Nitrification is a ubiquitous microbially mediated process in the environment and an essential process in engineered systems such as wastewater and drinking water treatment plants. However, nitrification also contributes to fertilizer loss from agricultural environments, increasing the eutrophication of downstream aquatic ecosystems, and produces the greenhouse gas nitrous oxide. As ammonia-oxidizing bacteria are the most dominant ammonia-oxidizing microbes in fertilized agricultural soils, understanding their responses to a variety of environmental conditions is essential for curbing the negative environmental effects of nitrification. Notably, oxygen limitation has been reported to significantly increase nitric oxide and nitrous oxide production during nitrification. Here, we investigate the physiology of the best-characterized ammonia-oxidizing bacterium, Nitrosomonas europaea, growing under oxygen-limited conditions.
KEYWORDS: ammonia and oxygen limitation, ammonia-oxidizing bacteria, chemostat, nitrification, Nitrosomonas europaea, transcriptome
ABSTRACT
Ammonia-oxidizing microorganisms perform the first step of nitrification, the oxidation of ammonia to nitrite. The bacterium Nitrosomonas europaea is the best-characterized ammonia oxidizer to date. Exposure to hypoxic conditions has a profound effect on the physiology of N. europaea, e.g., by inducing nitrifier denitrification, resulting in increased nitric and nitrous oxide production. This metabolic shift is of major significance in agricultural soils, as it contributes to fertilizer loss and global climate change. Previous studies investigating the effect of oxygen limitation on N. europaea have focused on the transcriptional regulation of genes involved in nitrification and nitrifier denitrification. Here, we combine steady-state cultivation with whole-genome transcriptomics to investigate the overall effect of oxygen limitation on N. europaea. Under oxygen-limited conditions, growth yield was reduced and ammonia-to-nitrite conversion was not stoichiometric, suggesting the production of nitrogenous gases. However, the transcription of the principal nitric oxide reductase (cNOR) did not change significantly during oxygen-limited growth, while the transcription of the nitrite reductase-encoding gene (nirK) was significantly lower. In contrast, both heme-copper-containing cytochrome c oxidases encoded by N. europaea were upregulated during oxygen-limited growth. Particularly striking was the significant increase in transcription of the B-type heme-copper oxidase, proposed to function as a nitric oxide reductase (sNOR) in ammonia-oxidizing bacteria. In the context of previous physiological studies, as well as the evolutionary placement of N. europaea’s sNOR with regard to other heme-copper oxidases, these results suggest sNOR may function as a high-affinity terminal oxidase in N. europaea and other ammonia-oxidizing bacteria.
IMPORTANCE Nitrification is a ubiquitous microbially mediated process in the environment and an essential process in engineered systems such as wastewater and drinking water treatment plants. However, nitrification also contributes to fertilizer loss from agricultural environments, increasing the eutrophication of downstream aquatic ecosystems, and produces the greenhouse gas nitrous oxide. As ammonia-oxidizing bacteria are the most dominant ammonia-oxidizing microbes in fertilized agricultural soils, understanding their responses to a variety of environmental conditions is essential for curbing the negative environmental effects of nitrification. Notably, oxygen limitation has been reported to significantly increase nitric oxide and nitrous oxide production during nitrification. Here, we investigate the physiology of the best-characterized ammonia-oxidizing bacterium, Nitrosomonas europaea, growing under oxygen-limited conditions.
INTRODUCTION
Nitrification is a microbially mediated aerobic process involving the successive oxidation of ammonia (NH3) and nitrite (NO2−) to nitrate (NO3−) (1). In oxic environments, complete nitrification is accomplished through the complementary metabolisms of ammonia-oxidizing bacteria (AOB)/archaea (AOA) and nitrite-oxidizing bacteria (NOB) or by comammox bacteria (2, 3). The existence of nitrite-oxidizing archaea (NOA) has been proposed but not yet confirmed (4). Although an essential process during wastewater and drinking water treatment, nitrification is also a major cause of nitrogen (N) loss from N-amended soils. Nitrifiers increase N loss through the production of NO3−, which is more susceptible to leaching from soils than ammonium (NH4+), serves as terminal electron acceptor for denitrifiers, and contributes to the eutrophication of downstream aquatic environments (5).
In addition, ammonia oxidizers produce and release nitrogenous gases such as nitric (NO) and nitrous (N2O) oxide during NH3 oxidation at a wide range of substrate and oxygen (O2) concentrations (6, 7). Nitrogenous gases are formed through enzymatic processes (8–13) but also by a multitude of chemical reactions that use the key metabolites of ammonia oxidizers, hydroxylamine (NH2OH) and NO2− (or its acidic form HNO2), as the main precursors (14, 15). AOB, in particular, release NO and N2O either during NH2OH oxidation (16–21) or via nitrifier denitrification—the reduction of NO2− to N2O via NO (22–25). The first pathway is the dominant process at atmospheric O2 levels, while the latter is more important under O2-limited (hypoxic) conditions (26, 27), where NO2− and NO serve as alternative sinks for electrons generated by NH3 oxidation.
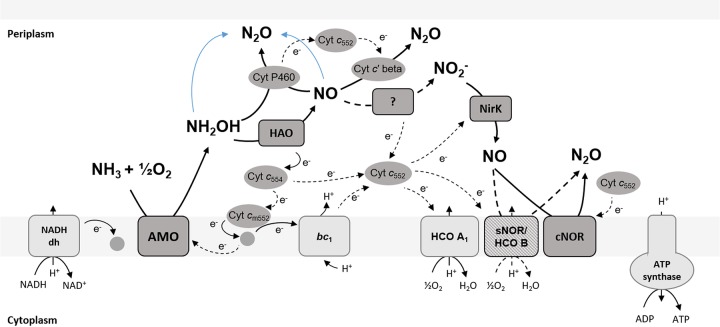
Nitrosomonas europaea strain ATCC 19718 was the first AOB to have its genome sequenced (28) and is widely used as a model organism in physiological studies of NH3 oxidation and NO/N2O production in AOB (27, 29–36). The enzymatic background of NO and N2O production in N. europaea is complex and involves multiple interconnected processes (Fig. 1). Most AOB harbor a copper-containing nitrite reductase, NirK, which is necessary for efficient NH3 oxidation by N. europaea at atmospheric O2 levels. NirK is also involved in but not essential for NO production during nitrifier denitrification in N. europaea (26, 27, 29, 35) and is upregulated in response to high NO2− concentrations (37). Moreover, two forms of membrane-bound cytochrome (cyt) c oxidases (cNOR and sNOR) and three cytochromes, referred to as cyt P460 (CytL), cyt c′ beta (CytS), and cyt c554 (CycA), have been implicated in N2O production in N. europaea and other AOB (12, 24, 32, 38–40). However, the involvement of cyt c554 in N2O production has recently been disputed (41). Finally, recent research has confirmed that the oxidation of NH3 to NO2− in AOB includes the formation of NO as an obligate intermediate, produced by NH2OH oxidation via the hydroxylamine dehydrogenase (HAO) (20). The enzyme responsible for the oxidation of NO to NO2− (the proposed nitric oxide oxidase) has not yet been identified (40).
FIG 1.
A simplified schematic of electron transport and NO/N2O-producing pathways in N. europaea. Solid lines indicate confirmed and dashed lines indicate postulated reactions or electron transfer processes. Abiotic N2O production is indicated in blue. NADH dh, NADH dehydrogenase (complex I); AMO, ammonia monooxygenase; HAO, hydroxylamine dehydrogenase; NirK, nitrite reductase; bc1, cytrochrome bc-I complex (complex III); HCO A1, heme-copper-containing cytochrome c oxidase A1-type (complex IV); sNOR/HCO B, heme-copper-containing NO reductase/heme-copper-containing cytochrome c oxidase B-type (complex IV); cNOR, heme-iron-containing nitric oxide reductase.
The production of NO and N2O by N. europaea, grown under oxic as well as hypoxic (oxygen-limited) conditions, was previously demonstrated and quantified in multiple batch and chemostat culture studies (11, 12, 34, 35, 42, 43). Furthermore, recent studies have investigated the instantaneous rate of NO and N2O production by N. europaea during the transition from oxic to oxygen-limited or anoxic conditions (12, 35, 36). Despite this large body of literature describing the effect of oxygen (O2) limitation on NH3 oxidation and NO/N2O production in N. europaea, little attention has been paid to the regulation of other processes under these conditions. Previous studies have utilized reverse transcription-quantitative PCR (RT-qPCR) assays to examine transcriptional patterns of specific mainly N cycle-related genes in AOB grown under O2-limited conditions (34, 36, 44). To date, no study has evaluated the global transcriptomic response of N. europaea to O2-limited growth. However, research on the effect of stressors other than reduced O2 tension have demonstrated the suitability of transcriptomics for the analysis of physiological responses in AOB (43, 45–48).
N. europaea utilizes the Calvin-Benson-Bassham (CBB) cycle to fix inorganic carbon (28, 49). Whereas all genome-sequenced AOB appear to use the CBB cycle, differences exist in the number of copies of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) genes encoded as well as the presence or absence of carbon dioxide (CO2)-concentrating mechanisms (50–52). N. europaea harbors a single form IA green-like (high-affinity) RuBisCO enzyme and two carbonic anhydrases but no carboxysome-related genes (28). RuBisCO is considered to function optimally in hypoxic environments, as it also uses O2 as a substrate and produces the off-path intermediate 2-phosphoglycolate (53, 54). However, the effects of O2 limitation on the transcription of RuBisCO-encoding genes and resulting growth yield in AOB are still poorly understood.
In this study, we expand upon previous work investigating the effects of O2 limitation on N. europaea by profiling the transcriptomic response to substrate (NH3) versus O2 limitation. N. europaea was grown under steady-state NH3- or O2-limited conditions, which allowed for the investigation of differences in transcriptional patterns between growth conditions. We observed a downregulation of genes associated with CO2 fixation as well as increased expression of two distinct heme-copper-containing cytochrome c oxidases (HCOs) during O2-limited growth. Our results provide new insights into how N. europaea physiologically adapts to thrive in O2-limited environments and identified putative key enzymes for future biochemical characterization.
RESULTS AND DISCUSSION
Growth characteristics.
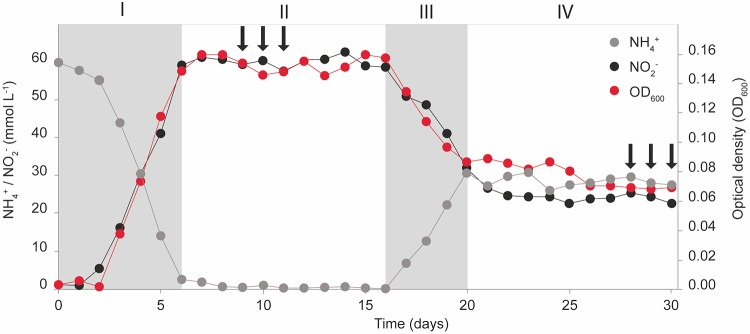
N. europaea was grown as a continuous steady-state culture under both NH3- and O2-limited growth conditions. During NH3-limited steady-state growth, the culture was kept oxic with a constant supply of filtered atmospheric air, was continuously stirred (400 rpm), and contained a standing NO2− concentration of ∼60 mmol liter−1. N. europaea grown under NH3-limited conditions consumed ∼98% of substrate provided; therefore, cultures were considered to have nonlimiting amounts of O2 (Table 1). In contrast, during O2-limited steady-state growth, no additional air inflow was provided, but the stirring was increased (800 rpm) to facilitate O2 transfer between the headspace and growth medium. As a consequence of O2 limitation, the medium contained standing concentrations (∼30 mmol liter−1) of both NH4+ and NO2− (Fig. 2; Table 1).
TABLE 1.
Comparison of N. europaea growth characteristics and NH4+ to NO2− conversion stoichiometry during NH3- and O2-limited steady-state growth
| Growth condition | Period (days) | OD600a | Input NH3b (mmol day−1) | NH3 consumeda (mmol day−1) | Steady-statea NH4+ (mmol liter−1) | Steady-statea NO2− (mmol liter−1) | N balancea,c,d (mmol) | Ammonia oxidation ratea,d (qNH3) (mmol g [dry cell weight]−1 h−1) | Apparent growth yielda,d (Y) (g [dry cell weight] mol−1 NH3) |
|---|---|---|---|---|---|---|---|---|---|
| NH3 limited | 7–16 | 0.15 ± 0.01 | 14.4 | 14.2 ± 0.1 | 0.9 ± 0.5 | 60.1 ± 1.4 | 61.0 ± 1.7 A | 24.04 ± 0.93 C | 0.42 ± 0.02 C |
| 9–11 | 0.15 ± 0.004 | 14.4 | 14.2 ± 0.1 | 0.9 ± 0.4 | 59.1 ± 1.4 | 60.0 ± 1.8 c | 24.73 ± 0.53 c | 0.40 ± 0.01 c | |
| O2 limited growth | 23–32 | 0.07 ± 0.01 | 14.4 | 7.5 ± 0.4 | 28.9 ± 1.5 | 24.1 ± 0.8 | 52.8 ± 1.8 B | 26.44 ± 2.28 D | 0.38 ± 0.03 D |
| 28–30 | 0.07 ± 0.0005 | 14.4 | 7.5 ± 0.3 | 28.6 ± 1.1 | 24.3 ± 1.4 | 52.9 ± 2.4 d | 28.51 ± 1.13 d | 0.35 ± 0.01 d |
a Average values from 3 sampling days or 10-day steady-state period, ± standard deviations (see Table S1 in the supplemental material).
b The NH4+ concentration of the influx medium (60 mmol liter−1) multiplied by the influx rate (0.24 liter day−1).
c Sum of effluent NH4+ and NO2− concentrations.
d Letters A and B represent highly significant differences (P ≤ 0.01), and letters C and D represent significant differences (P ≤ 0.05) within parameters. Capital letters represent comparisons between 10-day periods, whereas lowercase letters represent comparisons between 3-day periods.
FIG 2.
N. europaea culture dynamics and sampling scheme. N. europaea grown in a chemostat operated in batch mode (I), under steady-state NH3-limited conditions as a continuous culture (II), transitioning from NH3-limited to O2-limited steady-state growth as a continuous culture (III), and under steady-state O2-limited conditions as a continuous culture (IV). Arrows indicate transcriptome sampling points during NH3-limited (days 9, 10, and 11) and O2-limited (days 28, 29, and 30) steady-state growth.
N. europaea culture dynamics and NH3 oxidation activity calculations. Shaded areas represent non-steady-state growth phases. Transcriptome sampling days are indicated in bold; g (dry cell weight) in the chemostat; qNH3, NH3 consumption rate; Y, apparent growth yield. Download Table S1, PDF file, 0.05 MB (47.6KB, pdf) .
Copyright © 2020 Sedlacek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
During NH3-limited steady-state growth (days 7 to 16) (Fig. 2), N. europaea stoichiometrically oxidized all supplied NH4+ to NO2− (N balance = 61.0 ± 1.7 mmol liter−1) and maintained an optical density at 600 nm (OD600) of 0.15 ± 0.01 (Table 1). During O2-limited steady-state growth (days 23 to 32) (Fig. 2), N. europaea was able to consume on average 31.1 ± 1.5 mmol liter−1 (51.8%) of the supplied NH4+ and maintained an OD600 of 0.07 ± 0.01 (Table 1). A decrease in OD600 was expected, as the O2-limited culture oxidized less total substrate (NH4+), resulting in less biomass produced. The conversion of NH4+ to NO2− was not stoichiometric during O2-limited growth, as only 77.5% (24.1 ± 0.8 mmol liter−1) of the NH4+ oxidized was measured as NO2− in the effluent, resulting in an N balance of 52.8 ± 1.8 mmol liter−1 (Table 1). The significant difference (P ≤ 0.01) in the N balance between NH4+ consumed and NO2− formed during O2-limited growth is in accordance with previous reports and likely due to increased N loss in the form of NH2OH, NO, and N2O under O2-limited conditions (12, 35, 42, 55).
The dilution rate (0.01 h−1) of the chemostat was kept constant during both NH3- and O2-limited growth, and resulted in 14.4 mmol day−1 NH4+ delivered into the chemostat. On days 9, 10, and 11, which were sampled for NH3-limited growth transcriptomes, N. europaea consumed NH3 at a rate (qNH3) of 24.73 ± 0.53 mmol g (dry cell weight)−1 h−1 with an apparent growth yield (Y) of 0.40 ± 0.01 g (dry cell weight) mol−1 NH3. During days sampled for O2-limited growth transcriptomes (days 28, 29, and 30), the qNH3 was significantly higher (28.51 ± 1.13 mmol g [dry cell weight]−1 h−1; P ≤ 0.05), while Y was significantly lower (0.35 ± 0.01 g [dry cell weight] mol−1 NH3; P ≤ 0.05). When the whole 10-day NH3- and O2-limited steady-state growth periods were considered, the qNH3 and Y trends remained statistically significant (P ≤ 0.05) (Table 1). Overall, NH3 oxidation was less efficiently coupled to biomass production under O2-limited growth conditions.
Global transcriptomic response of N. europaea to growth under NH3- versus O2-limited conditions.
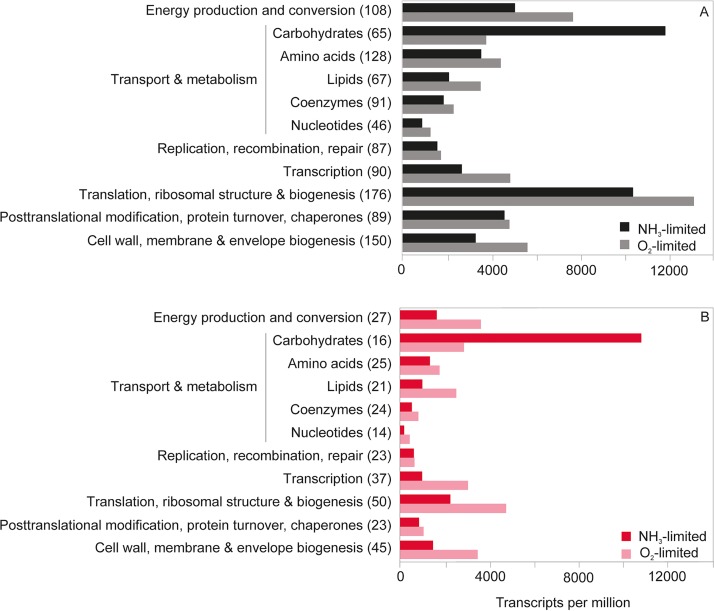
Under both NH3- and O2-limited growth conditions, transcripts mapping to 2,535 of 2,572 protein-coding genes (98.5%) and 3 RNA-coding genes (ffs, rnpB, and transfer-messenger RNA [tmRNA]) were detected. Many of the 37 genes not detected encode phage elements or transposases, some of which may have been excised from the genome in the >15 years of culturing since genome sequencing (see Data Set S1 in the supplemental material). In addition, no tRNA transcripts were detected. The high proportion of transcribed genes is in line with recent N. europaea transcriptomic studies, where similarly high fractions of transcribed genes were detected (43, 48). A significant difference in transcript levels between growth conditions was detected for 615 (∼24%) of transcribed genes (see Fig. S1). Of these 615 genes, 435 (∼71%) were present at higher levels, while 180 (∼29%) were present at lower levels during O2-limited growth. Genes encoding hypothetical proteins with no further functional annotation accounted for ∼21% (130) of the differentially transcribed genes (Data Set S1). Steady-state growth under O2-limited conditions mainly impacted the transcription of genes in clusters of orthologous groups (COGs) related to transcription and translation, ribosome structure and biogenesis, carbohydrate transport and metabolism, and energy production and conversion (Fig. 3).
FIG 3.
The sum of transcripts per million (TPM) for protein-coding genes transcribed in given COG categories (number of transcribed genes per category is given in parentheses) in the N. europaea transcriptomes. (A) Contributions and numbers of all transcribed genes in a given COG category. (B) Contributions and numbers of statistically significantly differentially transcribed genes in a given COG category.
Differential transcription (mean TPMs) of N. europaea genes during NH3- versus O2-limited growth. Transcripts differentially regulated by ≥1.5-fold change and a Welch’s t test P value threshold of ≤0.01 or ≤0.05 are shown in red or black, respectively. Download FIG S1, PDF file, 1.7 MB (1.7MB, pdf) .
Copyright © 2020 Sedlacek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential transcription (mean TPMs) and the corresponding fold changes of all the N. europaea genes that transcripts were detected during NH3- and/or O2-limited growth. P values were determined by a Welch’s t test. Download Data Set S1, XLSX file, 0.3 MB (292KB, xlsx) .
Copyright © 2020 Sedlacek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Universal and reactive oxygen stress.
The transcript levels of various chaperone proteins and sigma factors considered to be involved in the general stress response in N. europaea (45) differed between NH3- and O2-limited growth, with no discernible trend of regulation (see Table S2; Data Set S1). Overall, prolonged exposure to O2 limitation did not seem to induce a significantly increased general stress response in N. europaea. Key genes involved in oxidative stress defense (superoxide dismutase, catalase, peroxidases, and thioredoxins) were transcribed at lower levels during O2-limited growth, as expected (Table S2; Data Set S1). Surprisingly, rubredoxin (NE1426) and a glutaredoxin family protein-encoding gene (NE2328) did not follow this trend and were transcribed at significantly higher levels (2.8- and 1.8-fold, respectively) during O2-limited growth (Table S2). Although their role in N. europaea is currently unresolved, both have been proposed to be involved in cellular oxidative stress response (56, 57), iron homeostasis (58, 59), or both.
Differential expression of select genes during NH3-limited versus O2-limited growth. *, P ≤ 0.05; **, P ≤ 0.01. Download Table S2, PDF file, 0.1 MB (116.7KB, pdf) .
Copyright © 2020 Sedlacek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Carbon fixation and carbohydrate and storage compound metabolism.
There was a particularly strong effect of O2-limited growth on the transcription of several genes related to CO2 fixation (Fig. 3B). The four genes of the RuBisCO-encoding cbb operon (cbbOQSL) were among the genes displaying the largest decrease in detected transcript numbers (Fig. 4; Table S2). Correspondingly, the transcriptional repressor of the cbb operon (cbbR) was transcribed at 4.5-fold higher levels (Fig. 4; Table S2). This agrees with the previously reported decrease in transcription of the N. europaea cbbOQSL operon in O2-limited batch culture experiments (60). The reduced transcription of RuBisCO-encoding genes potentially reflects a decreased RuBisCO enzyme concentration needed to maintain an equivalent CO2 fixation rate during O2-limited growth. Since O2 acts as a competing substrate for the RuBisCO active site, the CO2-fixing carboxylase reaction proceeds more efficiently at lower O2 concentrations (53, 61, 62). When N. europaea is grown under CO2 limitation, the transcription of RuBisCO-encoding genes increases significantly (43, 60, 63). Due to the absence of carboxysomes, N. europaea appears to regulate CO2 fixation at the level of RuBisCO enzyme concentration.
FIG 4.
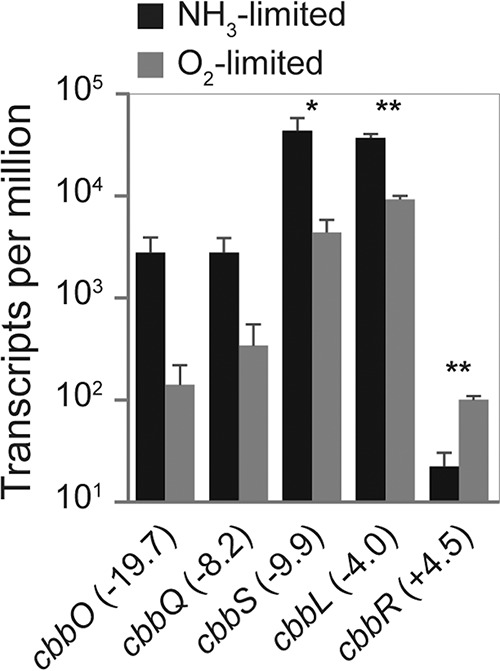
Mean TPMs of all RuBisCO-encoding genes (cbbOQSL) and the corresponding transcriptional regulator (cbbR) in N. europaea. The fold changes of gene transcription between NH3- versus O2-limited growth are given in parentheses. Error bars represent the standard deviations between replicate samples (n = 3) for each growth condition. A Welch’s t test was used to determine significantly differentially transcribed genes. *, P < 0.05; **, P < 0.01. For gene annotations, refer to Table S2 in the supplemental material.
Genes encoding the remaining enzymes of the CBB pathway and carbonic anhydrases were not significantly differentially regulated, with the exception of the transketolase-encoding cbbT gene (Table S2). Likewise, almost no differences in transcription were observed for the majority of genes in other central metabolic pathways (glycolysis/gluconeogenesis, tricarboxylic acid [TCA] cycle) (Data Set S1). As the specific growth rate of N. europaea was kept constant during both NH3- and O2-limited growth, it is not surprising that genes associated with these core catabolic pathways were transcribed at comparable levels.
Differential transcription of polyphosphate (PP) metabolism-related genes suggests an increased accumulation of PP storage during O2-limited growth. Transcripts of the polyphosphate kinase (ppk) involved in PP synthesis were detected in significantly higher numbers (2.1-fold), while transcription of the gene encoding the PP-degrading exopolyphosphatase (ppx) did not change (Table S2). Indeed, N. europaea was previously shown to accumulate PP when ATP generation (NH3 oxidation) and ATP consumption become uncoupled and surplus ATP is available (64). As the specific growth rate was kept constant throughout the experiment, PP accumulation could be a result of increased efficiency in ATP-consuming pathways, such as CO2 fixation or oxidative stress-induced repair. A decrease in the reaction flux through the energetically wasteful oxygenase reaction catalyzed by RuBisCO could result in surplus ATP being diverted to PP production.
Energy conservation.
Genes encoding the known core enzymes of the NH3 oxidation pathway in N. europaea were all highly transcribed during both NH3- and O2-limited growth (Table S2). These included ammonia monooxygenase (AMO; amoCAB operons and the singleton amoC gene) and the genes encoding HAO (haoBA) and the accessory cyt c554 (cycA) and cyt cm552 (cycX). Due to a high level of sequence conservation among the multiple AMO and HAO operons (65), it is not possible to decipher the transcriptional responses of paralogous genes in these clusters. Therefore, we report the regulation of AMO and HAO operons as single units (Table S2). The transcript numbers of genes in the AMO operons decreased up to 3.3-fold during O2-limited growth, while transcripts of the singleton amoC were present at 1.9-fold higher levels. However, these transcriptional differences were not statistically significant. The HAO cluster genes were also not significantly differentially transcribed (Table S2).
Previous research has shown that transcription of AMO, and to a lesser extent of HAO, is induced by NH3 in a concentration-dependent manner (66). In contrast, other studies have reported an increase in amoA transcription by N. europaea following substrate limitation (44, 67). Furthermore, N. europaea has been reported to increase amoA and haoA transcription during growth under low-O2 conditions (34). However, exposure to repeated transient anoxia did not significantly change amoA or haoA mRNA levels (36). As both NH3 and O2 limitation were previously shown to induce transcription of AMO- and HAO-encoding genes, the high transcription levels observed here under both NH3- and O2-limited steady-state growth conditions are not surprising.
The periplasmic red copper protein nitrosocyanin (NcyA) was among the most highly transcribed genes under both NH3- and O2-limited growth conditions (Table S2). Nitrosocyanin has been shown to be expressed at levels similar to those of other nitrification and electron transport proteins (68) and is among the most abundant proteins commonly found in AOB proteomes (47, 69). To date, the nitrosocyanin-encoding gene ncyA has been identified only in AOB genomes (24) and has been proposed as a candidate for the nitric oxide oxidase (40). However, as comammox Nitrospira do not encode ncyA (2, 3, 13), nor do all genome-sequenced AOB (70), nitrosocyanin cannot be the NO oxidase in all ammonia oxidizers. In this study, a slight (1.7-fold) but not statistically significantly higher number of ncyA transcripts was detected during O2-limited growth (Table S2). This agrees with a previous study comparing ncyA mRNA levels in N. europaea continuous cultures grown under high- and low-O2 conditions (44). However, N. europaea performing pyruvate-dependent NO2− reduction also significantly upregulated ncyA, while transcription of amoA and haoA decreased (44). Overall, there is evidence for an important role of nitrosocyanin in NH3 oxidation or electron transport in AOB, but further experiments are needed to elucidate its exact function.
Three additional cytochromes are considered to be involved in the ammonia-oxidizing pathway of N. europaea: (i) cyt c552 (cycB), essential for electron transfer; (ii) cyt P460 (cytL), responsible for N2O production from NO and hydroxylamine (39); and (iii) cyt c′-beta (cytS), hypothesized to be involved in N oxide detoxification and metabolism (24, 71). All three were among the most highly transcribed genes (top 20%) under both growth conditions (Table S2). In this study, cytS was transcribed at significantly lower levels (2.3-fold) during O2-limited growth. However, transcription levels of cycB and cytL were not significantly different (Table S2). While the in vivo function of cytS remains elusive, it is important to note that in contrast to ncyA, the cytS gene is present in all sequenced AOB and comammox Nitrospira genomes (12, 13, 52). The ubiquitous detection of cytS in genomes of all AOB, comammox Nitrospira, and in methane-oxidizing bacteria capable of NH3 oxidation (72) indicates that cyt c′-beta might play an important yet unresolved role in bacterial aerobic NH3 oxidation.
Nitrifier denitrification.
During O2-limited growth, N. europaea either performs nitrifier denitrification or experiences a greater loss of N intermediates such as NH2OH (73) or NO (20), which leads to the observed N imbalance between total NH4+ consumed and NO2− produced (Fig. 2; Table 1). The Cu-containing NO2− reductase NirK and the iron-containing membrane-bound cyt c-dependent NO reductase (cNOR; NorBC) are considered to be the main nitrifier denitrification enzymes (24, 35). N. europaea NirK plays an important role in both nitrifier denitrification and NH3 oxidation (27) and is known to be expressed during both O2-replete and -limited growth (29, 30, 35). However, under O2-limited conditions, nirK was among the genes with the largest decrease in transcript numbers (4.2-fold) observed in this study (Fig. 5; Table S2). In N. europaea, nirK transcription is regulated via the nitrite-sensitive transcriptional repressor nsrA (30). Thus, in contrast to the nirK of many denitrifiers (74), nirK transcription in N. europaea is regulated in response to NO2− concentration and not NO or O2 availability (31, 34, 48). The reduced O2 supply during O2-limited growth resulted in an ∼50% decrease in total NH3 oxidized and an ∼60% reduction in steady-state NO2− concentration (Fig. 2; Table 1). The decrease in NO2− concentration during O2-limited growth likely induced the transcription of nsrA, which was significantly (2.1-fold) upregulated (Fig. 5; Table S2). Therefore, the large decrease in nirK transcription observed here was likely due to the lower NO2− concentrations and not a direct reflection of overall nitrifier denitrification activity. In more natural nitrifying systems (e.g., agricultural soils or wastewater treatment plants [WWTPs]) changes in NO2− concentration could have a greater effect on AOB nirK expression than O2 availability. However, it should be noted that environmental NO2− concentrations are unlikely to reach those observed in this study (30 to 60 mmol liter−1 NO2−).
FIG 5.
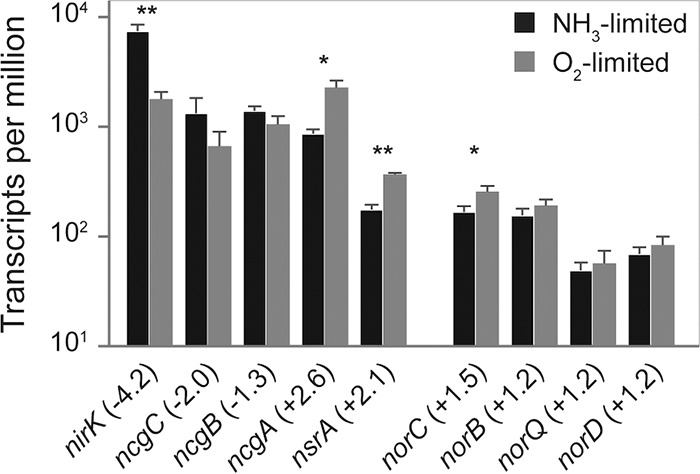
Mean TPMs of genes encoding the NirK and cNOR gene clusters in N. europaea. The fold changes of gene transcription between NH3- versus O2-limited growth are given in parentheses. Error bars represent the standard deviations between replicate samples (n = 3) for each growth condition. A Welch’s t test was used to determine significantly differentially transcribed genes. *, P < 0.05; **, P < 0.01. For gene annotations refer to Table S2.
Regulation of nirK transcription in response to primarily NO2− and not O2 concentration is consistent with the observation that NirK is not essential for NO2− reduction to NO in N. europaea. This supports the hypothesis that a not-yet-identified nitrite reductase is present in this organism. Previously, it was shown that N. europaea nirK knockout mutants are still able to enzymatically produce NO and N2O (29, 35), even if hydrazine is oxidized by HAO instead of hydroxylamine as an electron donor (35). In addition, NO and N2O formation have also been observed in the AOB Nitrosomonas communis that does not encode nirK (12). The other three genes in the NirK cluster (ncgCBA) were differentially transcribed, with ncgC and ncgB being transcribed at lower levels (2- and 1.3-fold, respectively), while ncgA was transcribed at a significantly higher level (2.6-fold) during O2-limited growth. The role of ncgCBA in N. europaea has not been fully elucidated, but all three genes were previously implicated in the metabolism or tolerance of N oxides and NO2− (31).
In contrast, transcripts of the norCBQD gene cluster, encoding the iron-containing cyt c-dependent cNOR-type NO reductase, were present at slightly higher (1.2- to 1.5-fold) but not significantly different levels during O2-limited growth (Fig. 5; Table S2). Previous research has demonstrated that in N. europaea, cNOR functions as the main NO reductase under anoxic and hypoxic conditions (35). Interestingly, all components of the proposed alternative heme-copper-containing NO reductase (sNOR), including the NO/low-oxygen sensor senC (24), were transcribed at significantly higher levels (2.7- to 10.8-fold) during O2-limited growth (Fig. 6; Table S2). Therefore, it is possible that the phenotype describing cNOR as the main NO reductase in N. europaea (35) was a product of short incubation times and that during longer term O2-limited conditions, sNOR contributes to NO reduction during nitrifier denitrification. Another possibility is that the increased transcription of sNOR observed here during O2-limited growth is primarily related to respiration and not NO reductase activity.
FIG 6.
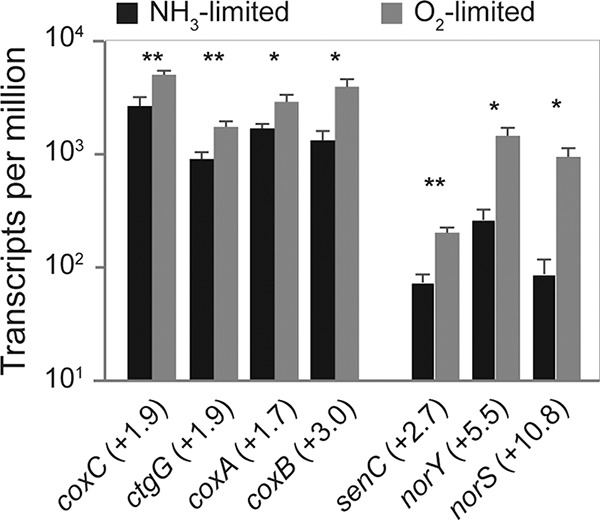
Mean TPMs of all genes encoding the A1-type and B-type HCO in N. europaea. The fold changes of gene transcription between NH3- versus O2-limited growth are given in parentheses. Error bars represent the standard deviations between replicate samples (n = 3) for each growth condition. A Welch’s t test was used to determine significantly differentially transcribed genes. *, P < 0.05; **, P < 0.01. For gene annotations, refer to Table S2.
Respiratory chain and terminal oxidases.
N. europaea harbors a low-affinity cyt c aa3 (A1 type) HCO but not a high-affinity cbb3-type (C type) cyt c HCO harbored by other AOB such as N. eutropha or Nitrosomonas sp. GH22 (28, 50, 52). Significantly higher numbers of transcripts (1.7- to 3.0-fold) of all three subunits of the cyt c aa3 HCO and the cyt c oxidase assembly gene ctaG were detected during O2-limited growth (Fig. 6; Table S2). Increased transcription of the terminal oxidase was expected, as it is a common bacterial response to O2 limitation (75). In addition, transcripts of all three subunits of the proton translocating cyt bc-I complex (complex III) were present in higher numbers (Table S2). The genes encoding NADPH dehydrogenase (complex I) and ATP synthase (complex V) were transcribed at similar levels during both growth conditions (Table S2).
As mentioned above, transcripts of both subunits of sNOR (norSY, previously called coxB2A2), and the NO/low-oxygen sensor senC were present at significantly higher numbers (2.7- to 10.8-fold) during O2-limited growth (Fig. 6; Table S2). The NO reductase function of the sNOR enzyme complex was proposed based on domain similarities between NorY and NorB (24, 32). Yet, norY phylogenetically affiliates with and structurally resembles B-type HCOs (76). In addition, NorY does not contain the five well-conserved and functionally important NorB glutamate residues (77), which are present in the canonical NorB of N. europaea. All HCOs studied thus far can reduce O2 to H2O and couple this reaction to proton translocation, albeit B- and C-type HCOs translocate fewer protons per mole O2 reduced than A-type HCOs (78). Notably, NO reduction to N2O is a known side reaction of the A2-, B-, and C-type but not A1-type HCOs (79–81). The transcriptional induction of sNOR during O2-limited growth reported here, as well as the high O2 affinity of previously studied B-type HCOs (82), indicates that sNOR might function as a high-affinity terminal oxidase in N. europaea and possibly other sNOR-harboring AOB. Furthermore, functionally characterized B-type HCOs display a lower NO turnover rate than the more widespread high-affinity C-type HCOs (79, 80). Taken together, these observations indicate that B-type HCOs, such as sNOR, are ideal for scavenging O2 during O2-limited growth conditions that coincide with elevated NO concentrations, which would impart a fitness advantage for AOB growing under these conditions. Lastly, the NOR of Roseobacter denitrificans structurally resembles cNOR but contains an HCO-like heme-copper center in place of the heme-iron center of canonical cNORs. Interestingly, this cNOR readily reduces O2 to H2O but displays very low NO reductase activity (83, 84). Therefore, in line with previous hypotheses (79, 83), the presence of a heme-copper center in NOR/HCO superfamily enzymes, such as the sNOR of N. europaea, may indicate O2 reduction as the primary enzymatic function. Notably, a recent study provided the first indirect evidence of NO reductase activity of sNOR in the marine NOB, Nitrococcus mobilis (85). However, further research is needed to resolve the primary function of sNOR in nitrifying microorganisms.
Conclusions.
In this study, we examined the transcriptional response of N. europaea to continuous growth under steady-state NH3- and O2-limited conditions. Overall, O2-limited growth resulted in a decreased growth yield but did not invoke a significant stress response in N. europaea. On the contrary, a reduced need for oxidative stress defense was evident. Interestingly, no clear differential regulation was observed for genes classically considered to be involved in aerobic NH3 oxidation. In contrast, a strong decrease in transcription of RuBisCO-encoding genes during O2-limited growth was observed, suggesting that control of CO2 fixation in N. europaea is exerted at the level of RuBisCO enzyme concentration. Furthermore, the remarkably strong increase in transcription of the genes encoding sNOR (B-type HCO) indicates this enzyme complex might function as a high-affinity terminal oxidase in N. europaea and other AOB. Overall, despite lower growth yield, N. europaea successfully adapts to growth under hypoxic conditions by regulating core components of its carbon fixation and respiration machinery.
MATERIALS AND METHODS
Cultivation.
N. europaea ATCC 19718 was cultivated at 30°C as a batch and continuous chemostat culture as previously described (43, 48). Briefly, N. europaea was grown in mineral medium containing 30 mmol liter−1 (NH4)2SO4, 0.75 mmol liter−1 MgSO4, 0.1 mmol liter−1 CaCl2, and trace minerals (10 μmol liter−1 FeCl3, 1.0 μmol liter−1 CuSO4, 0.6 μmol liter−1 Na2Mo4O4, 1.59 μmol liter−1 MnCl2, 0.6 μmol liter−1 CoCl2, 0.096 μmol liter−1 ZnCl2). After sterilization by autoclaving, the medium was buffered by the addition of 6 ml liter−1 autoclaved phosphate-carbonate buffer solution (0.52 mmol liter−1 NaH2PO4·H2O, 3.5 mmol liter−1 KH2PO4, 0.28 mmol liter−1 Na2CO3, pH adjusted to 7.0 with HCl).
For steady-state growth, a flowthrough bioreactor (Applikon Biotechnology) with a 1-liter working volume was inoculated with 2% (vol/vol) of an exponential-phase N. europaea batch culture. The bioreactor was set to “batch” mode until the NH4+ concentration reached <5 mmol liter−1 (6 days) (see Table S1 in the supplemental material). Subsequently, the bioreactor was switched to continuous flow “chemostat” mode, at a dilution rate/specific growth rate (μ) of 0.01 h−1 (doubling time = ∼70 h), which was controlled by a peristaltic pump (Thermo Scientific). The culture was continuously stirred at 400 rpm, and the pH was automatically maintained at 7.0 ± 0.1 by addition of sterile 0.94 mol liter−1 (10% [wt/vol]) Na2CO3 solution. Sterile-filtered (0.2 μm) air, at a rate of 40 ml min−1, was supplied during batch and NH3-limited steady-state growth. Once NH3-limited steady-state was reached (day 7), the chemostat was continuously operated under NH3-limited conditions for 10 days. To transition to O2-limited steady-state growth, after day 16, the air input was stopped, and the stirring speed was increased to 800 rpm to facilitate gas exchange between the medium and the headspace. The headspace was continuously replenished with O2 by the passive diffusion of atmospheric air into the chemostat through open air inlets containing a sterile filter (0.2 μm). O2-limited steady-state growth was achieved on day 23 as defined by the persistence of 26.4 to 31 mmol liter−1 NH4+ and the accumulation of 22.8 to 25.5 mmol liter−1 NO2− in the growth medium. The culture was continuously grown under these conditions for 10 days.
Sterile samples (∼5 ml) were taken on a daily basis. Culture purity was assessed by periodically inoculating ∼100 μl of culture onto lysogeny broth (Sigma-Aldrich) agar plates, which were incubated at 30°C for at least 4 days. Any observed growth on agar plates was considered contamination, and those cultures were discarded. NH4+ and NO2− concentrations were determined colorimetrically (86), and cell density was determined spectrophotometrically (Beckman) by making optical density measurements at 600 nm (OD600) (Table S1). Total biomass in grams (dry cell weight) per liter, substrate consumption rate (qNH3), and apparent growth yield (Y) were calculated as described in Mellbye et al. (43). To test for statistically significant differences in NH4+ to NO2− conversion stoichiometry, qNH3, and Y between NH3- and O2-limited steady-state growth, a Welch’s t test was performed.
RNA extraction and transcriptome sequencing.
For RNA extraction and transcriptome sequencing, three replicate samples (40 ml) were collected on three separate days during NH3-limited (days 9, 10, 11) and O2-limited (days 28, 29, 30) steady-state growth (Fig. 2). The samples were harvested by centrifugation (12,400 × g, 30 min, 4°C), resuspended in RNeasy RLT buffer with 2-mercaptoethanol, and lysed with an ultrasonication probe (3.5 output, pulse of 30 s on/30 s off for 1 min; Heatsystems Ultrasonic Processor XL). RNA was extracted using the RNeasy minikit (Qiagen) followed by the MICROBExpress-bacteria RNA Enrichment kit (Ambion/Life Technologies) according to the manufacturer’s instructions. Depleted RNA quality was assessed using the Bioanalyzer 6000 Nano Lab-Chip kit (Agilent Technologies). Sequencing libraries were constructed from at least 200 ng rRNA-depleted RNA with the TruSeq targeted RNA expression kit (Illumina), and 100-bp paired-end libraries were sequenced on a HiSeq 2000 (Illumina) at the Center for Genome Research and Biocomputing Core Laboratories (CGRB) at Oregon State University.
Transcriptome analysis.
Paired-end transcriptome sequence reads were processed and mapped to open reading frames (ORFs) deposited at NCBI for the N. europaea ATCC 19718 (NC_004757.1) reference genome using the CLC Genomics Workbench (CLC bio) under default parameters as previously described (43). Residual reads mapping to the rRNA operon were excluded prior to further analysis. An additive consensus read count was manually generated for all paralogous genes. Thereafter, mapped read counts for each gene were normalized to the gene length in kilobases, and the resulting read per kilobase (RPK) values were converted to transcripts per million (TPM) (87). To test for statistically significant differences between transcriptomes obtained from NH3- and O2-limited steady-state growth, TPMs of biological triplicate samples were used to calculate P values based on a Welch’s t test. The more stringent Welch’s rather than the Student’s t test was selected due to the limited number of biological replicates (88). Additionally, linear fold changes between average TPMs under both growth conditions for each expressed ORF were calculated. Transcripts with a P value of ≤0.05 and a transcription fold change of ≥1.5× between conditions were considered present at significantly different levels.
Data availability.
All retrieved transcriptome sequence data have been deposited in the European Nucleotide Archive (ENA) under the project accession number PRJEB31097.
ACKNOWLEDGMENTS
We thank the Center for Genome Research and Biocomputing at Oregon State University for the sequencing services. We also thank Fillipa Sousa for helpful discussions.
This work was funded by Department of Energy (DOE) award ER65192 (co-principal investigators, L.A.S.-S. and P.J.B.). C.J.S., H.D., and M.W. were supported by the Comammox Research Platform of the University of Vienna. In addition, M.W. and C.J.S. were supported by the European Research Council (ERC) via the Advanced Grant project NITRICARE 294343, and C.J.S. and H.D. were supported by Austrian Science Fund (FWF) grant 30570-B29. A.T.G. and D.W. were supported by the ERC Starting Grant 636928, under the European Union’s Horizon 2020 research and innovation program.
REFERENCES
- 1.Kuypers MMM, Marchant HK, Kartal B. 2018. The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 2.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–506. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Kessel M, Speth DR, Albertsen M, Nielsen PH, den Camp HJO, Kartal B, Jetten MSM, Lücker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitzinger K, Koch H, Lücker S, Sedlacek CJ, Herbold C, Schwarz J, Daebeler A, Mueller AJ, Lukumbuzya M, Romano S, Leisch N, Karst SM, Kirkegaard R, Albertsen M, Nielsen PH, Wagner M, Daims H. 2018. Characterization of the first “Candidatus Nitrotoga” isolate reveals metabolic versatility and separate evolution of widespread nitrite-oxidizing bacteria. mBio 9:e01186-18. doi: 10.1128/mBio.01186-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA. 2008. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320:889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- 6.Dundee L, Hopkins DW. 2001. Different sensitivities to oxygen of nitrous oxide production by Nitrosomonas europaea and Nitrosolobus multiformis. Soil Biol Biochem 33:1563–1565. doi: 10.1016/S0038-0717(01)00059-1. [DOI] [Google Scholar]
- 7.Shaw LJ, Nicol GW, Smith Z, Fear J, Prosser JI, Baggs EM. 2006. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ Microbiol 8:214–222. doi: 10.1111/j.1462-2920.2005.00882.x. [DOI] [PubMed] [Google Scholar]
- 8.Ahn JH, Kwan T, Chandran K. 2011. Comparison of partial and full nitrification processes applied for treating high-strength nitrogen wastewaters: microbial ecology through nitrous oxide production. Environ Sci Technol 45:2734–2740. doi: 10.1021/es103534g. [DOI] [PubMed] [Google Scholar]
- 9.Kool DM, Dolfing J, Wrage N, Van Groenigen JW. 2011. Nitrifier denitrification as a distinct and significant source of nitrous oxide from soil. Soil Biol Biochem 43:174–178. doi: 10.1016/j.soilbio.2010.09.030. [DOI] [Google Scholar]
- 10.Santoro AE, Buchwald C, McIlvin MR, Casciotti KL. 2011. Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science 333:1282–1285. doi: 10.1126/science.1208239. [DOI] [PubMed] [Google Scholar]
- 11.Stein LY. 2011. Surveying N2O-producing pathways in bacteria, p 131–152. In Klotz MG. (ed), Methods in enzymology. Academic Press, Waltham, MA. [DOI] [PubMed] [Google Scholar]
- 12.Kozlowski JA, Kits KD, Stein LY. 2016. Comparison of nitrogen oxide metabolism among diverse ammonia-oxidizing bacteria. Front Microbiol 7:1090. doi: 10.3389/fmicb.2016.01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kits KD, Jung MY, Vierheilig J, Pjevac P, Sedlacek CJ, Liu S, Herbold C, Stein LY, Richter A, Wissel H, Brüggemann N, Wagner M, Daims H. 2019. Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata. Nat Commun 10:1836. doi: 10.1038/s41467-019-09790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber F, Wunderlin P, Udert KM, Wells GF. 2012. Nitric oxide and nitrous oxide turnover in natural and engineered microbial communities: biological pathways, chemical reactions, and novel technologies. Front Microbiol 3:372. doi: 10.3389/fmicb.2012.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heil J, Vereecken H, Brüggemann N. 2016. A review of chemical reactions of nitrification intermediates and their role in nitrogen cycling and nitrogen trace gas formation in soil. Eur J Soil Sci 67:23–39. doi: 10.1111/ejss.12306. [DOI] [Google Scholar]
- 16.Hooper AB. 1968. A nitrite-reducing enzyme from Nitrosomonas europaea. Preliminary characterization with hydroxylamine as electron donor. Biochim Biophys Acta 162:49–65. doi: 10.1016/0005-2728(68)90213-2. [DOI] [PubMed] [Google Scholar]
- 17.Hooper AB, Terry KR. 1977. Hydroxylamine oxidoreductase from Nitrosomonas: inactivation by hydrogen peroxide. Biochemistry 16:455–459. doi: 10.1021/bi00622a018. [DOI] [PubMed] [Google Scholar]
- 18.Hooper AB, Terry KR, Maxwell PC. 1977. Hydroxylamine oxidoreductase of Nitrosomonas. Oxidation of diethyldithiocarbamate concomitant with stimulation of nitrite synthesis. Biochim Biophys Acta 462:141–152. doi: 10.1016/0005-2728(77)90196-7. [DOI] [PubMed] [Google Scholar]
- 19.Anderson IC, Poth M, Homstead J, Burdige D. 1993. A comparison of NO and N2O production by the autotrophic nitrifier Nitrosomonas europaea and the heterotrophic nitrifier Alcaligenes faecalis. Appl Environ Microbiol 59:3525–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caranto JD, Lancaster KM. 2017. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc Natl Acad Sci U S A 114:8217–8222. doi: 10.1073/pnas.1704504114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellbye BL, Giguere AT, Murthy GS, Bottomley PJ, Sayavedra-Soto LA, Chaplen FWR. 2018. Genome-scale, constraint-based modeling of nitrogen oxide fluxes during coculture of Nitrosomonas europaea and Nitrobacter winogradskyi. mSystems 3:e00170-17. doi: 10.1128/mSystems.00170-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poth M, Focht DD. 1985. 15N kinetic analysis of N2O production by Nitrosomonas europaea: an examination of nitrifier denitrification. Appl Environ Microbiol 49:1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrage N, Velthof GL, van Beusichem ML, Oenema O. 2001. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem 33:1723–1732. doi: 10.1016/S0038-0717(01)00096-7. [DOI] [Google Scholar]
- 24.Klotz MG, Stein LY. 2008. Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol Lett 278:146–156. doi: 10.1111/j.1574-6968.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- 25.Giguere AT, Taylor AE, Suwa Y, Myrold DD, Bottomley PJ. 2017. Uncoupling of ammonia oxidation from nitrite oxidation: impact upon nitrous oxide production in non-cropped Oregon soils. Soil Biol Biochem 104:30–38. doi: 10.1016/j.soilbio.2016.10.011. [DOI] [Google Scholar]
- 26.Schmidt I, van Spanning RJM, Jetten M. 2004. Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK- and NorB-deficient mutants. Microbiology 150:4107–4114. doi: 10.1099/mic.0.27382-0. [DOI] [PubMed] [Google Scholar]
- 27.Cantera JJL, Stein LY. 2007. Role of nitrite reductase in the ammonia-oxidizing pathway of Nitrosomonas europaea. Arch Microbiol 188:349–354. doi: 10.1007/s00203-007-0255-4. [DOI] [PubMed] [Google Scholar]
- 28.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, Sayavedra-Soto L, Arciero D, Hommes N, Whittaker M, Arp D. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol 185:2759–2773. doi: 10.1128/jb.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaumont HJ, Hommes NG, Sayavedra-Soto LA, Arp DJ, Arciero DM, Hooper AB, Westerhoff HV, van Spanning R. 2002. Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers tolerance to nitrite. J Bacteriol 184:2557–2560. doi: 10.1128/jb.184.9.2557-2560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beaumont HJ, Lens SI, Reijnders WN, Westerhoff HV, van Spanning RJ. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol Microbiol 54:148–158. doi: 10.1111/j.1365-2958.2004.04248.x. [DOI] [PubMed] [Google Scholar]
- 31.Beaumont HJ, Lens SI, Westerhoff HV, van Spanning RJ. 2005. Novel nirK cluster genes in Nitrosomonas europaea are required for NirK-dependent tolerance to nitrite. J Bacteriol 187:6849–6851. doi: 10.1128/JB.187.19.6849-6851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho CMH, Yan T, Liu X, Wu L, Zhou J, Stein LY. 2006. Transcriptome of a Nitrosomonas europaea mutant with a disrupted nitrite reductase gene (nirK). Appl Environ Microbiol 72:4450–4454. doi: 10.1128/AEM.02958-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellitteri-Hahn MC, Halligan BD, Scalf M, Smith L, Hickey WJ. 2011. Quantitative proteomic analysis of the chemolithoautotrophic bacterium Nitrosomonas europaea: comparison of growing-and energy-starved cells. J Proteomics 74:411–419. doi: 10.1016/j.jprot.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Yu R, Chandran K. 2010. Strategies of Nitrosomonas europaea 19718 to counter low dissolved oxygen and high nitrite concentrations. BMC Microbiol 10:70. doi: 10.1186/1471-2180-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozlowski JA, Price J, Stein LY. 2014. Revision of N2O-producing pathways in the ammonia-oxidizing bacterium, Nitrosomonas europaea ATCC 19718. Appl Environ Microbiol 80:4930–4935. doi: 10.1128/AEM.01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu R, Perez-Garcia O, Lu H, Chandran K. 2018. Nitrosomonas europaea adaptation to anoxic-oxic cycling: insights from transcription analysis, proteomics and metabolic network modeling. Sci Total Environ 615:1566–1573. doi: 10.1016/j.scitotenv.2017.09.142. [DOI] [PubMed] [Google Scholar]
- 37.Cua LS, Stein LY. 2011. Effects of nitrite on ammonia-oxidizing activity and gene regulation in three ammonia-oxidizing bacteria. FEMS Microbiol Lett 319:169–175. doi: 10.1111/j.1574-6968.2011.02277.x. [DOI] [PubMed] [Google Scholar]
- 38.Upadhyay AK, Hooper AB, Hendrich MP. 2006. NO reductase activity of the tetraheme cytochrome c554 of Nitrosomonas europaea. J Am Chem Soc 128:4330–4337. doi: 10.1021/ja055183+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caranto JD, Vilbert AC, Lancaster KM. 2016. Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission. Proc Natl Acad Sci U S A 113:14704–14709. doi: 10.1073/pnas.1611051113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lancaster KM, Caranto JD, Majer SH, Smith MA. 2018. Alternative bioenergy: updates to and challenges in nitrification metalloenzymology. Joule 2:421–441. doi: 10.1016/j.joule.2018.01.018. [DOI] [Google Scholar]
- 41.McGarry JM, Pacheco A. 2018. Upon further analysis, neither cytochrome c554 from Nitrosomonas europaea nor its F156A variant display NO reductase activity, though both proteins bind nitric oxide reversibly. J Biol Inorg Chem 23:861–878. doi: 10.1007/s00775-018-1582-4. [DOI] [PubMed] [Google Scholar]
- 42.Kester RA, De Boer W, Laanbroek HJ. 1997. Production of NO and N2O by pure cultures of nitrifying and denitrifying bacteria during changes in aeration. Appl Environ Microbiol 63:3872–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellbye BL, Giguere A, Chaplen F, Bottomley PJ, Sayavedra-Soto LA. 2016. Steady-state growth under inorganic carbon limitation increases energy consumption for maintenance and enhances nitrous oxide production in Nitrosomonas europaea. Appl Environ Microbiol 82:3310–3318. doi: 10.1128/AEM.00294-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beyer S, Gilch S, Meyer O, Schmidt I. 2009. Transcription of genes coding for metabolic key functions in Nitrosomonas europaea during aerobic and anaerobic growth. J Mol Microbiol Biotechnol 16:187–197. doi: 10.1159/000142531. [DOI] [PubMed] [Google Scholar]
- 45.Gvakharia BO, Permina EA, Gelfand MS, Bottomley PJ, Sayavedra-Soto LA, Arp DJ. 2007. Global transcriptional response of Nitrosomonas europaea to chloroform and chloromethane. Appl Environ Microbiol 73:3440–3445. doi: 10.1128/AEM.02831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park S, Ely RL. 2008. Genome-wide transcriptional responses of Nitrosomonas europaea to zinc. Arch Microbiol 189:541–548. doi: 10.1007/s00203-007-0341-7. [DOI] [PubMed] [Google Scholar]
- 47.Kartal B, Wessels HJ, van der Biezen E, Francoijs KJ, Jetten MS, Klotz MG, Stein LY. 2012. Effects of nitrogen dioxide and anoxia on global gene and protein expression in long-term continuous cultures of Nitrosomonas eutropha C91. Appl Environ Microbiol 78:4788–4794. doi: 10.1128/AEM.00668-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez J, Buchanan A, Mellbye B, Ferrell R, Chang JH, Chaplen F, Bottomley PJ, Arp DJ, Sayavedra-Soto LA. 2015. Interactions of Nitrosomonas europaea and Nitrobacter winogradskyi grown in co-culture. Arch Microbiol 197:79–89. doi: 10.1007/s00203-014-1056-1. [DOI] [PubMed] [Google Scholar]
- 49.Sayavedra-Soto LA, Arp DJ. 2011. Ammonia-oxidizing Bacteria: their biochemistry and molecular biology, p 11–38. In Ward BB, Arp DJ, Klotz MG (ed). Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 50.Stein LY, Arp DJ, Berube PM, Chain PS, Hauser L, Jetten MS, Klotz MG, Larimer FW, Norton JM, Op den Camp HJ, Shin M, Wei X. 2007. Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ Microbiol 9:2993–3007. doi: 10.1111/j.1462-2920.2007.01409.x. [DOI] [PubMed] [Google Scholar]
- 51.Berg IA. 2011. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sedlacek CJ, McGowan B, Suwa Y, Sayavedra-Soto LA, Laanbroek HJ, Stein LY, Norton JM, Klotz MG, Bollmann A. 2019. A physiological and genomic comparison of Nitrosomonas cluster 6a and 7 ammonia-oxidizing bacteria. Microb Ecol 78:985–994. doi: 10.1007/s00248-019-01378-8. [DOI] [PubMed] [Google Scholar]
- 53.Andrews TJ, Lorimer GH. 1978. Photorespiration—still unavoidable? FEBS Lett 90:1–9. doi: 10.1016/0014-5793(78)80286-5. [DOI] [Google Scholar]
- 54.Badger MR, Bek EJ. 2008. Multiple RuBisCo forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot 59:1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- 55.Goreau TJ, Kaplan WA, Wofsy SC, McElroy MB, Valois FW, Watson SW. 1980. Production of NO2− and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl Environ Microbiol 40:526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prieto-Alamo MJ, Jurado J, Gallardo-Madueno R, Monje-Casas F, Holmgren A, Pueyo C. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J Biol Chem 275:13398–13405. doi: 10.1074/jbc.275.18.13398. [DOI] [PubMed] [Google Scholar]
- 57.Coulter ED, Kurtz DM Jr. 2001. A role for rubredoxin in oxidative stress protection in Desulfovibrio vulgaris: catalytic electron transfer to rubrerythrin and two-iron superoxide reductase. Arch Biochem Biophys 394:76–86. doi: 10.1006/abbi.2001.2531. [DOI] [PubMed] [Google Scholar]
- 58.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 59.Rouhier N, Couturier J, Johnson MK, Jacquot JP. 2010. Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci 35:43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei X, Sayavedra-Soto LA, Arp DJ. 2004. The transcription of the cbb operon in Nitrosomonas europaea. Microbiology 150:1869–1879. doi: 10.1099/mic.0.26785-0. [DOI] [PubMed] [Google Scholar]
- 61.Lorimer GH. 1981. The carboxylation and oxygenation of ribulose 1,5-bisphosphate: the primary events in photosynthesis and photorespiration. Annu Rev Plant Physiol 32:349–382. doi: 10.1146/annurev.pp.32.060181.002025. [DOI] [Google Scholar]
- 62.McNevin D, von Caemmerer S, Farquhar G. 2006. Determining RuBisCO activation kinetics and other rate and equilibrium constants by simultaneous multiple non-linear regression of a kinetic model. J Exp Bot 57:3883–3900. doi: 10.1093/jxb/erl156. [DOI] [PubMed] [Google Scholar]
- 63.Jiang D, Khunjar WO, Wett B, Murthy SN, Chandran K. 2015. Characterizing the metabolic trade-off in Nitrosomonas europaea in response to changes in inorganic carbon supply. Environ Sci Technol 49:2523–2531. doi: 10.1021/es5043222. [DOI] [PubMed] [Google Scholar]
- 64.Terry KR, Hooper AB. 1970. Polyphosphate and orthophosphate content of Nitrosomonas europaea as a function of growth. J Bacteriol 103:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arp DJ, Sayavedra-Soto LA, Hommes NG. 2002. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol 178:250–255. doi: 10.1007/s00203-002-0452-0. [DOI] [PubMed] [Google Scholar]
- 66.Sayavedra-Soto LA, Hommes NG, Russell SA, Arp DJ. 1996. Induction of ammonia monooxygenase and hydroxylamine oxidoreductase mRNAs by ammonium in Nitrosomonas europaea. Mol Microbiol 20:541–548. doi: 10.1046/j.1365-2958.1996.5391062.x. [DOI] [PubMed] [Google Scholar]
- 67.Chandran K, Love NG. 2008. Physiological state, growth mode, and oxidative stress play a role in Cd (II)-mediated inhibition of Nitrosomonas europaea 19718. Appl Environ Microbiol 74:2447–2453. doi: 10.1128/AEM.01940-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whittaker M, Bergmann D, Arciero D, Hooper AB. 2000. Electron transfer during the oxidation of ammonia by the chemolithotrophic bacterium Nitrosomonas europaea. Biochim Biophys Acta 1459:346–355. doi: 10.1016/S0005-2728(00)00171-7. [DOI] [PubMed] [Google Scholar]
- 69.Zorz JK, Kozlowski JA, Stein LY, Strous M, Kleiner M. 2018. Comparative proteomics of three species of ammonia-oxidizing bacteria. Front Microbiol 9:938. doi: 10.3389/fmicb.2018.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bollmann A, Sedlacek CJ, Norton J, Laanbroek HJ, Suwa Y, Stein LY, Klotz MG, Arp D, Sayavedra-Soto L, Lu M, Bruce D, Detter C, Tapia R, Han J, Woyke T, Lucas SM, Pitluck S, Pennacchio L, Nolan M, Land ML, Huntemann M, Deshpande S, Han C, Chen A, Kyrpides N, Mavromatis K, Markowitz V, Szeto E, Ivanova N, Mikhailova N, Pagani I, Pati A, Peters L, Ovchinnikova G, Goodwin LA. 2013. Complete genome sequence of Nitrosomonas sp. Is79, an ammonia oxidizing bacterium adapted to low ammonium concentrations. Stand Genomic Sci 7:469–482. doi: 10.4056/sigs.3517166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elmore BO, Bergmann DJ, Klotz MG, Hooper AB. 2007. Cytochromes P460 and c′-beta; a new family of high-spin cytochromes c. FEBS Lett 581:911–916. doi: 10.1016/j.febslet.2007.01.068. [DOI] [PubMed] [Google Scholar]
- 72.Zahn JA, Arciero DM, Hooper AB, Dispirito AA. 1996. Cytochrome c′ of Methylococcus capsulatus Bath. Eur J Biochem 240:684–691. doi: 10.1111/j.1432-1033.1996.0684h.x. [DOI] [PubMed] [Google Scholar]
- 73.Liu S, Han P, Hink L, Prosser JI, Wagner M, Brüggemann N. 2017. Abiotic conversion of extracellular NH2OH contributes to N2O emission during ammonia oxidation. Environ Sci Technol 51:13122–13132. doi: 10.1021/acs.est.7b02360. [DOI] [PubMed] [Google Scholar]
- 74.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bueno E, Mesa S, Bedmar EJ, Richardson DJ, Delgado MJ. 2012. Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid Redox Signal 16:819–852. doi: 10.1089/ars.2011.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sousa FL, Alves RJ, Pereira-Leal JB, Teixeira M, Pereira MM. 2011. A bioinformatics classifier and database for heme-copper oxygen reductases. PLoS One 6:e19117. doi: 10.1371/journal.pone.0019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y. 2010. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 78.Sousa FL, Alves RJ, Ribeiro MA, Pereira-Leal JB, Teixeira M, Pereira MM. 2012. The superfamily of heme-copper oxygen reductases: types and evolutionary considerations. Biochim Biophys Acta 1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 79.Giuffre A, Stubauer G, Sarti P, Brunori M, Zumft WG, Buse G, Soulimane T. 1999. The heme-copper oxidases of Thermus thermophilus catalyze the reduction of nitric oxide: evolutionary implications. Proc Natl Acad Sci U S A 96:14718–14723. doi: 10.1073/pnas.96.26.14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forte E, Urbani A, Saraste M, Sarti P, Brunori M, Giuffrè A. 2001. The cytochrome cbb3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur J Biochem 268:6486–6491. doi: 10.1046/j.0014-2956.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 81.Pereira MM, Teixeira M. 2004. Proton pathways, ligand binding and dynamics of the catalytic site in haem-copper oxygen reductases: a comparison between the three families. Biochim Biophys Acta 1655:340–346. doi: 10.1016/j.bbabio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 82.Han H, Hemp J, Pace LA, Ouyang H, Ganesan K, Roh JH, Daldal F, Blanke SR, Gennis RB. 2011. Adaptation of aerobic respiration to low O2 environments. Proc Natl Acad Sci U S A 108:14109–14114. doi: 10.1073/pnas.1018958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuda Y, Inamori KI, Osaki T, Eguchi A, Watanabe A, Kawabata SI, Iba K, Arata H. 2002. Nitric oxide-reductase homologue that contains a copper atom and has cytochrome c-oxidase activity from an aerobic phototrophic bacterium Roseobacter denitrificans. J Biochem 131:791–800. doi: 10.1093/oxfordjournals.jbchem.a003167. [DOI] [PubMed] [Google Scholar]
- 84.Zumft WG. 2005. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J Inorg Biochem 99:194–215. doi: 10.1016/j.jinorgbio.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 85.Füssel J, Lücker S, Yilmaz P, Nowka B, van Kessel M, Bourceau P, Hach PF, Littmann S, Berg J, Spieck E, Daims H, Kuypers MMM, Lam P. 2017. Adaptability as the key to success for the ubiquitous marine nitrite oxidizer Nitrococcus. Sci Adv 3:e1700807. doi: 10.1126/sciadv.1700807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hood-Nowotny R, Umana NHN, Inselbacher E, Oswald-Lachouani P, Wanek W. 2010. Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Sci Soc Am J 74:1018–1027. doi: 10.2136/sssaj2009.0389. [DOI] [Google Scholar]
- 87.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. 2010. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Götz F, Pjevac P, Markert S, McNichol J, Becher D, Schweder T, Mussmann M, Sievert SM. 2019. Transcriptomic and proteomic insight into the mechanism of cyclooctasulfur- versus thiosulfate-oxidation by the chemolithoautotroph Sulfurimonas denitrificans. Environ Microbiol 21:244–258. doi: 10.1111/1462-2920.14452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
N. europaea culture dynamics and NH3 oxidation activity calculations. Shaded areas represent non-steady-state growth phases. Transcriptome sampling days are indicated in bold; g (dry cell weight) in the chemostat; qNH3, NH3 consumption rate; Y, apparent growth yield. Download Table S1, PDF file, 0.05 MB (47.6KB, pdf) .
Copyright © 2020 Sedlacek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential transcription (mean TPMs) of N. europaea genes during NH3- versus O2-limited growth. Transcripts differentially regulated by ≥1.5-fold change and a Welch’s t test P value threshold of ≤0.01 or ≤0.05 are shown in red or black, respectively. Download FIG S1, PDF file, 1.7 MB (1.7MB, pdf) .
Copyright © 2020 Sedlacek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential transcription (mean TPMs) and the corresponding fold changes of all the N. europaea genes that transcripts were detected during NH3- and/or O2-limited growth. P values were determined by a Welch’s t test. Download Data Set S1, XLSX file, 0.3 MB (292KB, xlsx) .
Copyright © 2020 Sedlacek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential expression of select genes during NH3-limited versus O2-limited growth. *, P ≤ 0.05; **, P ≤ 0.01. Download Table S2, PDF file, 0.1 MB (116.7KB, pdf) .
Copyright © 2020 Sedlacek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All retrieved transcriptome sequence data have been deposited in the European Nucleotide Archive (ENA) under the project accession number PRJEB31097.