Abstract
Background
Only a few microbial studies have conducted in IVF (in vitro fertilization), showing the high-variety bacterial contamination of IVF culture media to cause damage to or even loss of cultured oocytes and embryos. We aimed to determine the prevalence and counts of bacteria in IVF samples, and to associate them with clinical outcome.
Methods
The studied samples from 50 infertile couples included: raw (n = 48), processed (n = 49) and incubated (n = 50) sperm samples, and IVF culture media (n = 50). The full microbiome was analyzed by 454 pyrosequencing and quantitative analysis by real-time quantitative PCR. Descriptive statistics, t-, Mann-Whitney tests and Spearman’s correlation were used for comparison of studied groups.
Results
The study involved normozoospermic men. Normal vaginal microbiota was present in 72.0% of female partners, while intermediate microbiota and bacterial vaginosis were diagnosed in 12.0 and 16.0%, respectively. The decreasing bacterial loads were found in raw (35.5%), processed (12.0%) and sperm samples used for oocyte insemination (4.0%), and in 8.0% of IVF culture media. The most abundant genera of bacteria in native semen and IVF culture media were Lactobacillus, while in other samples Alphaproteobacteria prevailed. Staphylococcus sp. was found only in semen from patients with inflammation. Phylum Bacteroidetes was in negative correlation with sperm motility and Alphaproteobacteria with high-quality IVF embryos.
Conclusion
Our study demonstrates that IVF does not occur in a sterile environment. The prevalent bacteria include classes Bacilli in raw semen and IVF culture media, Clostridia in processed and Bacteroidia in sperm samples used for insemination. The presence of Staphylococcus sp. and Alphaproteobacteria associated with clinical outcomes, like sperm and embryo quality.
Keywords: Bacteria, Contamination, In vitro fertilization (IVF), Infertility, Sperm microbiota
Background
Assisted reproductive technologies (ART) are the cornerstone of contemporary infertility treatment. Despite considerable progress made in ART, the implantation rate of replaced embryos remains low, and has been shown to depend on numerous clinical and laboratory factors. The success and failure in ART have largely been attributed to variables such as the patient’s age, weight, endometrial receptivity, and embryo quality and the transfer technique used. The viability of IVF embryos, in turn, depends on the composition of embryo culture media and physical environmental factors applied in embryo culture. At the same time bacterial contamination of gamete samples used in ART may impair the embryo culture environment, causing damage to or even loss of cultured oocytes and embryos [1].
Semen is not sterile [2] and may contain microorganisms even after processing for ART. Although most of the microorganisms detected in semen samples are non-pathogenic commensals or contaminants, their presence has a great significance on in vitro fertilization (IVF), a treatment in which the natural defense of the female genital tract is largely bypassed [3]. Therefore, different approaches have been proposed to reduce the microbial contamination and load in IVF culture media by improving semen preparation and embryo culture protocols. The majority of IVF laboratories use culture media containing antibiotics to minimize the risks of microbial growth. This has been a common practice since the first successful IVF treatment in 1978, when it was suggested that contamination during IVF procedure could negatively affect treatment outcome [4].
Nevertheless, microorganisms may colonize culture dishes with oocytes and embryos; most likely originating from semen samples, as follicular fluid samples are largely sterile and good laboratory practice of IVF eliminates the possibility that embryo culture media will become contaminated by microbes during the procedure. However, the exact frequency of these microbial contaminations is unknown due to low number of investigations [5]. Moreover, there is very little information available on how to handle embryos derived from culture dishes with obvious bacterial contamination. Therefore, the better understanding of whether seminal-derived bacteria have a negative impact on IVF conception could lead to the adoption of more efficacious interventions that can improve the pregnancy and delivery rate in assisted conception [6].
In the current study we aimed to determine the prevalence and counts of bacteria in native semen samples used for IVF, processed semen samples and IVF culture media, and to associate them with IVF embryo quality and pregnancy rate.
Methods
Ethical considerations
Participation in the study was voluntary. Informed written consent was obtained from the patients. The study was approved by the Ethics Review Committee on Human Research of the University of Tartu (Permission No. 193/T-16).
Study group and lab standards
The study group included 50 infertile couples attending the Nova Vita clinic (Tallinn, Estonia) in 2012–2013 for IVF procedure. The average age of women and men were 33.4 ± 4.4 and 37.1 ± 6.3 years, respectively. Additional file 1: Tables S3, S4 provide the clinical and lifestyle background data for the study group. The patients had been suffering from infertility for at least 1 year being otherwise healthy. Only the couples undergoing IVF were recruited, while the couples requiring ICSI (intracytoplasmic sperm injection) were excluded.
Before IVF procedure, sexually transmitted infections were tested and treated whenever needed. Gram stained vaginal smears were microscopically examined to assess vaginal candidiasis as well as bacterial vaginosis according to standardized classification developed by Nugent [7]. Composite score was categorized into three categories, scores 0–3 being normal, 4–6 being intermediate, and 7–10 being definite bacterial vaginosis [7]. Inflammatory prostatitis was assessed by the neutrophil concentration in semen as described [8]. The involved IVF laboratory air quality corresponds to class D and cells were handled under the laminar where A class air quality is mandatory according to ISO 15189 standards. Air particle counting and microbiology measurements were done annually with no deviation.
IVF, sample collection and processing
The patients underwent the standard ovarian stimulation with exogenous gonadotrophins promoting the multi-follicular development. Transvaginal ultrasonography-guided follicle aspiration was performed under short full anaesthesia. The follicles with size > 16 mm were aspirated, follicular fluid was evaluated under the stereomicroscope and oocyte-cumulus complexes were isolated and washed several times in clean culture medium (Origio Universal IVF media). Oocyte-cumulus complexes were incubated for 4 hours at 37 °C, 6% CO2 conditions until the planned insemination with washed semen.
Semen samples were obtained after 2–7 days of sexual abstinence. Before sample collection to sterile container men were asked to urinate and wash their glans penis with soap and warm water [2]. After ejaculation, the sperm sample was shortly (for maximum of 10 min) incubated at 37 °C and left for 25–45 min in room temperature for liquefaction. The semen analysis was performed according to WHO guidelines [9] (Additional file 1: Table S2). Thereafter the semen samples were processed using a 40–80% (2 ml + 2 ml) discontinuous gradient centrifugation method (PureSperm, Nidacon); to separate motile spermatozoa from non-living sperm cells, immotile spermatozoa and seminal plasma. 1–2 ml of the sperm ejaculate was layered over the gradient and centrifuged at 500 g for 20 min. After centrifugation, the supernatant was removed, and the sperm pellet was transferred to the clean 15 ml tube and resuspended in 5 ml of fresh medium (Sperm preparation medium, Origio). Thereafter a washing at 300 g for 10 min was performed, and the supernatant was discarded. The sperm pellet was gently transferred to new 5 ml tube and resuspended in 0.5–1 ml sperm-washing medium and incubated at room temperature for 1 h before oocyte insemination.
In normal IVF, oocytes were inseminated 4–5 h after follicular aspiration with ~ 150,000–200,000 progressively motile spermatozoa per 4–6 oocytes in 1 ml of culture media (Origio Universal IVF media). Fertilization was checked 16–18 h after insemination and normally fertilized oocytes with two pronuclei and polar bodies were further cultured in Origio ISM1 media for 24–48 h before being transferred or cryopreserved. Cleavage stage embryo quality was evaluated daily considering the number of blastomeres, the degree of fragmentation, and the uniformity of blastomeres. Embryos with better quality were selected for embryo transfer or cryopreserved for future use. The following embryo grading system was used: Grade 1 embryos have equal size and symmetrically located blastomeres with < 10% fragments; Grade 2 embryos have 10–25% fragmentation; Grade 3 and 4 embryos exhibit 25–50 and > 50% fragmentation, respectively. Uneven size of blastomeres, multinucleation and other abnormal features of the embryos downgrade the quality of the embryos. Both Grade 1 and 2 embryos are considered as good quality embryos and are preferred for transfer or cryopreservation, while Grade 3 embryos are classified as moderate quality embryos with lower chance of pregnancy [10].
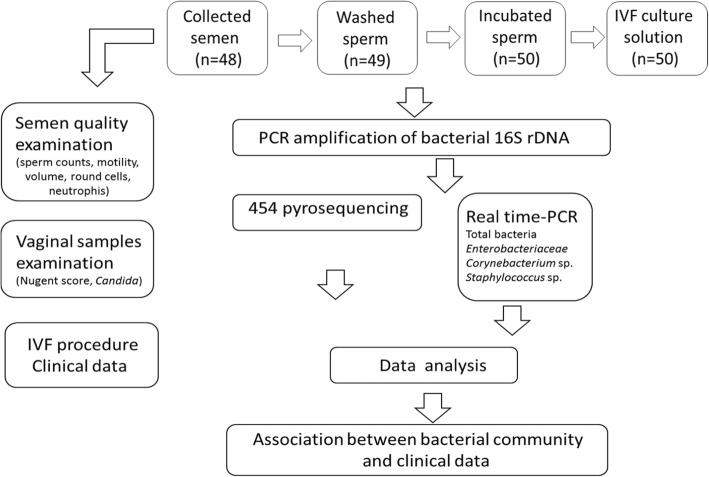
Altogether 197 samples were available for analyses, including: 1) 0.2 ml fresh ejaculate samples (n = 48) frozen before processing; 2) 0.1–0.2 ml washed/prepared sperm samples prepared for oocyte insemination (n = 49) and frozen immediately after processing; 3) 0.1–0.4 ml of leftover semen suspensions (n = 50) used for oocyte insemination, but incubated overnight at 37 °C and 6% CO2, and frozen thereafter; and 4) 1–3 ml of collected IVF culture media (n = 50) where 4–6 oocytes have been incubated together with ~ 150,000–200,000 progressively motile spermatozoa for 16–18 h and frozen thereafter (Fig. 1). Samples were frozen at − 20 °C for further DNA extraction and microbiological studies.
Fig. 1.
Overview of the study. Schematic information about samples and studies methods
Embryos were cultured usually for two or 3 days post fertilization and one or two embryos with better quality were selected for uterine transfer. A positive serum hCG test performed 2 weeks after embryo transfer confirmed biochemical pregnancy. The clinical pregnancy was documented by the presence of a positive fetal heart activity on transvaginal sonography at the sixth or seventh week of pregnancy.
Molecular methods
Bacterial DNA of type strains was extracted using QiaAmp DNA mini kit (Qiagen, Hilden, Germany) according to manufacture instructions. DNA extraction from samples was performed using QIAamp DNA Blood Mini Kit (Qiagen) with some modifications. The sequencing of the DNA library was performed on the Roche 454 FLX next generation sequencing platform. Real-time PCR was applied to quantify the total counts of bacteria, Enterobacteriaceae group and indicator species Staphylococcus and Corynebacterium. The details of molecular methods are presented in Table 1 and Additional file 1: Table S1 [13, 16–18].
Table 1.
Specific primers and probes used for 454 pyrosequencing and qPCR
| Study methods | Target groups (amplicon length, Tm, assay) | Primers/Probes | Sequence (5′-3′) | References |
|---|---|---|---|---|
| 454 pyrosequencing | 16S rRNA gene | TIBACB-ra | TTGGCAGTCTCAG (NNNNNNNN)AGAGTTTGATCCTGGCTCAG | Fabrice et al. 2009 [11] |
| TIBACA/TI357RA-fa | GTCTCCGACTCAG (NNNNNNNN)CTGCTGCCTYCCGTA | |||
| A | CCATCTCATCCCTGCGTGTCTCCGACTCAG | |||
| B | CCTATCCCCTGTGTGCCTTGGCAGTCTCAG | |||
| Real-time PCR |
Total bacteria (466 bp, 60 °C, TaqMan) |
Univ-f Univ-r Univ (probe) |
TGGAGCATGTGGTTTAATTCGA TGCGGGACTTAACCCAACA CACGAGCTGACGACA (AG)CCATGCA |
Ott et al. 2004 [12] |
|
Enterobacteriaceae (195 bp, 58 °C, SYBR) |
Eco1457-f Eco1652-r |
CATTGACGTTACCCGCAGAAGAAGC CTCTACGAGACTCAAGCTTGC |
Bartosch et al. 2004 [13] | |
|
Staphylococcus sp. (560 bp, 62 °C, SYBR) |
TstaG422 TstaG765 |
GGCCGTGTTGAACGTGGTCAAATCA TIACCATTTCAGTACCTTCTGGTAA |
Martineau et al. 2001 [14] | |
|
Corynebacterium sp. (516 bp, 60 °C, SYRR) |
CF-1 CF-2 |
CGTAGGGTGCGAGCGTTGTCCG CGTTGCGGGACTTAACCCAACACT |
Adderson et al. 2008 [15] |
aIn TIBACB and in TIBACA/TI357RA, A and B adapter sequences are underlined, the barcode is indicated by N-s in parentheses and specific primers’ sequences 8F and 357R are shown in italics. In reverse primer, the same barcode was incorporated to all samples
Statistical analysis
The statistical analysis of clinical and qPCR data was performed using SIGMASTAT 2.0 (Systat Software, Chicago, USA) statistic software package. According to the data descriptive statistics, t-, Mann-Whitney tests and Spearman’s correlation were applied to compare the differences in microbiological indices. Statistically significant difference was considered if P < 0.05.
Results
Altogether 50 couples attending IVF procedure participated in the study. For microbiological analyses two molecular approaches were combined, high-throughput sequencing that allows a global systemic view of the microbiome, and qPCR with specific primers that provide an accurate and sensitive method for quantification of individual bacteria in total bacterial count.
Clinical indices
Clinical and lifestyle data of study subjects are presented in Additional file 1: Tables S2, S3 and S4. Semen volume and sperm motility and concentration were normal in all males (Additional file 1: Table S2). According to the WHO threshold value [19], the subset of men with increased neutrophil concentration in semen was 20.0% (10/50). The sperm concentrations were higher before washing in comparison to after washing and sperm suspension used for insemination (p < 0.001, both). In contrast, the sperm motility (A + B) were increased after sperm washing (p < 0.001), where A and B are progressively moving sperm cells.
According to Nugent score data, normal vaginal microbiota was present in 72.0% (36/50) of women; additionally, intermediate microbiota and bacterial vaginosis were diagnosed in 12.0% (6.0/50) and 16.0% (8.0/50) of women, respectively (Additional file 1: Table S2). Biochemical pregnancy after IVF embryo transfer was recorded in 36.0% (18/50) of the couples, while ultrasound scan confirmed the clinical pregnancy in 28.0% (14/50) of the cases.
Microbiome of samples used for IVF procedure
We applied the pyrosequencing of 16S rRNA V2-V3 region to reveal the full microbiome of the investigated samples. Among all 197 samples, 35.5% (17/48) of raw semen, 12.0% (6.0/49) of washed sperm, 4.0% (2.0/50) of incubated semen samples, and 8.0% (4.0/50) of IVF culture media were positive by sequencing method. The number of sequences decreased in studied samples during sperm treatment (Table 2).
Table 2.
Average number (±SD) of sequences, phylotypes abundance (OTUs) and Shannon ‘H’ diversity index in the studied samples
| Samples | Number of sequences (gene copies/μl) | Phylotype abundance (OTUs) | Shannon ‘H’ index (diversity) |
|---|---|---|---|
| Semen | 7911 ± 3562a,b,c | 81.5 ± 44.5 | 33.6 ± 16.5 |
| Washed sperm | 4100 ± 3931a | 64.5 ± 38.5 | 35.6 ± 7.9 |
| Incubated sperm | 1692 ± 1296b | 30.5 ± 12.0 | 24.4 ± 4.3 |
| IVF culture media | 2572 ± 1080c | 36.2 ± 5.7 | 30.6 ± 6.9 |
ap = 0.038
bp = 0.027
cp = 0.008
In total, 188,983 sequences were obtained, with an average 7911 ± 3562 reads for each of the raw semen sample, an average of 4100 ± 3931 and 1692 ± 1296 reads for washed and incubated sperm samples, respectively; as well as an average of 2572 ± 1080 reads for IVF insemination media. The phylotype abundance and Shannon ‘H’ diversity index were also higher in semen and washed sperm than the incubated sperm and IVF culture media though the differences were slightly over significance level.
A principal coordinate analysis (PCoA) plot based on different taxonomic levels (phylum, classes and genera) was constructed to assess the relationships between the community structures of studied samples. Figure 2 showed that the microbiota of different studied samples clustered separately as expected. In native semen the phylum Firmicutes displayed the highest relative abundance (median 91.5%) (Fig. 3, Additional file 1: Table S5). The processed/washed sperm solution displayed more diverse composition of bacteria, in addition to Firmicutes also Proteobacteria and Bacteroidetes displayed remarkable proportions (medians 19.6 to 36.4%). Almost half of the bacteria in incubated semen and in IVF culture media were represented by Proteobacteria.
Fig. 2.
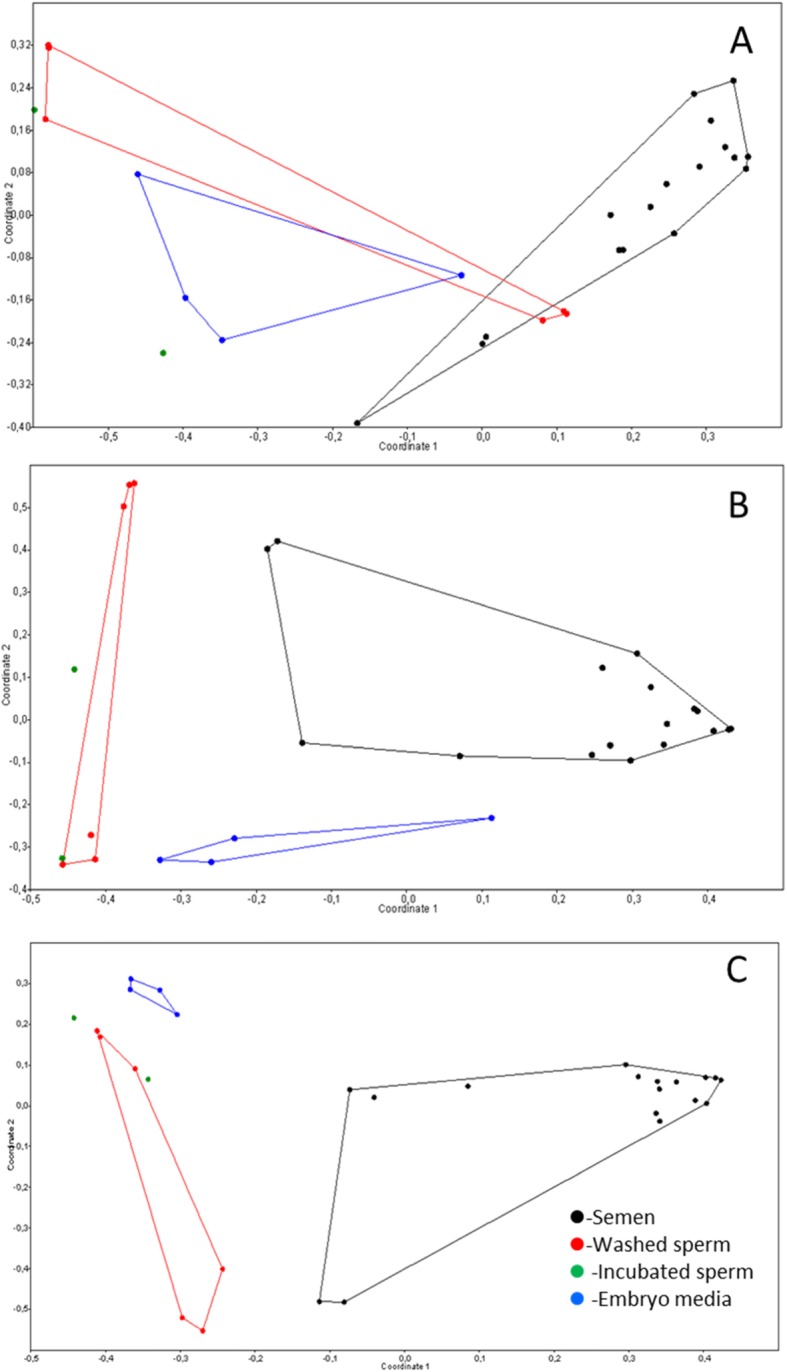
Principal coordinate analysis (PCoA) of bacterial communities in studied samples based on (a) phylum, (b) classes and (c) genera levels. A principal coordinate analysis plot demonstrates different clustering of different specimens (semen, washed sperm, incubated sperm and IVF culture solution)
Fig. 3.
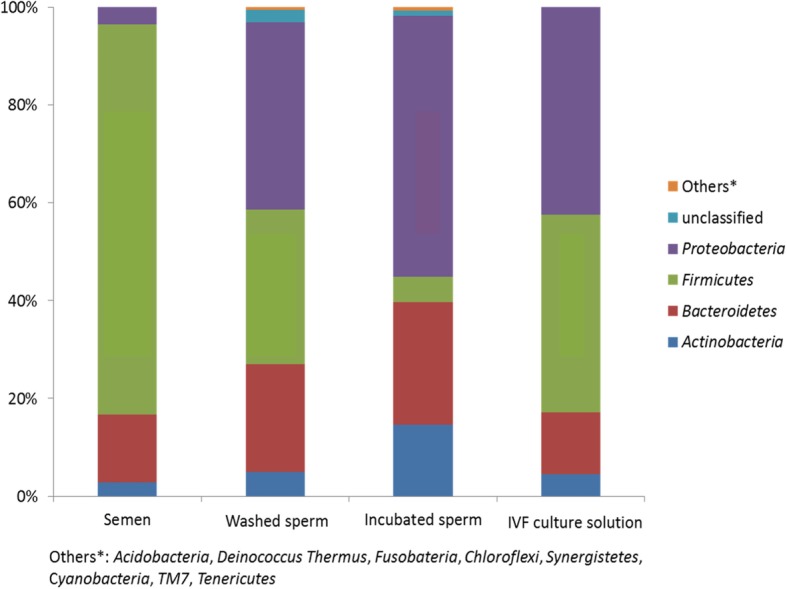
Relative abundance of different bacterial phyla in microbial communities of different samples. Bar charts showing mean values of 4 most abundant phyla in semen, washed and incubated sperm and IVF culture solution. Others: Acidobacteria, Deinococcus Thermus, Fusobacteria, Chloroflexi, Synergistetes, Cyanobacteria, TM7 and Tenericutes. Phylum Firmicutes displayed the highest relative abundance in semen. The processed/washed sperm solution displayed more diverse composition of bacteria, in addition to Firmicutes also Proteobacteria and Bacteroidetes displayed remarkable proportions. Almost half of the bacteria in incubated semen and in IVF culture media were represented by Proteobacteria
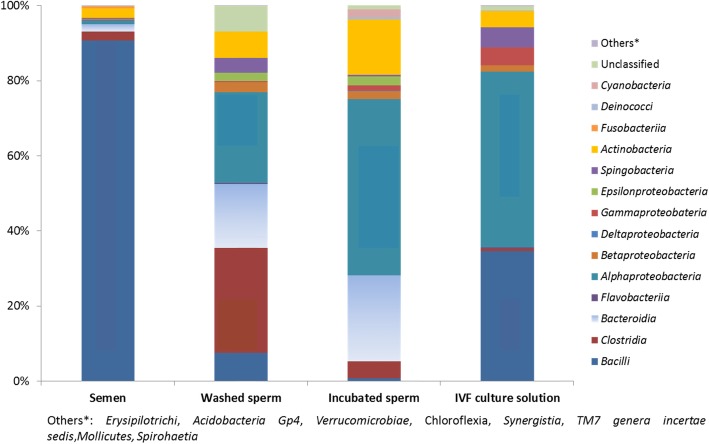
On class level, Bacilli displayed the highest relative abundance in sperm before washing (85.7%) and in IVF culture media (32.7%); Clostridia (20.6%) in washed sperm and Bacteroidia in both washed and incubated sperm (12.6 and 22.4%) (Fig. 4, Additional file 1: Table S6). Alphaproteobacteria displayed high proportions in incubated sperm and IVF culture media (45.7 and 44.1%).
Fig. 4.
Relative abundance of different bacterial classes in microbial communities of different samples. Bar charts showing mean values of most abundant classes in semen, washed and incubated sperm and IVF culture solution. Others: Erysipilotrichi, Acidobacteria Gp4, Verrucomicrobiae, Chloroflexia, Synergistia, TM7 eneta incertae sedis, Mollicutes and Spirohaetia. Bacilli displayed the highest relative abundance in sperm before washing and in IVF culture media; Clostridia in washed sperm and Bacteroidia in both washed and incubated sperm. Alphaproteobacteria displayed high proportions in incubated sperm and IVF culture media
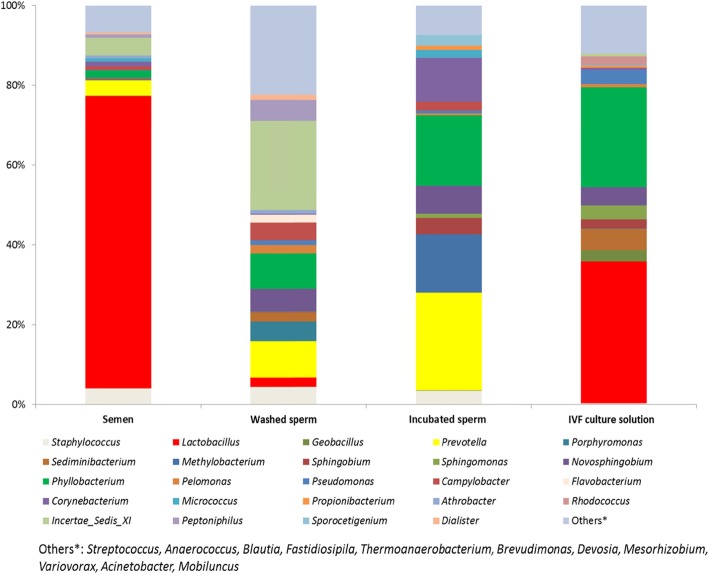
The most abundant genera of bacteria in the sperm before washing and IVF culture solution were Lactobacillus (73.3 and 35.5%, respectively), followed by Incertae sedis XI (4.5%), Staphylococcus (4%) and Prevotella (3.9%) in raw semen samples, while in other samples more heterogenous microbial composition was noted (Fig. 5, Additional file 1: Table S7).
Fig. 5.
Relative abundance of most frequent bacterial genera of microbial communities of different samples. Bar charts showing mean values of most abundant genera in semen washed and incubated sperm and IVF culture solution. Others: Streptococcus, Anaerococcus, Blautia, Fastidiosipila, Thermoanaerobacterium, Brevudimonas, Devosia, Msorhizobium, Variovorax, Acinetobacter and Mobiluncus. The most abundant genera of bacteria in the sperm before washing and IVF culture solution were Lactobacillus, followed by Incertae sedis XI, Staphylococcus and Prevotella in raw semen samples
Prevalence of common aerobic bacteria in IVF samples as revealed by qPCR method
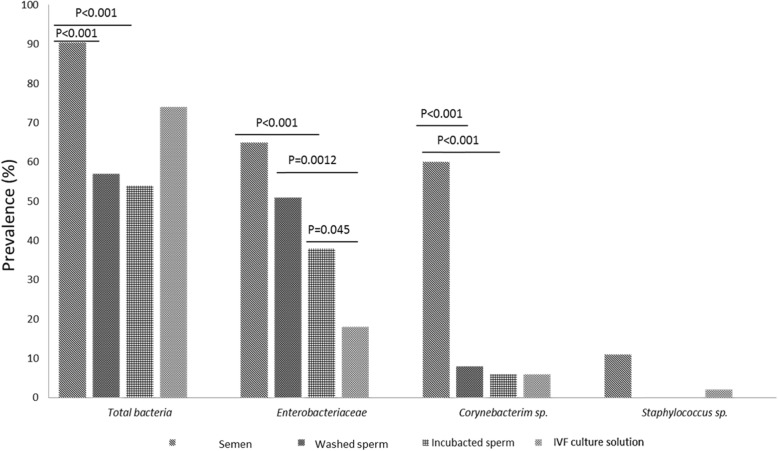
We additionally applied qPCR to detect prevalence and concentration of total bacteria as well as three common groups of bacteria in male semen – Enterobacteriaceae, Corynebacterium sp. and Staphylococcus sp. The prevalence of bacteria in the studied sperm samples significantly decreased after washing and incubation (Fig. 6); while the mean total counts of bacteria decreased during all treated procedures (Table 3). The prevalence of Enterobacteriaceae was lower in IVF culture media than in washed and incubated sperm (Fig. 6), while the counts were lowest in incubated sperm than in raw and washed sperm (Table 3). The counts of Corynebacterium sp. were higher in raw semen in comparison to both washed and incubated sperm as well as IVF insemination media (Table 3).
Fig. 6.
The prevalence (%) of total and three common groups of bacteria Enterobacteriaceae, Corynebacterium sp. and Staphylococcus sp. according to qPCR in study samples. The prevalence of bacteria in the studied sperm samples significantly decreased after washing and incubation. The prevalence of Enterobacteriaceae was lower in IVF culture media than in washed and incubated semen
Table 3.
The counts (log10 plasmid gene copies/ml sperm; mean ± SD) of total bacteria and three common groups of bacteria, Enterobacteriaceae, Corynebacterium sp. and Staphylococcus sp., in study samples as revealed by qPCR
| Bacterial groups | Semen | Washed sperm | Incubated sperm | IVF culture media | p |
|---|---|---|---|---|---|
| Total bacteria | 5.79 ± 0.44a,b,c | 5.17 ± 0.53a | 4.92 ± 0.36b | 4.85 ± 0.28c | a,b,c p < 0.001 |
| Enterobacteriaceae | 4.67 ± 0.89a,b | 3.92 ± 0.26a,c,d | 3.43 ± 0.31b,d | 3.95 ± 0.56c |
a p < 0.005 b,c p < 0.001 d p = 0.008 |
| Corynebacterium sp. | 4.51 ± 0.67a,b,c | 3.78 ± 1.01a | 4.04 ± 0.35b | 3.36 ± 0.49c | a,b,c p < 0.001 |
| Staphylococcus sp. | 3.55 ± 0.33 | 0 | 0 | 2.58 | NS |
a,b,c,d Indicate the compared pairs of values that resulted in statistically significant differences (p value less than 0.05, given in the last column)
Associations between bacteria detected both by sequencing and qPCR, and clinical data
Positive correlations between neutrophils and certain bacteria in raw semen (genus Staphylococcus, classes Erysipelotrichia and Bacteroidia) were found (Table 4).
Table 4.
Spearman’s rank-order correlation between bacteria presented in raw semen and washed sperm (*) detected by pyrosequencing (454), qPCR and clinical data
| Bacteria | Clinical data | R | P |
|---|---|---|---|
| Presence of Bacteroidia (454, class) | Neutrophils | 0.673 | 0.0028 |
| Presence of Erysipelotrichia (454, class) | 0.523 | 0.0309 | |
| Erysipelotrichia (454, OTU numbers) | 0.636 | 0.006 | |
| Counts of Staphylococcus spp. (qPCR) | 0.448 | 0.002 | |
| Total OTU numbers (454, phylum) | Sperm motility | −0.472 | 0.0551 |
| Bacteroidetes (454, phylum, OTU numbers) | −0.518 | 0.0328 | |
| Bacteroidia (454, class, OTU numbers) | −0.498 | 0.0411 | |
| Proteobacteria (454, phylum, OTU numbers) | −0.661 | 0.0036 | |
| Alphaproteobacteria (454, class, OTU numbers) | −0.471 | 0.0551 | |
| Sphingobacteria (454, class, OTU numbers) | −0.635 | 0.0006 | |
| Alphaproteobacteria (454, OTU numbers)* | Embryo quality (Grade 1, 2, 3) | 0.413 | 0.0260 |
Staphylococcus sp. was detected only in semen samples of patients with inflammation.
At the same time the class Bacteroidia, and the whole phylum Bacteroidetes of raw semen were in negative correlation with sperm motility, like also some other bacteria – Proteobacteria (phylum), Alphaproteobacteria (class), and Sphingobacteria (class).
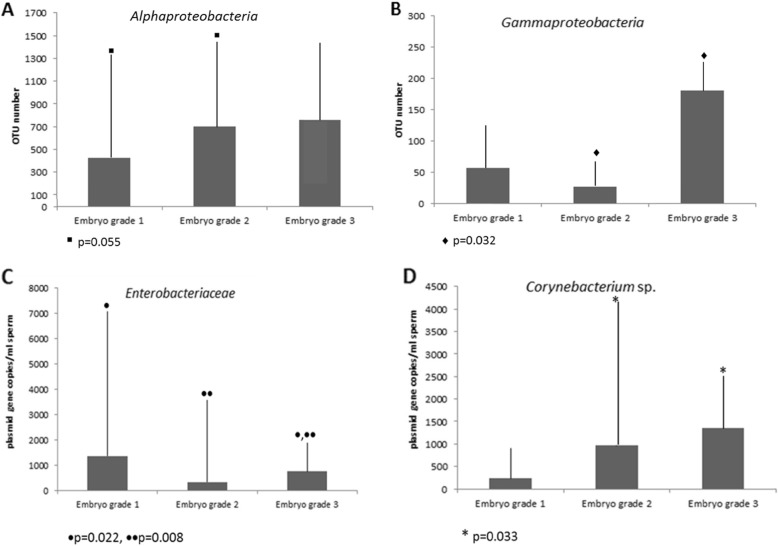
The positive correlation between Alphaproteobacteria (454 pyrosequencing) in washed sperm and low-quality embryos was found (Table 4). Additionally, the higher counts of Alphaproteobacteria and Gammaproteobacteria (454 pyrosequencing) in washed sperm, and Corynebacterium sp. (qPCR) in raw semen samples were found in patients with lower embryo quality (Fig. 7a, b, d). However, the mean proportion of Enterobacteriaceae group in raw semen was higher in couples with better embryo quality (Fig. 7c). No correlation between the prevalence or counts of bacteria presented in IVF culture media and pregnancy results was found.
Fig. 7.
The differences in the counts of Alphaproteobacteria (a) and Gammaproteobacteria (b); Enterobacteriaceae (qPCR) (c) and Corynebacterium sp. (qPCR) (d) in solutions with different embryo grades. The higher counts of Alphaproteobacteria (a) and Gammaproteobacteria (b) (454 pyrosequencing, mean + SD, OTU), in washed sperm, and Corynebacterium sp. (d) (qPCR, mean + SD, plasmid gene copies/ml sperm) in raw semen samples were found in patients with lower embryo quality (a, b, d). The mean proportion of Enterobacteriaceae (c) group (qPCR) in raw semen was higher in couples with better embryo quality (c)
Discussion
This study revealed the qualitative and quantitative bacterial composition of the samples used in IVF. We found that there are considerable bacterial changes in IVF samples with prevalence of classes Bacilli in raw semen and embryo culture media, Clostridia in washed sperm, Bacteroidia in incubated sperm and Alphaproteobacteria in incubated sperm and IVF culture media. The associations between certain clinical data (such as increased counts of neutrophils, sperm motility, embryo quality) and presence of some bacterial phyla and genera (Bacteroidetes, Proteobacteria, Staphylococcus, Corynebacterium spp.) were also found. Although our study is not the first research project describing the presence of some groups of microbes in raw and processed sperm and IVF culture media [2, 20, 21], our study is the first one giving a deep assessment of bacterial composition of IVF culture media based on 16S rRNA gene fragments (454 sequencing platform) to help monitor IVF culture conditions.
The goal of embryo culture in IVF is to keep gametes and embryos in as similar condition to their native environment. For that in IVF laboratory, the maintenance of gametes and embryos requires stringent culture conditions. A high standard of hygiene, cleaning and waste disposal must be followed in order to avoid infection of medical staff and patients, and contamination of the culture dishes and equipment. Every step in the laboratory procedures and manipulations must be carried out with a rigorous discipline of aseptic techniques [22]. Therefore, the sterile culture conditions should be pursued in the conditions, where semen samples and follicular fluid samples – obtained via transvaginal ultrasound-guided aspiration, are believed to contain polymicrobial communities. Indeed, the presence of bacteria in environment and patients’ bodies, like semen and follicular fluid samples, and cervical regions passed in egg retrieval and embryo transfer, have been associated with adverse pregnancy outcomes in IVF [23]. Similarly, a small number of researchers have reported isolating microorganisms from IVF culture media [24, 25]. Semen, technician contamination, for example from oil overlaying human embryo culture, is the most often quoted sources of contamination. The most common species identified are Escherichia coli, Aspergillus, Candida albicans and Gram-negative cocci [23].
We found bacterial load of embryo media in around 8% of the samples by 454 sequencing and more than 70% by real-time PCR method. Previously, Kastrop et al. examined > 14,000 and Ben-Chetrir et al. > 700 IVF cycles by cultivation and found that in both studies 0.7% of IVF cycles had isolated microorganisms [5, 26]. The differences in results may be explained by methods used for bacteria examination. In our study the lower limit of PCR amplicons for 454 sequencing was 0.5 ng/μl but for real-time PCR we used DNA of all 197 isolated samples. Additionally, qPCR specific primers for Enterobacteriaceae group (Gammaproteobacteria) may amplify some other bacterial species such as Moellerella, Morganella, Proteus, Leminorella and etc. [13] that was not found by 454 pyrosequencing. It may equally be explained by the fact that the qPCR used in the current study is more sensitive for specific groups of bacteria that agrees with the study of Al-Mously et al. [27].
The microbiome of semen has been studied mostly in connection with male infertility or prostatitis [28–30]. We found that the predominant bacterial genera in semen samples were Lactobacillus, Incertae sedis XI, Staphylococcus, Prevotella, Phyllobacterium and Corynebacterium. Previously, the high abundance of Lactobacillus in semen was also published [2, 21, 29]. Most abundant genera presented in semen by Hou et al. study were also identified abundant in our data, such as Lactobacillus, Prevotella, Corynebacterium, Staphylococcus and Veillonella [31]. Semen quality in Lactobacillus-predominant semen samples is higher than in the case of the other community types, as lactobacilli prevent sperm lipid peroxidation, thus preserving sperm motility and viability [28]. Some authors indicated that Gram-positive bacteria such as Lactobacillus and Corynebacterium, might protect against the negative influence of Gram-negative bacteria such as Prevotella, Aggregatibacter and Pseudomonas [21]. Prevotella is a genus of Gram-negative anaerobic bacteria [32], that is a member of both semen and vaginal microbial communities while its increased counts have been described in patients with low-quality semen [21, 33]. The clinical significance of strict anaerobes in sperm samples is a subject of dispute. Anaerobic bacteria are not routinely sought in sperm samples, because they are fastidious to cultivate. In fact, by using molecular methods Kiessling et al. detected and identified many anaerobes in the semen of men undergoing fertility evaluation [34]. Our study demonstrated correlation between presences of gram-negative bacteria (Bacteroidia, Sphingobacteria (class), Proteobacteria (phylum), Alphaproteobacteria (class)) with sperm motility. The gram-negative bacteria contain in their cell walls lipopolysaccharide that associated with more pro-inflammatory and oxidant environment and due to these mechnanism may disrupts sperm motility [35].
There is no data about the presence of Incertae sedis XI in semen samples. Previous publications indicated that Clostridiales Family XI Incertae Sedis bacteria are enriched in the colons of healthy adults and are also found on skin and genitals of women suffering from bacterial vaginosis [36–38].
Similar to semen, Lactobacillus genus was also dominating in embryo culture media. Next generation sequencing revealed that Lactobacillus sp. are present in endometrial and ovarian follicular microbiome [25, 39]. The authors associated it with embryo development, and difference in the microbiome between the left and right ovaries, which was attributed to differences in haematogenous spread, was also demonstrated [40]. In contrast, the presence of some other species, such as Propionibacterium and Actinomyces among others, has been associated with adverse IVF outcomes. In addition, E. coli and Streptococcus spp. in follicular fluid might inhibit follicle-stimulating hormone (FSH) from binding to its receptor on granulosa cells [41, 42]. To conclude, the follicular fluid bacteria have been associated with both positive and negative IVF outcomes [25, 40]. In our study, the presence of bacteria in IVF culture media did not influence the pregnancy rate. Also, we were unable to determine the origin of the microbiota in embryo culture media.
Since incubation temperature is a determining factor for bacterial growth, incubation of IVF media at 37 °C could influence the bacterial growth and activity. We found that washed and incubated sperm samples had quite heterogenous microbial composition with prevalence of genera Prevotella and Staphylococcus, and class Alphaproteobacteria. Interestingly, Alphaproteobacteria was the most prevailed class of bacteria in processed sperm samples without and with further incubation, including the highest prevalence of genera Phyllobacterium in all treated sperm samples as well as Methylobacterium in incubated sperm and Novosphingobium in washed sperm and embryo culture media. Presence of these genera in IVF culture media was not previously published. At the same time previous studies have indicated that coliform bacteria, including E. coli that belong to Alphaproteobacteria were found in higher concentration in semen and media used during IVF procedure [5, 21, 43]. Kala et al. demonstrated that inoculation of E. coli caused adhesion to the sperm membrane and subsequent destruction leading to reduced motility and viability in washed samples [44]. Presence of S. aureus and E. coli may induce apoptosis in human sperm with two possible putative mechanisms: a direct cytotoxic activity of bacterial toxins and the contact with pili and flagella [28].
The majority of IVF laboratories use culture media containing antibiotics to avoid the risks of microbial growth during IFV procedure. The most commonly used antibiotics are penicillin (β-lactam), streptomycin and gentamicin [45]. In our study, both semen incubation media and embryo culture media were supplemented with gentamicin sulphate. Gentamicin is a broad-spectrum bactericidal agent of aminoglycoside group that is effective against Gram-positive and Gram-negative aerobic bacteria. Gentamicin binds to four nucleotides of 16S rRNA and a single amino acid of protein S12. This leads to interference with the initiation complex and misreading of mRNA so that incorrect amino acids are inserted into the polypeptide leading to non-functional or toxic peptides and the breakup of polysomes into non-functional monosomes. Though the counts of Enterobacteriaceae decreased with treatment (qPCR), the abundance of some species (Methylobacterium, Phyllobacterium) belonging to Alphaproteobacteria classes increased. Some of these bacteria species may be resistant to gentamicin. The resistance of E. coli to both penicillin and streptomycin in culture media has been reported previously [5, 24]. Although, in the study of 70 bacterial strains isolated from contaminated culture media were subsequently found to be sensitive to gentamicin, we may support the view that antimicrobials in the culture media probably provide little inhibition to the potentially large numbers of bacteria, including anaerobic bacteria. Moreover, it has been shown that aminoglycosides have toxic effects on sperm motility [46]. A review analyzing randomized controlled trials investigating the effect of antibiotics on embryo transfer concluded that the administration of amoxycillin and clavulanic acid prior to embryo transfer reduced the upper genital tract microbial contamination but did not influence clinical pregnancy rates [47]. Furthermore, there are no data about randomised controlled trials to support or refute other antibiotic regimens in this setting [47, 48].
Our findings showed that the simple presence of bacteria might alter the sperm quality. In the present study the counts of Staphylococcus sp. were correlated with presence of neutrophils in semen. Previously, Moretti et al. demonstrated the sperm concentration and the percentage of progressive motility were significantly decreased in sperm samples containing S. epidermidis, S. aureus and E. coli [28]. In addition, we found that counts of Alphaproteobacteria and Enterobacteriaceae may influence embryo quality. In accordance with our results, it has been previously indicated that if the embryo culture dishes are contaminated with bacteria the quality of the developing embryos is poor [5].
Our study has some limitations. First, the number of samples was moderate. In addition, qPCR did not cover wide spectrum of bacteria.
Conclusion
In conclusion, our study demonstrated that IVF does not occur in a sterile environment. The prevalence and counts of bacteria in the IVF procedure decrease during the semen treatment. We demonstrated the prevalence of classes Bacilli (Lactobacillus genera) in raw semen and IVF culture media, Clostridia in washed sperm and Bacteroidia in incubated sperm samples. The presence of Staphylococcus sp. and Alphaproteobacteria are associated with clinical indicators such as sperm and embryo quality. Therefore, future research should focus on the methods aiding to reduce the adverse impact of these microorganisms on IVF embryo development and helping to avoid IVF failure.
Supplementary information
Additional file 1: Table S1. Details of molecular methods. Table S2. Overview of the semen and vaginal samples of the study subjects. Table S3. Overview of general and reproductive health of female partners. Table S4. Overview of general and reproductive health of male partners. Table S5. Relative abundance (%) of different bacterial phyla in microbial communities of different samples (mean±SD). Table S6. Relative abundance (%) of different bacterial classes in microbial communities of different samples (mean±SD). Table S7. Relative abundance (%) of most frequent bacterial genera of microbial communities of different samples (mean±SD).
Acknowledgments
We thank Irja Roots and Dagmar Hoidmets for technical assistance.
Abbreviations
- ART
Assisted reproductive technologies
- FSH
Follicle-stimulating hormone.
- hCG test
Human chorionic gonadotropin test
- ICSI
Intracytoplasmic sperm injection
- IVF
In vitro fertilization
- PCoA
Principal coordinate analysis
- qPCR
Quantitative polymerase chain reaction
Authors’ contributions
The authors’ responsibilities were as follows: J Š and J B were responsible for the study performance, molecular analyses, data analysis and manuscript drafting; J S was responsible for bioinformatic analysis; Ü P, T R and S S were responsible for molecular analysis; P K, M J, and K R were involved in clinical evaluation and the collection of samples; A S and R M were the principal investigators and participated in the design of the study, coordination and drafting the manuscript. All authors contributed to the interpretation of data and in writing and approval of the final manuscript.
Funding
This study was financially supported by Enterprise Estonia (grant No. EU48695), Estonian Ministry of Education and Research (grant No. KOGU-HUMB), Estonian Research Council (grant No. IUT34–19 and IUT34–16), Horizon 2020 innovation programme (WIDENLIFE, 692065) and MSCA-RISE-2015 project MOMENDO (691058).
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding.
author on reasonable request.
Ethics approval and consent to participate
Participation in the study was voluntary. Informed written consent was obtained from the patients. The study was approved by the Ethics Review Committee on Human Research of the University of Tartu (Permission No. 193/T-16).
Consent for publication
All authors agree to the submission of this article for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12958-019-0562-z.
References
- 1.Shu Y, Prokai D, Berga S, Taylor R, Johnston-MacAnanny E. Transfer of IVF-contaminated blastocysts with removal of the zona pellucida resulted in live births. J Assist Reprod Genet. 2016;33(10):1385–1388. doi: 10.1007/s10815-016-0776-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandar R, Punab M, Korrovits P, Turk S, Ausmees K, Lapp E, et al. Seminal microbiome in men with and without prostatitis. Int J Urol. 2017;24(3):211–216. doi: 10.1111/iju.13286. [DOI] [PubMed] [Google Scholar]
- 3.Palini S, Primiterra M, De Stefani S, Pedna MF, Sparacino M, Farabegoli P, et al. A new micro swim-up procedure for sperm preparation in ICSI treatments: preliminary microbiological testing. JBRA Assist Reprod. 2016;20(3):94–98. doi: 10.5935/1518-0557.20160023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czernobilsky B. Endometritis and infertility. Fertil Steril. 1978;30(2):119–130. doi: 10.1016/S0015-0282(16)43448-5. [DOI] [PubMed] [Google Scholar]
- 5.Kastrop PM, de Graaf-Miltenburg LA, Gutknecht DR, Weima SM. Microbial contamination of embryo cultures in an ART laboratory: sources and management. Hum Reprod. 2007;22(8):2243–2248. doi: 10.1093/humrep/dem165. [DOI] [PubMed] [Google Scholar]
- 6.Selman H, Mariani M, Barnocchi N, Mencacci A, Bistoni F, Arena S, et al. Examination of bacterial contamination at the time of embryo transfer, and its impact on the IVF/pregnancy outcome. J Assist Reprod Genet. 2007;24(9):395–399. doi: 10.1007/s10815-007-9146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991;29(2):297–301. doi: 10.1128/JCM.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Punab M, Loivukene K, Kermes K, Mandar R. The limit of leucocytospermia from the microbiological viewpoint. Andrologia. 2003;35(5):271–278. doi: 10.1111/j.1439-0272.2003.tb00856.x. [DOI] [PubMed] [Google Scholar]
- 9.WHO . WHO laboratory manual for the examination of human semen and sperm-cervical mucus interaction (4th Edition) Cambridge: Cambridge University Press; 2010. [Google Scholar]
- 10.Nasiri N, Eftekhari-Yazdi P. An overview of the available methods for morphological scoring of pre-implantation embryos in in vitro fertilization. Cell J. 2015;16(4):392–405. doi: 10.22074/cellj.2015.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabrice AD, R. Exploring microbial diversity using 16S rRNA high-throughput methods. J Comp Sci Syst Biol. 2009;2:074–092. [Google Scholar]
- 12.Ott SJ, Musfeldt M, Ullmann U, Hampe J, Schreiber S. Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric flora. J Clin Microbiol. 2004;42(6):2566–2572. doi: 10.1128/JCM.42.6.2566-2572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartosch S, Fite A, Macfarlane GT, McMurdo ME. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol. 2004;70(6):3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martineau F, Picard FJ, Ke D, Paradis S, Roy PH, Ouellette M, et al. Development of a PCR assay for identification of staphylococci at genus and species levels. J Clin Microbiol. 2001;39(7):2541–2547. doi: 10.1128/JCM.39.7.2541-2547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adderson EE, Boudreaux JW, Cummings JR, Pounds S, Wilson DA, Procop GW, et al. Identification of clinical coryneform bacterial isolates: comparison of biochemical methods and sequence analysis of 16S rRNA and rpoB genes. J Clin Microbiol. 2008;46(3):921–927. doi: 10.1128/JCM.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fite A, Macfarlane GT, Cummings JH, Hopkins MJ, Kong SC, Furrie E, et al. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut. 2004;53(4):523–529. doi: 10.1136/gut.2003.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang QG, Garrity M, Tiedje JM, Cole JR. Naıve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO . Laboratory manual for the examination and processing of human semen. 5. Geneva: WHO; 2011. [Google Scholar]
- 20.Abeysundara PK, Dissanayake D, Wijesinghe PS, Perera R, Nishad A. Efficacy of two sperm preparation techniques in reducing non-specific bacterial species from human semen. J Hum Reprod Sci. 2013;6(2):152–157. doi: 10.4103/0974-1208.117169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng SL, Chiu CM, Lin FM, Huang WC, Liang C, Yang T, et al. Bacterial communities in semen from men of infertile couples: metagenomic sequencing reveals relationships of seminal microbiota to semen quality. PLoS One. 2014;9(10):e110152. doi: 10.1371/journal.pone.0110152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elder K, Baker D, Ribes J. Infections, infertility and assisted reproduction. Part II: infections in reproductive medicine & part III: infections and the assisted reproductive laboratory. Cambridge: Cambridge University Press; 2005.
- 23.Pomeroy KO. Contamination of human IVF cultures by microorganisms: a review. J Clin Embryo. 2010;13:11–30. [Google Scholar]
- 24.Cottell E, Lennon B, McMorrow J, Barry-Kinsella C, Harrison RF. Processing of semen in an antibiotic-rich culture medium to minimize microbial presence during in vitro fertilization. Fertil Steril. 1997;67(1):98–103. doi: 10.1016/S0015-0282(97)81863-8. [DOI] [PubMed] [Google Scholar]
- 25.Pelzer ES, Allan JA, Cunningham K, Mengersen K, Allan JM, Launchbury T, et al. Microbial colonization of follicular fluid: alterations in cytokine expression and adverse assisted reproduction technology outcomes. Hum Reprod. 2011;26(7):1799–1812. doi: 10.1093/humrep/der108. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Chetrit A, Shen O, Haran E, Brooks B, Geva-Eldar T, Margalioth EJ. Transfer of embryos from yeast-colonized dishes. Fertil Steril. 1996;66(2):335–337. doi: 10.1016/S0015-0282(16)58463-5. [DOI] [PubMed] [Google Scholar]
- 27.Al-Mously N, Cross NA, Eley A, Pacey AA. Real-time polymerase chain reaction shows that density centrifugation does not always remove chlamydia trachomatis from human semen. Fertil Steril. 2009;92(5):1606–1615. doi: 10.1016/j.fertnstert.2008.08.128. [DOI] [PubMed] [Google Scholar]
- 28.Moretti E, Capitani S, Figura N, Pammolli A, Federico MG, Giannerini V, et al. The presence of bacteria species in semen and sperm quality. J Assist Reprod Genet. 2009;26(1):47–56. doi: 10.1007/s10815-008-9283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandar R, Punab M, Borovkova N, Lapp E, Kiiker R, Korrovits P, et al. Complementary seminovaginal microbiome in couples. Res Microbiol. 2015;166(5):440–447. doi: 10.1016/j.resmic.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Monteiro C, Marques PI, Cavadas B, Damiao I, Almeida V, Barros N, et al. Characterization of microbiota in male infertility cases uncovers differences in seminal hyperviscosity and oligoasthenoteratozoospermia possibly correlated with increased prevalence of infectious bacteria. Am J Reprod Immunol. 2018;79(6):e12838. doi: 10.1111/aji.12838. [DOI] [PubMed] [Google Scholar]
- 31.Hou D, Zhou X, Zhong X, Settles ML, Herring J, Wang L, et al. Microbiota of the seminal fluid from healthy and infertile men. Fertil Steril. 2013;100(5):1261–1269. doi: 10.1016/j.fertnstert.2013.07.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah HN, Collins DM. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Bacteriol. 1990;40(2):205–208. doi: 10.1099/00207713-40-2-205. [DOI] [PubMed] [Google Scholar]
- 33.Donders G. Diagnosis and management of bacterial vaginosis and other types of abnormal vaginal bacterial flora: a review. Obstet Gynecol Surv. 2010;65(7):462–473. doi: 10.1097/OGX.0b013e3181e09621. [DOI] [PubMed] [Google Scholar]
- 34.Kiessling AA, Desmarais BM, Yin HZ, Loverde J, Eyre RC. Detection and identification of bacterial DNA in semen. Fertil Steril. 2008;90(5):1744–1756. doi: 10.1016/j.fertnstert.2007.08.083. [DOI] [PubMed] [Google Scholar]
- 35.O'Doherty AM, Di Fenza M, Kölle S. Lipopolysaccharide (LPS) disrupts particle transport, cilia function and sperm motility in an ex vivo oviduct model. Sci Rep. 2016;6:24583. doi: 10.1038/srep24583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett E, Guinane CM, Ryan CA, Dempsey EM, Murphy BP, O'Toole PW, et al. Microbiota diversity and stability of the preterm neonatal ileum and colon of two infants. Microbiologyopen. 2013;2(2):215–225. doi: 10.1002/mbo3.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert JA, Kalra A, Dodge CT, John S, Sobel JD, Akins RA. Novel PCR-based methods enhance characterization of vaginal microbiota in a bacterial vaginosis patient before and after treatment. Appl Environ Microbiol. 2013;79(13):4181–4185. doi: 10.1128/AEM.01160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hugon P, Ramasamy D, Robert C, Couderc C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Kallipyga massiliensis gen. Nov., sp. nov., a new member of the family Clostridiales Incertae Sedis XI. Stand Genomic Sci. 2013;8(3):500–515. doi: 10.4056/sigs.4047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno I, Codoner FM, Vilella F, Valbuena D, Martinez-Blanch JF, Jimenez-Almazan J, et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol. 2016;215(6):684–703. doi: 10.1016/j.ajog.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 40.Pelzer ES, Allan JA, Waterhouse MA, Ross T, Beagley KW, Knox CL. Microorganisms within human follicular fluid: effects on IVF. PLoS One. 2013;8(3):e59062. doi: 10.1371/journal.pone.0059062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sluss PM, Reichert LE., Jr Secretion of an inhibitor of follicle-stimulating hormone binding to receptor by the bacteria Serratia, including a strain isolated from porcine follicular fluid. Biol Reprod. 1984;31(3):520–530. doi: 10.1095/biolreprod31.3.520. [DOI] [PubMed] [Google Scholar]
- 42.Sluss PM, Fletcher PW, Reichert LE., Jr Inhibition of 125I-human follicle-stimulating hormone binding to receptor by a low molecular weight fraction of bovine follicular fluid: inhibitor concentration is related to biochemical parameters of follicular development. Biol Reprod. 1983;29(5):1105–1113. doi: 10.1095/biolreprod29.5.1105. [DOI] [PubMed] [Google Scholar]
- 43.Dissanayake DM, Amaranath KA, Perera RR, Wijesinghe PS. Antibiotics supplemented culture media can eliminate non-specific bacteria from human semen during sperm preparation for intra uterine insemination. J Hum Reprod Sci. 2014;7(1):58–62. doi: 10.4103/0974-1208.130859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kala S, Singh A, Prabha V, Singh R, Sharma P. Escherichia coli attaches to spermatozoa: affecting sperm parameters. Arh Appl Sci Res. 2011;3(5):618–23.
- 45.Malik S, Agarwal A. Understanding male infertility global practices & Indian perspectives. Gurgaon: Elsevier India; 2014. [Google Scholar]
- 46.Lemeire K, Van Merris V, Cortvrindt R. The antibiotic streptomycin assessed in a battery of in vitro tests for reproductive toxicology. Toxicol in Vitro. 2007;21(7):1348–1353. doi: 10.1016/j.tiv.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Kroon B, Hart RJ, Wong BM, Ford E, Yazdani A. Antibiotics prior to embryo transfer in ART. Cochrane Database Syst Rev. 2012;(3):CD008995. 10.1002/14651858.CD008995.pub2 [DOI] [PubMed]
- 48.Brook N, Khalaf Y, Coomarasamy A, Edgeworth J, Braude P. A randomized controlled trial of prophylactic antibiotics (co-amoxiclav) prior to embryo transfer. Hum Reprod. 2006;21(11):2911–2915. doi: 10.1093/humrep/del263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Details of molecular methods. Table S2. Overview of the semen and vaginal samples of the study subjects. Table S3. Overview of general and reproductive health of female partners. Table S4. Overview of general and reproductive health of male partners. Table S5. Relative abundance (%) of different bacterial phyla in microbial communities of different samples (mean±SD). Table S6. Relative abundance (%) of different bacterial classes in microbial communities of different samples (mean±SD). Table S7. Relative abundance (%) of most frequent bacterial genera of microbial communities of different samples (mean±SD).
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding.
author on reasonable request.