Abstract
Endometrial cancer (EC) is a common form of cancer in women. Metastasis is the main cause of EC treatment failure. Eukaryotic translation initiation factor 4E (eIF4E) is an oncogene that is overexpressed in a variety of malignancies and their distant metastases. The present study analyzed microarray data from the Oncomine database and revealed that high eIF4E expression was associated with poor prognosis and high pathological grade of EC. The expression of eIF4E was higher in EC tissues compared with in adjacent normal tissues. In addition, microRNA (miR)-320a and miR-340-5p expression levels were downregulated in EC tissues compared with those in adjacent normal tissues, which suggested that these microRNAs may serve as EC tumor suppressor genes. miR-320a and miR-340-5p could bind to the 3′-UTR of eIF4E mRNA, thus downregulating the expression of eIF4E and phosphorylated (p)-eIF4E in EC cells. Overexpression of miR-320a or miR-340-5p effectively suppressed HEC-1A cell migration and invasion. The downregulation of eIF4E and p-eIF4E following miR-320a or miR-340-5p transfection reduced the invasiveness and metastatic capability of EC cells in a manner associated with decreased expression of matrix metallopeptidase (MMP)-3 and MMP-9. In addition, one of the effects of transforming growth factor β1 (TGF-β1), which is to induce the phosphorylation of eIF4E, was suppressed by miR-320a and miR-340-5p overexpression. These two microRNAs also attenuated the features of TGF-β1-induced epithelial-mesenchymal transition (EMT). In conclusion, the results of the present study demonstrated that eIF4E was upregulated in EC, whereas miR-320a and miR-340-5p were downregulated in EC compared with adjacent normal tissues. In vitro, miR-320a and miR-340-5p inhibited the migratory capability of EC cells by downregulating MMP-3 and MMP-9 and prevented TGF-β1-induced EMT through p-eIF4E.
Keywords: microRNA, endometrial cancer, eukaryotic translation initiation factor 4E, cell metastasis, epithelial-mesenchymal transition, transforming growth factor β1
Introduction
Endometrial cancer (EC) is the most common malignancy of the female reproductive system, causing 76,000 deaths every year worldwide (1). Despite major advances in EC diagnosis and treatment strategies, metastasis is a significant clinical challenge and represents the main cause of EC mortality; in patients with EC without metastatic disease, the 5-year overall survival ranges between 74 and 91% (2), whereas patients with stage III or IV EC exhibit 5-year overall survival rates of 57–65 and 20–26%, respectively (1). Thus, understanding the underlying mechanisms of metastatic disease may help in the development of more effective therapeutic strategies for EC.
Eukaryotic translation initiation factor 4E (eIF4E) is the most important component of the eukaryotic translation initiation complex eIF4F. At the initiation of translation, eIF4E binds to the 5′-7-methylguanosine cap structure of an mRNA, connects it to the ribosome and enables translation (3). In addition to cap-dependent protein synthesis, eIF4E also contributes to malignancy as mRNAs regulated by eIF4E generally encode key proteins involved in cell proliferation, angiogenesis, survival and malignant transformation [e.g. cyclin D1, c-MYC, vascular endothelial growth factor and matrix metallopeptidase 9 (MMP-9)] (4). As an oncogene, eIF4E has been demonstrated to be upregulated in a variety of malignancies, such as breast (5), colorectal (6) and prostate (4) cancer, but its role in EC remains to be elucidated. A previous study has revealed that eIF4E is more frequently upregulated in EC extending outside the uterus (FIGO stage III/IV vs. I/II, and downregulation of eIF4E by small interfering (si)RNA significantly reduced the proliferation of HEC-1A cells (7). These findings suggested that eIF4E may serve an important role in the metastasis of EC and may represent a potential anti-metastatic therapeutic target.
The epithelial-mesenchymal transition (EMT) is an important mechanism in tumor metastasis that is triggered by the activation of transcription factors such as Snail family transcriptional repressor 1 (Snail), Twist-related protein 1, Snail family transcriptional repressor 2, forkhead box C2, SOX4 and zinc finger E-box-binding homeobox (8). These activated transcription factors downregulate the epithelial marker E-cadherin, as well as polarity-related proteins, such as lethal giant larvae 2 (9,10), and upregulate mesenchymal markers such as N-cadherin and vimentin (11). The transcriptional events controlling EMT are well characterized, but the associated post-transcriptional mechanisms have not been clearly elucidated (12). A previous study has demonstrated that phosphorylation of eIF4E promotes transforming growth factor β1 (TGF-β1)-mediated EMT via Snail and MMP-3 translation activation of (9), suggesting that eIF4E may serve an important role in the translational control of EMT.
MicroRNAs (miRNAs) are an abundant class of small regulatory RNAs in animals and plants that serve important regulatory roles by interacting with the 3′-untranslated region (3′-UTR) of target mRNAs for cleavage or translational repression (13). A number of studies have demonstrated that miRNAs serve important roles in cell proliferation, apoptosis, migration, chemosensitivity and radio-resistance (14–18), and recent findings have revealed that miRNAs also regulate EMT in various tumor cells. In hepatocellular carcinoma, miRNA (miR)-199b-5p attenuates TGF-β1-induced EMT by directly targeting N-cadherin (19), whereas miR-190 suppresses TGF-β1-induced EMT by targeting SMAD2 in breast cancer (20). Although several miRNAs have been demonstrated to regulate EMT and metastasis of EC, the roles of miR-320a and miR-340-5p in EC have not yet been fully elucidated.
The present study aimed to investigate the expression of eIF4E, miR-320a and miR-340-5p in EC and identify their interactions. The effects of miR-320a and miR-340-5p on cell metastatic potential and EMT were further investigated in EC cells.
Materials and methods
eIF4E gene expression data from patients with EC in the Oncomine database
The Oncomine database (https://www.oncomine.org/resource/login.html) was used to mine the data of eIF4E gene expression in EC using the key words ‘eIF4E’ and ‘endometrial carcinoma’. For the Kaplan-Meier survival analysis, patients with eIF4E expression values below the 20th percentile were classified as having low eIF4E levels.
Endometrial cancer tissues
Between August 2016 and July 2017, eight pairs of EC and adjacent normal tissues (≥2 cm from the tumor edge), were collected from eight patients who underwent hysterectomy at the Affiliated Hospital of Binzhou Medical University. All samples were diagnosed by surgical-pathology or biopsy. The tissues were snap-frozen in liquid nitrogen and stored at −80°C for later experiments, including RNA extraction and western blotting. This study was approved by the Medical Ethics Committee of Binzhou Medical University (approval no. 2016-21). Informed consent was obtained from all patients prior to the collection of samples.
Cell culture
The human EC cell line HEC-1A was purchased from the Shanghai Institute of Cell Biology. The human EC cell lines Ishikawa and RL95-2 were obtained from Dalian Medical University. HEC-1A, RL-952 and Ishikawa cells were cultured in McCoy's 5A (Beijing Macgene Biotechnology Co., Ltd.), DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) and RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) medium, respectively. The media were supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin-streptomycin (Sigma-Aldrich; Merck KGaA) at 37°C with 5% CO2 and saturated humidity.
miRNA transfections
EC cells (HEC-1A and RL95-2) at the logarithmic growth phase were seeded in 6-well plates at 3×105 cells/well. Transfection was performed in triplicate at 50–60% confluency using 1 µg miRNA mimics, mutation mimics (mu-320a or mu-340-5p), siRNA or an eIF4E-encoding vector (Sino Biological, Inc.) in 2.5 µl Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The sequences of the miRNA mimics were as follows: miR-320a sense, 5′-AAAAGCUGGGUUGAGAGGGCG-3′ and antisense, 5′-GCCCUCUCAACCCAGCUUUUUU-3′; miR-340-5p sense, 5′-UUAUAAGCAAUGAGACUGAUU-3′ and antisense, 5′-UCAGUCUCAUUGCUUUAUAAUU-3′. The mimics were synthesized by Shanghai GenePharma Co., Ltd. The sequences of si-eIF4E were sense, 5′-GCUUCUGUAUUCUAAUCUAAU-3′ and antisense, 5′-UAGAUUAGAAUACAGAAGCUU-3′, synthesized by Shanghai GeneChem Co., Ltd. Transfection was performed according to the manufacturer's instructions at room temperature; the transfection complex were replaced with complete medium 6–8 h post-transfection, and the cells were incubated for 24–48 h at 37°C prior to subsequent experiments. For experiments involving TGF-β1 treatment, various concentrations of TGF-β1 (5, 10 and 20 ng/ml; Sino Biological, Inc.) were added to treat EC cells for 48 h.
Reverse transcription-quantitative PCR (RT-qPCR)
Total miRNA of endometrial adenocarcinoma cells (HEC-1A and RL95-2) was isolated by RNAiso Plus (Takara Bio, Inc.), and polyA was added using a polyA polymerase (Ambion; Thermo Fisher Scientific, Inc.). cDNA was synthesized using PrimeScriptÔ RT reagent Kit with gDNA Eraser (Takara Bio, Inc.) with the RT primer 5′-AACATGTACAGTCCATGGATGd(T)30N(A, G, C or T)-3′ at 42°C for 15 min, and qPCR was performed to detect miR-320a and miR-340-5p. Primers used for amplification were as follows: miR-320a forward, 5′-AAAAGCTGGGTTGAGAGG-3′ and reverse, 5′-AACATGTACAGTCCATGGATG-3′; miR-340-5p forward, 5′-AAGCAATGAGACTGATT-3′ and reverse, 5′-AACATGTACAGTCCATGGATG-3′; human 5S rRNA forward, 5′-GCCATACCACCCTGAACG-3′ and reverse, 5′-AACATGTACAGTCCATGGATG-3′. A SYBR® Premix Ex Taq kit (Takara Bio, Inc.) was used according to the manufacturer's instructions. The expression levels of the two miRNAs were measured by the RG3000 system (Corbett Life Science; Qiagen, Inc.) using the following thermocycling conditions: Initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 20 sec, annealing at 56°C for 20 sec and an extension at 72°C for 20 sec. Fluorescence was detected at 585 nm, and the cycle threshold (Ct) was recorded. Human 5S rRNA served as a control. The relative expression of miR-320a and miR-340-5p was normalized to that of 5S rRNA, and the experiments were repeated three times in triplicate. The results ware quantified using the 2−ΔΔCq method (21).
Wound-healing assay
HEC-1A cells were seeded into 12-well plates at 1.5×105 cells/well and cultured to 90% confluency the next day. Subsequently, these cells were subjected to an in vitro wound-healing assay; a sterile 10 µl pipette tip was used to scratch the confluent cell monolayer, the cells were washed, suspended in using PBS and incubated in serum-free McCoy's 5A medium at 37°C. Images were captured using an inverted light microscope (×100 magnification; Leica Microsystems GmbH) at 0, 24 and 48 h of incubation. The rate of migration was measured by quantifying the distance that the HEC-1A cells moved from the edge of the scratch toward the center of the scratch (marked by dotted lines).
Transwell cell migration assays
HEC-1A or RL-952 cells were treated with miRNA mimics for 24 h. A total of 100 µl cell suspension was added to the upper chamber of the Transwell insert (Corning, Inc.) at a concentration of 5×105 cells/ml diluted with serum-free McCoy's 5A medium, whereas medium with 20% fetal calf serum was added to the lower chamber. At 24 h, the liquid in the upper chamber was removed, the surface was washed with PBS, the non-migrated cells were removed with a cotton swab, 600 µl 4% methanol was added to fix the cells (20 min at room temperature), and 600 µl 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) was added to stain the cells (15 min at room temperature). The number of migrated cells was counted under an inverted light microscope (×200 magnification; Leica Microsystems GmbH); the average number of migrated cells was determined by quantification in five random fields. The migratory ability of the cells was determined based on the number of transmembrane cells.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
For the MTT assay, 1×104 HEC-1A and RL95-2 cells/well were cultured in 96-well plates. The following day, cells were treated with the miR-320a or miR-340-5p mimics and control oligomers for 48 h. Each group was tested in six replicates. Subsequently, 10 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA) was added to each well and incubated for 4 h, followed by the addition of 100 µl DMSO (Sigma-Aldrich; Merck KGaA). The optical density (OD) was measured using an auto-microplate reader (Thermo Fisher Scientific, Inc.) at 490 nm.
Detection of apoptosis
Apoptosis was measured by fluorescence-activated cell sorting (FACS). Cells (HEC-1A and RL95-2) were cultured in 6-well plates at 3×105 cells/well and treated with miRNA mimics or control oligomers when the confluency reached 70% the next day. Detection of apoptosis was performed at 48 h using an Annexin V-FITC/PI apoptosis detection kit (BD Biosciences) according to the manufacturer's instructions. The cells were analyzed using a flow cytometer (Beckman Coulter, Inc.), and the CytExpert 1.2.11.0 software (Beckman Coulter, Inc.) were used for data analysis.
Construction of the pcDNA-GFP-eIF4E-3′UTR vector
The sequence of the eIF4E 3′-UTR was obtained from GenBank and was amplified by PCR from human genomic DNA (extracted from whole human blood). The primer sequences were as follows: eIF4E 3′-UTR forward, 5′-CCCAAGCTTTCATTCGCCTTTGTCTTGTA-3′ and reverse, 5′-CGGGGTACCTGGCAGGTGCTTGTAGTC-3′. The eIF4E 3′-UTR was then inserted into a pcDNA3.1-GFP-neo (+) (GenScript Biotech, Inc.) expression vector.
Western blotting
Cells (HEC-1A or RL95-2) were lysed with RIPA lysis buffer containing a protease inhibitor cocktail (cat. no. S8820; Sigma-Aldrich; Merck KGaA) for 30 min on ice. The protein concentrations were measured using the bicinchoninic acid assay, and the protein (35 µg/lane) was subjected to SDS-PAGE (10%) and transferred onto PVDF membranes. Subsequently, the membranes were blocked with 7% fat-free milk and were immunoblotted overnight at 4°C with antibodies against eIF4E (1:1,000; cat. no. BS3432), p-eIF4E (1:1,000; cat. no. BS5015), α-smooth muscle actin (α-SMA; 1:1,000; cat. no. BS70000; all from Biogot Technology Co., Ltd.), MMP-3 (1:400; cat. no. bs-0413R; Bioss), MMP-9 (1:400; cat. no. bs-4593R; Bioss), E-cadherin (1:1,000; cat. no. 20874-1-AP; Proteintech Group, Inc) and Snail (1:1,000; cat. no. 13099-1-AP; Proteintech Group, Inc). GAPDH (1:3,000; cat. no. AP0063; Biogot Technology Co., Ltd.) was used as a control. Following washing with TBS + Tween-20 (0.1%), the membranes were incubated with horseradish peroxidase-labeled goat anti-rabbit IgG (1:5,000; cat. no. ZB-2301; Beijing Zhongshan Golden Bridge Technology Co., Ltd.) for the detection of primary antibodies. The membranes were visualized with ECL (Shanghai Novland Co., Ltd.), and images were captured using an automatic chemiluminescence image analysis system (Tanon Science and Technology Co., Ltd.). Densitometric analysis of the blots was performed using Gel Image System 4.2 software (Tanon Science and Technology Co. Ltd.).
miRNA prediction
The online miRNA analysis software TargetScan (http://www.targetscan.org/vert_72/) was used to identify the miRNAs with potential binding sites in the eIF4E 3′-UTR.
Statistical analysis
Statistical significance of experimental data was evaluated with GraphPad Prism 5 (GraphPad Software, Inc.). Quantitative results are presented as the mean ± standard deviation. Paired Student's t-test was used to compare two groups. Differences among three or more groups were compared using one-way ANOVA followed by a Tukey's test. Correlations were calculated with a Spearman rank test. Array data of eIF4E were obtained from the Oncomine database (https://www.oncomine.org/resource/main.html; TCGA Endometrium 2 dataset). Survival rates were analyzed using Kaplan-Meier survival analysis by Gehan-Breslow-Wilcoxon tests. P<0.05 was considered to indicate a statistically significant difference.
Results
eIF4E is upregulated in EC tissues and is associated with poor clinical outcomes, whereas miR-320a and miR-340-5p are downregulated in EC tissues
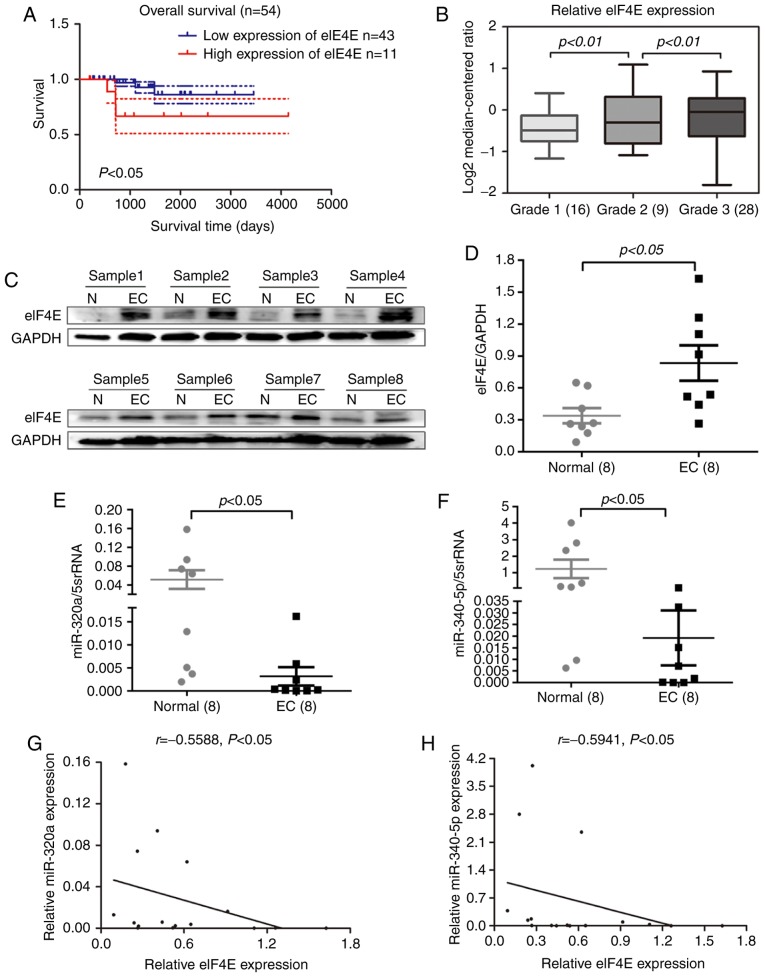
The expression profile of eIF4E in human EC was investigated using patient datasets from the Oncomine database. Data analysis revealed that high eIF4E expression levels in EC tissues were associated with reduced overall survival (Fig. 1A) and a high pathological grade (Fig. 1B). To validate this result, eIF4E protein expression was determined in eight pairs of EC and normal adjacent tissues by Western blotting. The results demonstrated that eIF4E expression levels in EC tissues were significantly higher compared with those in normal adjacent tissues (Fig. 1C and D). To explore their potential role in EC, the levels of miR-320a and miR-340-5p were measured by RT-qPCR in eight paired EC and adjacent tissues. Of note, miR-320a and miR-340-5p expression levels were significantly decreased in EC tissues compared with those in adjacent normal tissues (Fig. 1E and F). Correlation analysis indicated that the expression levels of miR-320a and miR-340-5p were inversely correlated with eIF4E expression (Fig. 1G and H). Collectively, these results suggested that eIF4E may function as a tumor promoter in EC.
Figure 1.
Relative expression of eIF4E, miR-320a and miR-340-5p in EC tissues. (A) Overall survival rate was determined through Kaplan-Meier survival analysis. Kaplan-Meier survival curves demonstrated that patients with EC with high eIF4E expression exhibited a poorer prognosis compared with those with low expression based on the data in the Oncomine database. (B) The expression of eIF4E was associated with the grade of EC in the Oncomine database. (C and D) Western blotting analysis demonstrated that eIF4E protein levels were higher in EC tissues compared with those in adjacent normal tissues. (E and F) miR-320a and miR-340-5p expression levels were decreased in EC tissues compared with those in adjacent normal tissues, as indicated by reverse transcription-quantitative PCR. (G and H) Expression levels of miR-320a and miR-340-5p were negatively correlated with eIF4E in EC. EC, endometrial carcinoma; N, adjacent normal tissue; eIF4E, eukaryotic translation initiation factor 4E; miR, microRNA.
eIF4E is overexpressed, whereas miR-320a and miR-340-5p are downregulated in HEC-1A and RL95-2 EC cell lines
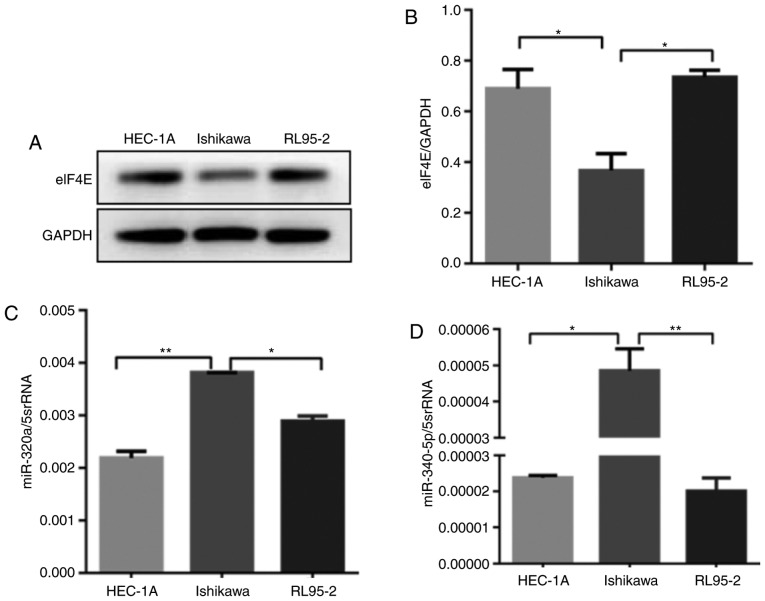
eIF4E expression was analyzed by western blotting in three endometrial cancer cell lines: HEC-1A, Ishikawa and RL95-2 cells. The expression of eIF4E was high in HEC-1A and RL95-2 cells, but low in Ishikawa cells (Fig. 2A and B). The expression levels of miR-320a and miR-340-5p in the three human EC cell lines were measured by RT-qPCR; miR-320a and miR-340-5p were downregulated in HEC-1A and RL95-2 compared with Ishikawa cells (Fig. 2C and D). Based on these results, it was hypothesized that this heterogeneity was due to the degree of differentiation in the three cell lines.
Figure 2.
eIF4E is upregulated, whereas miR-320a and miR-340-5p are downregulated in HEC-1A and RL95-2 cells. (A and B) eIF4E expression levels in HEC-1A, Ishikawa, and RL95-2 cells were determined by western blotting. (C) miR-320a expression levels in HEC-1A, Ishikawa and RL95-2 cells were determined by RT-qPCR. (D) miR-340-5p expression levels in HEC-1A, Ishikawa and RL95-2 cells were determined by RT-qPCR. *P<0.05 and **P<0.01. eIF4E, eukaryotic translation initiation factor 4E; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR.
eIF4E is a direct target of miR-320a and miR-340-5p
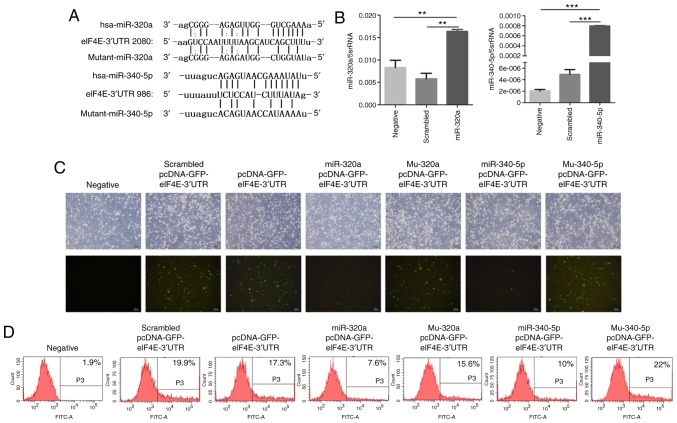
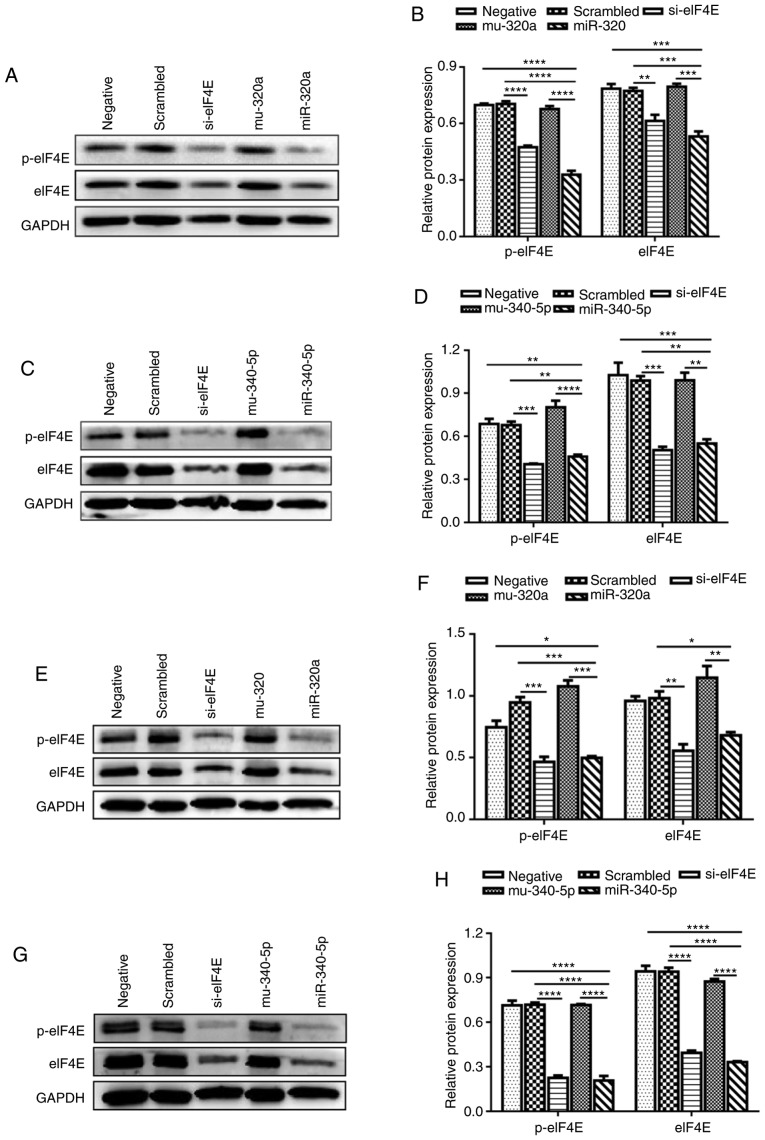
To investigate whether miR-320a and miR-340-5p were involved in regulating eIF4E expression, the potential miRNAs that target the 3′-UTR of eIF4E mRNA were determined. Based on the results of the online miRNA analysis software TargetScan, target sites for miR-320a and miR-340-5p were identified in the 3′-UTR of eIF4E (Fig. 3A). Treatment of HEC-1A cells with miR-320a or miR-340-5p mimics significantly increased their corresponding miRNA levels, as determined by RT-qPCR (Fig. 3B). Subsequently, a pcDNA expression vector encoding GFP-eIF4E-3′-UTR was co-transfected with miR-320a or miR-340-5p mimics into HEC-1A cells. Fluorescence microscopy revealed that the fluorescence intensity was significantly decreased in miR-320a and miR-340-5p mimic-treated cells compared with that in controls (Fig. 3C). Flow cytometry also demonstrated that the fluorescence decreased significantly in miR-320a and miR-340-5p mimic-treated cultures compared with that in the control groups (Fig. 3D). Western blotting confirmed that miR-320a and miR-340-5p mimics reduced the protein expression levels of not only eIF4E, but also p-eIF4E in HEC-1A cells (Fig. 4A-D). These experiments were repeated in RL95-2 cells, which confirmed the results obtained in HEC-1A cells (Fig. 4E-H). These results indicated that the eIF4E-3′UTR was directly targeted by miR-320a and miR-340-5p.
Figure 3.
eIF4E is a direct target of miR-320a and miR-340-5p. (A) The site of eIF4E 3′-UTR that was predicted to be targeted by miR-320a and miR-340-5p and the sequences of mutant-miR-320a and mutant-miR-340-5p. (B) Transfection of HEC-1A cells with miR-320a and miR-340-5p mimics significantly increased their expression levels. (C) GFP fluorescent analysis demonstrated that the fluorescence intensity decreased in miR-320a or miR-340-5p mimic-transfected groups compared with that in the control group. (D) Flow cytometry revealed that GFP levels were decreased in the miR-320a or miR-340-5p mimic-transfected groups compared with those in the control group. **P<0.01 and ***P<0.001 vs. negative or scrambled control. eIF4E, eukaryotic translation initiation factor 4E; miR, microRNA; 3′-UTR, 3′-untranslated region; negative, mock transfection; scrambled, cells treated with scrambled control microRNA mimics; miR-320a or miR-340-5p, cells treated with miR-320a or miR-340-5p mimics; pcDNA-GFP-eIF4E-3′UTR, cells treated with a pcDNA-GFP-eIF4E-3′UTR vector; Mu-320a or Mu-340-5p, cells treated with mutated sequences of miR-320a or miR-340-5p; GFP, green fluorescent protein.
Figure 4.
miR-320a and miR-340-5p mimics regulate the expression of eIF4E and p-eIF4E. (A and B) eIF4E and p-eIF4E levels were decreased in miR-320a-transfected HEC-1A cells. (C and D) eIF4E and p-eIF4E expression levels were decreased in miR-340-5p-transfected HEC-1A cells. (E and F) eIF4E and p-eIF4E levels were decreased in miR-320a mimic-transfected RL95-2 cells. (G and H) eIF4E and p-eIF4E expression levels were decreased in miR-340-5p mimic-transfected RL95-2 cells compared with the control. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. eIF4E, eukaryotic translation initiation factor 4E; miR, microRNA; negative, mock transfections; scrambled, cells treated with scrambled control miRNA mimics; miR-320a or miR-340-5p, cells treated with miR-320a or miR-340-5p mimics; mu-320a or mu-340-5p, cells treated with mutation sequence of mutant-miR-320a or mutant-miR-340-5p; siRNA-eIF4E, small interfering RNA targeting eIF4E.
Overexpression of miR-320a or miR-340-5p inhibits EC cell viability and migration
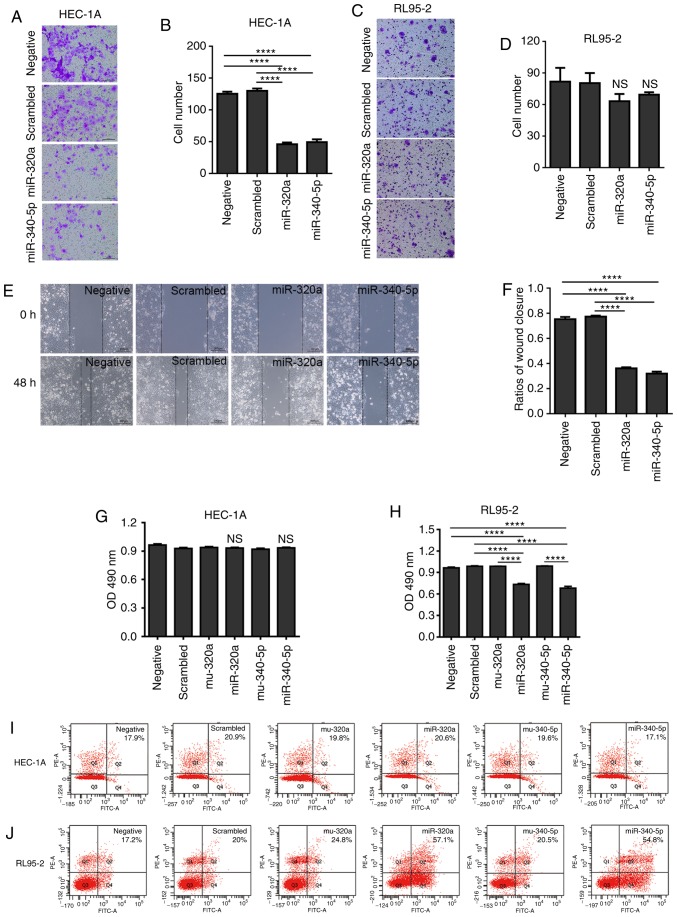
Considering that high expression of eIF4E is associated with the prognosis and grade of EC, the present study investigated whether miR-320a and miR-340-5p may reduce the metastatic capability of EC cells. Either miR-320a or miR-340-5p mimic treatment in HEC-1A cells reduced the number of cells that migrated to the lower chamber in the Transwell assay; however, no effect was observed in RL95-2 cells (Fig. 5A-D). The results of the wound-healing assay also demonstrated that miR-320a or miR-340-5p mimics significantly decreased the migration in HEC-1A cells (Fig. 5E and F).
Figure 5.
miR-320a and miR-340-5p mimics regulate HEC-1A cell viability and migration. (A) Treatment with miR-320a or miR-340-5p mimics reduced HEC-1A migration determined by Transwell assay. (B) Quantitative analysis of the migration rates in (A). (C) Treatment with miR-320a or miR-340-5p mimics did not affect RL95-2 migration determined by Transwell assays. (D) Quantitative analysis of the migratory rates in (C). (E) The migratory ability of HEC-1A cells transfected with miR-320a and mir-340-5p mimics was detected by wound-healing assay. (F) Quantitative analysis of the wound closure ratios in (E). (G) HEC-1A cell proliferation according to the MTT assay. (H) RL95-2 cell proliferation according to the MTT assay; the OD at 490 nm was lower in either the miR-320a or miR-340-5p mimic-treated RL95-2 cells compared with that in the negative, scrambled or mutant control groups. (I) Flow cytometry analysis of either miR-320a or miR-340-5p mimic-induced apoptosis in HEC-1A cells. (J) Flow cytometry analysis of either miR-320a or miR-340-5p mimic-induced late apoptosis in RL95-2 cells. The number of apoptotic cells was increased in miR-320a- or miR-340-5p mimic-treated cells compared with those in the negative, scrambled or mutant control groups. ****P<0.0001; NS, no significant difference. miR, microRNA; negative, mock transfections; scrambled, cells treated with scrambled-oligomer control RNA; miR-320a or miR-340-5p, cells treated with miR-320a or miR-340-5p mimics; mu-320a or mu-340-5p, cells treated with mutated sequences of miR-320a or miR-340-5p; OD, optical density.
To further investigate the proliferation inhibitory effect of miR-320a and miR-340-5p in EC cells, MTT and apoptosis detection assays were performed. miR-320a and miR-340-5p mimics inhibited RL95-2 cell proliferation, but did not affect HEC-1A cells (Fig. 5G and H). In addition, flow cytometric analysis of apoptosis indicated that miR-320a and miR-340-5p mimics induced apoptosis in RL95-2 cells, but had no effect on apoptosis in HEC-1A cells (Fig. 5I and J).
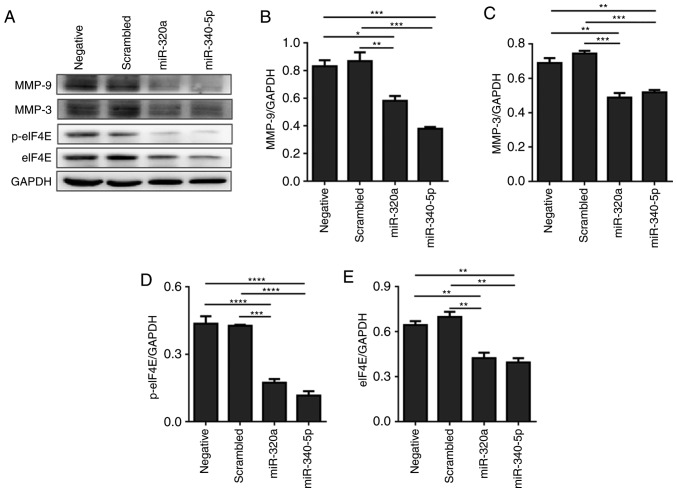
miR-320a and miR-340-5p mimics suppress MMP-3 and MMP-9 expression in HEC-1A cells
MMP-3 and MMP-9 expression levels were determined to explore the mechanisms involved in reduced cell migration and invasion following either miR-320a or miR-340-5p mimic treatment. As demonstrated in Fig. 6, MMP-3 and MMP-9 protein levels were attenuated, suggesting that the reduced level of MMP-3 and MMP-9 following miR-320a or miR-340-5p mimic treatment may account, at least in part, for the anti-migratory effects of miR-320a and miR-340-5p mimics.
Figure 6.
miR-320a and miR-340-5p mimic treatment suppresses the expression of MMP-3 and MMP-9 in HEC-1A cells. (A) Western blotting analysis indicated that the expression levels of MMP-3 and MMP-9 were downregulated following miR-320a or miR-340-5p mimic treatment. (B-E) Quantification of (B) MMP-3, (C) MMP-9, (D) p-eIF4E and (E) eIF4E relative expression levels (n=3). *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. eIF4E, eukaryotic translation initiation factor 4E; p, phosphorylated; miR, microRNA; MMP, matrix metallopeptidase; negative, mock transfections; scrambled, cells treated with scrambled-oligomer control RNA; miR-320a or miR-340-5p, cells treated with miR-320a or miR-340-5p mimics.
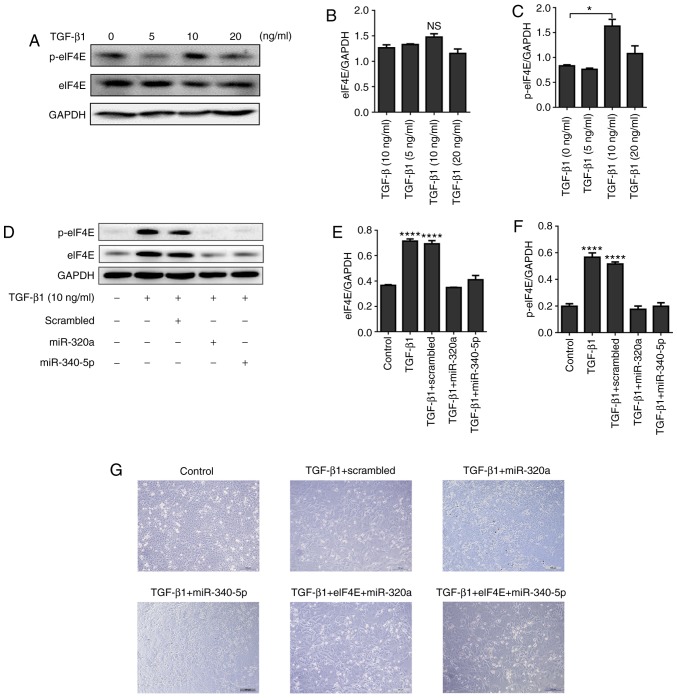
miR-320a and miR-340-5p mimics suppress TGF-β1-induced EMT and change p-eIF4E expression in EC cells
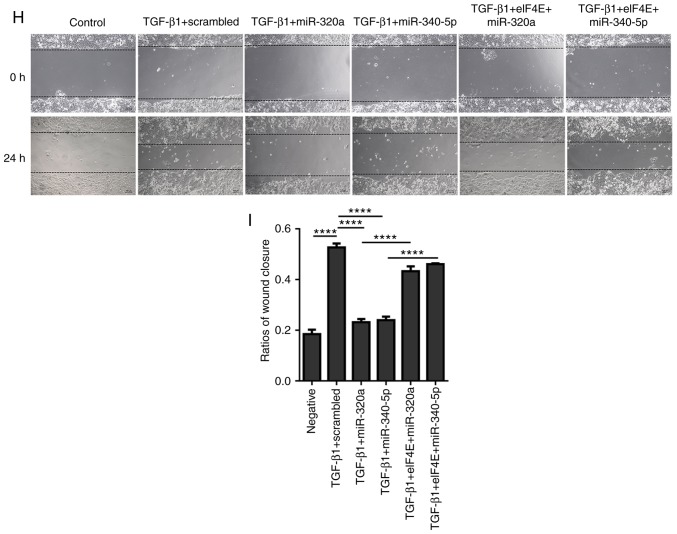
HEC-1A cells were treated with different concentrations of TGF-β1 (0, 5, 10 and 20 ng/ml) for 48 h. The expression of p-eIF4E was significantly enhanced by 10 ng/ml TGF-β1 (Fig. 7A-C), but this upregulation was suppressed when the cells were treated with miR-320a or miR-340-5p mimics (Fig. 7D-F). In terms of cell morphology, following 10 ng/ml TGF-β1 treatment for 48 h, HEC-1A cells exhibited fibroblast-like features; by contrast, a cobblestone-like appearance was observed in the control, miR-320a mimic + TGF-β1 and miR-340-5p mimic + TGF-β1 groups (Fig. 7G). An eIF4E-encoding vector co-transfected with either miR-320a or miR-340-5p into HEC-1A cells blocked the effects of miR-320a and miR-340-5p on cell morphology (Fig. 7G). To further assess the effects of miR-320a and miR-340-5p on the biological outcomes of EMT in HEC-1A cells, a wound-healing assay was performed; TGF-β1 promoted EC cell migration, whereas treatment with either miR-320a or miR-340-5p mimics prevented TGF-β1-induced cell migration (Fig. 7H and I).
Figure 7.
miR-320a and miR-340-5p mimic treatment inhibits TGF-β1-induced EMT. (A) Western blotting was performed to analyze eIF4E and p-eIF4E expression levels in HEC-1A cells following treatment with different concentrations of TGF-β1 for 48 h. (B and C) Quantification of eIF4E and p-eIF4E relative expression levels. (D) miR-320a or miR-340-5p mimic treatment suppressed p-eIF4E expression induced by TGF-β1 in HEC-1A cells. (E and F) Quantification of (E) eIF4E and (F) p-eIF4E relative expression. (G) Morphological characteristics of HEC-1A cells. TGF-β1-treated HEC-1A cells exhibited morphological changes typical to EMT (conversion from an epithelial to a fibroblastic phenotype), whereas miR-320a and miR-340-5p mimics reversed this transition. Cells treated with 10 ng/ml TGF-β1 for 72 h are presented. miR-320a and miR-340-5p mimic treatment inhibits TGF-β1-induced EMT. (H and I) The migratory ability of HEC-1A cells was enhanced in TGF-β1-treated HEC-1A cells compared with that of the controls and was inhibited by either miR-320a or miR-340-5p mimic treatment. *P<0.05 and ****P<0.0001. TGF-β1, transforming growth factor β1; scrambled, cells treated with scrambled control RNA; miR-320 or miR-340-5p; cells treated with miR-320a or miR-340-5p mimics; eIF4E, eukaryotic translation initiation factor 4E or cells treated with pCMV3-eIF4E; p, phosphorylated; EMT, epithelial-mesenchymal transition.
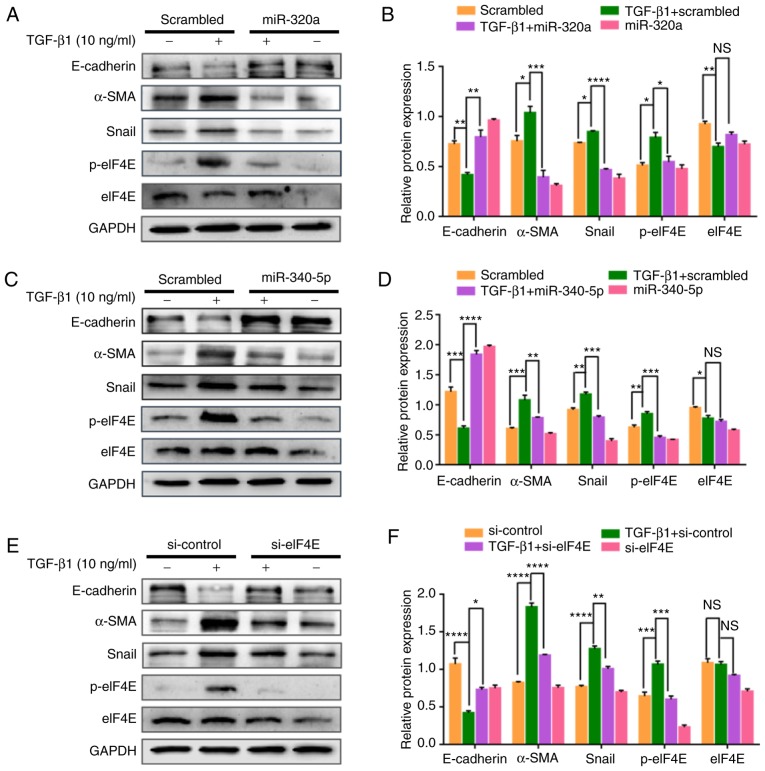
miR-320a and miR-340-5p mimics attenuate the TGF-β1- induced EMT marker expression in EC cells
To further explore the effects of miR-320a and miR-340-5p on the inhibition of EMT, HEC-1A cells transfected with either miR-320a or miR-340-5p mimics were exposed to TGF-β1 for 48 h, and EMT markers were subsequently assessed by western blotting. TGF-β1 suppressed the expression of the epithelial marker E-cadherin and enhanced the expression of the mesenchymal markers Snail and α-SMA (Fig. 8A-D). A siRNA targeting eIF4E exhibited similar results to those observed following miR-320a and miR-340-5p mimic treatments (Fig. 8E and F).
Figure 8.
miR-320a and miR-340-5p mimic treatment suppresses TGF-β1-induced epithelial-mesenchymal transition through the inhibition of p-eIF4E. (A, C and E) Western blotting analysis was performed to detect E-cadherin, α-SMA, Snail, p-eIF4E and eIF4E expression levels in HEC-1A cells transfected with the (A) miR-320a mimic, (B) miR-340-5p mimic or (C) si-eIF4E and treated with 10 ng/ml TGF-β1 for 48 h. (B, D and F) Quantification of E-cadherin, α-SMA, Snail, p-eIF4E and eIF4E relative expression levels in HEC-1A cells transfected with the (B) miR-320a mimic, (D) miR-340-5p mimic or (E) si-eIF4E and treated with 10 ng/ml TGF-β1 for 48 h (n=3). *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001; NS, not significant. TGF-β1, transforming growth factor β1; eIF4E, eukaryotic translation initiation factor 4E; miR, microRNA; α-SMA, α-smooth muscle actin; p, phosphorylated; scrambled, cells treated with scrambled control RNA; miR-320a or miR-340-5p, cells treated with miR-320a or miR-340-5p mimics; si-eIF4E or si-control, cells treated with small interfering RNA specific to eIF4E or siRNA-control.
Discussion
EC is the most common type of female reproductive system cancer and accounts for 4.8% of all cancers diagnosed in women (1). The mechanisms underlying metastasis, which is the main cause of EC treatment failure, have not been elucidated. Thus, a clearer understanding of metastasis is critical for the development of new therapeutic strategies for treating EC.
The miRNA-mediated regulation has complex cellular outcomes, as miRNAs can be involved in cell proliferation, apoptosis, invasion and migration (22,23). First identified as a potential modulator of aquaporin 1 and aquaporin 4, miR-320a also serves a role in cerebral ischemia (24). In metastatic colon cancer tissues and cells within the liver, miR-320a is downregulated, and its levels are associated with tumor progression in colorectal cancer (25). Additionally, miR-320a inhibits the proliferation of human colon cancer cells by directly targeting β-catenin (26). In non-small cell lung cancer, miR-320a suppresses cell migration and invasion through the PI3K/Akt signaling pathway by inhibiting the expression of E74-like ETS transcription factor 3 (16). Another miRNA that plays a role in a variety of tumors is miR-340-5p (also termed miR-340). In cervical cancer, miR-340 expression is downregulated compared with that in normal tissues, and miR-340 inhibits cervical cancer metastasis through targeting Ephrin type-A receptor 3 (27). In breast cancer, miR-340-5p inhibits the proliferation and drug resistance and increases the apoptosis of breast cancer cells by downregulating the expression of leucine-rich repeat-containing G protein-coupled receptor 5 via the Wnt/β-catenin pathway (28). In the present study, miR-320a and miR-340-5p expression levels were downregulated in EC tissues compared with those in adjacent normal tissues, and miR-320a or miR-340-5p mimics inhibited the migration and invasion of EC cells in vitro by downregulating eIF4E. In addition, miR-320a and miR-340-5p mimics exhibited different effects on different EC cell lines; HEC-1A migration was significantly affected by miR-320a or miR-340-5p, whereas RL95-2 cell migration was not. By contrast, miR-320a or miR-340-5p mimics exhibited effects on RL95-2 cell proliferation and apoptosis, which may have been a result of the different degrees of differentiation and different phenotypic characteristics of the two EC cell lines. Upregulation of eIF4E, a translation initiation factor, has been previously detected in many human tumors. Specifically, eIF4E has been associated with disease progression, cellular transformation, tumorigenesis and metastasis in experimental models (29). Choi et al (7) first reported that the positive rate of eIF4E expression is higher in metastatic EC and promotes the proliferation of HEC-1A cells in vitro. Additionally, another study demonstrated that eIF4E levels were higher in EC specimens compared with those in hyperplastic or normal endometrial tissue specimens (30). In the present study, analysis of the Oncomine database revealed that high eIF4E expression was associated with a poor prognosis and a high pathological grade of EC.
The results of the present study demonstrated that either miR-320a or miR-340-5p mimics downregulated the expression levels of MMP-3 and MMP-9 by targeting eIF4E. MMP-3 has been reported to be involved in vascular invasion and metastasis of EC through EMT (31). The translation of MMP-9 mRNA has been demonstrated to be exceptionally dependent on elevated eIF4F activity (32). MMP-9 is a target of eIF4E, and its upregulation is associated with vascular and lymphatic invasion in EC (33). Considering the results of the present study, it may be hypothesized that the inhibition of MMP-3 and MMP-9 expression may represent one of the mechanisms by which miR-320 and miR-340-5p prevent the invasion and migration of EC cells. This process may also be related to other mechanisms; in EMT, benign tumor cells acquire the capacity of invasion and metastasis and can infiltrate the surrounding tissues and eventually move to distant regions (34). During EMT and tumor invasion, eIF4E has been identified to serve important roles, as it has been demonstrated to regulate the expression of a number of proteins involved in EMT and metastasis, such as MMP-3 and Snail (35). Smith et al (9) demonstrated that overexpression of eIF4E induced EMT in lung epithelial cells. TGF-β is a major regulator of the EMT (34). In the present study, exogenous TGF-β1-induced EMT of HEC-1A cells was accompanied by an upregulation of p-eIF4E. These results suggested that eIF4E may promote a metastatic phenotype of EC, in part by regulating EMT. It was thus predicted that suppression of eIF4E may inhibit EMT in EC; transfection with either miR-320a or miR-340-5p mimics in EC cells prevented TGF-β1-induced changes in cell morphology and the upregulation of p-eIF4E. In addition, the expression of Snail was attenuated by miR-320a and miR-340-5p mimics. Snail, which is a key regulator of EMT, induces epithelial cells with migratory and invasive properties during tumor progression (36), and several studies have confirmed that Snail stimulates invasion and metastasis of EC (37,38). Robichaud et al (35) have demonstrated that Snail is regulated by p-eIF4E. The results of the present study demonstrated that downregulation of Snail by either miR-320a/p-eIF4E or miR-40-5p/p-eIF4E may represent a part of the mechanism underlying the prevention of TGF-β1-induced EMT. In addition, the TGF-β1-induced downregulation of E-cadherin and upregulation of α-SMA were prevented in miR-320a or miR-340-5p mimic-treated cells.
In conclusion, the results of the present study demonstrated that in EC, eIF4E was upregulated, whereas miR-320a and miR-340-5p were downregulated. Specifically, miR-320a and miR-340-5p mimics inhibited the proliferation and migration of EC cells in vitro by downregulating MMP-3 and MMP-9 expression and prevented the TGF-β1-induced EMT by targeting p-eIF4E. These results suggested that miR-320a and miR-340-5p may be potential therapeutic targets for EC treatment.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- miRNA
microRNA
- EC
endometrial cancer
- eIF4E
eukaryotic translation initiation factor 4E
- 3′-UTR
3′-untranslated region
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5diphenyltetrazolium bromide
- TGF-β1
transforming growth factor β1
- EMT
epithelial- mesenchymal transition
Funding
This study was supported by National Natural Scientific Grants (grant no. 31570798 and 31971209), Liaoning Excellent Talents in University (grant no. LR2017042), Liaoning Provincial Program for Top Discipline of Basic Medical Sciences and the Shandong Science and Technology Committee (grant no. 2018GSF118056).
Availability of data and materials
The datasets used or analyzed in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
HHZ and YK conceived and designed the experiments. HHZ, RL, YJL, XXY, QNS, and AYL performed the experiments. HHZ and YK analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All experiments with human specimens were performed in accordance with the relevant guidelines and were approved by the Medical Ethics Committee of Binzhou Medical University (Yantai, China). Prior to study inclusion, written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Van Nyen T, Moiola CP, Colas E, Annibali D, Amant F. Modeling endometrial cancer: Past, present, and future. Int J Mol Sci. 2018;19:E2348. doi: 10.3390/ijms19082348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387:1094–1108. doi: 10.1016/S0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 5.Pettersson F, Yau C, Dobocan MC, Culjkovic-Kraljacic B, Retrouvey H, Puckett R, Flores LM, Krop IE, Rousseau C, Cocolakis E, et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin Cancer Res. 2011;17:2874–2884. doi: 10.1158/1078-0432.CCR-10-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkel HJ, Turbat-Herrera EA, Shi R, de Benedetti A. Expression of the translation initiation factor eIF4E in the polyp-cancer sequence in the colon. Cancer Epidemiol Biomarkers Prev. 2001;10:663–666. [PubMed] [Google Scholar]
- 7.Choi CH, Lee JS, Kim SR, Lee YY, Kim CJ, Lee JW, Kim TJ, Lee JH, Kim BG, Bae DS. Direct inhibition of eIF4E reduced cell growth in endometrial adenocarcinoma. J Cancer Res Clin Oncol. 2011;137:463–469. doi: 10.1007/s00432-010-0902-z. [DOI] [PubMed] [Google Scholar]
- 8.Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2014;33:1755–1763. doi: 10.1038/onc.2013.128. [DOI] [PubMed] [Google Scholar]
- 9.Smith KA, Zhou B, Avdulov S, Benyumov A, Peterson M, Liu Y, Okon A, Hergert P, Braziunas J, Wagner CR, et al. Transforming growth factor-β1 induced epithelial mesenchymal transition is blocked by a chemical antagonist of translation factor eIF4E. Sci Rep. 2015;5:18233. doi: 10.1038/srep18233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Xie Y, Cui D, Ma Y, Sui L, Zhu C, Kong H, Kong Y. Osteopontin promotes invasion, migration and epithelial-mesenchymal transition of human endometrial carcinoma cell HEC-1A through AKT and ERK1/2 signaling. Cell Physiol Biochem. 2015;37:1503–1512. doi: 10.1159/000438518. [DOI] [PubMed] [Google Scholar]
- 12.Aparicio LA, Abella V, Valladares M, Figueroa A. Posttranscriptional regulation by RNA-binding proteins during epithelial-to-mesenchymal transition. Cell Mol Life Sci. 2013;70:4463–4477. doi: 10.1007/s00018-013-1379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.He M, Xue Y. MicroRNA-148a suppresses proliferation and invasion potential of non-small cell lung carcinomas via regulation of STAT3. Onco Targets Ther. 2017;10:1353–1361. doi: 10.2147/OTT.S123518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang HH, Pang M, Dong W, Xin JX, Li YJ, Zhang ZC, Yu L, Wang PY, Li BS, Xie SY. miR-511 induces the apoptosis of radioresistant lung adenocarcinoma cells by triggering BAX. Oncol Rep. 2014;31:1473–1479. doi: 10.3892/or.2014.2973. [DOI] [PubMed] [Google Scholar]
- 16.Zhao W, Sun Q, Yu Z, Mao S, Jin Y, Li J, Jiang Z, Zhang Y, Chen M, Chen P, et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene. 2018;670:31–37. doi: 10.1016/j.gene.2018.05.100. [DOI] [PubMed] [Google Scholar]
- 17.Ge X, Cui H, Zhou Y, Yin D, Feng Y, Xin Q, Xu X, Liu W, Liu S, Zhang Q. miR-320a modulates cell growth and chemosensitivity via regulating ADAM10 in gastric cancer. Mol Med Rep. 2017;16:9664–9670. doi: 10.3892/mmr.2017.7819. [DOI] [PubMed] [Google Scholar]
- 18.Wu X, Tang H, Liu G, Wang H, Shu J, Sun F. miR-448 suppressed gastric cancer proliferation and invasion by regulating ADAM10. Tumour Biol. 2016;37:10545–10551. doi: 10.1007/s13277-016-4942-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhou SJ, Liu FY, Zhang AH, Liang HF, Wang Y, Ma R, Jiang YH, Sun NF. MicroRNA-199b-5p attenuates TGF-β1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Br J Cancer. 2017;117:233–244. doi: 10.1038/bjc.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y, Luo W, Yang ZJ, Chi JR, Li YR, Ding Y, Ge J, Wang X, Cao XC. miR-190 suppresses breast cancer metastasis by regulation of TGF-β-induced epithelial-mesenchymal transition. Mol Cancer. 2018;17:70. doi: 10.1186/s12943-018-0818-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sepramaniam S, Armugam A, Lim KY, Karolina DS, Swaminathan P, Tan JR, Jeyaseelan K. MicroRNA 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia. J Biol Chem. 2010;285:29223–29230. doi: 10.1074/jbc.M110.144576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P, Zhang Q, Dong L, Liu Y, Dong J. microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol Rep. 2012;27:685–694. doi: 10.3892/or.2011.1561. [DOI] [PubMed] [Google Scholar]
- 26.Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ, et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem Biophys Res Commun. 2012;420:787–792. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 27.Xiao H, Yu L, Li F, Wang H, Li W, He X. MiR-340 suppresses the metastasis by targeting EphA3 in cervical cancer. Cell Biol Int. 2018;42:1115–1123. doi: 10.1002/cbin.10974. [DOI] [PubMed] [Google Scholar]
- 28.Shi S, Chen X, Liu H, Yu K, Bao Y, Chai J, Gao H, Zou L. LGR5 acts as a target of miR-340-5p in the suppression of cell progression and drug resistance in breast cancer via Wnt/β-catenin pathway. Gene. 2019;683:47–53. doi: 10.1016/j.gene.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–634. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 30.Shi ZM, Liu YN, Fu B, Shen YF, Li LM. Expression profile of eukaryotic translation initiation factor and matrix metalloproteinase 9 in endometrial cancer tissue. J Biol Regul Homeost Agents. 2017;31:1053–1059. [PubMed] [Google Scholar]
- 31.Mannelqvist M, Stefansson IM, Bredholt G, Hellem Bø T, Oyan AM, Jonassen I, Kalland KH, Salvesen HB, Akslen LA. Gene expression patterns related to vascular invasion and aggressive features in endometrial cancer. Am J Pathol. 2011;178:861–871. doi: 10.1016/j.ajpath.2010.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konicek BW, Dumstorf CA, Graff JR. Targeting the eIF4F translation initiation complex for cancer therapy. Cell Cycle. 2008;7:2466–2471. doi: 10.4161/cc.7.16.6464. [DOI] [PubMed] [Google Scholar]
- 33.Karahan N, Guney M, Baspinar S, Oral B, Kapucuoglu N, Mungan T. Expression of gelatinase (MMP-2 and MMP-9) and cyclooxygenase-2 (COX-2) in endometrial carcinoma. Eur J Gynaecol Oncol. 2007;28:184–188. [PubMed] [Google Scholar]
- 34.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015;34:2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vega S, Morales AV, Ocaña OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dragomirescu M, Stepan AE, Margaritescu C, Simionescu CE. The immunoexpression of p53 and snail in endometrioid endometrial carcinomas. Rom J Morphol Embryol. 2018;59:131–137. [PubMed] [Google Scholar]
- 38.Xiong S, Klausen C, Cheng JC, Leung PC. Activin B promotes endometrial cancer cell migration by down-regulating E-cadherin via SMAD-independent MEK-ERK1/2-SNAIL signaling. Oncotarget. 2016;7:40060–40072. doi: 10.18632/oncotarget.11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used or analyzed in the present study are available from the corresponding author upon reasonable request.