Abstract
The aim of the present study was to investigate whether miR-203 can inhibit transforming growth factor-β (TGF-β)-induced epithelial-mesenchymal transition (EMT), and the migration and invasion ability of non-small cell lung cancer (NSCLC) cells by targeting SMAD3. In the present study, the expression levels of miR-203, SMAD3 mRNA and protein in NSCLC tissues were examined, as well as their corresponding paracancerous samples. The miR-203 mimics and miR-203 inhibitor were transfected into the H226 cell line. RT-qPCR was used to assess the expression levels of E-cadherin, Snail, N-cadherin and vimentin mRNA, and western blotting was performed to detect the expression levels of p-SMAD2, SMAD2, p-SMAD3, SMAD3 and SMAD4. The cell migration and invasion abilities were detected by Transwell assays. The target site of SMAD3 was predicted by the combined action between miR-203 and dual luciferase. The results revealed that the RNA levels of miR-203, compared with paracancerous tissues, were decreased in NSCLC tissues, while SMAD3 mRNA and protein levels were upregulated, and miR-203 inhibited SMAD3 expression. Induction of TGF-β led to decreased E-cadherin mRNA levels, upregulation of Snail, N-cadherin and vimentin mRNA levels (P<0.05), and significant increase in cell migration and invasion, whereas transfection of miR-203 mimics reversed the aforementioned results (P<0.05). Conversely, miR-203 inhibitor could further aggravate the aforementioned results (P<0.05). Western blot results revealed that transfection of miR-203 mimics significantly reduced the protein expression of SMAD3 and p-SMAD3 (P<0.05). Furthermore, the results of the Dual-Luciferase assay revealed that miR-203 inhibited SMAD3 expression by interacting with specific regions of its 3′-UTR. Overall, a novel mechanism is revealed, in which, miR-203 can inhibit SMAD3 by interacting with specific regions of the 3′-UTR of SMAD3, thereby restraining TGF-β-induced EMT progression and migration and invasion of NSCLC cells.
Keywords: miR-203, epithelial-mesenchymal transition, non-small cell lung cancer, SMAD3
Introduction
Malignant tumors are one of the most important causes of death worldwide. In most countries, including China, lung cancer has the highest morbidity and mortality and is still increasing yearly. Smoking, diet, air pollution and genetic factors are all related to the occurrence of lung cancer (1–3). According to histopathology, lung cancer can be divided into non-small cell lung cancer (NSCLC) and small-cell lung cancer (SCLC), of which NSCLC accounts for ~85% of all lung cancer (4). Despite the continuous improvement of medical advancements, the diagnosis and treatment of NSCLC has not progressed significantly over the past few decades, and the overall 5-year survival rate remains at ~15% (5,6). Since there are no specific clinical symptoms in the early stage of lung cancer, most lung cancer patients are already in the advanced stage at the time of diagnosis, which means that tumor cells have already metastasized or have the characteristics of invasion and metastasis (7). Therefore, identifying key factors and elucidating their molecular mechanisms in invasion and metastasis of NSCLC are particularly important for reducing patient mortality and improving the quality of life of patients.
The invasion and metastasis of malignant tumors, is an extremely complicated process coordinated by multiple steps and factors. Studies have revealed that epithelial-mesenchymal transition (EMT) plays an important role in lung cancer invasion and metastasis, occurring in the early stage of lung cancer invasion and metastasis (8). EMT refers to the biological process by which polar epithelial cells are transformed into mesenchymal cells under certain specific physiological or pathological conditions. EMT is one of the most important mechanisms for malignant tumor cells to acquire the ability of migration and invasion, and plays a vital part in the occurrence and development of cancer (9). In the process of EMT, the epithelial cells are less exposed to surrounding cells and matrix, and cell adhesion is weakened. The epithelial cells obtain the ability of higher migration, invasion, anti-apoptosis and degradation of the extracellular matrix, exhibiting strong mesenchymal characteristics and transforming into mesenchymal cells. In addition, the related gene expression profile of the cells changes (10,11). The morphology of the cells changes significantly after EMT. For example, the cells become narrow and long, and form pseudopods. The intercellular gaps significantly increase, and the cells become more mobile. EMT is the result of the coordination of many factors, involving the effects of transcription factors, microRNA regulation, the TGF-signaling pathway as well as other chemical signaling pathways.
Transforming growth factor-β (TGF-β) is a multifunctional cytokine that triggers diverse cellular processes including tissue fibrosis, growth arrest and EMT (12). Previous studies have revealed that TGF-β can promote EMT progression by activating kinase-dependent signaling processes and is currently the classical inducing factor for the induction of EMT in lung cancer cells (10,13). The EMT process induced by TGF-β can be expressed as changes in the expression of some molecular markers, such as a loss of the epithelial phenotype indicated by downregulation of E-cadherin and the gain of the mesenchymal phenotype indicated by upregulation of N-cadherin and vimentin (14). TGF-β inhibits epithelial cell proliferation in the early stage of cancer, but promotes tumor growth and metastasis in the later stage. Studies have revealed that patients with higher TGF-β levels have a poor prognosis (15). Therefore, it was hypothesized that inhibition of TGF-β or its receptors can suppress the activation of the EMT pathway, thereby impairing the ability of tumor cell invasion and metastasis. TGF-β mainly exerts its biological function through the SMAD protein family. Specifically, binding of TGF-β to its receptor leads to phosphorylation of SMAD2/3, which binds to SMAD4 and enters the nucleus, where the SMAD transcription complex regulates the expression of specific target genes (16,17). Previous studies have reported that SMAD3 plays a key role in the occurrence and development of various cancers (18,19), and that SMAD3 can promote the invasion and metastasis of tumor cells through its mediated EMT (20,21). Loss or lack of SMAD3 will impede the EMT process and may alleviate epithelial deterioration (18,19). In addition, studies have revealed that silencing of SMAD3 can inhibit the migration and invasion of nasopharyngeal carcinoma cells (22), and the downregulated expression of N-cadherin can also be detected (14); miR-140 inhibited the migration and invasion of colorectal cancer cells by targeting SMAD3 (23). These data indicated that SMAD3 plays an essential role in TGF-β-induced EMT progression, however, in this process, the regulatory mechanisms of miRNA are still unclear.
miRNAs are a class of endogenous non-coding RNA with a wide distribution and a length of 19–25 nucleotides. They mainly form partial complementary sequences by binding to the 3′-UTR, 5′-UTR or coding region of a specific target gene, thereby affecting the transcriptional stability and post-transcriptional translation process of the target gene (24). The expression of ~30–50% of protein-coding genes in humans may be regulated by targeted miRNAs (25). Therefore, miRNAs play an important role in the fine regulation of a variety of biological processes, including cell proliferation, differentiation, apoptosis, migration and tumor formation (26,27). In addition, miRNAs must also regulate TGF-signaling by targeting the expression of key members in the TGF- pathway (28), known as TGF-β pathway-associated miRNAs. Several studies have revealed that miR-203 can inhibit the cell invasion of NSCLC and nasopharyngeal carcinoma, and is frequently downregulated in NSCLC (26,29,30). Ding et al revealed that miR-203 plays an important role in TGF-β-induced EMT progression and is downregulated in highly metastatic breast cancer cells (9). These studies indicated that miR-203 may regulate the process of EMT in NSCLC by regulating the TGF-β signaling pathway, and the mechanism of miR-203 in this process remains to be further elucidated.
In the present study, miR-203 was transfected into NSCLC cells to verify the hypothesis that SMAD3 is a target gene for miR-203, and miR-203 regulates the hypothesis that SMAD3 inhibits TGF-β-induced EMT and tumor invasion and metastasis. The present results clarified that miR-203 in NSCLC cell line can suppress the expression of SMAD3, affect the TGF-β-induced EMT process, inhibit the invasion and metastasis of tumor cells, and provide a new experimental basis for the diagnosis and treatment of NSCLC.
Materials and methods
Human tissue samples
Fresh NSCLC tissue samples from 10 patients (32–61 years old) and their corresponding paracancerous samples were collected in the study (n=10). The patients were diagnosed with NSCLC based on pathology and did not receive any chemotherapy and/or radiotherapy before surgery. There were 6 males and 4 females with an average age of 48.70±11.25 years. All of the specimens were examined and evaluated by two independent pathologists. Clinicopathological data were collected from the patient medical records and are presented in Table I. All patients provided their written informed consent and ethics approval was obtained from the Ethics Committees of the First Affiliated Hospital of Wenzhou Medical University (2017063).
Table I.
Clinicopathological characteristics of the NSCLC patients.
| Characteristics | Total (10, %) |
|---|---|
| Sex | |
| Male | 6 (60.0) |
| Female | 4 (40.0) |
| Age (years) | |
| <40 | 4 (40.0) |
| ≥40 | 6 (60.0) |
| Histologic type | |
| ADC | 6 (60.0) |
| SQCC | 4 (40.0) |
| Lymph node metastasis | |
| Yes | 3 (30.0) |
| No | 7 (70.0) |
| TNM stage | |
| I–II | 7 (70.0) |
| III–IV | 3 (30.0) |
NSCLC, non-small cell lung cancer; ADC, adenocarcinoma; SQCC, squamous cell carcinoma.
Cell lines and cell cultures
Human NSCLC cell line H226 cells (Institute of Cell Sciences, Chinese Academy of Sciences) were cultured in modified RPMI-1640 medium (Hyclone, GE Healthcare Life Sciences), supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and a mixture of antibiotics (penicillin, Sigma-Aldrich; streptomycin, Invitrogen), and the cells were incubated in a humidified 5% CO2 incubator at 37°C.
Real time-qPCR (RT-qPCR)
RNA, according to the manufacturer's protocol, was extracted from the cells using the RNAiso Plus kit (Thermo Fisher Scientific, Inc.). Synthesis of cDNA with reverse transcriptase was performed using the M-MLV First Strand kit (Invitrogen; Thermo Fisher Scientific, Inc.). The primer sequences used for RT-qPCR were as follows: miR-203-F, ACACTCCAGCTGGGAGTGGTTCTTAACAGTTC and miR-203-R, TGGTGTCGTGGAGTCG; SMAD3-F, TGGACGCAGGTTCTCCAAAC and SMAD3-R, CCGGCTCGCAGTAGGTAAC; SMAD2-F, ATCTTGCCATTCACTCCGCC and SMAD2-R, CTGTTCTCCACCACCTGCTC; E-cadherin-F, ATTTTTCCCTCGACACCCGAT and E-cadherin-R, TCCCAGGCGTAGACCAAGA; N-cadherin-F, TGCGGTACAGTGTAACTGGG and N-cadherin-R, GAAACCGGGCTATCTGCTCG; vimentin-F, AGTCCACTGAGTACCGGAGAC and vimentin-R, CATTTCACGCATCTGGCGTTC; Snail-F, ACTGCAACAAGGAATACCTCAG and Snail-R, GCACTGGTACTTCTTGACATCTG; U6-F, CTCGCTTCGGCAGCACA and U6-R, AACGCTTCACGAATTTGCGT; GAPDH-F, CTGGGCTACACTGAGCACC and GAPDH-R, AAGTGGTCGTTGAGGGCAATG. The cycling parameters (31) were as follows: One cycle at 94°C for 5 min, 35 cycles of a denaturing step at 94°C for 30 sec, an annealing step at 55°C for 30 sec, an extension step at 72°C for 1 min and, lastly, one cycle of an additional extension at 72°C for 10 min. PCR products were analyzed by 3% (w/v) agarose gel electrophoresis. All reactions were carried out in triplicate using SYBR-Green on the ABI StepOnePlus Real-time PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.), using standard cycling parameters. Standard SYBR-Green PCR conditions were used, with an annealing temperature at 59°C and 40 cycles. The 2−ΔΔCq method (32) was used to calculate the relative expression of miR-203, SMAD3, SMAD2, E-cadherin, N-cadherin, vimentin and Snail mRNA. As an internal control, mRNAs of U6 and GAPDH were measured under the same reaction conditions. All samples were tested in triplicate.
Immunohistochemistry
The paraffin-embedded tissue pieces were cut into 4-µm-thick sections, mounted on the slides and heated. The sections were dewaxed in xylene, and rehydrated in a gradient of ethanol through a series of alcohol gradient solutions. Digestion with 3 mol/l urea for 30 min was performed to expose target antigens in the tissue. After antigen addition to the citrate solution, the sections were treated with 3% H2O2 solution for 10 min, light was avoided, and then the sections were treated with 5% bovine serum albumin (BSA) for 30 min. The sections were then incubated with the monoclonal rabbit anti-human Smad3 antibody (dilution 1:100; product code ab40854; Abcam), placed in a refrigerator at 4°C overnight, and reacted for 45 min in a 37°C water temperature chamber the next day. Visualization of antibody binding was performed using DAB staining. The nuclei were stained with hematoxylin for 90 sec, then soaked in hydrochloric acid alcohol differentiation solution for 7 sec, and the gelatin was sealed after dehydration. The results of immunostaining were independently assessed by two pathologists.
The IHC score was based on staining intensity and percentage of positive cells, as follows: The intensity score was 0 (no staining), 1 (weak staining), 2 (moderate staining) and 3 (strong staining); the proportion score was 0 (<5% positive cells), 1 (6–25% positive cells), 2 (26–50% positive cells), 3 (51–75% positive cells) and 4 (>75% positive cells). The final staining fraction was obtained by multiplying strength and proportion fraction: 0 (negative), + (1–4), ++ (5–8) and +++ (9–12). For statistical analysis, negative or positive final staining scores were combined into the low-expression group, while ++ or +++ final staining scores were combined into the high-expression group.
All the tissue points on the tissue chip were positively scored with the same criteria, specifically, the product of the staining intensity of the target cells and the percentage of positive cells. Positive cells were distinguished from background or non-specific staining. The staining intensity was scored according to the staining characteristics of the target cells: 0 for non-staining, 1 for light yellow, 2 for brownish yellow and 3 for brown.
Western blot analysis
Protein markers used in western blot analysis were as follows: p-SMAD2 (cat. no. ARG55037), SMAD2 (cat. no. ARG54942), p-SMAD3 (cat. no. ARG51797), SMAD3 (cat. no. ARG53570), SMAD4 (cat. no. ARG54741, all from arigo Biolaboratories) and GAPDH (product code ab8245; Abcam). After H226 cells were treated with ice-cold RIPA buffer (Beyotime Institute of Biotechnology, Inc.), the supernatant was collected by centrifugation at 14000 × g. The concentration of protein was determined by the BCA method. The total protein concentration of the extracted sample was adjusted to 2 µg/ml. Total protein (20 µg), was transferred to the polyvinylidene fluoride (PVDF) membrane by 10% SDS-PAGE gel electrophoresis, and was blocked with 5% non-fat milk for 1 h at room temperature, and then incubated overnight at 4°C after treatment with a primary antibody (SMAD2, 1:1,000; SMAD3, 1:1,000; SMAD4, 1:1,000; p-SMAD2, 1:1,000; p-SMAD3, 1:1,000; GAPDH, 1:10,000). After washing away the excess primary antibody, TBST was used to dilute the corresponding HRP-labeled secondary antibody, so that the PVDF membrane could be immersed in the secondary antibody incubation solution (goat anti-mouse IgG-HRP, cat. no. sc-2058, dilution 1:3,000; goat anti-rabbit IgG-HRP, cat. no. sc-2301, dilution 1:3,000; Santa Cruz Biotechnology, Inc.), and incubation followed for 2 h at 37°C on a shaking table. Next, the PVDF membrane was washed 5–6 times with TBST, and the color was developed by ECL chemiluminescence (Beyotime Institute of Biotechnology). The gel images were obtained with an Alpha Gel imager (Alpha Innotech Co.), and absorbance analysis was performed using Quantity One v4.62 software (Bio-Rad Laboratories). The expression level of the target protein was expressed by the absorbance ratio between its absorbance and the corresponding GAPDH. The experiment was repeated 3 times.
Cell transfection
H226 cells were seeded at 2×105 cells/well in 6-well plates and transfected with miR-203 mimics, miR-203 inhibitors, and scrambled controls (miR-NC) (Shanghai GenePharma CO., Ltd.) using Invitrogen™ Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.). The sequences of these miRNAs were: miR-203 mimics, 5′-TGCTTTGGCCACTGACTGTCC-3′; miR-203 inhibitors, 5′-ACGAAACCGGTGACTGACAGG-3′; miR-NC, 5′-TCGCCACATGATCGCCTAAGT-3′. The expression levels of all transfected genes were confirmed with RT-PCR. Cells were transfected with appropriate miRNA, siRNA oligonucleotides and plasmids using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. The medium was replenished 6 h after transfection.
Transwell assays
For the invasion assay, the upper Transwell chamber (Corning, Inc.) was coated with 50 ml of 20 mg/ml Matrigel for filtering. The invasive chamber (Corning, Inc.) with an 8-µm pore polycarbonate membrane was pretreated for 2 h in serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.) medium at 37°C, 5% CO2. After the removal of the medium, 0.25% trypsin-digested monolayer cells were added in an amount of 1 ml/25 cm2 of surface area. The cells were resuspended and 100–200 µl were used for counting after pouring out the excess trypsin. The passaged cells were digested with 0.25% trypsin, washed with PBS, and added to the culture solution to prepare a suspension of the cells to be tested, and cell density was counted and calculated. H226 cells were plated into 6-well plates, and transfected with miR-NC, miR-203 mimics, and miR-203 inhibitors 12 h later. After 12 h of transfection, the cells in the 6-well plate were re-plated into the Transwell chamber with 1.5×104 cells/well. After transfection for 18 h, the cells in the upper chamber were replaced after the cells were attached, and the cells were treated with TGF (10 ng/ml). Cell status was observed after treatment with TGF for 48 h, cell samples were collected, and cells on the lower surface of the chamber were fixed with 4% paraformaldehyde for 30 min. After washing 3 times with PBS, the samples were stained with crystal violet (0.5 mM) dye for 30 min, and observed and photographed under the microscope. For the migration assay, a procedure similar to invasion assay was performed using the migration chamber without Matrigel.
Dual luciferase assay
When the cell density in the cell culture flask reached 70–80%, the H226 cells were digested with trypsin. Cells were collected by centrifugation at 40 × g, and then the cells were placed in the 24-well plate to make the density reach ~60% after resuspended. After mixture, the cells continued to be cultured in an incubator at 37°C. Plasmid transfection was performed using the TurboFect™ transfection kit (Fermentas, Inc.), and fresh medium was replaced 12 h after transfection. After 72 h of transfection, the cells were washed twice with PBS, and 250 µl of 1X PLB lysis buffer (Thermo Fisher Scientific, Inc.) was then added to each well. LAR II reagent (100 µl) and lysate (20 µl) were added into a 96-well plate, and then 100 µl of Stop & Glo substrate (Promega Corp.) was added within 10 sec to measure the luciferase activity. After exporting the data, the results were analyzed and plotted using GraphPad Prism 5 software (GraphPad Software, Inc.).
Statistical analysis
Statistical analysis was conducted by SPSS 17.0 (SPSS, Inc.) statistical software. Measured data were compared using t-tests. The count data were analyzed by Chi-square test. The rank-sum test was employed to compare the clinical grade data sets. The course of disease and score were statistically expressed as the mean ± standard deviation. Student's t-test analysis was used for comparison between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-203 and SMAD3 in human NSCLC tissues
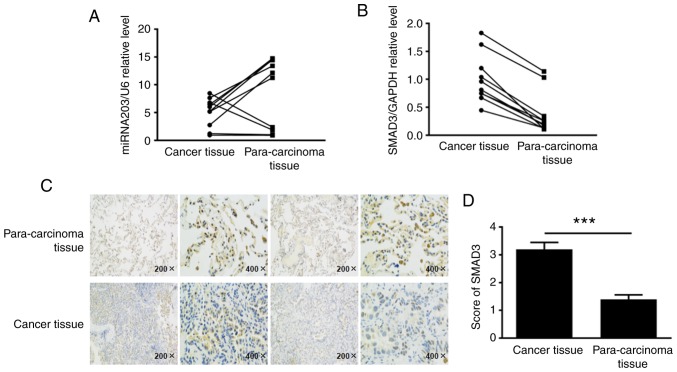
To elucidate the expression levels of miR-203 and SMAD3 in non-small cell lung cancer tissues, ~10 fresh NSCLC tissue samples and their corresponding paracancerous samples were collected, total RNA was extracted, and the expression levels of miR-203 and SMAD3 mRNA were detected by RT-PCR. The results revealed that miR-203 RNA levels were decreased in human NSCLC tissues compared with paracancerous tissues, while SMAD3 mRNA levels were upregulated (Fig. 1A and B). Tissue paraffin-embedded sections were used to detect the expression of SMAD3 protein by immunohistochemistry, according to the cell positive ratio, the score was 0 for ~0–5%, 1 for ~6–25%, 2 for ~26–50%, 3 for ~51–75%, and 4 for >75% (Fig. 1C and D). Collectively, these findings indicated that the SMAD3 protein was highly expressed in cancer tissues (P<0.001).
Figure 1.
In the NSCLC tissue samples and their corresponding paracancerous samples, the mRNA expression levels of (A) miR-203 and (B) SMAD3 were detected by RT-qPCR. (C and D) Immunohistochemistry was used to detect the relative expression of SMAD3 protein. ***P<0.001, compared with the para-carcinoma tissue. NSCLC, non-small cell lung cancer.
miR-203 affects the expression of SMAD3 in NSCLC cell lines
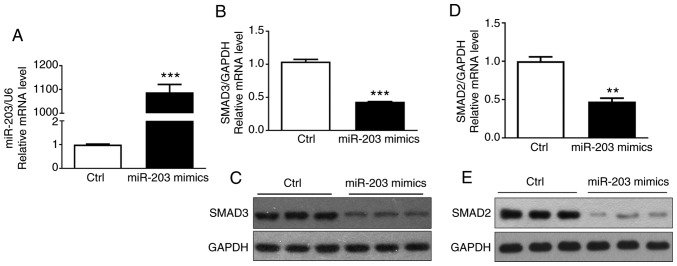
To determine that miR-203 affected the expression of SMAD3 in NSCLC cells, miR-203 mimics were transfected into H226 cells, and the transfection efficiency of miR-203 and the expression of SMAD3 and SMAD2 mRNA were then detected by RT-PCR. The results indicated that miR-203 mRNA expression was significantly increased (P<0.05) (Fig. 2A), while the mRNA expression of SMAD3 and SMAD2 was significantly decreased (P<0.05) (Fig. 2B and D). In addition, the western blotting assay confirmed that the protein expression of SMAD3 and SMAD2 was also significantly reduced (Fig. 2C and E), consistent with the aforementioned results. These results demonstrated that miR-203 inhibited the expression of SMAD3 and SMAD2.
Figure 2.
Effect of miR-203 on the expression of SMAD3 is detected by RT-qPCR and western blotting. (A) Relative expression of miRNA203 mRNA. (B) Relative expression of SMAD3 mRNA. (C) Western blot results of SMAD3 protein. (D) Relative expression of SMAD2 mRNA. (E) Western blot results of SMAD2 protein. **P<0.01, ***P<0.001, compared with the control group.
Effect of miR-203 on the TGF-β-induced EMT process
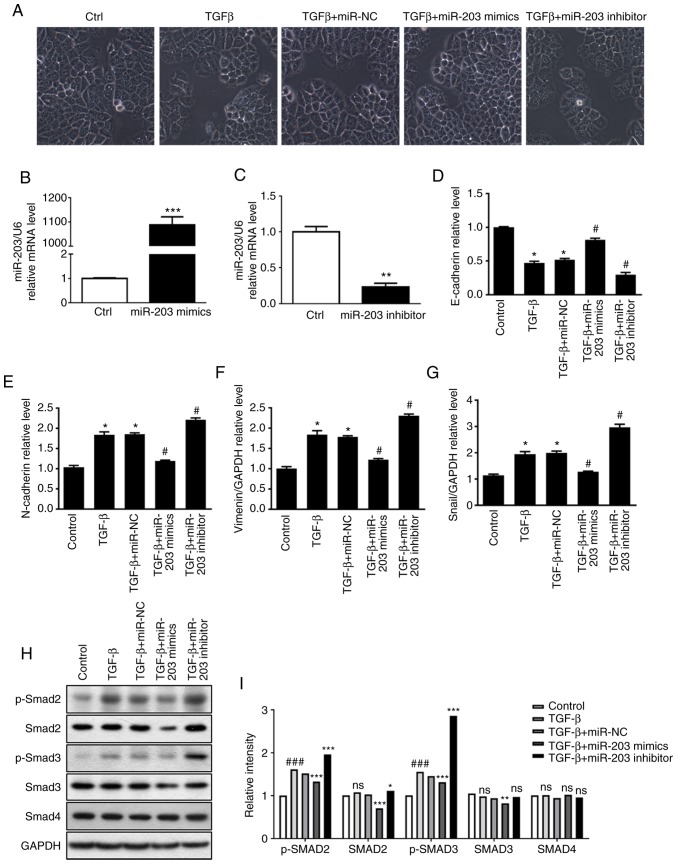
In order to clarify the role of miR-203 in TGF-β-induced EMT, miR-203 mimics and miR-203 inhibitor were synthesized according to miR-203 sequence, and then they were transiently transfected into the H226 cell line, respectively. In addition, the random mimics fragment miR-NC was also synthesized as a control. The RT-PCR results revealed that the miR-203 mRNA expression was significantly increased after the miR-203 mimics were transfected (P<0.001) (Fig. 3B), while it was significantly decreased after the miR-203 inhibitor was transfected (P<0.01) (Fig. 3C). The experimental groups were divided into a control group, TGF-β group, TGF-β+miR-NC group, TGF-β+miR-203 mimics group, and TGF-β+miR-203 inhibitor group. The changes in cell morphology of H226 cells induced by TGF-β were first identified, and it was revealed that the intercellular gap was significantly increased. When cells were transfected with miR-203 mimics, the intercellular gap was significantly reduced. After the inhibition of miR-203, the intercellular space was significantly larger compared with the TGF-β+miR-NC group (Fig. 3A). RT-PCR was used to detect the mRNA expression of several related factors during EMT. The results revealed that the mRNA expression of the epithelial marker E-cadherin was decreased after TGF-β induction, while the expression of mesenchymal markers Snail, N-cadherin and vimentin was upregulated, indicating significant differences (P<0.05). However, transfection of miR-203 mimics significantly reversed the aforementioned effects. Compared with the TGF-β+miR-NC group, the mRNA levels of E-cadherin in the TGF-β+miR-203 mimics group was significantly increased (P<0.05), whereas the mRNA levels of Snail N-cadherin and vimentin were significantly decreased (P<0.05). Conversely, after the transfection of miR-203 inhibitor, the mRNA level of E-cadherin in the TGF-β+miR-203 inhibitor group was significantly decreased (P<0.05), while the mRNA levels of Snail, N-cadherin and vimentin were significantly increased (P<0.05) (Fig. 3D-G). Western blotting was used to detect the expression of p-SMAD2, SMAD2, p-SMAD3, SMAD3 and SMAD4 in the TGF-β pathway under different TGF-β stimulation conditions. These results indicated that in the presence of TGF-β, the protein expression of SMAD3 and p-SMAD3 were significantly reduced after transfection with miR-203 mimics compared with the TGF-β-miR-NC group (P<0.05). Notably, after transfection with miR-203 inhibitor, there was no significant difference in SMAD3, while p-SMAD3 expression was significantly upregulated compared with the TGF-β-miR-NC group (P<0.05) (Fig. 3H and I). Collectively, these findings indicated that miR-203 may inhibit TGF-β-induced EMT progression by blocking SMAD3 phosphorylation.
Figure 3.
(A) The effect of miR-203 expression on H226 cell morphology and EMT phenomenon. RT-qPCR was used to detect (B) the transfection efficiency of miR-203 mimics, (C) the transfection efficiency of miR-203 inhibitor, the expression of (D) E-cadherin, (E) N-cadherin, (F) vimentin, (G) Snail mRNA during EMT. (H and I) Protein expression of p-SMAD2, SMAD2, p-SMAD3, SMAD3 and SMAD4. *P<0.5, **P<0.01, ***P<0.001, compared with the control group. #P<0.5, ###P<0.01, compared with the TGF-β+miR-NC group. EMT, epithelia-mesechymal transition; TGF-β, transforming growth factor.
Effect of miR-203 on the migration and invasion of NSCLC cells
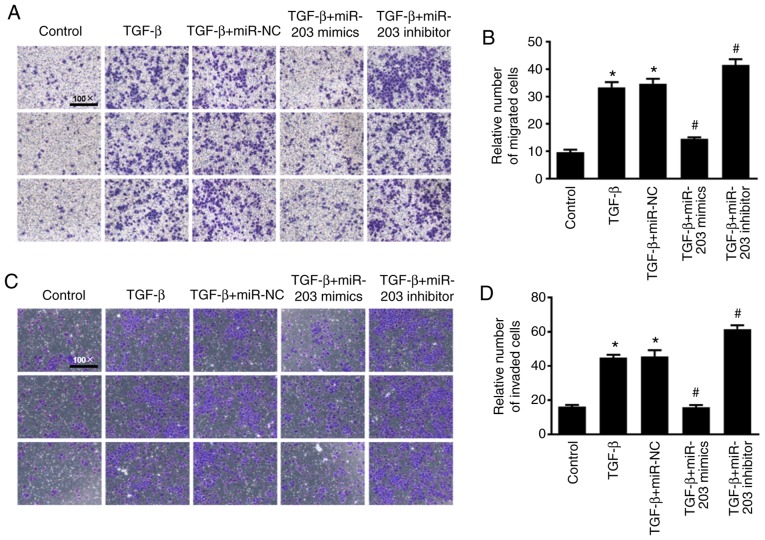
To investigate the effect of miR-203 on the migration and invasion of NSCLC cells, Transwell cell migration and invasion assays were performed and the results were quantified using ImageJ software to count the relative number of cells. As revealed in Fig. 4, under the induction of TGF-β, the migration and invasion abilities of the cells were significantly enhanced, while the migration and invasion abilities of H226 cells were significantly decreased after overexpression of miR-203 (P<0.05). In contrast, the migration and invasion abilities in the miR-203 inhibitor group were significantly increased (P<0.05).
Figure 4.
(A) Transwell cell migration assay. (B) ImageJ quantified the relative number of migrating cells. (C) Transwell cell invasion assay. (D) ImageJ quantified the relative number of invading cells. *P<0.5, compared with the control group, #P<0.5, compared with the TGF-β+miR-NC group. TGF-β, transforming growth factor.
miR-203 inhibits SMAD3 expression by targeting specific sites of SMAD 3′-UTR
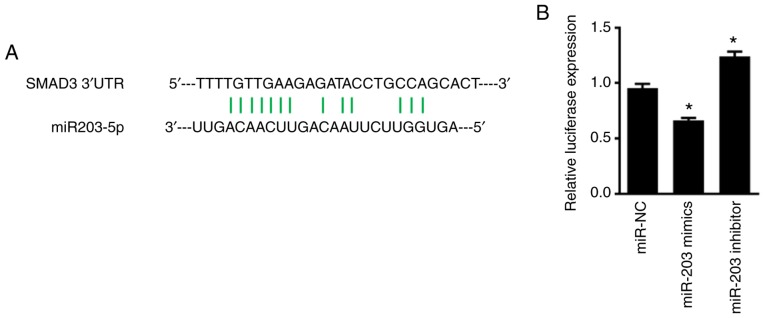
To determine whether miR-203 directly binds to SMAD, the luciferase plasmid of SMAD 3′-UTR was synthesized, and co-transfected NSCLC cells with miR-203. As revealed in Fig. 5, miR-203 significantly suppressed the fluorescence activity of the luciferase plasmid of SMAD 3′-UTR in H226 cells (P<0.05). When the expression of miR-203 was inhibited, however, the fluorescence activity of the luciferase plasmid of SMAD 3′-UTR was significantly increased (P<0.05). Overall, these findings indicated that the inhibitory effect of miR-203 on SMAD3 expression was exerted by interaction with a specific region of its 3′-UTR.
Figure 5.
(A) The binding site of miR-203 to SMAD. (B) Dual luciferase assay for the fluorescence activity of the luciferase plasmid of SMAD 3′-UTR. *P<0.05, compared with the miR-NC group.
Discussion
In the present study, the regulatory mechanism of miR-203 on SMAD3 in TGF-β-induced EMT in NSCLC and cell migration and invasion abilities were investigated. The results revealed that miR-203 inhibited SMAD3 by interacting with specific regions of the 3′-UTR of SMAD3, thereby inhibiting TGF-β-induced EMT progression and migration and invasion of NSCLC cells.
EMT occurs early in the process of epithelial-derived tumor metastasis (9), which involves changes in cell phenotype, the cytoskeleton, and some protein expression, and ultimately leads to the ability of tumor cells to migrate and invade. Previous studies have revealed that EMT is not only associated with tumor migration and invasion, but is also related with resistance of NSCLC (33,34). For advanced lung cancer cases, ionizing radiation is one of the main treatment methods. However, EMT promotes radioactive fibrosis and related metastasis after radiotherapy (35–37), and plays an important role in tumor development and follow-up treatment. Downregulated expression of epithelial marker E-cadherin and upregulated expression of mesenchymal markers N-cadherin, vimentin and Snail are the main molecular biological markers of EMT (38,39). As an essential factor in the induction of EMT, TGF-β plays an important role in promoting the migration and invasion of NSCLC cells. Many studies have revealed that TGF-β is highly expressed in many types of tumors (40,41), and its signaling pathway can induce the progression of EMT (21,42–44). As a significant mediator of the TGF-β pathway in tumor cells, SMAD3 plays different roles in the development and progression of cancer by regulating different transcriptional reactions, depending on the type of tumor cells and the clinical stage of cancer (15). Studies have suggested that SMAD3 plays a key role in promoting EMT (45,46), and downregulation of SMAD3 or downregulation of its activated form p-SMAD3 significantly inhibited the TGF-β-mediated EMT and slowed the progression of pulmonary fibrosis (47). Yang et al (14) also revealed that TGF-β/SMAD3 can directly transcribe and activate the expression of N-cadherin, thereby promoting the EMT process of NSCLC cells. In the present study, after TGF-β induced H226 cells, p-SMAD3 protein expression was significantly increased, the mRNA levels of E-cadherin were decreased, Snail, N-cadherin and vimentin mRNA expression was upregulated, and these changes were statistically significant. In addition, the migration and invasion abilities of the cells were significantly enhanced. The aforementioned results indicated that TGF-β promoted SMAD3 activation, thereby stimulating the occurrence of EMT and enhancing the migration and invasion abilities of tumor cells, which was consistent with previous studies.
More than 500 miRNAs have been identified through current research, and miRNAs can participate in the regulation of various biological processes, including proliferation, differentiation, and apoptosis (48,49). Evidence has demonstrated that miRNAs regulate cancer metastasis by targeting different key proteins (50). During regulation, the target gene is silenced or degraded mainly by binding to the 3-UTR region of the target gene mRNA (51,52). The miR-203 gene sequence is located on chromosome 14q32.33 and encodes ~12% of the miRNA known to humans, and has been revealed to express abnormalities in many types of tumors (53). Zhou et al revealed that miR-203 could directly target the LIN28B gene to enhance the biosynthesis of the tumor suppressor let-7 in lung cancer and exert its anticancer effect (54). Wang et al revealed that miR-203 inhibited the expression of SRC as well as the proliferation and migration of lung cancer cells and promoted apoptosis of lung cancer cells (30). The signaling pathway between TGF-β and the transcription factor SNAI2 was revealed to inhibit the expression of miR-203 and promote EMT and tumor metastasis (9). The aforementioned studies have revealed that the miR-203 regulatory mechanism and target genes are diverse, suggesting that it may play a role in multiple signaling pathways and target genes. Notably, further identification and clarification of the target genes of miR-203 are particularly important for understanding the entire regulatory network of miR-203 and then carrying out targeted intervention. The 3′-UTR region of SMAD3 has a complementary pairing region with miR-203, indicating that SMAD3 may be a target gene for miR-203. Therefore, the luciferase plasmid of SMAD 3′-UTR was synthesized and NSCLC cells were co-transfected with miR-203. The results revealed that miR-203 significantly inhibited its fluorescence activity, and the fluorescence activity of the luciferase plasmid was significantly increased after inhibition of the expression of miR-203. In addition, studies have revealed that miR-203 can inhibit the migration of lung cancer cells by directly targeting PKCα (26). Chen et al also demonstrated that miR-203 inhibits the migration and invasion of NSCLC cells by targeting Bmi1 (29). In the present study, it was revealed that miR-203 significantly inhibited TGF-β-induced EMT and protein expression of SMAD3 and p-SMAD3, and the migration and invasion abilities of NSCLC H226 cells were significantly attenuated (P<0.05). Conversely, after inhibition of miR-203, the EMT process was further aggravated, and the expression of p-SMAD3 was significantly upregulated, and the migration and invasion abilities of the H226 cell line was significantly enhanced (P<0.05). These results indicated that miR-203 targeted the 3′-UTR region of SMAD3 and blocked SMAD3 phosphorylation to inhibit TGF-β-induced EMT progression as well as migration and invasion of NSCLC cells.
In NSCLC tissues, it was revealed that miR-203 was significantly downregulated, while SMAD3 mRNA and protein expression were significantly upregulated, which confirmed that miR-203 played a tumor suppressor role in the development of NSCLC, clinically. Additionally, the possibility that other miRNAs may affect the expression of SMAD3 cannot be excluded. Yang et al revealed that miR-136 can target SMAD3, thereby inhibiting the migration and invasion of lung adenocarcinoma cells, accompanied by increased epithelial marker expression and decreased mesenchymal marker expression (55). In a recent study on pancreatic ductal adenocarcinoma, it was reported that miR-323-3p could also target the inhibition of SMAD3 expression, thereby inhibiting the invasion and metastasis of cancer cells (56). These findings demonstrated that SMAD3 plays an important role in tumor EMT and migration and invasion (19). In addition to exploring the effects of miR-203 and SMAD3, the present study also investigated the mechanism of action between miR-203 and EMT marker molecules induced by TGF-β, which has not been reported previously.
Considering the important role of EMT in tumorigenesis and subsequent treatment, the present study demonstrated that miR-203 can directly target SMAD3 and inhibit TGF-β-induced migration and invasion of EMT and NSCLC cells, providing a theoretical basis for the development of new drugs for tumor invasion and metastasis. Moreover, the present study also offered insights into targeted treatment strategies to solve problems related to lung cancer resistance.
Acknowledgements
Not applicable.
Funding
The present study was supported by Zhejiang Natural Science Fund Youth Project (LQ18H010004) and the Wenzhou Science and Technology Bureau (Y20160047).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WH was responsible for the conception and design of the study, the data analysis and interpretation and the manuscript revision. YW was responsible for the collection and assembly of data, data analysis and interpretation, and manuscript writing. DC was responsible for the collection and assembly of data, and revised the manuscript. ZH carried out the collection and assembly of data, wrote and reviewed the manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients provided their written informed consent and ethics approval was obtained from the Ethics Committees of the First Affiliated Hospital of Wenzhou Medical University (2017063). The study adhered to the ethical standards of the Helsinki Declaration.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zhang S, Zou X. Estimation and projection of lung cancer incidence and mortality in China. Zhongguo Fei Ai Za Zhi. 2010;13:488–493. doi: 10.3779/j.issn.1009-3419.2010.05.20. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson KW, Sandler AB. The role of MET receptor tyrosine kinase in non-small cell lung cancer and clinical development of targeted anti-MET agents. Oncologist. 2013;18:115–122. doi: 10.1634/theoncologist.2012-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosell R, Karachaliou N. Lung cancer: Maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol. 2013;10:549–550. doi: 10.1038/nrclinonc.2013.152. [DOI] [PubMed] [Google Scholar]
- 6.Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, Kunkler I, EUROCARE-4 Working Group Recent cancer survival in Europe: A 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8:784–796. doi: 10.1016/S1470-2045(07)70246-2. [DOI] [PubMed] [Google Scholar]
- 7.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gal A, Sjöblom T, Fedorova L, Imreh S, Beug H, Moustakas A. Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene. 2008;27:1218–1230. doi: 10.1038/sj.onc.1210741. [DOI] [PubMed] [Google Scholar]
- 9.Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J Biol Chem. 2013;288:10241–10253. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 11.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 12.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang HJ, Wang HY, Zhang HT, Su JM, Zhu J, Wang HB, Zhou WY, Zhang H, Zhao MC, Zhang L, Chen XF. Transforming growth factor-β1 promotes lung adenocarcinoma invasion and metastasis by epithelial-to-mesenchymal transition. Mol Cell Biochem. 2011;355:309–314. doi: 10.1007/s11010-011-0869-3. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, Wang L, Zhao J, Chen Y, Lei Z, Liu X, Xia W, Guo L, Zhang HT. TGF-β-activated SMAD3/4 complex transcriptionally upregulates N-cadherin expression in non-small cell lung cancer. Lung Cancer. 2015;87:249–257. doi: 10.1016/j.lungcan.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Millet C, Zhang YE. Roles of Smad3 in TGF-beta signaling during carcinogenesis. Crit Rev Eukaryot Gene Expr. 2007;17:281–293. doi: 10.1615/CritRevEukarGeneExpr.v17.i4.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 17.Yang G, Yang X. Smad4-mediated TGF-beta signaling in tumorigenesis. Int J Biol Sci. 2010;6:1–8. doi: 10.7150/ijbs.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Roberts AB, Tian F, Byfield SD, Stuelten C, Ooshima A, Saika S, Flanders KC. Smad3 is key to TGF-beta-mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev. 2006;17:19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, Feng XH, Sawaya R, Medema RH, Hung MC, Huang S. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer metastasis. J Clin Invest. 2014;124:564–579. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, Pietras K, Virtanen I, Philipson L, Leopold PL, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11:943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, Sun P, Lei Z, Li M, Wang Y, Zhang HT, Liu J. miR-145 inhibits invasion and metastasis by directly targeting Smad3 in nasopharyngeal cancer. Tumour Biol. 2015;36:4123–4131. doi: 10.1007/s13277-015-3046-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhao W, Zou J, Wang B, Fan P, Mao J, Li J, Liu H, Xiao J, Ma W, Wang M, et al. microRNA-140 suppresses the migration and invasion of colorectal cancer cells through targeting Smad3. Zhonghua Zhong Liu Za Zhi. 2014;36:739–745. (In Chinese) [PubMed] [Google Scholar]
- 24.Huang S, Wu S, Ding J, Lin J, Wei L, Gu J, He X. MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res. 2010;38:7211–7218. doi: 10.1093/nar/gkq564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Wang X, Liang H, Wang T, Yan X, Cao M, Wang N, Zhang S, Zen K, Zhang C, Chen X. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PLoS One. 2013;8:e73985. doi: 10.1371/journal.pone.0073985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider MR. MicroRNAs as novel players in skin development, homeostasis and disease. Br J Dermatol. 2012;166:22–28. doi: 10.1111/j.1365-2133.2011.10568.x. [DOI] [PubMed] [Google Scholar]
- 28.Butz H, Rácz K, Hunyady L, Patócs A. Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol Sci. 2012;33:382–393. doi: 10.1016/j.tips.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Chen T, Xu C, Chen J, Ding C, Xu Z, Li C, Zhao J. MicroRNA-203 inhibits cellular proliferation and invasion by targeting Bmi1 in non-small cell lung cancer. Oncol Lett. 2015;9:2639–2646. doi: 10.3892/ol.2015.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang N, Liang H, Zhou Y, Wang C, Zhang S, Pan Y, Wang Y, Yan X, Zhang J, Zhang CY, et al. miR-203 suppresses the proliferation and migration and promotes the apoptosis of lung cancer cells by targeting SRC. PLoS One. 2014;9:e105570. doi: 10.1371/journal.pone.0105570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickie LJ, Aziz AM, Savic S, Lucherini OM, Cantarini L, Geiler J, Wong CH, Coughlan R, Lane T, Lachmann HJ, et al. Involvement of X-box binding protein 1 and reactive oxygen species pathways in the pathogenesis of tumour necrosis factor receptor-associated periodic syndrome. Ann Rheum Dis. 2012;71:2035–2043. doi: 10.1136/annrheumdis-2011-201197. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, Pham TQ, Soriano R, Stinson J, Seshagiri S, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 34.Shintani Y, Okimura A, Sato K, Nakagiri T, Kadota Y, Inoue M, Sawabata N, Minami M, Ikeda N, Kawahara K, et al. Epithelial to mesenchymal transition is a determinant of sensitivity to chemoradiotherapy in non-small cell lung cancer. Ann Thorac Surg. 2011;92:1794–1804. doi: 10.1016/j.athoracsur.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 35.Jung JW, Hwang SY, Hwang JS, Oh ES, Park S, Han IO. Ionising radiation induces changes associated with epithelial-mesenchymal transdifferentiation and increased cell motility of A549 lung epithelial cells. Eur J Cancer. 2007;43:1214–1224. doi: 10.1016/j.ejca.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 36.Zhou YC, Liu JY, Li J, Zhang J, Xu YQ, Zhang HW, Qiu LB, Ding GR, Su XM, Mei-Shi, Guo GZ. Ionizing radiation promotes migration and invasion of cancer cells through transforming growth factor-beta-mediated epithelial-mesenchymal transition. Int J Radiat Oncol Biol Phys. 2011;81:1530–1537. doi: 10.1016/j.ijrobp.2011.06.1956. [DOI] [PubMed] [Google Scholar]
- 37.Theys J, Jutten B, Habets R, Paesmans K, Groot AJ, Lambin P, Wouters BG, Lammering G, Vooijs M. E-Cadherin loss associated with EMT promotes radioresistance in human tumor cells. Radiother Oncol. 2011;99:392–397. doi: 10.1016/j.radonc.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat Cell Biol. 2007;9:1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 41.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Yang H, Lei Z, Zhao J, Chen Y, Chen P, Li C, Zeng Y, Liu Z, Liu X, Zhang HT. Repression of TIF1γ by SOX2 promotes TGF-β-induced epithelial-mesenchymal transition in non-small-cell lung cancer. Oncogene. 2016;35:867–877. doi: 10.1038/onc.2015.141. [DOI] [PubMed] [Google Scholar]
- 43.Cho HJ, Baek KE, Saika S, Jeong MJ, Yoo J. Snail is required for transforming growth factor-beta-induced epithelial-mesenchymal transition by activating PI3 kinase/Akt signal pathway. Biochem Biophys Res Commun. 2007;353:337–343. doi: 10.1016/j.bbrc.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 44.Shintani Y, Maeda M, Chaika N, Johnson KR, Wheelock MJ. Collagen I promotes epithelial-to-mesenchymal transition in lung cancer cells via transforming growth factor-beta signaling. Am J Respir Cell Mol Biol. 2008;38:95–104. doi: 10.1165/rcmb.2007-0071OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruna A, Darken RS, Rojo F, Ocaña A, Peñuelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, Seoane J. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 46.Liu RY, Zeng Y, Lei Z, Wang L, Yang H, Liu Z, Zhao J, Zhang HT. JAK/STAT3 signaling is required for TGF-β-induced epithelial-mesenchymal transition in lung cancer cells. Int J Oncol. 2014;44:1643–1651. doi: 10.3892/ijo.2014.2310. [DOI] [PubMed] [Google Scholar]
- 47.Wang C, Song X, Li Y, Han F, Gao S, Wang X, Xie S, Lv C. Low-dose paclitaxel ameliorates pulmonary fibrosis by suppressing TGF-β1/Smad3 pathway via miR-140 upregulation. PLoS One. 2013;8:e70725. doi: 10.1371/journal.pone.0070725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao H, Yang CS, Rana TM. Evolutionary emergence of microRNAs in human embryonic stem cells. PLoS One. 2008;3:e2820. doi: 10.1371/journal.pone.0002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakayama K, Nakayama N, Katagiri H, Miyazaki K. Mechanisms of ovarian cancer metastasis: Biochemical pathways. Int J Mol Sci. 2012;13:11705–11717. doi: 10.3390/ijms130911705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuesta R, Martínez-Sánchez A, Gebauer F. miR-181a regulates cap-dependent translation of p27(kip1) mRNA in myeloid cells. Mol Cell Biol. 2009;29:2841–2851. doi: 10.1128/MCB.01971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, Xu ZM, Yin YB. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 53.Abella V, Valladares M, Rodriguez T, Haz M, Blanco M, Tarrío N, Iglesias P, Aparicio LA, Figueroa A. miR-203 regulates cell proliferation through its influence on Hakai expression. PLoS One. 2012;7:e52568. doi: 10.1371/journal.pone.0052568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Y, Liang H, Liao Z, Wang Y, Hu X, Chen X, Xu L, Hu Z. miR-203 enhances let-7 biogenesis by targeting LIN28B to suppress tumor growth in lung cancer. Sci Rep. 2017;7:42680. doi: 10.1038/srep42680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y, Liu L, Cai J, Wu J, Guan H, Zhu X, Yuan J, Chen S, Li M. Targeting Smad2 and Smad3 by miR-136 suppresses metastasis-associated traits of lung adenocarcinoma cells. Oncol Res. 2013;21:345–352. doi: 10.3727/096504014X14024160459285. [DOI] [PubMed] [Google Scholar]
- 56.Wang C, Liu P, Wu H, Cui P, Li Y, Liu Y, Liu Z, Gou S. MicroRNA-323-3p inhibits cell invasion and metastasis in pancreatic ductal adenocarcinoma via direct suppression of SMAD2 and SMAD3. Oncotarget. 2016;7:14912–14924. doi: 10.18632/oncotarget.7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.