Abstract
Niclosamide is an FDA-approved anthelmintic drug, and may elicit antineoplastic effects through direct STAT3 inhibition, which has been revealed in numerous human cancer cells. Chemotherapy is the standard treatment for advanced esophageal cancers, but also causes severe systemic side effects. The present study represents the first study evaluating the anticancer efficacy of niclosamide in esophageal cancers. Through western blot assay, it was demonstrated that niclosamide suppressed the STAT3 signaling pathway in esophageal adenocarcinoma cells (BE3) and esophageal squamous cell carcinoma cells (CE48T and CE81T). In addition, niclosamide inhibited cell proliferation as determined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay and soft agar colony forming assay, and induced cell apoptosis as determined by Annexin V and PI staining. The induction of p21 and G1 arrest of the cell cycle also was revealed in niclosamide-treated CE81T cells by qPCR and flow cytometric assays, respectively. Furthermore, in the combination analysis of niclosamide and chemotherapeutic agents by MTS assay, low IC50 values were detected in cells co-treated with niclosamide, with the exception of cisplatin-treated CE81T cells. To confirm the results using an apoptosis assay, the apoptotic enhancement of niclosamide was only demonstrated in CE48T cells co-treated with 5-FU, cisplatin, or paclitaxel, and in BE3 cells co-treated with paclitaxel, but not in CE81T cells. These findings indicate a future clinical application of niclosamide in esophageal cancers.
Keywords: esophageal cancer, niclosamide, chemotherapy, apoptosis, cell cycle arrest
Introduction
In 2018, esophageal cancer ranked seventh for cancer incidence and sixth as the leading cause of cancer-related mortality in the world, and ~70% of cases occurred in men. In men, the rates were also 2-fold higher in Human Development Index (HDI) countries, with mortality rates ranking fifth in these countries (1). Esophageal squamous cell carcinomas (ESCC) and esophageal adenocarcinomas (EAC) account for >95 % of the various esophageal malignancies. Moreover, the incidence of EAC is rapidly increasing in the United States and Western Europe (2). Esophagectomy alone is associated with a high rate of recurrence and low 5-year survival rates. A recent clinical trial revealed that neoadjuvant chemotherapy significantly improved survival in patients with resectable tumors (3). Conventional chemotherapeutic agents can effectively kill actively proliferating cells, however, the non-selective targeting can cause the destruction of both healthy and cancerous cells. Ultimately, chemotherapy leads to multiple side effects and the occurrence of drug resistance in patients. Therefore, an important approach is to identify low-toxicity and non-cancer agents for combination therapy in order to reduce the dose requirement of chemotherapeutic agents (4). Drug repurposing is an efficient approach, using clinically-approved drugs for different disease treatments bypassing the long streamline of the drug discovery process (5). In order to enhance the killing efficacy of cancer cells and ameliorate the side effects caused by cancer treatments, it is significant to study and evaluate both the feasibility and efficacy of combination therapy with chemotherapeutic agents plus repurposed non-cancer drugs targeting similar pathways found in cancer (6). Based on this concept, searching for non-cancer clinical medications and evaluating the feasibility of combination therapy of these non-cancer clinical medications and chemotherapeutic agents for esophageal cancers is the focus of this research.
Niclosamide is an FDA-approved chewable tablet consumed orally against tapeworms and schistosomiasis. The mechanism of niclosamide action is through the inhibition of glucose uptake, oxidative phosphorylation, and anaerobic metabolism in the target worms (7). Except for antiprotozoal function, the antineoplastic effects of niclosamide has been revealed in many human cancer cells, including adrenocortical carcinoma (8), breast (9–11), colorectal (12,13) and cervical cancer (14), glioblastoma (15), hepatocellular carcinoma (16,17), head and neck (18,19) and lung cancer (20,21), leukemia (22,23), nasopharyngeal cancer (24), osteosarcoma (25), oral squamous cell carcinoma (26,27), ovarian (28–30), prostate (31–33), renal (34) and thyroid cancer (35). Niclosamide treatment was reported to induce apoptosis of cancer cells in vitro (7,10,23) and suppress tumor size in animal studies (11,29). Moreover, the combination of anticancer agents with niclosamide synergistically suppressed cell proliferation of acute myelogenous leukemia, head and neck, ovarian, prostate and non-small lung cancer (19,21,23,30,31). However, whether niclosamide is effective against esophageal cancer has not been investigated yet.
The molecular mechanisms underlying the antineoplastic effect of niclosamide have been explored in many human malignant cancers, indicating that niclosamide exhibits anticancer activity by suppressing many oncogenic signaling pathways concurrently (7,13,17,23,27,28,30,36). For instance, niclosamide has been identified as a direct inhibitor of signal transducer and activator of transcription 3 (STAT3) through interaction with the DNA-binding domain (37). In ovarian cancer, niclosamide significantly decreased the expression of proteins in the wingless/integrated (Wnt), mammalian target of rapamycin (mTOR) and STAT3 pathways and caused significant inhibition of proliferation of cells (28). In acute myeloid leukemia, niclosamide could induce apoptosis of AML blast cells through inhibition of the nuclear factor-κB (NF-κB) pathway and increasing the production of reactive oxygen species (23). In lung and head and neck cancers, niclosamide suppressed erlotinib-induced STAT3 phosphorylation, and a combination of erlotinib and niclosamide decreased tumor size in animal model experiments (19,21). In advanced prostate cancer, niclosamide blocked the interleukin 6 (IL6)/STAT3/androgen receptor (AR) pathway to overcome enzalutamide resistance and inhibit migration and invasion (31).
In the present study, the antineoplastic effects of niclosamide on esophageal cancer cells were investigated and it was revealed that niclosamide suppressed the STAT3 signaling pathway and inhibited cell proliferation in esophageal cancer cells. Niclosamide also induced cell apoptosis and G1-phase arrest of the cell cycle. Furthermore, the combination treatment of niclosamide and chemotherapeutic drugs selectively reduced the dose requirement of the chemotherapeutic drugs in order to obtain the IC50 efficacy. These findings indicated that niclosamide may be used as a single or combined drug treatment for esophageal cancer.
Materials and methods
Reagents
Niclosamide (product no. N3510), 5-fluorouracil (5-FU) (product no. F6627), cisplatin (P4394), and paclitaxel (T7402) were purchased from Sigma-Aldrich (Merk KGaA). Cisplatin was dissolved in ddH2O, whereas, niclosamide, 5-FU and paclitaxel were dissolved in dimethyl sulfoxide (DMSO). The solvent was routinely used in the control group of the experiment.
Cell culture
Esophageal cancer cell lines, BE3 (adenocarcinoma), CE48T/VGH and CE81T/VGH (squamous cell carcinoma) were courtesy of Dr Yen (38) and Dr Lee (39), respectively. BE3 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) and CE48T and CE81T were cultured in Roswell Park Memorial Institute (RPMI) 1640 (Gibco BRL; Thermo Fisher Scientific, Inc.), supplemented with 10% heat-inactivated fetal bovine serum, 1% penicillin/streptomycin solution (Gibco-BRL; Thermo Fisher Scientific, Inc.). The cells were grown in a humidified incubator containing 5% CO2 at 37°C.
MTS assay
To determine the cytotoxicity of niclosamide and the combined effect of niclosamide and chemotherapeutic agents, cells were seeded in 96-well plates overnight and treated for 72 h with different concentrations of niclosamide (1.25–20 µM) or co-treated with 2.5 µM niclosamide and serial doses of chemotherapeutic drugs, or DMSO as a vehicle control. The cell viability was then evaluated by MTS assay. Twenty microliters of the MTS CellTiter 96 Cell Proliferation Assay reagent (Promega Corporation) were added to each well. After 2 h of incubation at 37°C, the absorbance of the colored product was measured on an EPOCH2 microplate reader (BioTek Instruments, Inc.).
Colony-forming assay
Esophageal cancer cell lines to be tested were trypsinized and dissociated into single-cell suspensions for plating in 6-well plates. An agar suspension (0.3% agar) containing colony-forming cells (2×104 cells/well) is plated over an agar underlay (0.6% agar). Niclosamide (2.5 µM) was delivered to the cells. Tissue culture medium was replenished every 3 days. After 21 days of culture, the cells were washed twice with phosphate-buffered saline (PBS) and incubated in fresh medium with MTS reagent for 2 h at 37°C. The living colony formation efficiency was determined by formation of dark-colored formazan dye by reduction of the tetrazolium salt MTS by metabolically active cells. Digital images of the plates were captured by Nikon camera.
Western blot analysis
To evaluate the protein expression levels after niclosamide treatment, western blot analysis was performed. The cells were dissolved in a lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA, 5 mM sodium orthovanadate, 25 mM sodium fluoride, 5 mM sodium pyrophosphate decahydrate and 5 mM β-glycerophosphate. Protein concentration was quantified by Bio-Rad protein assay. Fifty micrograms of protein was fractionated via 10 or 12% SDS-PAGE and transferred to Immobilon Transfer Membranes (EMD Millipore), and then the membranes were blocked in 5% skim milk at room temperature for 1 h. Primary antibodies used in the present study included those against β-actin (GTX109639) (dilution 1:10,000; GeneTex, Inc.), poly(ADP-ribose) polymerase (PARP) (9542S), β-catenin (8480S), STAT3 (4904S), phosphor-STAT3 (9145S) (dilution 1:1,000; Cell Signaling Technology, Inc.), cyclin D1 (H295), cyclin E1 (HE-12), cyclin A (H432), and cyclin B (GNS1) (dilution 1:1,000; Santa Cruz Biotechnology, Inc.). The HRP-conjugated secondary antibodies recognizing mouse-IgG (7076) or rabbit-IgG (7074) were purchased from Cell Signaling Technology, Inc. and those recognizing goat-IgG (SC-2020) were purchased from Santa Cruz Biotechnology, Inc. (dilution, ~1:5,000–10,000). The chemiluminescence signal was developed with Immobilon Western Chemiluminescent HRP Substrate (WBKLS0500; Millipore) and visualized and recorded using the BioSpectrum UVP810 Imaging System with VisionWorks software (GE Healthcare Life Sciences).
Real-time quantitative RT-PCR (RT-qPCR) analysis
To quantify the endogenous mRNA expression levels, qRT-PCR was performed. Total RNA of cells was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. One microgram of total RNA was used to synthesize the cDNA using SuperScript III reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) with Oligo-dT primer. Quantitative PCR was performed using Kapa SYBR fast qPCR master mix (Kapa Biosystems, Inc.). The primer sequences for qPCR were as follows: GAPDH forward, 5′-GAAGGTGAAGGTCGGAGT-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′; p21 forward, 5′-GAAGACCATGTGGACCTGTC-3′ and reverse, 5′-TCCTCTTGGAGAAGATCAGC-3′; p27 forward, 5′-GTTTCAGACGGTTCCCCAAA-3′ and reverse, 5′-CCTTGCTTCATCAAGCAGTG-3′. The qPCR reaction was run in a final volume of 20 µl containing 1 µl of reverse transcriptase product, 10 µl of 2X Kapa SYBR fast qPCR master mix, and 0.6 µl of each primer (10 µM). The qPCR mixtures were pre-incubated at 95°C for 3 min, followed by 40 cycles at 95°C for 3 sec and 60°C for 30 sec. All qPCR was performed in triplicate using an iQ5 Real-Time PCR detection system (Bio-Rad Laboratories, Inc.). Relative expression was calculated using the 2−ΔΔCq method (40).
Annexin V and propidium iodide (PI) double staining
Following drug treatment, the cells were collected and washed twice with cold PBS, and stained with 5 µl of Annexin V-FITC and 10 µl of PI (5 g/ml) in 0.5 ml of binding buffer (10 mM HEPES, pH 7.4; 140 mM NaOH; 2.5 mM CaCl2) for 15 min at room temperature in the dark. The apoptotic cells were determined using a BD FACSVerse Flow Cytometer (BD Biosciences).
Cell cycle analysis
After niclosamide treatments, the cells were collected and fixed with cold 80% ethanol at −20°C for 1 h. The cells were then washed twice with PBS and stained with 0.5 ml PI/RNase at RT for 30 min. The samples were analyzed by BD FACSVerse Flow Cytometer.
Statistical analysis
All values were expressed as the mean ± standard error. The Student's t-test was used to analyze the results, and a P-value of <0.05 was considered to indicate a statistically significant difference.
Results
Niclosamide suppresses the STAT3 signaling pathway and inhibits the cell growth of esophageal cancer cell lines
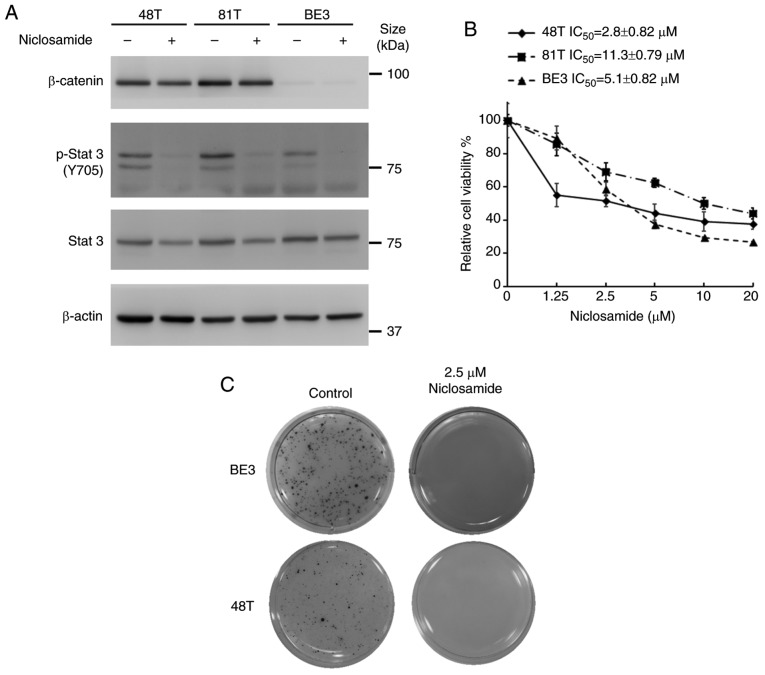
Niclosamide, a direct STAT3 inhibitor, had been reported to exhibit the ability to suppress multiple oncogenic signaling transduction pathways. In order to explore the effect of niclosamide on STAT3 and β-catenin signaling pathways of cultured esophageal cancer cells, CE48T, CE81T and BE3 cells were treated with 10 µM niclosamide for 24 h, and the total forms of STAT3 and β-catenin and phosphorylated (p)-STAT3 (Y705) were investigated by western blot analysis. As revealed in Fig. 1A, there were different expression levels of β-catenin in these cell lines. The protein levels of endogenous β-catenin could only be detected in CE48T and CE81T cells, but not in BE3 cells. Moreover, niclosamide treatment did not markedly affect the protein level of β-catenin. Conversely, the STAT3 signaling pathway was constitutively active in these cancer cells, and niclosamide treatment markedly suppressed STAT3 (Y705) phosphorylation. Furthermore, to explore the effect of niclosamide on the proliferation of esophageal cancer cells, these cells were treated with niclosamide at different concentrations for 72 h and then cell viability was assessed by MTS assay. As revealed in Fig. 1B, the cell viabilities of these three cancer cell lines were dose-dependently decreased by niclosamide treatment. The IC50 concentrations of niclosamide were 2.8, 11.3, and 5.1 µM for CE48T, CE81T and BE3, respectively. Moreover, a clonogenic assay was conducted to investigate the long-term growth inhibitory effect of niclosamide on esophageal cancer cells. Since CE81T cells did not grow on the soft agar to form a colony, only BE3 and CE48T cells were repeated and long-term treated with a physiological concentration of niclosamide (2.5 µM) for 21 days and living colony formation efficiency was determined by formation of dark-colored formazan dye by reduction of the tetrazolium salt MTS by metabolically active cells. As revealed in Fig. 1C, niclosamide completely inhibited colony formation in BE3 and CE48T cells, indicating that niclosamide may be considered as a potential inhibitor for esophageal cancer cell growth.
Figure 1.
Niclosamide treatment blocks the STAT3 signaling pathway and inhibits the cell growth of human esophageal cancer cell lines. (A) CE48T, CE81T and BE3 cells were treated with 10 µM niclosamide for 24 h, and the phosphorylation of Y705 and the total form of STAT3, as well as β-catenin, were investigated by western blot assay using β-actin as a loading control. (B) Cancer cells were seeded in 96-well plates and treated with a concentration series of niclosamide (0, 1.25, 2.5, 5, 10 or 20 µM) for 72 h, and then the relative cell viabilities were determined by MTS assay. Error bars represent the standard deviation. The viability of control cells was designated as 100%, and that of the other groups were expressed as the percent of the control. (C) Colony-forming ability of CE48T and BE3 cells analyzed by clone formation assay. Images of living cell clones indicated by the formation of dark-colored formazan dye by reduction of the tetrazolium salt MTS by metabolically active cells. STAT3, signal transducer and activator of transcription 3; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
Niclosamide treatment induces apoptosis of esophageal cancer cell lines
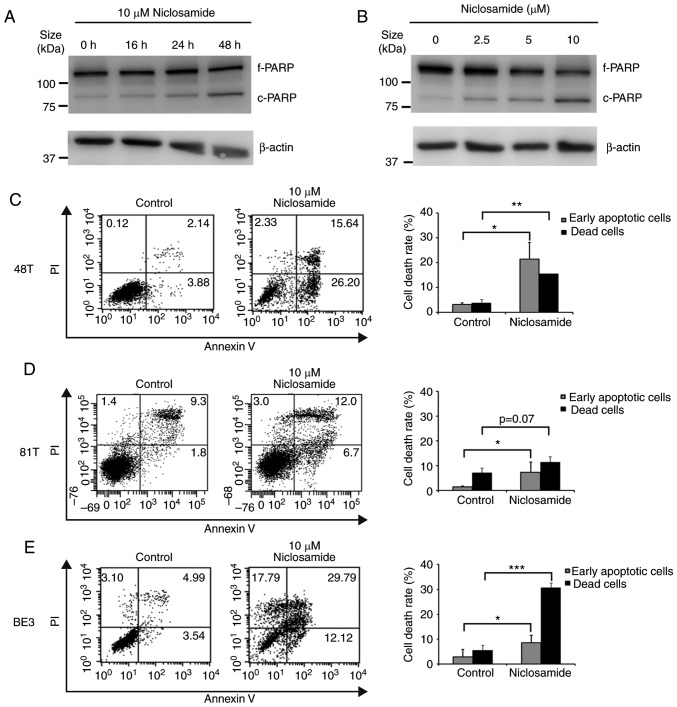
To further explore if the decrease in esophageal cancer cell viability was a consequence of niclosamide-induced apoptosis, BE3 cells treated with niclosamide were subjected to western blot analysis of cleaved PARP. As revealed in Fig. 2A and B, the highest level of cleaved PARP product was detected at 10 µM niclosamide and 48 h of treatment, indicating that the apoptotic level was positively associated with the niclosamide dose and exposure time in BE3 cells. To further demonstrate the induction of apoptosis by niclosamide, CE48T, CE81T and BE3 cells were treated with niclosamide for 48 h, and subjected to Annexin V and PI double-staining assay. The early apoptotic and dead cells were quantified by flow cytometric analysis. As revealed in Fig. 2C-E, niclosamide significantly (P<0.05) increased the proportion of early apoptotic cells in the three cell lines, but only significantly increased the proportion of dead cells in CE48T (P<0.005) and BE3 (P<0.0005) cells. The results indicated that the reduction of cell viabilities of CE48T and BE3 cells was mainly caused by niclosamide-induced cell apoptosis, but was partial to that of CE81T cells.
Figure 2.
Niclosamide treatment induces apoptosis in esophageal cancer cell lines. (A) BE3 cells were treated with 10 µM niclosamide, then harvested at different time-points after treatment, or (B) treated with various doses of niclosamide for 48 h. Treated cells were lysed for western blot analysis of PARP. The position of the native PARP (f-PARP) and the cleaved fragment (c-PARP) is indicated. (C) CE48T, (D) CE81T and (E) BE3 cells were treated with 10 µM niclosamide for 48 h and the apoptosis levels were detected by Annexin V/PI staining. DMSO treatment was used as the control. The proportion of Annexin V+/PI− and Annexin V+/PI+ cells indicated the early apoptotic and dead cells, respectively. Results were expressed as the means ± SD of three independent experiments. *P<0.05, **P<0.005, and ***P<0.0005. PARP, poly(ADP-ribose) polymerase.
Niclosamide treatment causes G1-phase arrest in CE81T cells
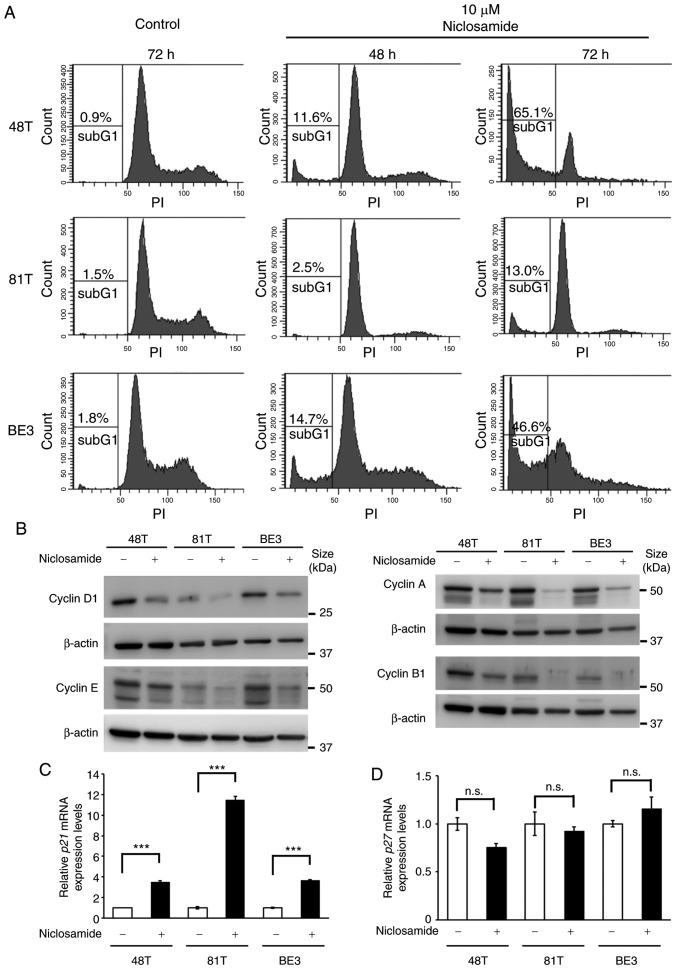
Given that only a small proportion of apoptotic cells were detected in CE81T cells after treatment with 10 µM niclosamide for 48 h (Fig. 2D), cell cycle dynamics were analyzed by flow cytometric analysis to investigate why niclosamide suppressed cell growth of CE81T cells. The cell cycle analysis revealed that after treatment with 10 µM niclosamide for 72 h a high proportion of sub-G1 (cell debris) was only detected in CE48T and BE3 cells, while a low proportion of sub-G1 was revealed in CE81T cells. Moreover, an apparent G1 peak was only revealed in CE81T cells, as well as absence of S and G2/M phases (Fig. 3A). In addition, the physiological concentration of niclosamide (2.5 µM) was also used to conduct cell cycle analysis in CE48T, CE81T and BE3 cells. The proportion of sub-G1 cells was increased in a time-dependent manner in CE48T and BE3 cells after 48 and 72 h treatment. In addition, at 72 h of treatment, the proportion of G1-phase cells was decreased and the cells of the S and G2/M phases were still detectable. In contrast, in CE81T cells, the proportion of sub-G1 cells did not significantly increase, the proportion of G1-phase cells was increased and the proportion of S-phase cells was decreased after 72 h of treatment (Fig. S1). This finding indicated that niclosamide induced G1-phase arrest in CE81T cells. Cyclin is a family of proteins that control the progression of cells through the cell cycle. The protein levels of various cyclins in CE48T, CE81T and BE3 cells were assessed by western blot assay, and it was revealed that all of the expression levels of cyclin D1, E, A and B1 were almost completely lost in CE81T cells treated with niclosamide for 48 h (Fig. 3B). In addition, the expression of cell cycle inhibitor genes, p21 and p27, were also examined in these cells treated with or without 10 µM niclosamide for 24 h. As revealed in Fig. 3C and D, an ~11-fold increase in p21 mRNA expression was detected in niclosamide-treated CE81T cells, as well as an ~3.5-fold increase in niclosamide-treated CE48T and BE3 cells. In contrast, niclosamide treatment did not affect p27 mRNA expression in these cells.
Figure 3.
Niclosamide treatment causes CE81T cell cycle arrest. (A) CE48T, CE81T and BE3 cells were treated with 10 µM niclosamide for 48 or 72 h. Cells were stained with PI for DNA content and analyzed by flow cytometry. The relative proportion of the sub-G1 cells are indicated. (B) The protein expression levels of cyclin D1, E, A and B1 in CE48T, CE81T and BE3 were determined with western blot assay after cells were treated with 10 µM niclosamide for 48 h. Protein loading was normalized based on β-actin. (C and D) These cells were treated with or without 10 µM niclosamide for 24 h and total RNA was collected for p21 and p27 assessment by RT-qPCR assays. The data were normalized by GAPDH expression and presented as the mean ± SEM (n=3). ***P<0.001; n.s., not significant. PI, propidium iodide.
Niclosamide co-treatment selectively reduces the dosage of anticancer drugs for achieving IC50 in esophageal cancer cells
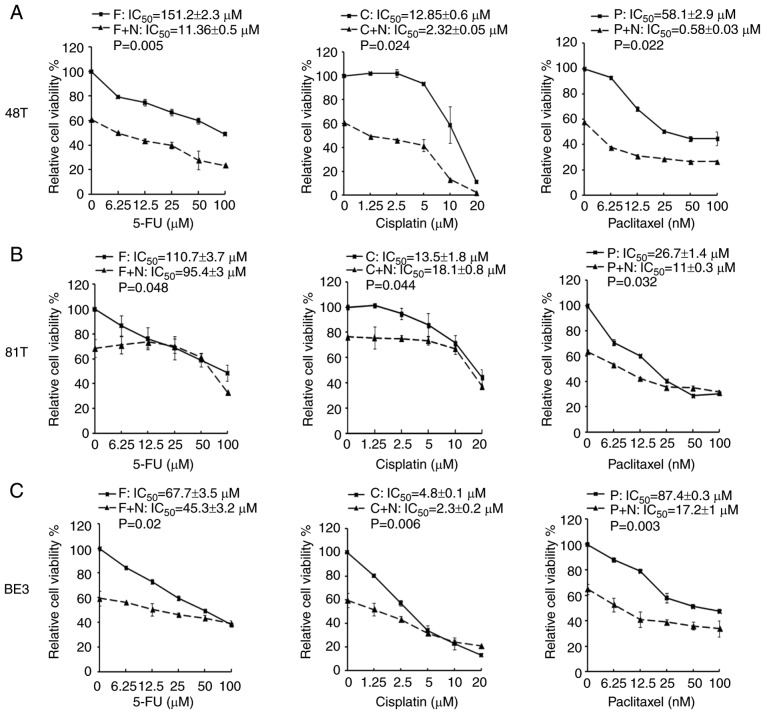
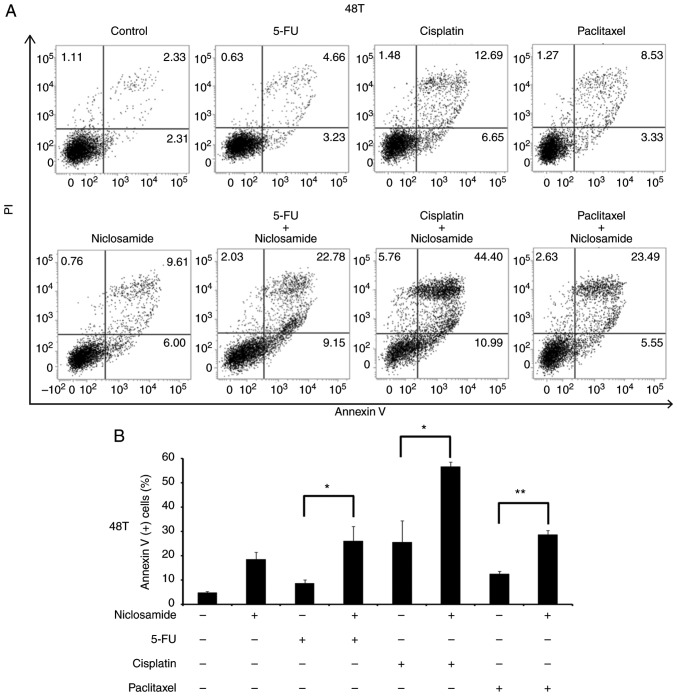
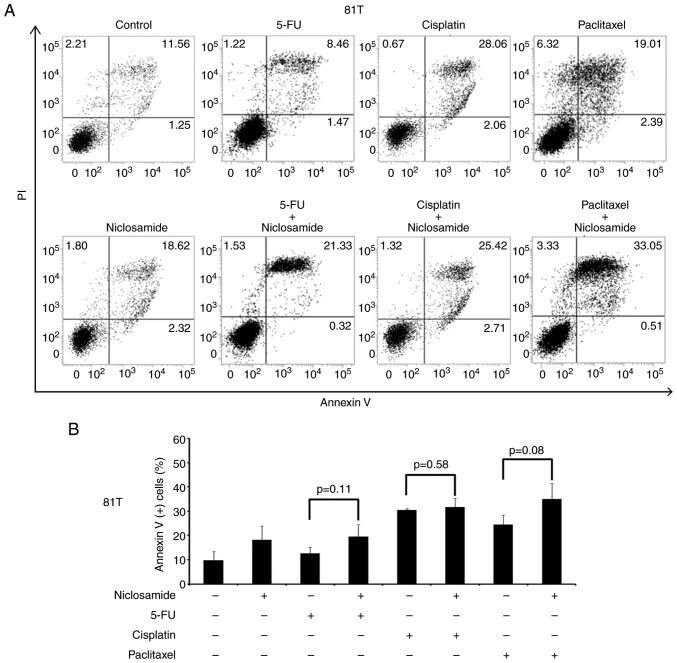
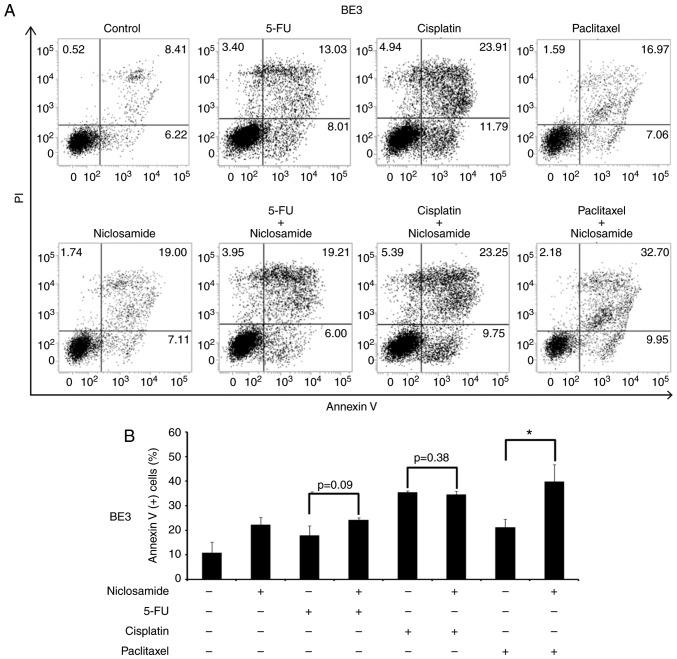
A previous study reported that the combination of niclosamide and anticancer drugs resulted in a synergistic antitumor efficacy against xenografted AML tumor cells (23). 5-FU, cisplatin, and paclitaxel are three frontline chemotherapeutic drugs currently used in clinical esophageal therapy (41–43). Therefore, they were used in the present experiment to explore whether the treatment of these anticancer drugs combined with niclosamide could have an additive benefit of antitumor efficacy for esophageal cancer cells. Niclosamide appears to be minimally absorbed from the gastrointestinal tract and a pharmacokinetics study revealed that the oral dosage of niclosamide for adults in cytocidal treatment was 2 g as a single dose, leading to maximal serum concentration of 0.76 to 18.35 µM (44). In the present study, the esophageal cancer cell lines were treated with the combination of 2.5 µM niclosamide plus different chemotherapeutic drugs for 72 h, and cell viability was assessed by MTS assay. In CE48T and BE3 cells, the IC50 values of 5-FU, cisplatin, and paclitaxel were reduced in the combination groups of niclosamide, when compared to the anticancer drug administrated alone (Fig. 4A and C). In CE81T cells, the niclosamide combination only reduced the IC50 values of 5-FU and paclitaxel as compared with anticancer drug treatment alone (Fig. 4B). To further validate the benefit of combination treatment with niclosamide, the proportion of apoptotic cells (Annexin V+ cells) induced by the anticancer drug alone, or in combination with niclosamide was investigated by flow cytometric analysis. As revealed in Fig. 5, the treatment of 2.5 µM niclosamide for 48 h induced 18.46±3.06% of apoptotic cells in CE48T cells, and the combination of niclosamide and 5-FU (50 µM) caused a higher proportion of apoptotic cells than 5-FU treatment alone (26.14±5.89% vs. 8.62±1.47%; P<0.05). Moreover, the combination of niclosamide and cisplatin (10 µM) resulted in a higher proportion of apoptotic cells than cisplatin treatment alone (56.68±1.82% vs. 25.57±8.81%; P<0.05), as well as the combination of niclosamide and paclitaxel (10 nM) which induced a higher proportion of apoptotic cells than paclitaxel treatment alone (28.69±1.75% vs. 12.54±1.12%; P<0.005). In CE81T cells, there was no statistically significant difference detected in the proportion of apoptotic cells between treatment with these three chemotherapeutic agents alone and the combination of niclosamide with 5-FU, cisplatin, or paclitaxel (as revealed in Fig. 6). In BE3 cells, the combination of niclosamide and paclitaxel induced a higher proportion of apoptotic cells than paclitaxel treatment alone (39.88±6.83% vs. 21.26±3.26%; P<0.05) (as revealed in Fig. 7). The results of the cell apoptosis assays were similar with the findings of the cell viability assays, indicating that niclosamide selectively reduced the dosage requirement of anticancer drug for obtaining the IC50 in esophageal cancer cells.
Figure 4.
MTS assays of esophageal cancer cells treated with a chemotherapeutic drug alone or in combination with niclosamide. Esophageal cancer cells (A) CE48T, (B) CE81T and (C) BE3 cells were seeded in 96-well plates and treated for 72 h with serial doses of 5-FU, cisplatin, paclitaxel, or in combination with 2.5 µM of niclosamide. Cell viability was then analyzed by MTS assay. F, C, and P indicate the treatment of 5-FU, cisplatin, or paclitaxel, respectively. F+N, C+N, and P+N indicate the combination treatment of niclosamide with 5-FU, cisplatin, or paclitaxel, respectively. 5-FU, 5-fluorouracil.
Figure 5.
Cell apoptosis assay of CE48T cells using Annexin V/PI double-staining method. (A) CE48T cells were treated with niclosamide (2.5 µM), 5-FU (50 µM), cisplatin (10 µM), paclitaxel (10 nM) alone, or in combination with niclosamide and each of these chemotherapeutic drugs for 48 h. The apoptotic levels were detected by Annexin V/PI staining and analyzed by flow cytometric assay. DMSO treatment was used as the control. The proportion of Annexin V (+) cells indicate the stage of apoptosis. (B) The percentage of Annexin V (+) cells were expressed as the means ± SD of three independent experiments. *P<0.05 and **P<0.005. 5-FU, 5-fluorouracil.
Figure 6.
Cell apoptosis assay of CE81T cells using Annexin V/PI double-staining method. (A) CE81T cells were treated with niclosamide (2.5 µM), 5-FU (50 µM), cisplatin (10 µM), paclitaxel (10 nM) alone, or in combination with niclosamide and each of these chemotherapeutic drugs for 48 h. The apoptotic levels were detected by Annexin V/PI staining and analyzed by flow cytometric assay. DMSO treatment was used as the control. The proportion of Annexin V (+) cells indicate the stage of apoptosis. (B) The percentage of Annexin V (+) cells were expressed as the means ± SD of three independent experiments. 5-FU, 5-fluorouracil.
Figure 7.
Cell apoptosis assay of BE3 cells using Annexin V/PI double-staining method. (A) BE3 cells were treated with niclosamide (2.5 µM), 5-FU (50 µM), cisplatin (10 µM), paclitaxel (10 nM) alone, or in combination with niclosamide and each of these chemotherapeutic drugs for 48 h. The apoptotic levels were detected by Annexin V/PI staining and analyzed by flow cytometric assay. DMSO treatment was used as the control. The proportion of Annexin V (+) cells indicated the stage of apoptosis. (B) The percentage of Annexin V (+) cells were expressed as the means ± SD of three independent experiments. *P<0.05. 5-FU, 5-fluorouracil.
Discussion
The current chemotherapeutic strategy is inevitably accompanied by a variety of dose-dependent and sometimes lethal adverse effects. Thus, it is worth exploring cost-effective drug repurposing and combinations to ameliorate unpleasant adverse effects following traditional chemotherapy. Niclosamide is an FDA-approved anthelmintic drug which exerts antineoplastic activity. The antineoplastic efficacy of niclosamide in numerous human cancer studies (8–35) has been demonstrated. However, whether niclosamide is active against esophageal cancer is unknown. Herein, the present study for the first time demonstrated that niclosamide exhibited antineoplastic effects in esophageal cancer cell lines.
Niclosamide has been identified as a direct STAT3 inhibitor through interaction with the DNA-binding domain (37). In addition to its role in STAT3 inhibition, niclosamide has also been revealed to concurrently inhibit multiple intracellular signaling pathways, including the Wnt, Notch 1, mTOR and NF-κB signaling cascades (10,45). In many solid and hematological cancers, STAT3 is constitutively active, promoting tumor survival and progression. For example, a previous study by Huang et al reported that positive p-STAT3 expression was observed in 47.7% of patients with esophageal squamous cell carcinoma. Patients with p-STAT3 positive disease had significantly worse clinical outcomes in terms of median progression-free survival (5.0 vs. 6.9 months, P<0.001) and median overall survival (8.9 vs. 9.9 months, P=0.026) (46). In addition, Wang et al reported that constitutively activated STAT3 in normal human esophageal epithelium cells (HET-1A) promoted proliferation and decreased apoptosis. Moreover, STAT3 activated HET-1A cells to form tumors in vivo (47), suggesting that aberrant STAT3 expression plays a crucial role in esophageal carcinogenesis. Conversely, STAT3 knockdown significantly reduced cell proliferation of OE21 (ESCC) and OE33 (EAC) cells, and a reduced STAT3 level was revealed to be associated with significant downregulation of cell cycle genes in both cell lines (48), indicating that STAT3 plays a pivotal role in cell proliferation of esophageal cancer cells. Through western blot analysis, it was revealed that the STAT3 signaling pathway, but not the Wnt pathway, was constitutively active in the three esophageal cancer cell lines. In addition, niclosamide treatment did not only markedly suppress the phosphorylation of Y705 of STAT3, but also slightly decreased the total protein levels of STAT3. Similar results were also obtained by Yoshikawa et al in a niclosamide-treated ovarian clear cell line (KK) (49). However, the mechanism resulting in the decrease of STAT3 total protein form by niclosamide treatment is still unclear. The IC50 values of niclosamide were 2.8, 11.3, and 5.1 µM for CE48T, CE81T and BE3 cells, respectively. Notably, ESCC CE81T cells were more resistant than ESCC CE48T cells to niclosamide. Similar resistant responses were revealed in a previous study in which CE81T cells were resistant to photofrin-induced inhibition of EGFR as well as to photofrin-mediated dark toxicity, compared with CE48T cells (39). Since niclosamide is minimally absorbed from the gastrointestinal tract, 2.5 µM niclosamide was used, a concentration less than the IC50 value, to investigate the long-term growth inhibitory effect of niclosamide in esophageal cancer cells by colony-forming assay. Niclosamide completely inhibited colony formation in BE3 and CE48T cells after 21 days of treatment, indicating that niclosamide may be considered as a potential inhibitor of esophageal cancer cell growth, as demonstrated in other types of cancer cells.
According to the results of western blot analysis of PARP and the Annexin V/PI staining assay, it was demonstrated that the inhibitory effect of niclosamide on cell growth in esophageal adenocarcinoma cell line BE3 was caused by niclosamide-induced apoptosis. By contrast, in esophageal squamous cell carcinoma cell lines, niclosamide significantly (P<0.05) increased the proportion of early apoptotic cells in both CE48T and CE81T cells, however only significantly increased the proportion of dead cells in CE48T (P<0.005), but not in CE81T cells. After 48 h of treatment of niclosamide, only a small proportion of Annexin V-positive cells were detected in CE81T, compared with CE48T and BE3 cells. The detailed mechanism of the different response of CE81T cells to niclosamide compared to CE48T and BE3 cells remains unclear and further investigation is required.
Previous studies in oral squamous cell carcinoma (26), adrenocortical carcinoma (8) and head and neck cancer (18) and osteosarcoma (25) revealed that niclosamide could induce G1-phase arrest of the cell cycle. Through flow cytometric analysis using DNA dye PI staining, it was revealed that a high proportion of sub-G1 cells were detected in CE48T and BE3 cells after 10 µM niclosamide treatment for 72 h, while only a small proportion of sub-G1 was detected in CE81T cells. Moreover, an evident G1 peak was revealed in CE81T cells. Similar experiments were also performed using the physiological concentration of niclosamide (2.5 µM) and the results were consistent with the high dose treatment. In addition, the protein levels of the various cyclins were explored by western blot assay, and it was revealed that the protein levels of cyclin D1, E, A and B1 almost completely disappeared in CE81T cells after niclosamide treatment for 48 h. As G1-phase arrest, cell cycle checkpoint proteins, such as p21 and p27, may mediate assembly and activation of Cdk4(6)-cyclin D in the cytoplasm, as well as inhibiting their activity in the nucleus. Ectopic induction of p21 or p27 expression completely arrested cells at the G1 phase of the cell cycle by p27 and at G1 and G2 phases by p21 (50). Therefore, in the present study, it was explored whether p21, p27 or both were involved in niclosamide-induced G1-phase arrest in CE81T cells. As revealed, niclosamide after 24 h of treatment significantly increased p21 mRNA expression in CE81T cells, but not p27 mRNA. These findings demonstrated that niclosamide induced G1-phase arrest in ESCC CE81T cells, however, the detailed mechanism is not clear yet.
Niclosamide may be beneficial not only as a monotherapy but also in combination therapy. Niclosamide has been demonstrated to increase the sensitivity of cancer cells for radiation therapy in lung cancer (51), nasopharyngeal cancer (24) and breast cancer (9). Furthermore, niclosamide was also revealed to exert a synergistic effect with numerous cancer drugs in human cancer cells. Compared with cancer drug treatment alone, the combination treatment of niclosamide with bicalutamide, cisplatin, paclitaxel, carboplatin, erlotinib, dasatinib, or sorafenib exhibited an improved antineoplastic efficacy in prostate (32), lung (52), cervical (14), ovarian (30), colon (53), chronic myeloid leukemia (22) and renal cancer (34), respectively. To date, the frontline chemotherapeutic agents for advanced esophageal cancer cells are 5-FU, cisplatin, and paclitaxel. In the present study, the antineoplastic efficacy of combination treatment of niclosamide (2.5 µM) plus 5-FU, cisplatin, or paclitaxel was assessed in esophageal cancer cells, and it was revealed that niclosamide co-treatment selectively reduced the dosage of anticancer drugs for obtaining the IC50. In CE48T and BE3 cells, the IC50 values of 5-FU, cisplatin, or paclitaxel were reduced in the combination groups with niclosamide, when compared to the anticancer drug administered alone. In CE81T cells, niclosamide combination treatment only reduced the IC50 values of 5-FU and paclitaxel as compared with the anticancer drug treatments alone. The beneficial effects of niclosamide combination on inhibition of cancer cell proliferation were further validated in the cell apoptosis assays. These findings indicated that niclosamide may be used as a repurposing drug to reduce the dosage requirement of anticancer drug for achieving the IC50 in esophageal cancer cells.
Sustained inhibition of tumor colony formation by long-term niclosamide treatment with potentially achievable serum concentration is indicative of a novel maintenance strategy for esophageal cancers adjunctive to current curatively local treatments or palliative chemotherapy with limited choices and survival benefits but well-known side effects. Due to approved indication of niclosamide as an antiparasitic, there is little clinical data with regard to the adverse effects of prolonged niclosamide treatment. Andrews et al reviewed associated results in laboratory animals (including mice, rats and dogs) and reported few cumulative or long-term toxicities across species. They mentioned that there was no indication that a higher incidence of tumors occurred. From all long-term experiments it is evident that the closely-related substances niclosamide ethanolamine salt and niclosamide are not high-risk substances (44). To date, at least three clinical trials for niclosamide in prostate and colon cancers are registered, due to cumulative encouraging laboratory results. There is no reported early termination due to unexpected/unacceptable adverse effects. It is anticipated that if the combination of niclosamide and chemotherapeutic drugs was used clinically it would offer two important advantages: A reduction in the dose requirement of chemotherapeutic drugs, and amelioration of the side effects caused by aggressive treatment with high doses of chemotherapy.
Supplementary Material
Acknowledgements
Esophageal adenocarcinoma cell line (BE3) was generously provided by Dr Chia-Jui Yen (Division of Hematology and Oncology, Department of Internal Medicine, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan, Taiwan) and esophageal squamous cell carcinoma cell lines (CE48T and CE81T) were kindly provided from Dr Jang-Ming Lee (Department of Surgery, National Taiwan University Hospital and National Taiwan University College of Medicine, Taipei, Taiwan). We thank the staff of the Second Core Lab, Department of Medical Research, National Taiwan University Hospital for technical support during the study.
Glossary
Abbreviations
- AML
acute myelogenous leukemia
- AR
androgen receptor
- EAC
esophageal adenocarcinoma
- EGFR
epidermal growth factor receptor
- ER
estrogen receptor
- ESCC
esophageal squamous cell carcinoma
- 5-FU
5-fluorouracil
- FDA
Food and Drug Administration
- IC50
half maximal inhibitory concentration
- IL6
interleukin 6
- LRP6
lipoprotein receptor-related protein 6
- mTOR
mammalian target of rapamycin
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- NF-κB
nuclear factor-κB
- PI
propidium iodide
- PI3K
phosphoinositide 3 kinase
- ROS
reactive oxygen species
- STAT3
signal transducer and activator of transcription 3
- WNT
wingless and int-1
Funding
The present study was financially supported partly from NTUH grants i) NTUH 99S-1309, ii) NTUH 101-S1867, iii) 107-14, and iv) 108-13, and partly from the National Science Council of the Republic of China grants i) NSC 98-2314-B-002-091-MY3 and ii) NSC 101-2320-B-002-010-MY3). We also thank Dr Jau-Min Wong, Dr Mei-Lin Wu and ‘Liver Disease Prevention & Treatment Research Foundation, Taiwan’ for partial funding support.
Availability of data and materials
All data used during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MCL and BRL conceived and designed the experiments. MCL and YJH performed the experiments. MCL, BRL and YKC collected and analyzed the data, and interpreted the findings. MCL wrote the manuscript. BRL and YKC edited the manuscript. All authors have read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
There authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wang K, Johnson A, Ali SM, Klempner SJ, Bekaii-Saab T, Vacirca JL, Khaira D, Yelensky R, Chmielecki J, Elvin JA, et al. Comprehensive genomic profiling of advanced esophageal squamous cell carcinomas and esophageal adenocarcinomas reveals similarities and differences. Oncologist. 2015;20:1132–1139. doi: 10.1634/theoncologist.2015-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu B, Bo Y, Wang K, Liu Y, Tang X, Zhao Y, Zhao E, Yuan L. Concurrent neoadjuvant chemoradiotherapy could improve survival outcomes for patients with esophageal cancer: A meta-analysis based on random clinical trials. Oncotarget. 2017;8:20410–20417. doi: 10.18632/oncotarget.14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayat Mokhtari R, Homayouni TS, Baluch N, Morgatskaya E, Kumar S, Das B, Yeger H. Combination therapy in combating cancer. Oncotarget. 2017;8:38022–38043. doi: 10.18632/oncotarget.16723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Huang WT, Wu HY, He RQ, Ma J, Liu AG, Chen G. Novel drug candidate for the treatment of several softtissue sarcoma histologic subtypes: A computational method using survivalassociated gene signatures for drug repurposing. Oncol Rep. 2019;41:2241–2253. doi: 10.3892/or.2019.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun W, Sanderson PE, Zheng W. Drug combination therapy increases successful drug repositioning. Drug Discov Today. 2016;21:1189–1195. doi: 10.1016/j.drudis.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan JX, Ding K, Wang CY. Niclosamide, an old antihelminthic agent, demonstrates antitumor activity by blocking multiple signaling pathways of cancer stem cells. Chin J Cancer. 2012;31:178–184. doi: 10.5732/cjc.011.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satoh K, Zhang L, Zhang Y, Chelluri R, Boufraqech M, Nilubol N, Patel D, Shen M, Kebebew E. Identification of niclosamide as a novel anticancer agent for adrenocortical carcinoma. Clin Cancer Res. 2016;22:3458–3466. doi: 10.1158/1078-0432.CCR-15-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu L, Dong J, Wang L, Xia Q, Zhang D, Kim H, Yin T, Fan S, Shen Q. Activation of STAT3 and Bcl-2 and reduction of reactive oxygen species (ROS) promote radioresistance in breast cancer and overcome of radioresistance with niclosamide. Oncogene. 2018;37:5292–5304. doi: 10.1038/s41388-018-0340-y. [DOI] [PubMed] [Google Scholar]
- 10.Ye T, Xiong Y, Yan Y, Xia Y, Song X, Liu L, Li D, Wang N, Zhang L, Zhu Y, et al. The anthelmintic drug niclosamide induces apoptosis, impairs metastasis and reduces immunosuppressive cells in breast cancer model. PLoS One. 2014;9:e85887. doi: 10.1371/journal.pone.0085887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang YC, Chao TK, Chang CC, Yo YT, Yu MH, Lai HC. Drug screening identifies niclosamide as an inhibitor of breast cancer stem-like cells. PLoS One. 2013;8:e74538. doi: 10.1371/journal.pone.0074538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suliman MA, Zhang Z, Na H, Ribeiro AL, Zhang Y, Niang B, Hamid AS, Zhang H, Xu L, Zuo Y. Niclosamide inhibits colon cancer progression through downregulation of the Notch pathway and upregulation of the tumor suppressor miR-200 family. Int J Mol Med. 2016;38:776–784. doi: 10.3892/ijmm.2016.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Ren XR, Piao H, Zhao S, Osada T, Premont RT, Mook RA, Jr, Morse MA, Lyerly HK, Chen W. Niclosamide-induced Wnt signaling inhibition in colorectal cancer is mediated by autophagy. Biochem J. 2019;476:535–546. doi: 10.1042/BCJ20180385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Wang L, Shen H, Lin H, Li D. Anthelminthic drug niclosamide sensitizes the responsiveness of cervical cancer cells to paclitaxel via oxidative stress-mediated mTOR inhibition. Biochem Biophys Res Commun. 2017;484:416–421. doi: 10.1016/j.bbrc.2017.01.140. [DOI] [PubMed] [Google Scholar]
- 15.Cheng B, Morales LD, Zhang Y, Mito S, Tsin A. Niclosamide induces protein ubiquitination and inhibits multiple pro-survival signaling pathways in the human glioblastoma U-87 MG cell line. PLoS One. 2017;12:e0184324. doi: 10.1371/journal.pone.0184324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Zhou X, Xu H, Shi X, Zhao J, Yang M, Zhang L, Jin X, Hu Y, Li X, et al. Niclosamide inhibits cell growth and enhances drug sensitivity of hepatocellular carcinoma cells via STAT3 signaling pathway. J Cancer. 2018;9:4150–4155. doi: 10.7150/jca.26948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomizawa M, Shinozaki F, Motoyoshi Y, Sugiyama T, Yamamoto S, Sueishi M, Yoshida T. Niclosamide suppresses Hepatoma cell proliferation via the Wnt pathway. Onco Targets Ther. 2013;6:1685–1693. doi: 10.2147/OTT.S50065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Z, Li Q, Wang Y, Wang L, Li X, Ge N, Wang Y, Guo C. Niclosamide induces cell cycle arrest in G1 phase in head and neck squamous cell carcinoma through Let-7d/CDC34 axis. Front Pharmacol. 2019;9:1544. doi: 10.3389/fphar.2018.01544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R, You S, Hu Z, Chen ZG, Sica GL, Khuri FR, Curran WJ, Shin DM, Deng X. Inhibition of STAT3 by niclosamide synergizes with erlotinib against head and neck cancer. PLoS One. 2013;8:e74670. doi: 10.1371/journal.pone.0074670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MO, Choe MH, Yoon YN, Ahn J, Yoo M, Jung KY, An S, Hwang SG, Oh JS, Kim JS. Antihelminthic drug niclosamide inhibits CIP2A and reactivates tumor suppressor protein phosphatase 2A in non-small cell lung cancer cells. Biochem Pharmacol. 2017;144:78–89. doi: 10.1016/j.bcp.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Li R, Hu Z, Sun SY, Chen ZG, Owonikoko TK, Sica GL, Ramalingam SS, Curran WJ, Khuri FR, Deng X. Niclosamide overcomes acquired resistance to erlotinib through suppression of STAT3 in non-small cell lung cancer. Mol Cancer Ther. 2013;12:2200–2212. doi: 10.1158/1535-7163.MCT-13-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Li Y, Lv C, Wang L, Song H. Anthelmintic drug niclosamide enhances the sensitivity of chronic myeloid leukemia cells to dasatinib through inhibiting Erk/Mnk1/eIF4E pathway. Biochem Biophys Res Commun. 2016;478:893–899. doi: 10.1016/j.bbrc.2016.08.047. [DOI] [PubMed] [Google Scholar]
- 23.Jin Y, Lu Z, Ding K, Li J, Du X, Chen C, Sun X, Wu Y, Zhou J, Pan J. Antineoplastic mechanisms of niclosamide in acute myelogenous leukemia stem cells: Inactivation of the NF-kappaB pathway and generation of reactive oxygen species. Cancer Res. 2010;70:2516–2527. doi: 10.1158/0008-5472.CAN-09-3950. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Li H, Zhan D, Xiang M, Yang J, Zuo Y, Yu Y, Zhou H, Jiang D, Luo H, et al. Niclosamide sensitizes nasopharyngeal carcinoma to radiation by downregulating Ku70/80 expression. J Cancer. 2018;9:736–744. doi: 10.7150/jca.20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Z, Nan G, Yan Z, Zeng L, Deng Y, Ye J, Zhang Z, Qiao M, Li R, Denduluri S, et al. The anthelmintic drug niclosamide inhibits the proliferative activity of human osteosarcoma cells by targeting multiple signal pathways. Curr Cancer Drug Targets. 2015;15:726–738. doi: 10.2174/1568009615666150629132157. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Ding R, Han Z, Ma Z, Wang Y. Targeting of cell cycle and let-7a/STAT3 pathway by niclosamide inhibits proliferation, migration and invasion in oral squamous cell carcinoma cells. Biomed Pharmacother. 2017;96:434–442. doi: 10.1016/j.biopha.2017.09.149. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Yang Z, Han Z, Wen Y, Ma Z, Wang Y. Niclosamide acts as a new inhibitor of vasculogenic mimicry in oral cancer through upregulation of miR-124 and downregulation of STAT3. Oncol Rep. 2018;39:827–833. doi: 10.3892/or.2017.6146. [DOI] [PubMed] [Google Scholar]
- 28.Arend RC, Londono-Joshi AI, Gangrade A, Katre AA, Kurpad C, Li Y, Samant RS, Li PK, Landen CN, Yang ES, et al. Niclosamide and its analogs are potent inhibitors of Wnt/β-catenin, mTOR and STAT3 signaling in ovarian cancer. Oncotarget. 2016;7:86803–86815. doi: 10.18632/oncotarget.13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yo YT, Lin YW, Wang YC, Balch C, Huang RL, Chan MW, Sytwu HK, Chen CK, Chang CC, Nephew KP, et al. Growth inhibition of ovarian tumor-initiating cells by niclosamide. Mol Cancer Ther. 2012;11:1703–1712. doi: 10.1158/1535-7163.MCT-12-0002. [DOI] [PubMed] [Google Scholar]
- 30.Arend RC, Londoño-Joshi AI, Samant RS, Li Y, Conner M, Hidalgo B, Alvarez RD, Landen CN, Straughn JM, Buchsbaum DJ. Inhibition of Wnt/β-catenin pathway by niclosamide: A therapeutic target for ovarian cancer. Gynecol Oncol. 2014;134:112–120. doi: 10.1016/j.ygyno.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Liu C, Lou W, Armstrong C, Zhu Y, Evans CP, Gao AC. Niclosamide suppresses cell migration and invasion in enzalutamide resistant prostate cancer cells via Stat3-AR axis inhibition. Prostate. 2015;75:1341–1353. doi: 10.1002/pros.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C, Armstrong CM, Lou W, Lombard AP, Cucchiara V, Gu X, Yang JC, Nadiminty N, Pan CX, Evans CP, Gao AC. Niclosamide and bicalutamide combination treatment overcomes enzalutamide- and bicalutamide-resistant prostate cancer. Mol Cancer Ther. 2017;16:1521–1530. doi: 10.1158/1535-7163.MCT-16-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer MT, Haugk K, McKiernan JS, Gulati R, Cheng HH, Maes JL, Dumpit RF, Nelson PS, Montgomery B, McCune JS, et al. A phase I study of niclosamide in combination with enzalutamide in men with castration-resistant prostate cancer. PLoS One. 2018;13:e0198389. doi: 10.1371/journal.pone.0202709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X, Liu F, Zeng L, He F, Zhang R, Yan S, Zeng Z, Shu Y, Zhao C, Wu X, et al. Niclosamide exhibits potent anticancer activity and synergizes with sorafenib in human renal cell cancer cells. Cell Physiol Biochem. 2018;47:957–971. doi: 10.1159/000490140. [DOI] [PubMed] [Google Scholar]
- 35.Yu K, Wang T, Li Y, Wang C, Wang X, Zhang M, Xie Y, Li S, An Z, Ye T. Niclosamide induces apoptosis through mitochondrial intrinsic pathway and inhibits migration and invasion in human thyroid cancer in vitro. Biomed Pharmacother. 2017;92:403–411. doi: 10.1016/j.biopha.2017.05.097. [DOI] [PubMed] [Google Scholar]
- 36.Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- 37.Furtek SL, Matheson CJ, Backos DS, Reigan P. Evaluation of quantitative assays for the identification of direct signal transducer and activator of transcription 3 (STAT3) inhibitors. Oncotarget. 2016;7:77998–78008. doi: 10.18632/oncotarget.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen CJ, Izzo JG, Lee DF, Guha S, Wei Y, Wu TT, Chen CT, Kuo HP, Hsu JM, Sun HL, et al. Bile acid exposure up-regulates tuberous sclerosis complex 1/mammalian target of rapamycin pathway in Barrett's-associated esophageal adenocarcinoma. Cancer Res. 2008;68:2632–2640. doi: 10.1158/0008-5472.CAN-07-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang PW, Hung MC, Hsieh CY, Tung EC, Wang YH, Tsai JC, Lee JM. The effects of Photofrin-mediated photodynamic therapy on the modulation of EGFR in esophageal squamous cell carcinoma cells. Lasers Med Sci. 2013;28:605–614. doi: 10.1007/s10103-012-1119-y. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C, Ma Q, Shi Y, Li X, Wang M, Wang J, Ge J, Chen Z, Wang Z, Jiang H. A novel 5-fluorouracil-resistant human esophageal squamous cell carcinoma cell line Eca-109/5-FU with significant drug resistance-related characteristics. Oncol Rep. 2017;37:2942–2954. doi: 10.3892/or.2017.5539. [DOI] [PubMed] [Google Scholar]
- 42.Wu J, Wang L, Du X, Sun Q, Wang Y, Li M, Zang W, Liu K, Zhao G. α-solanine enhances the chemosensitivity of esophageal cancer cells by inducing microRNA-138 expression. Oncol Rep. 2018;39:1163–1172. doi: 10.3892/or.2018.6187. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Wu X, Zhang F, Mo S, Lu Y, Wei W, Chen X, Lan L, Lu B, Liu Y. Paclitaxel induces apoptosis of esophageal squamous cell carcinoma cells by downregulating STAT3 phosphorylation at Ser727. Oncol Rep. 2017;37:2237–2244. doi: 10.3892/or.2017.5503. [DOI] [PubMed] [Google Scholar]
- 44.Andrews P, Thyssen J, Lorke D. The biology and toxicology of molluscicides, bayluscide. Pharmacol Ther. 1982;19:245–295. doi: 10.1016/0163-7258(82)90064-X. [DOI] [PubMed] [Google Scholar]
- 45.Wieland A, Trageser D, Gogolok S, Reinartz R, Höfer H, Keller M, Leinhaas A, Schelle R, Normann S, Klaas L, et al. Anticancer effects of niclosamide in human glioblastoma. Clin Cancer Res. 2013;19:4124–4136. doi: 10.1158/1078-0432.CCR-12-2895. [DOI] [PubMed] [Google Scholar]
- 46.Huang C, Wang L, Yang X, Lai L, Chen D, Duan C. Expression of activated signal transducer and activator of transcription-3 as a predictive and prognostic marker in advanced esophageal squamous cell carcinoma. World J Surg Oncol. 2015;13:314. doi: 10.1186/s12957-015-0726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Zhu S, Shen M, Liu J, Wang M, Li C, Wang Y, Deng A, Mei Q. STAT3 is involved in esophageal carcinogenesis through regulation of Oct-1. Carcinogenesis. 2013;34:678–688. doi: 10.1093/carcin/bgs361. [DOI] [PubMed] [Google Scholar]
- 48.Timme S, Ihde S, Fichter CD, Waehle V, Bogatyreva L, Atanasov K, Kohler I, Schöpflin A, Geddert H, Faller G, et al. STAT3 expression, activity and functional consequences of STAT3 inhibition in esophageal squamous cell carcinomas and Barrett's adenocarcinomas. Oncogene. 2014;33:3256–3266. doi: 10.1038/onc.2013.298. [DOI] [PubMed] [Google Scholar]
- 49.Yoshikawa T, Miyamoto M, Aoyama T, Soyama H, Goto T, Hirata J, Suzuki A, Nagaoka I, Tsuda H, Furuya K, Takano M. JAK2/STAT3 pathway as a therapeutic target in ovarian cancers. Oncol Lett. 2018;15:5772–5780. doi: 10.3892/ol.2018.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon MK, Mitrea DM, Ou L, Kriwacki RW. Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochem Soc Trans. 2012;40:981–988. doi: 10.1042/BST20120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang M, Chen Z, Yang D, Li H, Zuo Y, Li J, Zhang W, Zhou H, Jiang D, Xu Z, Yu Z. Niclosamide enhances the antitumor effects of radiation by inhibiting the hypoxia-inducible factor-1α/vascular endothelial growth factor signaling pathway in human lung cancer cells. Oncol Lett. 2017;14:1933–1938. doi: 10.3892/ol.2017.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuo Y, Yang D, Yu Y, Xiang M, Li H, Yang J, Li J, Jiang D, Zhou H, Xu Z, Yu Z. Niclosamide enhances the cytotoxic effect of cisplatin in cisplatin-resistant human lung cancer cells via suppression of lung resistance-related protein and c-myc. Mol Med Rep. 2018;17:3497–3502. doi: 10.3892/mmr.2017.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi L, Zheng H, Hu W, Zhou B, Dai X, Zhang Y, Liu Z, Wu X, Zhao C, Liang G. Niclosamide inhibition of STAT3 synergizes with erlotinib in human colon cancer. Onco Targets Ther. 2017;10:1767–1776. doi: 10.2147/OTT.S129449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used during the current study are available from the corresponding author on reasonable request.