Abstract
There are no reliable criteria to assess risk of progression of non-muscle invasive bladder cancer to muscle invasive bladder cancer. The aim of the present study was to identify potential markers based on gene expression profiling to improve predictive power of disease progression and prognosis in patients with bladder cancer. In the present study, we screened seventy-three differentially expressed genes by analyzing bladder cancer samples with or without progression. Forty-seven prognosis-related genes were screened, 13 of which were identified to build a progression-associated gene signature using the LASSO regression method. Based on this 13-mRNA signature, patients were divided into high- and low-risk groups, with different prognostic outcomes. The gene signature was an independent prognostic factor for overall survival. Receiver operating characteristic analysis suggested that the signature performed well in the validation cohort and its predictive power outperformed other several published signatures. CTHRC1, MMP11, AEBP1, SNCAIP, COL1A1 and S100A8 were identified as hub genes and their expression levels were detected using reverse transcriptase-quantitative polymerase chain reaction. The expression of CTHRC1 was elevated in aggressive bladder cancer compared with non-invasive type, which suggests CTHRC1 may be a valuable biomarker for prediction of prognosis and progression of bladder cancer. Collectively, this 13-mRNA signature may be useful in predicting disease progression and prognosis, thereby contributing to individualized management of patients with bladder cancer.
Keywords: bladder cancer, mRNA signature, progression, prognosis, gene set enrichment analysis, nomogram
Introduction
Bladder cancer is the tenth most commonly diagnosed carcinoma, with an estimated 549,000 new cases and 200,000 deaths reported globally in 2018 (1). Approximately 75% newly diagnosed cases present with disease confined to the mucosa, namely non-muscle invasive bladder cancer (NMIBC), with a favorable long-term survival but a higher relapse rate after transurethral resection of bladder tumor (TURBT) and intravesical therapy. However, 10–15% of patients with NMIBC still suffer from disease progression to the deadlier muscle invasive bladder cancer (MIBC), the 5-year survival of which is often <50% (2–4). The undesirable outcome of bladder cancer is partly due to inadequate knowledge of the biological mechanisms of disease recurrence and progression. Thus, identification and development of novel molecular biomarkers based on genomic profiling are needed for predicting disease progression and improving prognosis of patients diagnosed as bladder cancer at a very early stage.
Rapidly emerging high-throughput sequencing technology has revealed genomic profiling and epigenetic alternations of diseases, which have provided significant insight into the molecular characteristics of various human cancers. Since the progressive accumulation of epigenetic variations can drive the tumorigenesis and development of bladder cancer, a genetic disorder, a series of array-based gene signatures has been established to better predict the risk of recurrence and progression and distinguish the prognosis of patients with bladder cancer beyond clinicopathologic parameters (5–7). However, few studies have focused on disease progression and prognosis of patients with NMIBC simultaneously. Establishing related gene markers is of great significance for clinical decision-makers in diagnosis and treatment selection.
In the present study, we aimed to explore and establish a multigene signature for predicting the risk of muscle invasion and survival prognosis in patients with bladder cancer by analyzing the transcriptome profiles using the Gene Expression Omnibus (GEO) data. A robust 13-mRNA signature was constructed with the use of univariate Cox regression analysis and the Least Absolute Shrinkage and Selection Operator logistic regression method (LASSO). The diagnostic and prognostic value of this mRNA-based risk score model was further validated using an independent microarray of GEO as well as The Cancer Genome Atlas (TCGA) cohort. In addition, to better assess its clinical significance, we also constructed a predictive nomogram with other clinicopathologic factors and compared the performance between the 13-gene model and other published biomarkers regarding prediction of progression to MIBC. Gene set enrichment analysis (GSEA) was later performed to identify underlying biological functions and molecular mechanisms associated with the occurrence and development of bladder cancer. Finally, hub genes identified from the co-expression network of the 13 genes were detected using experimental reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR), and we found that CTHRC1 may be a novel potential biomarker in the prediction of the progression and survival of patients with bladder cancer.
Materials and methods
Data collection
The mRNA expression profile matrix files of GSE13507 and GSE120736 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE13507 performed on the platform of Illumina human-6 v2.0 expression beadchip contained 68 normal bladder tissues and 188 bladder cancer tissues (8). GSE120736 calculated on the platform of Illumina HumanHT-12 V4.0 expression beadchip contained 145 bladder cancer tissues. Only progression-free and recurrence-free primary NMIBC and those with NMIBC with progression were enrolled in our study; thus 98 samples from GSE13507 (67 non-progressive and 31 progressive) and 83 samples from GSE120736 (67 non-progressive and 16 progressive) were selected for further analysis.
The mRNA sequencing expression profile and clinical data of patients with bladder cancer were obtained from the Cancer Genome Atlas (TCGA) data portal (https://gdc-portal.nci.nih.gov/). The inclusion criteria were set as follows: i) Diagnosis of bladder cancer; ii) the samples were recorded with complete RNAseq data; iii) the samples were recorded with detailed clinicopathological data including pathological stage, grade, overall survival (OS), recurrence-free survival (RFS) and corresponding follow-up time information. In total, 279 cases of bladder cancer were included for further study. In addition, our research follows the access rules and publication guidelines of TCGA.
Preprocessing of microarray data and differentially expressed mRNA screening
Raw microarray datasets from GEO database were normalized using Robust Multichip Average (RMA) and transformed to Log2 pattern for processed signals. Probes were annotated by using the Affymetrix annotation files (9). Then, the differentially expressed genes (DEGs) between primary NMIBC and progressive NMIBC specimens from training set (GSE13507) were screened with the use of DESeq R package. When adjusted P-value <0.05 and |Log2 (FC)|>1, the genes were regarded as DEGs, the expression profiles of which are listed in Table SI.
Development and validation of the gene signature
Univariate Cox regression analysis was utilized to investigate the prognosis-related DEGs. The gene was considered significant when P-value <0.05, and 47 candidates qualified, through which the LASSO Cox regression was employed to build mRNAs signature model for prediction of disease progression and prognosis. The optimal value of the penalty parameter λ was determined via 10-times cross-validations. 13-mRNA model was then constructed based on the optimal λ value and the risk score for each patient was calculated according to the expression of genes and corresponding weighted coefficient. The samples in each cohort were classified into a high-risk group and low-risk group based on the optimum cut-off value determined using the X-tile software (version 3.6.1, Yale University).
Construction of predictive nomogram
The predictive nomogram based on mRNAs signature and clinical related factors was plotted using ‘rms’ package of R software (version 3.5.1). Calibration curve was plotted to assess the performance of the nomogram. In the calibration graph, nomogram-predicted progression and observed outcome were presented on the x-axis and y-axis, respectively; the 45-degree dotted line indicated the ideal prediction.
Gene set enrichment analysis (GSEA)
To identify 13 mRNAs-related biological processes and pathways, GSEA was performed using TCGA dataset including 279 patients with bladder cancer divided into high- and low-risk groups according to the cut-off value, and executed using GSEA software 3.0 from the Broad Institute (10). The Hallmark gene sets (h.all.v6.1.symbols.gmt) representing specific well-defined biological processes and the gene sets of canonical pathways (c2.all.v6.0.symbols.gmt) were obtained from the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). Gene set permutations were performed 1,000 times for each analysis to obtain normalized enrichment score (NES) used for sorting pathways enriched in each phenotype. A result was regarded as significant when nominal P-value <0.05 and false discovery rate (FDR)<0.2.
Proteins network construction
Analysis of 13 gene-related networks was performed using GeneMANIA (http://www.genemania.org/), a website-based database and tool for predicting interactions and functions of genes and gene sets on the basis of multiple networks (11). To screen hub genes in regulatory network, node degree and betweenness were considered for analysis of protein-protein interaction from a Cytoscape plugin. The interaction networks for the gene signature were rebuilt and visualized by Cytoscape (ver. 3.5.1).
Collection of clinical samples
Twelve NMIBC tissues and 12 MIBC tissues were collected from patients diagnosed as primary bladder cancer between September, 2017 and October, 2018 in the Department of Urology of the First Affiliated Hospital of Chongqing Medical University. Once we obtained the specimens, they were frozen and stored at −80°C until used for RNA extraction. However, another 49 paraffin sections of bladder cancer (including 34 MIBC and 15 NMIBC tissues) were obtained from the Department of Pathology of Chongqing Medical University (specimens were collected from January, 2015 to December, 2017) for detection of proteins by immunohistochemistry assay. Notably, there was no cross-over between the two groups of patients. The study was approved by the Ethics Committees of the First Affiliated Hospital of Chongqing Medical University and informed consent was obtained from each patient involved.
RNA isolation and reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from bladder cancer tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized using 1 µg of total RNA and the PrimeScript RT reagent kit (Takara). RT-qPCR was performed using SYBR-Green assay (Takara) and executed by ABI 7500 Real-Time PCR system (Applied Biosystems). Gene expression was normalized to β-actin. Primers used for mRNAs (Invitrogen; Thermo Fisher Scientific, Inc.,) are summarized in Table SII. Relative quantification values of mRNAs were calculated with the use of the 2−ΔΔCq method (12). In addition, bladder cancer cells were also used for the isolation of RNA for RT-qPCR.
Immunohistochemistry (IHC)
The protocol of immunohistochemical staining was as previously described (13). Briefly, sections of formalin-fixed paraffin-embedded (FFPE) were deparaffinized and rehydrated, immersed in sodium citrate buffer for antigen retrieval, incubated with 3% H2O2 to remove endogenous peroxidase activity, blocked with normal goat serum, incubated with anti-CTHRC1 (Abcam, ab85739) at 4°C overnight, incubated with biotinylated goat anti-mouse IgG and streptavidin-biotin-conjugated horseradish peroxidase (HRP) at 37°C. Then signals were visualized using a diaminobenzidine kit (ZSGB-BIO) and slides were counterstained with hematoxylin. The IHC scores were assessed by two experienced pathologists and calculated based on the staining intensity and extent; the score criterion was described specifically in previous study (13).
Cell culture
Human bladder cancer cell lines (5637 and TCCSUP) were purchased from the American Type Culture Collection (ATCC). The 5637 and TCCSUP cells were cultured in RPMI-1640 medium (Corning) with 10% fetal bovine serum (FBS), 100 mg/ml penicillin and 100 mg/ml streptomycin at 37°C in 5% CO2.
Cell transfection
Transfections were performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.,) according to the manufacturer's protocol. The 5637 and TCCSUP cells were seeded in 6-well plates at density of 1×106 cells/well and cultured at 37°C with 5% CO2 for 24 h. Cells were transfected with CTHRC1 siRNAs (siRNA-1: Sense, 5′-GCCAAUGGCAUUCCGGGUATT-3′; Antisense, 5′-UACCCGGAAUGCCAUUGGCTT-3′. siRNA-2: Sense, 5′-CCUCUUCCCAUUGAAGCUATT-3′; Antisense, 5′-UAGCUUCAAUGGGAACAGGTT-3′.) or negative control siRNA (Sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; Antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) (GenePharma Inc.) at a concentration of 50 nM for 4 h. After 48 h, the treated cells were collected for subsequent experiments.
Cell migration assay
Cell migration assay was detected using Transwell chambers (Corning Inc.). After transfection with siRNA for 48 h, 5637 and TCCSUP cells were digested with trypsin and suspended (5×105 cells/ml) with serum-free RPMI-1640 medium. Cell suspension (200 µl) was added to the upper chamber, and 700 µl RPMI-1640 medium containing 10% FBS was added to the lower chamber. After 48 h, the migrated cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 15 min each at room temperature. Subsequently, the cells from five random fields were counted under a microscope (×100 magnification; Leica Microsystems GmbH).
Statistical analysis
Statistical analyses were carried out using R software (version 3.5.1), SPSS software (version 22; SPSS Inc.) and Graphpad 5.0. LASSO Cox regression analysis was conducted in the discovery dataset using the ‘glmnet’ package. Comparisons between the two groups were examined using the two-tailed Student's t-test. The prediction accuracy of the model was analyzed by receiver operating characteristic (ROC) or time-dependent ROC and evaluated by area under curve (AUC). The survival difference between high- and low-risk groups was assessed by the Kaplan-Meier curve and compared by the log-rank test. The prognosis significance of mRNA-based signature was analyzed by univariate and multivariate Cox proportional hazard regression model. One-way analysis of variance (ANOVA) was used for comparisons among multiple groups, followed by the Newman-Keuls post hoc test. P<0.05 was regarded as statistically significant.
Results
Preparation of bladder cancer datasets
Two gene expression datasets (GSE13507 and GSE120736) with disease progression information about patients with NMIBC were screened for establishing and validation of the model after a thorough search of the GEO database. A total of 279 cases of bladder cancer samples with complete survival data were downloaded from TCGA database for prognostic verification. The baseline data of these cohorts are summarized in Table I.
Table I.
Summary of baseline characteristics of patients with bladder cancer in the three datasets.
| Characteristic | GSE13507 (n=98) | GSE120736 (n=83) | TCGA (n=279) |
|---|---|---|---|
| Age | |||
| ≤65 | 42 | N/A | 171 |
| >65 | 56 | N/A | 108 |
| Sex | |||
| Male | 82 | 71 | 213 |
| Female | 16 | 12 | 66 |
| Grade | |||
| Low | 69 | 35 | 15 |
| High | 29 | 48 | 262 |
| Unkown | 0 | 0 | 2 |
| Tumor stage | |||
| Ta | 20 | 34 | 2 |
| T1 | 58 | 33 | 2 |
| T2 | 7 | 5 | 107 |
| T3–4 | 13 | 11 | 164 |
| Tx | 0 | 0 | 4 |
| Lymph node stage | |||
| N0 | 94 | N/A | 202 |
| N1–3 | 3 | N/A | 75 |
| Nx | 1 | N/A | 2 |
| Progression | |||
| No | 67 | 67 | N/A |
| Yes | 31 | 16 | N/A |
| Recurrence | |||
| No | 67 | 67 | 212 |
| Yes | 31 | 16 | 67 |
| Survival status | |||
| Alive | 54 | N/A | 194 |
| Deceased | 44 | N/A | 85 |
| Mean follow-up time (month) | 51.9 | N/A | 24.9 |
N/A, not applicable.
Development of a 13-mRNA-based classifier
A detailed flow-chart of the procedure of analysis is depicted in Fig. 1 to aid in better understanding the study. GSE13507 dataset (with 67 patients with confined NMIBC and 31 patients with progressive NMIBC) was used to construct the training cohort, the samples in which were divided into a non-progressive group and a progressive group. Seventy-three differentially expressed mRNAs were found between the two groups (Fig. 2A), including 24 downregulated mRNAs and 49 overexpressed mRNAs (Table SI). Among them we conducted univariate Cox regression to investigate the prognosis-related genes and identified 47 genes, which were significantly related to survival for further analysis (P<0.05) (Table SIII).
Figure 1.
Overall workflow diagram for the development and validation of prognosis-related mRNA signature.
Figure 2.
Identification and construction of NMIBC progression-related 13-mRNA classifier. (A) The heat map of the expression levels of 73 differentially expressed genes in the training dataset. (B) LASSO coefficient profiles of the 47 prognosis-associated mRNAs. Ten-time cross-validation was exhibited for tuning parameter selection and a vertical line was drawn at the value of the minimal λ used for establishment of 13-mRNA classifier. (C) The optimum cut-off value of the gene signature based risk scores was produced using the X-tile program in the GSE13507 training cohort. Red shows inverse association, while green indicates direct association between risk score and progression-free survival.
LASSO regression model was utilized to develop progression-related gene signatures and 13 of the 47 genes were selected in the training dataset (Fig. 2B). Risk score for each patient was accumulated based on the expression levels of the 13 mRNAs and corresponding weighted coefficients: Risk score=(0.17027* expression level of S100A8)+(0.06678* expression level of CTHRC1)+(0.10548* expression level of C15orf48)+(−0.16639* expression level of MPPED2)+(0.25600* expression level of CKAP2L)+(0.17159* expression level of SNCAIP)+(0.06363* expression level of SALL4)+(0.01503* expression level of MMP11)+(0.00057* expression level of COL1A1)+(0.22674* expression level of AEBP1)+(−0.08764* expression level of C10orf99)+(0.22416* expression level of KIF1A)+(−0.00367* expression level of ALDH1L1).
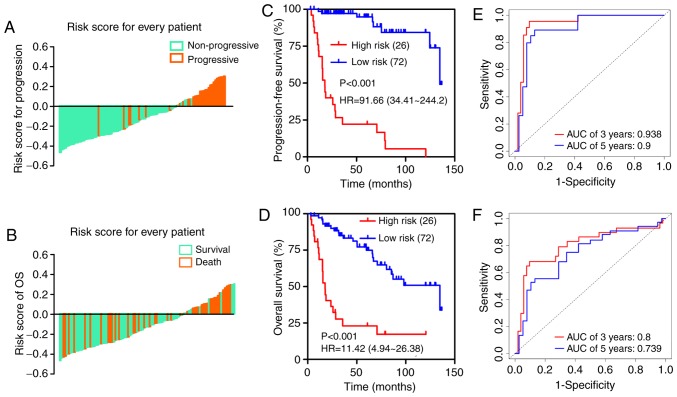
The optimum cut-off value of the mRNA classifier was determined as 3.3 according to the X-tile diagrams (Fig. 2C). We classified patients in the training dataset into high- and low-risk groups based on the established optimum cut-off point. The distribution of adjusted risk score (risk score minus cut-off value) for each patient is shown in Fig. 3A and B, which suggests that patients in the high-risk group (above the x-axis) had a higher possibility of disease progression and poorer survival than those in the low-risk group (below the x-axis). Kaplan-Meier survival curves demonstrated that the two groups had significantly different progression-free survival (PFS) time [hazard ratio (HR)=91.66, 95% confidence interval (CI)=34.41–244.2, P<0.001; Fig. 3C] and overall survival time (HR=11.42, 95% CI=4.94–26.38, P<0.001; Fig. 3D). Time-dependent ROC analysis indicated that the areas under the curve (AUCs) at 3- and 5-year were 0.938 and 0.9 for PFS (Fig. 3E) and 0.8 and 0.739 for OS (Fig. 3F), respectively.
Figure 3.
Analysis of the 13-mRNA signature in the training cohort. The distribution of each patient's adjusted risk score for (A) disease progression and (B) overall survival. Kaplan-Meier curves of (C) PFS and (D) OS in high- and low-risk groups. Time-dependent ROC curves based on the predictive model for (E) 3- and 5-year PFS and (F) OS probability.
Performance of the 13-mRNA signature in the validation dataset
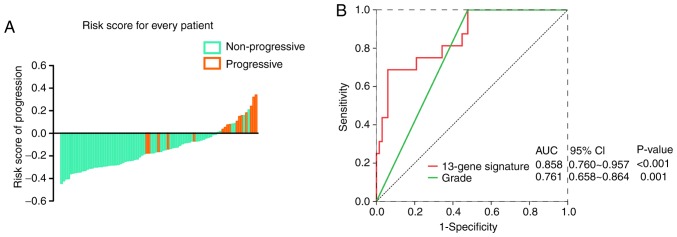
To further confirm the predictive value of the 13-mRNA signature in different datasets, we employed another qualified GEO microarray (GSE120736). A total of 83 patients in the validation cohort were divided into a high-risk group [n=16 (19.3%)] and a low-risk group [(n=67 (80.7%)] based on the same risk score model and cut-off point. Consistent with the findings described before, the proportion of disease progression of patients in the low-risk group was significantly lower than that of patients in the high-risk group (Fig. 4A). In Fig. 4B, The AUC in the ROC analyses for the 13-mRNA signature and grade were statistically significant (AUC=0.8581, 95% CI=0.760–0.957; P<0.001; and AUC=0.761, 95% CI=0.658–0.864; P=0.001, respectively).
Figure 4.
Performance of the 13-mRNA signature in an independent validation cohort. (A) The distribution of each patient's adjusted risk score for disease progression. (B) ROC analyses for the 13-mRNA signature and pathologic grade in the GSE120736 dataset.
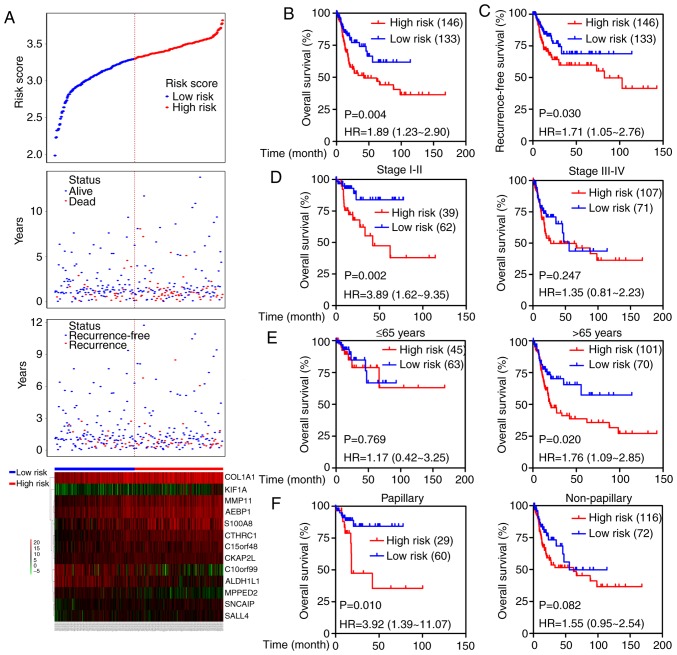
Additionally, the prognostic value of the 13-mRNA classifier was further tested in TCGA cohort. Based on the established cut-off point above, 146 (52.3%) patients were divided into the high-risk group, and 133 (47.7%) were in the low-risk group. The distribution of each patient's risk score, survival status, recurrence status, and expression profiles of the 13 genes were ranked and are shown in Fig. 5A. Kaplan-Meier curves showed that patients with high-risk scores had higher mortality and recurrence rates than patients with low-risk scores (OS: HR=1.89, 95% CI=1.23–2.90, P=0.004; RFS: HR=1.71, 95% CI=1.05–2.76, P=0.030; Fig. 5B and C). We next stratified the patients into different subgroups according to the American Joint Committee on Cancer (AJCC) stage (stage I–II versus stage III–IV), age (≤65 vs. >65 years) and tumor subtypes (papillary versus non-papillary). Prognosis of patients in the high-risk group was evidently worse than that in the low-risk group in stage I–II subgroup (HR=3.89; 95% CI=1.62–9.35, P=0.002) but not in the stage III–IV group (Fig. 5D). High-risk scores suggested a poor prognosis in the elderly subgroup (HR=1.76; 95% CI=1.09–2.85, P=0.020) but not in the young group (Fig. 5E), and predicted an unfavorable outcome in patients in the papillary subgroup (HR=3.92; 95% CI=1.39–11.07, P=0.010) but not in the non-papillary subgroup (Fig. 5F). Moreover, the prognostic value of the 13-mRNA signature combined with clinical factors was further analyzed by univariate and multivariate Cox regression. As shown in Table II, age, lymph-vascular invasion (LVI), and 13-mRNA-based risk score significantly correlated with OS even when adjusted for other covariates.
Figure 5.
Prognostic significance of the 13-mRNA signature in the TCGA cohort. (A) The distribution of risk score, survival status, recurrence status and the 13-mRNA expression profiles for each patient. Kaplan-Meier survival curves depicting (B) OS and (C) recurrence-free survival in high- and low-risk patients. Kaplan-Meier curves of OS in different subgroups stratified by (D) AJCC stage, (E) age and (F) tumor subtype.
Table II.
Univariate and multivariate Cox regression analysis for overall survival of patients with bladder cancer.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variates | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (≥65 vs. ≤65) | 3.23 (1.85–5.65) | <0.001c | 4.57 (1.72–12.11) | 0.002b |
| Sex (male vs. female) | 1.21 (0.72–2.05) | 0.465 | — | — |
| Subtype (non-papillary vs. papillary) | 1.60 (0.94–2.72) | 0.086 | — | — |
| LVI (yes vs. no) | 2.47 (1.45–4.23) | 0.001b | 2.45 (1.19–5.07) | 0.016a |
| pT stage (T3–4 vs. T1–2) | 1.89 (1.17–3.06) | 0.01a | — — | 0.909 |
| pN stage (N1–3 vs. N0) | 2.18 (1.41–3.37) | <0.001c | 1.08 (0.46–2.50) | 0.863 |
| pM stage (M1 vs. M0) | 4.12 (1.64–10.38) | 0.003b | 3.23 (0.38–27.46) | 0.283 |
| AJCC stage (III–IV vs. I–II) | 1.81 (1.10–2.96) | 0.019a | — — | 0.909 |
| 13-mRNA signature (high vs. low) | 3.40 (1.47–7.87) | 0.004b | 5.23 (1.14–24.06) | 0.034a |
The ‘—’ indicates that there is no information of patients in this cohort. The ‘——’ indicates that the value is not available.
P<0.05
P<0.01
P<0.001. HR, hazards ratio; CI, confidence interval; LVI, lymphovascular invasion.
Establishment of a nomogram to predict progression of NMIBC
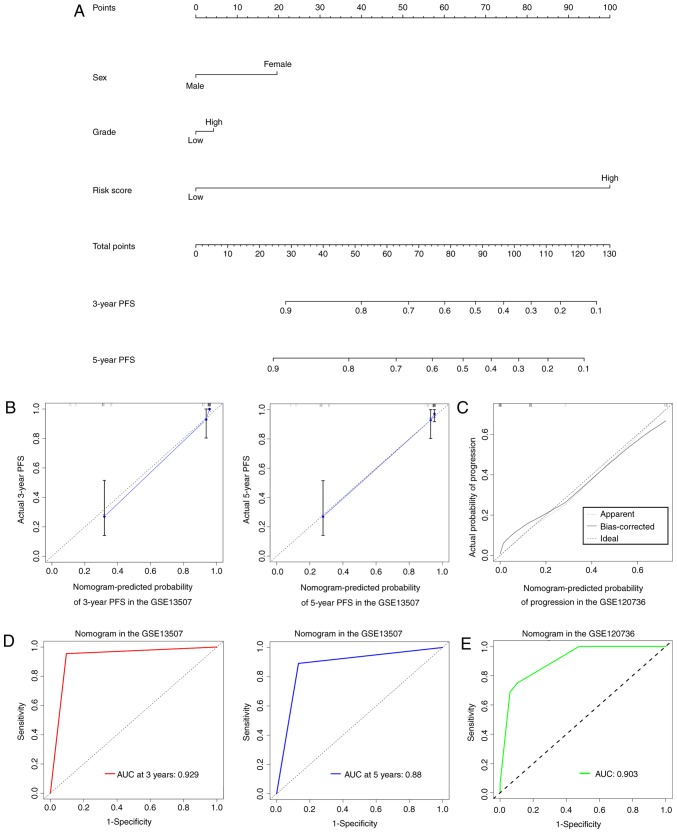
To provide a clinically practical method for clinicians to predict the risk of progression in patients with NMIBC, a nomogram was developed by integrating the 13-mRNA signature and other clinical factors including sex and grade that were available both in GSE13507 and GSE120736 datasets (Fig. 6A). In comparison to the ideal model, the calibration plots for 3- and 5-year PFS were predicted well in the training dataset (Fig. 6B), and the performance of the nomogram on prediction accuracy was also excellent in the validation dataset (Fig. 6C). Time-dependent ROC curves indicated that the AUCs at 3- and 5-year were 0.929 and 0.88 for the nomogram in the training dataset (Fig. 6D). ROC analysis showed a 0.903 of AUC for the nomogram in the validation dataset (Fig. 6E).
Figure 6.
The established nomogram to predict the risk of NMIBC progression. (A) The nomogram was generated in the training dataset, with gender, grade and 13-mRNA signature incorporated. (B) Calibration curves of the nomogram to predict PFS at 3- and 5-year in the training dataset. (C) Calibration curves of the nomogram in the validation dataset. The actual diagnosed disease progression is plotted on the y-axis; nomogram predicted probability is plotted on the x-axis. (D) Time-dependent ROC curves based on the nomogram for 3- and 5-year PFS in the training dataset. (E) ROC curves based on the nomogram in the validation dataset.
Comparison of the 13-mRNA signature with other progression-related biomarkers for NMIBC
Several studies have attempted to develop biomarkers for predicting disease progression in NMIBC during the past couple of years. We compared the diagnostic efficacy of the 13-gene classifier to that of other published biomarkers in the GSE13507 and GSE120736 cohorts to comprehensively evaluate the model's practical applicability. The published biomarkers included mRNA signatures and single genes, including a 5-gene signature to predict progression in T1G3 bladder cancer (14), a prognostic model based on a 12-gene progression score (15), and FOXM1 (16), HYAL-1 (17) and STAG2 (18) that were used for prediction of recurrence or progression in NMIBC.
The AUC and P-value for each possible signature or biomarker were calculated using the corresponding weighted formula algorithms. As shown in Fig. 7 and Table III, the 13-mRNA signature had a favorable performance in the two independent datasets, with an AUC of 0.905 in GSE13507 and 0.857 in GSE120736. Similar performance was observed in the 12-gene progression score model, with an AUC of 0.827 in GSE13507 and 0.902 in GSE120736. However, the 5-gene classifier exhibited inadequate performance, with an AUC of 0.689 in GSE13507 and 0.811 in GSE120736. In terms of the predictive value of several independent biomarkers in NMIBC progression, the diagnostic performance of FOXM1 was better than that of HYAL-1 and STAG2 in the two cohorts.
Figure 7.
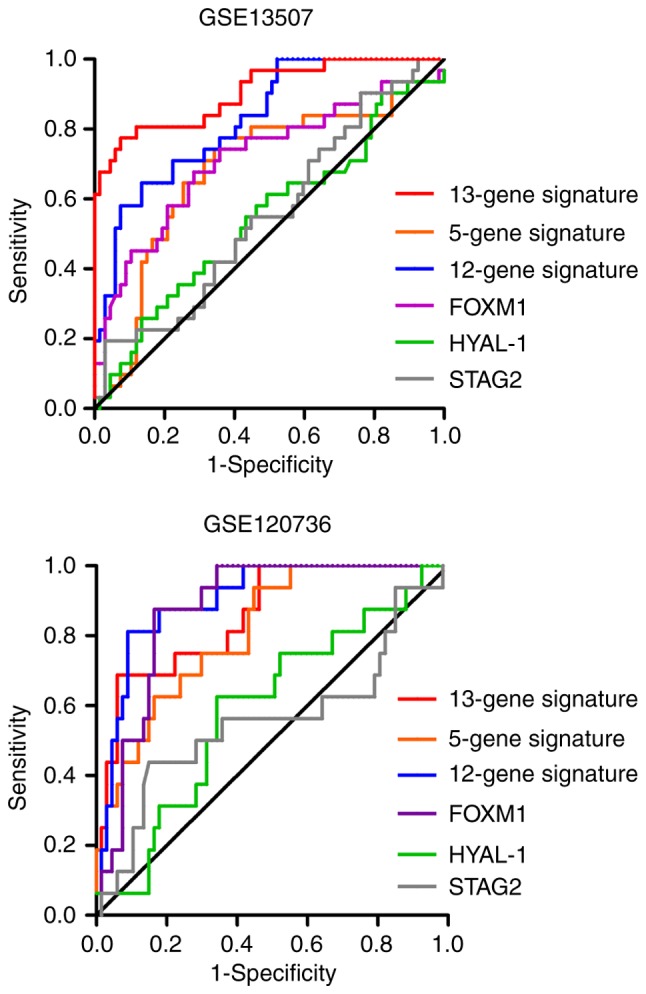
ROC analysis of 13-mRNA signature and other published single gene or multi-gene models in GSE13507 and GSE120736 datasets. Integration and comparison the differences of areas under curve in the six predictive biomarkers.
Table III.
AUCs of the signatures and biomarkers in the training and validation datasets.
| GSE13507 | GSE120736 | |||
|---|---|---|---|---|
| Approaches | AUC | P-values | AUC | P-values |
| 13-gene signature | 0.905 | P<0.0001 | 0.857 | P<0.0001 |
| 5-gene signature | 0.689 | P=0.003 | 0.811 | P=0.0001 |
| 12-gene signature | 0.827 | P<0.0001 | 0.902 | P<0.0001 |
| FOXM1 | 0.717 | P=0.001 | 0.874 | P<0.0001 |
| HYAL-1 | 0.545 | P=0.475 | 0.593 | P=0.2484 |
| STAG2 | 0.556 | P=0.378 | 0.557 | P=0.4778 |
Analysis of biological processes and signaling pathways
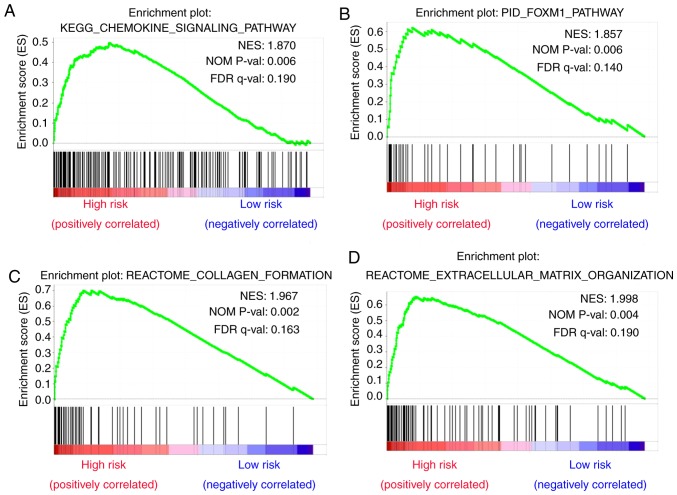
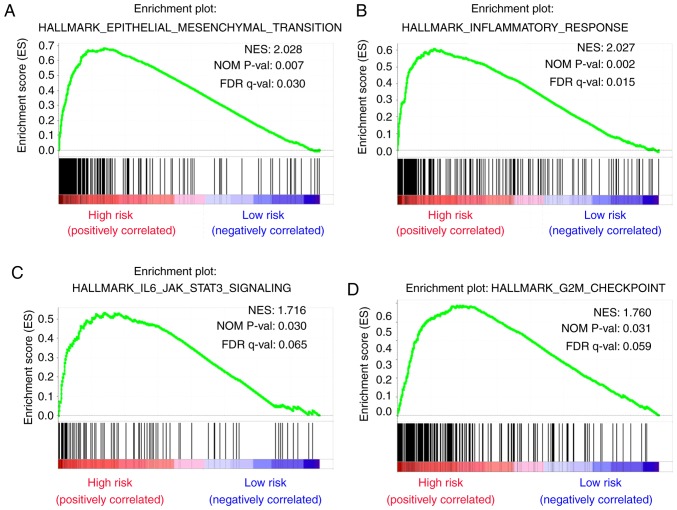
To identify dysregulated biological states and signaling pathways involved in the development of bladder cancer, GSEA was conducted with the use of 279 cases of subjects from TCGA cohort that were classified into a high-risk group [n=146 (52.3%)] and a low-risk group [n=133 (47.7%)] based on the established cut-off value. Gene sets were considered significantly enriched on the basis of NES, nominal P-value and FDR q-value. GSEA results showed that several canonical pathways, such as ‘chemokine signaling pathway’, ‘foxm1 pathway’, ‘collagen formation’ and ‘extracellular matrix organization’ that are involved in extracellular signal transduction, were highly enriched in the high-risk phenotype (Fig. 8A-D). Additionally, gene sets of ‘Hallmark’ involving ‘epithelial mesenchymal transition’, ‘inflammatory response’, ‘IL6-JAK-STAT3 signaling’ and ‘G2M checkpoint’ that are associated with biological processes of cancers were also enriched in the high-risk group (Fig. 9A-D).
Figure 8.
Gene sets of canonical pathways and Hallmark that are associated with risk score were performed by GSEA. Results of GSEA for gene sets of canonical pathways showing that (A) chemokine signaling pathway, (B) FOXM1 pathway, (C) collagen formation and (D) extracellular matrix organization were highly enriched in the high-risk group, with nominal P-value <0.05 and FDR<0.2.
Figure 9.
Gene sets of Hallmark that are associated with risk score were performed by GSEA. Results of GSEA for gene sets of Hallmark showing that (A) epithelial mesenchymal transition, (B) inflammatory response, (C) IL6-JAK-STAT3 SIGNALING and (D) G2M checkpoint were highly enriched in high-risk group, with nominal P-value <0.05 and FDR<0.2.
Identification and experimental validation of hub genes in the 13-mRNA signature
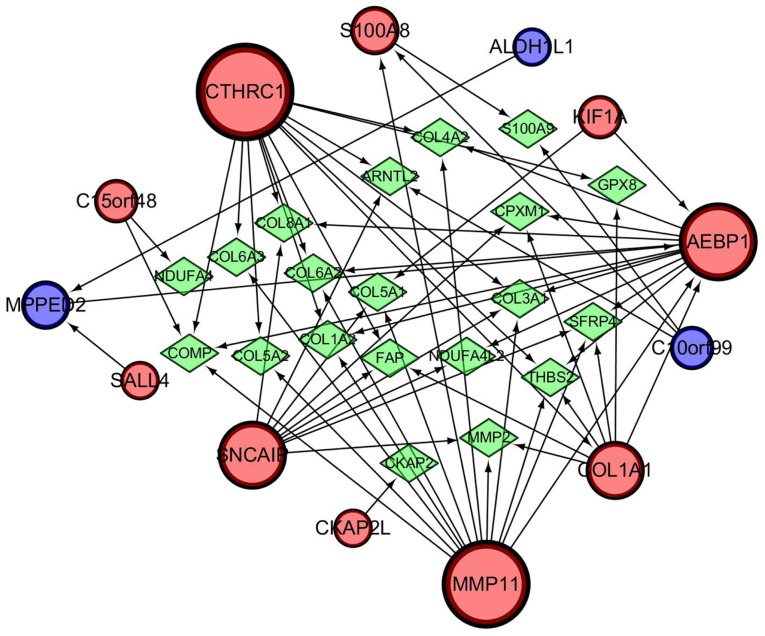
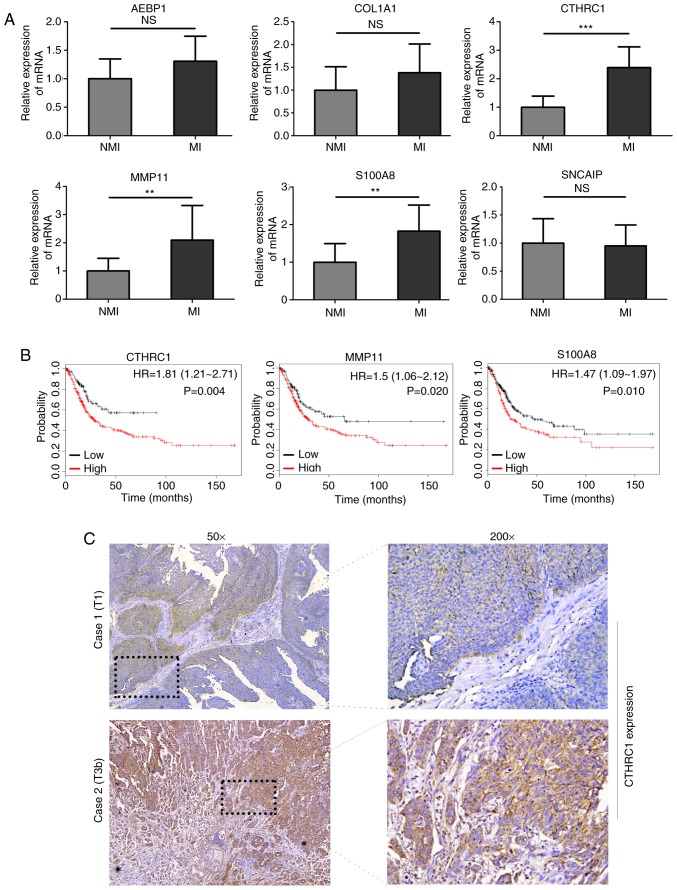
A gene regulatory network was established by GeneMANIA and visualized using Cytoscape software (ver. 3.5.1) to determine the interrelationships between the 13 genes related to progression and prognosis. The interaction network consisted of 33 genes, including 13 identified target genes and 20 additional genes spontaneously pulled by GeneMANIA (Fig. 10). The hub genes identified with the top six-node degree and betweenness were CTHRC1, MMP11, AEBP1, SNCAIP, COL1A1, and S100A8. We then measured the difference in expression levels of the six hub genes between NMIBC and MIBC tissues using RT-qPCR and found that the levels of expression of CTHRC1, S100A8, and MMP11 in MIBC were higher than that in NMIBC (Fig. 11A). A large TCGA cohort containing 404 patients with bladder cancer in Kaplan-Meier plotter database was employed to assess the prognostic significance of the three DEGs mentioned above. The result suggested that high expression of CTHRC1, MMP11, and S100A8 was correlated with poor OS (Fig. 11B). Considering the good performance of CTHRC1 on the gene regulatory network and prognosis, and the fact that few studies have focused on its role in bladder cancer, the present study focused on this gene. Compared with NMIBC samples, cytoplasmic and membranous CTHRC1 immunostaining was markedly enhanced in MIBC tissues (Fig. 11C), and high CTHRC1 expression was observed in the majority of MIBC tissues (22/34) while in part of non-invasive tumors (5/15), which was consistent with the result of RT-qPCR and the trend seen in the GEO datasets. Detailed information about clinical specimens is listed in Table IV. Furthermore, functional assay indicated that CTHRC1 knockdown inhibited migration ability of 5637 and TCCSUP cells (Fig. S1), suggesting CTHRC1 may be involved in the development and progression of bladder cancer.
Figure 10.
Protein-protein interaction network associated with the 13-mRNA classifier identified by GeneMANIA. Red circles indicate upregulated genes, whereas blue ones depict downregulated genes in 13-mRNA signature. Green rhombi represent additional genes spontaneously predicted by GeneMANIA.
Figure 11.
Validation of hub genes identified from the 13-mRNA signature. (A) RT-qPCR results showing levels of expression profiles of the six hub genes in NMIBC tissues (n=12) vs. MIBC tissues (n=12); **P<0.01 and ***P<0.001. NMI: non-muscle invasive bladder cancer, MI: muscle invasive bladder cancer. (B) Survival curves generated by Kaplan-Meier plotter (http://kmplot.com/analysis/) indicated that high expression of CTHRC1, MMP11 and S100A8 can predict unfavorable prognosis. (C) Immunohistochemical analysis of CTHRC1 protein expression in NIMBC and MIBC tissues. Scale bar, 100 µm.
Table IV.
Clinicopathological parameters of patients enrolled in the cohort.
| Characteristics | Used for RT-qPCR | Used for IHC |
|---|---|---|
| Number of patients | 24 | 49 |
| Age (mean ± SD) | 69.21±7.40 | 68.92±9.62 |
| Sex (male/female) | 6/18 | 40/9 |
| Pathology grade (low/high) | 7/17 | 11/38 |
| Tumor size (≤3 cm/>3 cm) | 15/9 | 26/23 |
| Subtype (papillary/non-papillary) | 11/13 | 20/29 |
| T stage (Ta-T1/T2-T4) | 12/12 | 15/34 |
| Lymph node metastasis (no/yes) | 24/0 | 41/8 |
| TNM stage (I/II/III/IV) | 12/9/3/0 | 15/18/9/7 |
IHC, immunohistochemistry; SD, standard deviation; TNM, tumor-node-metastasis.
Discussion
Comprehensive genomic studies on high-throughput RNA sequence and microarray profiles have attracted considerable attention for the prognostic prediction and exploration of molecular mechanisms of a variety of diseases in recent years. Progression of NMIBC to MIBC has been an unsolved life-threatening problem, and cannot be accurately predicted with the use of traditional clinicopathological parameters (15). Thus, identification of good sensitive and specific novel biomarkers for predicting disease progression of patients with NMIBC is of utmost clinical significance in treatment decisions and follow-up regimens. In the current study, a 13-mRNA signature was developed for both prediction of disease progression and survival prediction of patients with bladder cancer. Moreover, this model outperformed other published gene signatures and biomarkers in two GEO datasets included in the present study, demonstrating its considerable reliability and robustness in predictive accuracy.
In total, 73 DEGs were identified between patients with primary NMIBC and patients with progressive NMIBC, and univariate Cox regression analysis was used to screen 47 prognostic genes from those DEGs. However, this method is not suitable for such high-dimensional microarray data due to the familiar limitation of overfitting in selecting survival-related genes (19). Therefore, the LASSO Cox regression method that can eliminate the above limitation was applied and a 13-mRNA-based risk score model that can separate patients into low- and high-risk groups was constructed in the training dataset and then verified in another independent GEO dataset (for prediction of disease progression) and TCGA cohort (for prediction of survival), which indicated its favorable predictive prognostic performance.
Several novel multi-gene-based signatures and single biomarkers for prediction of progression of NMIBC to MIBC have been reported in recent years and comparison of the 13-mRNA signature with them was conducted in our study (14–18). The ROC curves indicated that the performance of the 13-mRNA signature was similar to that of the 12-gene progression score model, and superior to that of the 5-gene model and three other single-gene biomarkers in two GEO datasets. Since the 12-gene model was based on a large-scale multicenter prospective study, it had a robust predictive power, and we paid particular attention to it. The progression score model based on the expression levels of 12 genes detected by RT-qPCR assay correlated significantly with outcome and had independent prognostic power when the model was analyzed in combination with the European Organisation for Research and Treatment of Cancer (EORTC) risk score. Moreover, the 12-gene model was also significantly related to previous molecular classes identified with basal- and luminal-like characteristics (20). Unfortunately, there are no overlapping genes between our 13-mRNA signature and the 12-gene progression score model, which may be attributed to the high heterogeneity of bladder cancer and different statistical methods used for constructing the signatures. Pearson's correlation coefficient, ROC analysis, Wilcoxon signed-rank test and Cox regression analysis were employed to identify the 12 genes for inclusion in the ideal PCR signature by comparing normalized cycle threshold (Ct) values and clinical outcomes (21), whereas DEGs in the primary NMIBC and progressive NMIBC groups based on absolute fold change and adjusted t-test P-value were screened for developing the 13-mRNA model using univariate Cox regression and LASSO Cox regression methods. Notably, the current histopathological system of evaluation is the conventional system used to determine the prognosis of and to stratify treatment of patients with bladder cancer. Both the 1973 and 2004 World Health Organization grading systems provide independent clinical information for predicting disease progression in patients with NMIBC (2). The ROC curves in the present study showed that the 13-mRNA signature was superior to the grading system with respect to prediction of progression in the training and validation cohorts. Moreover, a predictive nomogram, which combined the 13-mRNA signature and available clinically related risk factors (gender and grade), was developed to help guide prediction of prognosis and enable clinicians to introduce an individualized therapeutic strategy for patients with NMIBC.
Gene set enrichment analysis revealed the differences in distinctive biological processes and signaling pathways between the high- and low-risk subgroups stratified by the 13-gene signature in TCGA cohort. Several cancer-related pathways, including the ‘FOXM1 pathway’, ‘extracellular matrix organization’, ‘epithelial mesenchymal transition’, ‘inflammatory response’ and ‘IL-6-JAK-STAT3’, which are involved in diverse functions that promote carcinogenesis or metastasis, were significantly enriched in the high-risk subgroup. The FOXM1 transcription factor, a member of the Fox transcription factor family, plays vital role in carcinogenesis via regulation of proliferation, cell cycle, transformation and apoptosis (22). FOXM1 facilitated to cell viability, migration and invasion of bladder cancer cells through reducing p27 level and increasing VEGF expression (23). High expression of FOXM1 was associated with adverse clinical features, including pathological grade, TNM stage, concomitant carcinoma in situ and multifocal tumors, and predicted a risk of luminal subtype with worse outcomes in patients with stage pT1 NMIBC (24). Emerging evidence supports the hypothesis that inflammation is one of the hallmark characteristics of cancer development and progression. Inflammatory cytokines, such as IL-6, S100A8, and TGF-β, play an essential role in cancer progression by directly interacting with tumor cells, promoting epithelial-to-mesenchymal transition and driving distant metastasis (25). As common markers of systemic inflammatory response, neutrophil-to-lymphocyte ratio (NLR) and C-reactive protein (CRP) were associated with disease progression in patients with NMIBC (26).
There were 6 hub genes screened from the 13-mRNA signature and 3 of 6 of them that were detected by RT-qPCR were further identified as target genes, which were differentially expressed between NMIBC and MIBC tissues. CTHRC1, a secreted glycosylated protein, has been found to be aberrantly upregulated in various human malignant cancers and involved in cellular processes related to cancer development and metastasis. Forced expression of CTHRC1 is closely associated with carcinogenesis and bone metastasis in breast cancer (27). Upregulation of CTHRC1 driven by N-glycosylation contributes to cell migration in oral squamous cell carcinoma through the non-canonical Wnt/planar cell polarity (PCP) signaling pathway (28). However, the expression patterns and biological functions of CTHRC1 in bladder cancer have not been well investigated and remain largely unclear. Here we found that expression of CTHRC1 was higher in invasive bladder cancer than the non-invasive subtype at mRNA and protein levels, and the Kaplan-Meier analysis showed that patients with high CTHRC1 expression had an unfavorable prognosis. In addition, Transwell assay has revealed that CTHRC1 deletion by specific small interfering RNA (siRNA) markedly reduced migration and invasion activities of 5637 and TCCSUP cells (data not shown), which may be attributed to aberrant MEK/ERK phosphorylation stimulated by rCTHRC1 (data not shown). S100A8 is one of the family members of calcium-binding proteins that regulates inflammatory responses and can drive proliferation and invasion of cancer cells by binding to the receptor for advanced glycation end-products (RAGE) to activate mitogen-activated protein kinase (MAPK) and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling pathways (29). Elevated S100A8 expression has been observed in multiple cancers, including bladder cancer. Several studies have devoted to explore the value of S100A8 being a biomarker for diagnosis and prediction of prognosis in bladder cancer. A prognostic signature based on S100A8-correlated genes can predict disease progression, and patients with NMIBC with high S100A8 cluster are more likely to progress to MIBC (30), indicating that S100A8 might be a novel promising biomarker of bladder cancer outcome. Using multiple comprehensive approaches of protein detection, Bansal et al (31) found that S100A8 and S100A9 were able to precisely discriminate over 80% of bladder cancer patients compared to healthy controls, with high sensitivity and specificity (ROC, 0.946). In addition, with a comparable tactic, S100A8 and S100A4 were able to distinguish 92% of low-grade cases from high-grade, with utmost sensitivity and specificity (ROC, 0.941). Immunohistochemistry assay was performed to test S100A8 expression in NMIBC, data showed that S100A8 expression was significantly higher in pT1 and high-grade tumors than in pTa and low-grade, and it remained an independent factor for RFS (P=0.024, HR 2.43) and PFS (P=0.002, HR 5.92) according to the multivariate Cox regression model (32). Those results suggest that S100A8 is a promising marker for identification of bladder cancer patients at high risk of recurrence and progression, which supports our main viewpoint that S100A8 as a member of 13-mRNA signature can be a novel potential biomarker for diagnosis and prediction of prognosis in patients with bladder cancer. As for the role and mechanism of S100A8 in bladder cancer, we think the relative research is necessary but independent of this manuscript. Recent studies have reported MMP11 as a marker of lymph node metastasis and predictor of poor survival in bladder cancer. Li et al found increased MMP-11 protein expression was related to increments of pathologic stage status, a stepwise increment of MMP11expression from normal urothelium and non-invasive urothelial carcinoma to superficially invasive subtype, and to high stage urothelial carcinoma. The expression of MMP-11 is positively related to high cancer-specific mortality and metastasis in bladder transitional cell carcinoma (33). Coincidentally, MMP11 was identified as a member of a five-mRNA-based classifier developed for predicting lymph node metastasis in bladder cancer (34). Those results suggest that MMP11 is a promising biomarker for prediction of patients with bladder cancer.
This study, however, has several limitations. First, this study is retrospective in nature, which may have resulted in a selection bias, and the sample size was insufficient in the training and validation datasets. The results of our study, therefore, need further validation in large prospective clinical trials. Secondly, information on clinical factors, like age, intravesical therapy, survival status and follow-up time was not available in the validation dataset; thus, only two parameters (sex and grade) that exist in both datasets were collected and combined with the 13-mRNA signature to construct a nomogram. This requires further verification in different cohorts in the future. Finally, more detailed studies are still needed to explore the biological roles and molecular mechanisms of the genes incorporated in the integrated signature in bladder cancer.
In conclusion, we have developed an integrated 13-mRNA signature for prediction of progression of NMIBC to MIBC and prognostication. The prognostic value of the 13-mRNA signature for PFS and OS, and a nomogram based on the model might be useful for clinicians to select personalized therapy for patients with bladder cancer. Furthermore, the gene signature could also provide insights into the underlying molecular mechanisms of development and progression of bladder cancer.
Supplementary Material
Acknowledgements
The authors would like to thank the Chongqing Key Laboratory of Molecular Oncology and Epigenetics (Chongqing, China) for technical guidance.
Funding
This study was supported by National Natural Science Foundation of China (No. 81874092), National Natural Science Foundation of China Youth Fund (No. 81801482) and Chongqing Science & Technology Commission (No. cstc2017shmsA130098 and No. cstc2019jcyj-msxmX0126).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CZ and DG conceived and designed the study. HY performed the procedures of bioinformatics analysis and wrote the initial manuscript. XG and WH were involved in the conception of the study and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (Chongqing, China), and written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Burger M, van der Aa MN, van Oers JM, Brinkmann A, van der Kwast TH, Steyerberg EC, Stoehr R, Kirkels WJ, Denzinger S, Wild PJ, et al. Prediction of progression of non-muscle-invasive bladder cancer by WHO 1973 and 2004 grading and by FGFR3 mutation status: A prospective study. Eur Urol. 2008;54:835–843. doi: 10.1016/j.eururo.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 3.Lamm D, Persad R, Brausi M, Buckley R, Witjes JA, Palou J, Böhle A, Kamat AM, Colombel M, Soloway M. Defining progression in nonmuscle invasive bladder cancer: It is time for a new, standard definition. J Urol. 2014;191:20–27. doi: 10.1016/j.juro.2013.07.102. [DOI] [PubMed] [Google Scholar]
- 4.Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, Newling DW, Kurth K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: A combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466–465. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 5.Le Goux C, Vacher S, Pignot G, Sibony M, Barry Delongchamps N, Terris B, Piaggio E, Zerbib M, Damotte D, Bieche I. mRNA Expression levels of genes involved in antitumor immunity: Identification of a 3-gene signature associated with prognosis of muscle-invasive bladder cancer. Oncoimmunology. 2017;6:e1358330. doi: 10.1080/2162402X.2017.1358330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 7.Yang L, Taylor J, Eustace A, Irlam JJ, Denley H, Hoskin PJ, Alsner J, Buffa FM, Harris AL, Choudhury A, West CML. A gene signature for selecting benefit from hypoxia modification of radiotherapy for high-risk bladder cancer patients. Clin Cancer Res. 2017;23:4761–4768. doi: 10.1158/1078-0432.CCR-17-0038. [DOI] [PubMed] [Google Scholar]
- 8.Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB, Kim SK, Kim YJ, Kim WJ, Chu IS. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol. 2010;28:2660–2667. doi: 10.1200/JCO.2009.25.0977. [DOI] [PubMed] [Google Scholar]
- 9.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 10.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Yin H, Yang X, Gu W, Liu Y, Li X, Huang X, Zhu X, Tao Y, Gou X, He W. HMGB1-mediated autophagy attenuates gemcitabine-induced apoptosis in bladder cancer cells involving JNK and ERK activation. Oncotarget. 2017;8:71642–71656. doi: 10.18632/oncotarget.17796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Heijden AG, Mengual L, Lozano JJ, Ingelmo-Torres M, Ribal MJ, Fernández PL, Oosterwijk E, Schalken JA, Alcaraz A, Witjes JA. A five-gene expression signature to predict progression in T1G3 bladder cancer. Eur J Cancer. 2016;64:127–136. doi: 10.1016/j.ejca.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Dyrskjøt L, Reinert T, Algaba F, Christensen E, Nieboer D, Hermann GG, Mogensen K, Beukers W, Marquez M, Segersten U, et al. Prognostic impact of a 12-gene progression score in non-muscle-invasive bladder cancer: A prospective multicentre validation study. Eur Urol. 2017;72:461–469. doi: 10.1016/j.eururo.2017.05.040. [DOI] [PubMed] [Google Scholar]
- 16.Rinaldetti S, Wirtz R, Worst TS, Hartmann A, Breyer J, Dyrskjot L, Erben P. FOXM1 predicts disease progression in non-muscle invasive bladder cancer. J Cancer Res Clin Oncol. 2018;144:1701–1709. doi: 10.1007/s00432-018-2694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer MW, Golshani R, Merseburger AS, Knapp J, Garcia A, Hennenlotter J, Duncan RC, Soloway MS, Jorda M, Kuczyk MA, et al. HYAL-1 hyaluronidase: A potential prognostic indicator for progression to muscle invasion and recurrence in bladder cancer. Eur Urol. 2010;57:86–93. doi: 10.1016/j.eururo.2009.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lelo A, Prip F, Harris BT, Solomon D, Berry DL, Chaldekas K, Kumar A, Simko J, Jensen JB, Bhattacharyya P, et al. STAG2 is a biomarker for prediction of recurrence and progression in papillary non-muscle-invasive bladder cancer. Clin Cancer Res. 2018;24:4145–4153. doi: 10.1158/1078-0432.CCR-17-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon R, Altman DG. Statistical aspects of prognostic factor studies in oncology. Br J Cancer. 1994;69:979–985. doi: 10.1038/bjc.1994.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedegaard J, Lamy P, Nordentoft I, Algaba F, Høyer S, Ulhøi BP, Vang S, Reinert T, Hermann GG, Mogensen K, et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell. 2016;30:27–42. doi: 10.1016/j.ccell.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Dyrskjøt L, Reinert T, Novoradovsky A, Zuiverloon TC, Beukers W, Zwarthoff E, Malats N, Real FX, Segersten U, Malmström PU, et al. Analysis of molecular intra-patient variation and delineation of a prognostic 12-gene signature in non-muscle invasive bladder cancer; technology transfer from microarrays to PCR. Br J Cancer. 2012;107:1392–1398. doi: 10.1038/bjc.2012.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai J, Yang L, Wang J, Xiao Y, Ruan Q. Prognostic value of FOXM1 in patients with malignant solid tumor: A meta-analysis and system review. Dis Markers. 2015;2015:352478. doi: 10.1155/2015/352478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Shi Y, Yan J, Fan H. Downregulation of FoxM1 inhibits cell growth and migration and invasion in bladder cancer cells. Am J Transl Res. 2018;10:629–638. [PMC free article] [PubMed] [Google Scholar]
- 24.Breyer J, Wirtz RM, Erben P, Rinaldetti S, Worst TS, Stoehr R, Eckstein M, Sikic D, Denzinger S, Burger M, et al. FOXM1 overexpression is associated with adverse outcome and predicts response to intravesical instillation therapy in stage pT1 non-muscle-invasive bladder cancer. BJU Int. 2019;123:187–196. doi: 10.1111/bju.14525. [DOI] [PubMed] [Google Scholar]
- 25.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–e503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 26.Mbeutcha A, Shariat SF, Rieken M, Rink M, Xylinas E, Seitz C, Lucca I, Mathieu R, Rouprêt M, Briganti A, et al. Prognostic significance of markers of systemic inflammatory response in patients with non-muscle-invasive bladder cancer. Urol Oncol. 2016;34:483.e17–483.e24. doi: 10.1016/j.urolonc.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Kharaishvili G, Cizkova M, Bouchalova K, Mgebrishvili G, Kolar Z, Bouchal J. Collagen triple helix repeat containing 1 protein, periostin and versican in primary and metastatic breast cancer: An immunohistochemical study. J Clin Pathol. 2011;64:977–982. doi: 10.1136/jclinpath-2011-200106. [DOI] [PubMed] [Google Scholar]
- 28.Liu G, Sengupta PK, Jamal B, Yang HY, Bouchie MP, Lindner V, Varelas X, Kukuruzinska MA. N-glycosylation induces the CTHRC1 protein and drives oral cancer cell migration. J Biol Chem. 2013;288:20217–20227. doi: 10.1074/jbc.M113.473785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller P, Kidwell KM, Thomas D, Sabel M, Rae JM, Hayes DF, Hudson BI, El-Ashry D, Lippman ME. Elevated S100A8 protein expression in breast cancer cells and breast tumor stroma is prognostic of poor disease outcome. Breast Cancer Res Treat. 2017;166:85–94. doi: 10.1007/s10549-017-4366-6. [DOI] [PubMed] [Google Scholar]
- 30.Kim SK, Kim EJ, Leem SH, Ha YS, Kim YJ, Kim WJ. Identification of S100A8-correlated genes for prediction of disease progression in non-muscle invasive bladder cancer. BMC Cancer. 2010;10:21. doi: 10.1186/1471-2407-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bansal N, Gupta A, Sankhwar SN, Mahdi AA. Low- and high-grade bladder cancer appraisal via serum-based proteomics approach. Clin Chim Acta. 2014;436:97–103. doi: 10.1016/j.cca.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Nicklas AP, Kramer MW, Serth J, Hennenlotter J, Hupe MC, Reimer DU, Stenzl A, Merseburger AS, Kuczyk MA, von Klot CJ. Calgranulin A (S100A8) immunostaining: A future candidate for risk assessment in patients with non-muscle-invasive bladder cancer (NMIBC) Adv Ther. 2018;35:2054–2068. doi: 10.1007/s12325-018-0789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li WM, Wei YC, Huang CN, Ke HL, Li CC, Yeh HC, Chang LL, Huang CH, Li CF, Wu WJ. Matrix metalloproteinase-11 as a marker of metastasis and predictor of poor survival in urothelial carcinomas. J Surg Oncol. 2016;113:700–707. doi: 10.1002/jso.24195. [DOI] [PubMed] [Google Scholar]
- 34.Wu SX, Huang J, Liu ZW, Chen HG, Guo P, Cai QQ, Zheng JJ, Qin HD, Zheng ZS, Chen X, et al. A genomic-clinicopathologic nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. EBioMedicine. 2018;31:54–65. doi: 10.1016/j.ebiom.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.