/j.ajhg.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 71. Dabrowska M, Juzwa W, Krzyzosiak WJ, et al. : Precise Excision of the CAG Tract from the Huntingtin Gene by Cas9 Nickases. Front Neurosci. 2018;12:75. 10.3389/fnins.2018.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Rath J: Safety and Security Risks of CRISPR/Cas9. In: Ethics Dumping Springer. Cham.2018;107–113. 10.1007/978-3-319-64731-9_13 [DOI] [Google Scholar]
- 73. Maulik PK, Mascarenhas MN, Mathers CD, et al. : Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil. 2011;32(2):419–436. 10.1016/j.ridd.2010.12.018 [DOI] [PubMed] [Google Scholar]
- 74. Westerinen H, Kaski M, Virta LJ, et al. : The nationwide register-based prevalence of intellectual disability during childhood and adolescence. J Intellect Disabil Res. 2017;61(8):802–809. 10.1111/jir.12351 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Ilyas M: Replication Data for: The Genetics of Intellectual Disability Review Tables. Harvard Dataverse, V1.2019. 10.7910/DVN/MMUNLR [DOI]
- 76. Ilyas M: The Genetics of Intellectual Disability.Harvard Dataverse, V1.2019. 10.7910/DVN/AEKQOL [DOI]
- 77. Ilyas M: Replication Data for: The Genetics of Intellectual Disability. Harvard Dataverse, V1.2019. 10.7910/DVN/9BJDI6 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ilyas M: Replication Data for: The Genetics of Intellectual Disability Review Tables. Harvard Dataverse, V1.2019. 10.7910/DVN/MMUNLR [DOI]
- Ilyas M: The Genetics of Intellectual Disability.Harvard Dataverse, V1.2019. 10.7910/DVN/AEKQOL [DOI]
- Ilyas M: Replication Data for: The Genetics of Intellectual Disability. Harvard Dataverse, V1.2019. 10.7910/DVN/9BJDI6 [DOI]
Data Availability Statement
Extended data
Harvard Dataverse: Replication Data for: The Genetics of Intellectual Disability Review Tables, https://doi.org/10.7910/DVN/MMUNLR 75
This project contains the following extended data:
Table 1: Estimated total number of genes involved in Intellectual Disability Genes from Literature search and from genome England Database
Harvard Dataverse: The Genetics of Intellectual Disability, https://doi.org/10.7910/DVN/AEKQOL 76
This project contains the following extended data:
Table 2: List of Genes causing Autosomal Recessive Intellectual Disability
Harvard Dataverse: Replication Data for: The Genetics of Intellectual Disability, https://doi.org/10.7910/DVN/9BJDI6 77
This project contains the following extended data:
Table 3: List of Autosomal Dominant Genes in Omim and PubMed DataBase
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).