ABSTRACT
In the era of immunotherapies there is an urgent need to implement the use of circulating biomarkers in clinical practice to facilitate personalized therapy and to predict treatment response. We conducted a prospective study to evaluate the usefulness of circulating exosomal-PD-L1 in melanoma patients’ follow-up. We studied the dynamics of exosomal-PD-L1 from 100 melanoma patients by using an enzyme-linked immunosorbent assay. We found that PD-L1 was secreted through exosomes by melanoma cells. Exosomes carrying PD-L1 had immunosuppressive properties since they were as efficient as the cancer cell from which they derive at inhibiting T-cell activation. In plasma from melanoma patients, the level of PD-L1 (n= 30, median 64.26 pg/mL) was significantly higher in exosomes compared to soluble PD-L1 (n= 30, 0.1 pg/mL). Furthermore, exosomal-PD-L1 was detected in all patients whereas only 67% of tumour biopsies were PD-L1 positive. Although baseline exosomal-PD-L1 levels were not associated with clinic-pathologic characteristics, their variations after the cures (ΔExoPD-L1) correlated with the tumour response to treatment. A ΔExoPD-L1 cut-off of> 100 was defined, yielding an 83% sensitivity, a 70% specificity, a 91% positive predictive value and 54% negative predictive values for disease progression. The use of the cut-off allowed stratification in two groups of patients statistically different concerning overall survival and progression-free survival. PD-L1 levels in circulating exosomes seem to be a more reliable marker than PD-L1 expression in tumour biopsies. Monitoring of circulating exosomal-PD-L1 may be useful to predict the tumour response to treatment and clinical outcome.
KEYWORDS: Melanoma, exosome, PD-L1/PD-1, immune checkpoint, follow-up
Introduction
Immune checkpoint inhibitors such as antibodies targeting PD-L1 (programmed death-ligand) are revolutionizing cancer patients’ management, particularly in melanoma and non-small cell lung cancer (NSCLC) treatment. Two monoclonal antibodies have already been approved by the FDA (Nivolumab and Pembrolizumab) in both indications [1–3]. In advanced melanoma patients, around 40% of objective response rate is observed whereas in NSCLC patients, Nivolumab response rate is approximately 20%. Therefore, there is an urgent need to validate a biomarker allowing selection of patients who might benefit from an immunotherapeutic approach. With this goal, the first studies aimed to identify PD-L1 expression in patients’ biopsies but this test has proven to be unreliable due to the heterogeneity of PD-L1 expression within the tumour [4,5]. Additionally, different studies attempted to measure PD-L1 level in blood samples. However, this protein is very unstable and not easy to detect. Recent studies report the interest of using circulating nanovesicles, such as exosomes, as a diagnostic tool.
Exosomes are 40–150 nm lipid bilayer membrane-bound particles generated and released by most kinds of cells through a defined intracellular trafficking route [6,7]. Exosomes carry nucleic acids, lipids and proteins, potential biomarkers that are protected by an exosome’s cholesterol-rich membrane. Among the proteins, exosomes contain membrane proteins (flotillin-1, tetraspanins, annexinV, ICAM-1), immunostimulatory molecules (MHC I/II) and ESCRT proteins (Alix, TSG101) [8–10]. In the setting of cancer, exosomes are released by all tumour cells and they can be isolated from circulating peripheral blood [6,11,12]. Exosome-based diagnostics provide higher sensitivity and specificity over conventional biopsies or liquid biomarkers due to their stability in biofluids. In addition, exosomal markers are readily available from most biofluids and recent isolation technologic advances make exosome-based diagnostics cost and labour effective [13]. Recent studies indicate that PD-L1 expression in extracellular vesicles isolated from cancer patients’ blood samples can correlate with tumour features [14–18]. Additionally, studies of tumour-derived exosomes, performed in cultured cells, have demonstrated that these exosomes represent an effective mechanism of immunosuppression [17,19–21].
In this work, we have performed a prospective clinical study to investigate whether PD-L1 levels in exosomes isolated from melanoma patients’ plasma could predict a patient’s response to immunotherapy.
Material and methods
Cell culture
Non-cancerous human lung cells (MRC-5) human lung fibroblasts, A549 human lung cancer cells, B16F10 mouse melanoma cells and SK-MEL-2 human melanoma cells were purchased from the ATCC. All cell lines were cultured at 37°C, 5% CO2 in RPMI 1640 medium (Dutscher) supplemented with 10% foetal bovine serum (Dutscher) depleted in exosomes and were tested weekly for mycoplasma contamination.
Exosomes purification from the cell’s supernatants
Cells were cultured in medium depleted from serum-derived exosomes. Supernatants were collected from cell lines and sequentially centrifuged at 300g for 10 min (4°C), at 2000g for 20 min and at 10,000g for 40 min. Then, exosomes were ultracentrifugated at 100,000g for 70 min and washed in PBS (Beckman Coulter, Optima XPN-100). Supernatants were carefully removed and exosome pellets were suspended either in 50 µl of 1% RIPA lysis buffer or in 50 µl of PBS.
Lymphocyte isolation and co-culture experiment
PBMC were isolated from buffy coats (EFS, Besançon) by centrifugation using a density gradient (Lymphocytes separation medium, Eurobio). Red blood cells were lysed and T cells were enriched (RosetteSep, Stem Cell). Then, purified T cells (105) were activated with CD2/CD3/CD28 beads (Miltenyi Biotec) and incubated with either human melanoma cells (105) (SK-MEL-2) or human lung cancer cells (A549) or exosomes derived from these cell lines (106). After 48 h of culture, T cells were collected and analysed by flow cytometry for the expression of membrane-bound PD-1 (46-1799-42, eBioscience), and intracellular Ki67 (48-5699-42, eBioscience), IFNϒ (506516, Biolegend) after fixation and permeabilization (BD Perm/Wash Buffer, Biosciences).
EXOMEL clinical study
This prospective study was conducted between January 2016 and December 2018 in the Department of Dermatology, University Hospital of Besançon, France. The study design was approved by the local research ethics committee and a written informed consent was provided before enrolment. Study adhered to the Declaration of Helsinki Principles. Patients with melanoma were included at different clinical stages graded according to the latest American Joint Committee on Cancer staging classification (8th edition, Balch 2018).
Study objectives and endpoints
The primary objective of this study was to quantify exosomal PD-L1 in the blood of melanoma patients. The secondary objectives were to determine whether the amount of ExoPD-L1 could be associated to: (i) stages diseases, (ii) response to treatment, (iii) overall survival (OS) and progression-free survival (PFS), (iv) clinical variable and (v) PD-L1 expression in tumours or soluble in the plasma. The primary endpoint of the study was the blood concentration of PD-L1-exosomes at different time points using ELISA. The secondary endpoints were to compare: (i) the baseline concentration of ExoPD-L1 with clinic/pathologic features (age, gender, primary melanoma histology subtype and location, tumour burden, prior therapy and disease status, biologic data-lactate dehydrogenase, lymphopenia, soluble PD-L1 using ELISA dosage and tumour PD-L1 using immunohistochemistry staining, (ii) the concentration of ExoPD-L1 and their variation of in patients having a complete response (CR), partial response (PR), progressive disease (PD) (based on immune-related irRECIST).
Blood collection and storage
Peripheral blood was drawn into sodium heparin tubes. First sampling at inclusion was labelled S1. Second sampling was labelled S2. Change in ExoPD-L1 from S1 to S2 was labelled ΔExoPD-L1. To ensure exosomes integrity, blood was centrifuged at 2400g for 15 min to remove cell debris and dead cells. Plasma samples were stored in 1 mL aliquots at −18°C.
Exosomes isolation from plasma samples
Thawed plasma samples of 5 mL were differentially centrifuged 300g for 5 min at 4°C and then 17,000g for 10 min at 4°C. Next, supernatants obtained in the previous step were ultra-centrifuged at 200,000g for 1 h at 4°C (Beckman Coulter, Optima XPN-100). Supernatants were carefully removed and exosome pellets suspended either in 50 µl of 1% RIPA lysis buffer or in 50 µl of PBS.
Exosomes characterization
Exosomes size and concentration were determined by nanoparticle tracking analysis using a NS300 Instrument (Nanosight, Amesbury, UK).
To determine PD-L1 expression at the exosome’s surface, protein–protein interaction experiments were conducted with an Octet Red instrument FortéBio, Menlo Park, CA). The ligand (PD-1) was biotinylated using EZ-Link NHS-PEG4-biotin (2 nM, 30 min, RT, Thermo Fisher Scientific, Germany) and immobilized on streptavidin sensors (black 96-well plate, FortéBio, USA). Functionalized sensors were incubated in PBS (10 min) then incubated with exosomes (106, isolated from either the supernatant of cancer cell lines, normal cells, melanoma patients’ plasma, lung cancer patients’ plasma or healthy donors’ plasma). All sensorgrams were corrected for baseline drift by subtracting a control sensor exposed to running buffer only (FortéBio, Data analysis software version 7.1.0.89).
Isolated exosomes were tested for the expression of exosomal markers. Nanovesicles were lysed and separated on SDS/PAGE gels. Proteins were transferred onto a polyvinylidene fluoride membrane (Amersham GE Healthcare Life Sciences) for western-blotting analysis. After transferring, membranes were blocked with 5% bovine serum albumin for 1 h and incubated overnight at 4°C with antibodies specific for Alix (2171s, Cell Signalling), TSG101 (sc-7964, Santacruz), PD-L1 (sc-50298), CD9 (ab92726, Abcam), CD63 (NBP2-4225, BioTechne), GRP94 (ab2791, Abcam), β-actin (A3854, Sigma Aldrich). Horse radish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch) was added and immunoreactive proteins revealed using ECL detection reagents (34095, ThermoFisher Scientific). Band intensities were captured using Chemidoc XRS+ system and images were analysed using Image Lab software (Bio-Rad Laboratories).
Determination of PD-L1 concentration in plasma and circulating exosomes
Soluble PD-L1 and PD-L1 in exosomes levels were measured using an enzyme-linked immunosorbent assay (PD-L1 Human ELISA Kit, Invitrogen), according to the manufacturer’s instructions. Protein concentrations were determined according to standard curves.
Immunohistochemistry of PD-L1 in tumour tissue samples
Formalin-fixed paraffin-embedded tumour tissue samples were obtained for correlation analysis of PD-L1 expression between tissue and circulating exosomes. All IHC procedures were performed using Benchmark apparatus (Ventana). PD-L1 expression was evaluated with 22C3 clone (Dako). PD-L1 immunostaining on tumour tissue samples (blind coded) was assessed by a dedicated pathologist.
Tumour response evaluation
Tumour response was based on immune-related irRECIST using unidimensional measurements on contrast-enhanced computed tomography scan or PET-CT (Positron Emission Tomography) [19]. CR, complete response; PR, partial response. Stable disease (SD) was labelled SD. High tumour burden was defined as 10 lesions or more, or only 1 lesion bigger than 3 cm. Imaging re-evaluation was performed in blind-coded samples.
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics v24.0 and the Graphpad Prism 6 program. Data presented are from at least three independent experiments. Error bars shown in graphical data represent mean ± SEM. When data are presented as box plots, the bar indicates the median, the box shows the interquartile range (25–75%) and the whiskers extend to 1.5 the interquartile range. For normally distributed data, significance of mean differences was determined using two-tailed paired or unpaired Student t-test. For data not normally distributed, non-parametric Kruskal–Wallis and Wilcoxon tests were used for unpaired and paired analysis, respectively. To test the relationship between variables, Spearmann correlation coefficients were calculated. Dependence was ruled out with a correlation coefficient (absolute q-value) of <0.7. Kaplan–Meier survival curve, Log-rank test and Cox proportional hazards model were used to analyse survival data. Results from Cox proportional hazards models were reported as hazard ratios (HRs) with 95% confidence interval (CIs). A two-tailed value of p < 0.05 was considered statistically significant. Quantitative data are expressed as mean ± SEM from at least three independent experiments. Quantitative results were compared using the Mann–Whitney test according to their distribution. The cut-off value for the change in PD-L1-Exo was calculated by the Receiver Operating Characteristic (ROC) curve analysis to identify melanoma patients with high probability to display disease control.
Results
Cancer cells release exosomes expressing membrane PD-L1
We first studied whether PD-L1 could be secreted through exosomes. We purified exosomes from different cell lines such as human and murine melanoma (SK-MEL-2, B16F10), human lung cancer (A549) and MRC-5. TSG101, Alix, CD9 and CD63 were used as markers of exosomes and Grp94 served as a negative control. As shown in Figure 1(a), PD-L1 was expressed in exosomes from the three cancer cell lines analysed but not in the non-cancerous cell line. We then analysed PD-L1 expression in exosomes (ExoPD-L1) isolated from pooled blood samples of melanoma patients (n = 6) or lung cancer patients (n = 6) or exosomes isolated from healthy donors (n = 5). As previously observed for cell lines, we only detected PD-L1 in the exosomes isolated from cancer patients (Figure 1(b)). While there was a difference between cancer patients and healthy donors in the detection of PD-L1 expressing exosomes, no difference was found in the total number and size of the exosomes (Supplementary Figure 1A-B).
Figure 1.
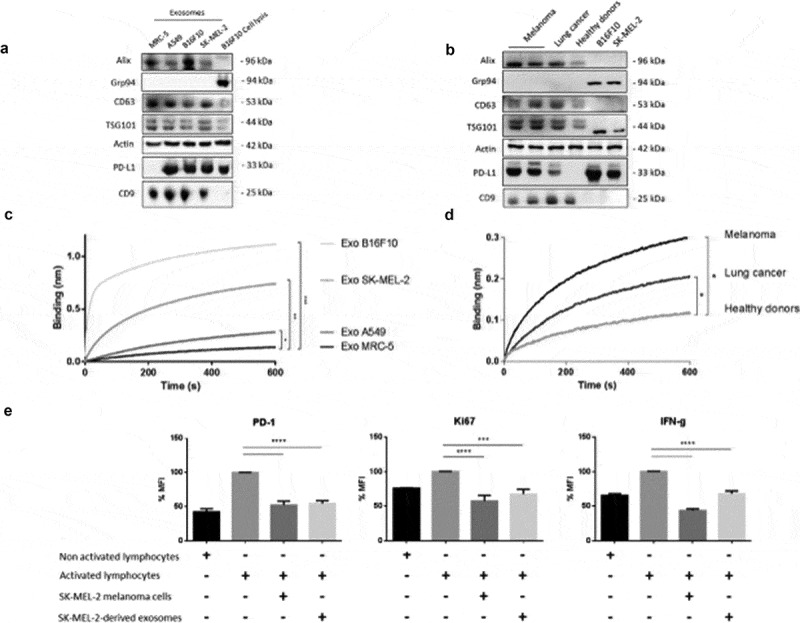
PD-L1 expression in tumour-derived exosomes. (A/B) Representative immunoblots showing expression of Alix, CD63, TSG101, PD-L1 and CD9 in exosomes derived from cell lines (a) MRC-5 (normal human lung cells), A549 (human lung cancer), B16F10 (mouse melanoma) or SK-MEL-2 (human melanoma) or (b) plasma-derived exosomes from melanoma patients, lung cancer patients and healthy donors. Grp94 is used here as an exosomal negative control and actin as a loading control. Cell lysates are also included. (C/D) Binding of exosomes (nm) derived from cell lines (MRC-5, A549, SK-MEL-2, B16F10) or from plasma samples of melanoma patients (n = 6) or lung cancer patients (n = 6) and healthy donors (n = 5) to immobilized biotinylated PD-1 determined by biolayer interferometry. Binding curves represent mean signal of triplicate measurements for each sample. Mann–Whitney. (c), ***p < 0.001, **p = 0.0081, *p = 0.0041. (d) *p = 0.0039 melanoma, *p = 0.0031 lung cancer). (e) Percentage of PD-1, Ki67 and IFNϒ mean fluorescence intensity in lymphocytes cultured 24 h in the presence or absence of SK-MEL-2 or SK-MEL-2-derived exosomes, determined by flow cytometry (***p = 0.0006, ****p <0.0001).
To determine whether PD-L1 was expressed at the membrane of the vesicles, we used an interference biolayer approach to capture exosomes with a PD-1 recombinant protein. As shown in Figure 1(c–d), exosomes could be captured both from cancer cell lines and cancer patients’ blood samples (melanoma, lung cancer), indicating the presence of PD-L1 in their membrane. In contrast, no ExoPD-L1 (or negligible amounts) could be captured from the non-cancerous cells’ supernatant or from the blood of healthy donors. These results indicate that PD-L1 is present at the membrane of tumour-derived exosomes.
Next, we determined whether isolated PD-L1-exosomes were functional. Primary T cells were activated (CD3, CD28) and then co-cultured for 24 h with either human melanoma SK-MEL-2 cells (positive control) or with exosomes purified from those cells’ supernatant (ExoPD-L1). As shown in Figure 1(e), the exosomes were as efficient as cancer cells in inhibiting T-cell activation, as determined by the expression of PD-1, Ki67 and IFNϒ. Indeed, we observed an almost two-fold decrease in the percentage of PD-1, Ki67 and IFNϒ in the T cells cultured in the presence of SK-MEL-2 cells and SK-MEL-2-derived exosomes (respective comparisons: PD-1: 52.43% ± 7.56 and 54.84% ± 7.60 vs 100%; 57.54% ± 5.95, Ki67: 67.51% ± 6.20 vs 100%; 43.71% ± 3.45 and IFNϒ: 68.01% ± 3.44 vs 100%) (Figure 1(e)). Similar results were observed with lung cancer cells (Supplementary Figure 1C).
Altogether, these results show that PD-L1 is secreted in exosomes derived from cancer cells and that this molecule is functional since it is able to mediate T-cell immunosuppression.
Patient characteristics: the EXOMEL cohort
To study ExoPD-L1 as potential biomarkers, we opened a prospective clinical study. Over the study period, 100 melanoma patients were included. Among these patients, 43% received anti-PD-1 antibodies, 10% ipilimumab and 18% BRAF and MEK inhibitors. Treatment information and clinical outcomes are detailed in Supplemental Table 1. Overall, 12 had a CR, 28 had a PR, 10 had SD, and 46 had PD). In the cohort, 46 patients were evaluated after a median follow-up of 16 months. Among them 5 had a CR, 23 had a PR, 8 had SD, and 10 had PD, giving an overall risk ratio of 60.8%. Mean age was 64 years old (SD, 13.7) and 39% of patients were female. Most primary melanomas were cutaneous (84%) and carried a BRAF mutation (51%). At the time of inclusion, most patients were AJCC stage IV (74%) with M1c and M1d disease (80%) (Table 1).
Table 1.
Clinical characteristics of melanoma patients with available ∆ExoPD-L1 (n = 46).
| Patients N = 46 |
||
|---|---|---|
| Age, years | ||
| <70 | 28 (60.9%) | |
| =70 | 18 (39.1%) | |
| Gender | ||
| Male | 28 (60.9%) | |
| Female | 18 (39.1%) | |
| ECOG performance status | ||
| 0 | 36 (78.3%) | |
| =1 | 10 (21.7%) | |
| Anatomic location of primary melanoma | ||
| Cutaneous | 36 (78.3%) | |
| Head and neck | 7 | |
| Lower limb | 8 | |
| Upper limb | 6 | |
| Trunk | 14 | |
| Hand and foot | 1 | |
| Mucosal | 4 (8.7%) | |
| Uveal | 3 (6.5%) | |
| Unknown | 3 (6.5%) | |
| Histological subtype* | ||
| SSM | 14 (30.4%) |
|
| NM | 8 (17.4%) | |
| LMN | - | |
| ALM | 1 (2.2%) | |
| Desmoplastic | 1 (2.2%) | |
| Unknown | 12 (26.1%) | |
| NA | 10 (21.7%) | |
| Breslow, mm* | ||
| 0–1 mm | 7 (15.2%) | |
| 1.01–2 mm | 7 (15.2%) | |
| 2.01–4 mm | 12 (26.1%) | |
| =4 mm | 9 (19.6%) | |
| Unknown | 1 (2.2%) | |
| NA | 10 (21.7%) |
Expression of PD-L1 in plasma or in circulating exosomes versus tumour biopsies
First, we compared the expression of PD-L1 within exosomes in blood samples versus PD-L1 tumour expression in biopsies by IHC. Then, in plasma samples, we compared by ELISA the levels of soluble PD-L1 with PD-L1 in exosomes (Figure 2(a)). As expected, the level of ExoPD-L1 (n = 30, median 64.26 pg/mL) was significantly higher compared with soluble PD-L1 in the plasma, which was barely detectable (n = 30, 0.1 pg/mL). Interestingly, ExoPD-L1 was detected in all patients (100%) (Figure 2(b)) whereas, only 67% were PD-L1 positive in tumour biopsies (Figure 2(c–d)). We conclude that PD-L1 was much easier to detect and quantify in circulating exosomes rather than in biopsies or when soluble in plasma.
Figure 2.
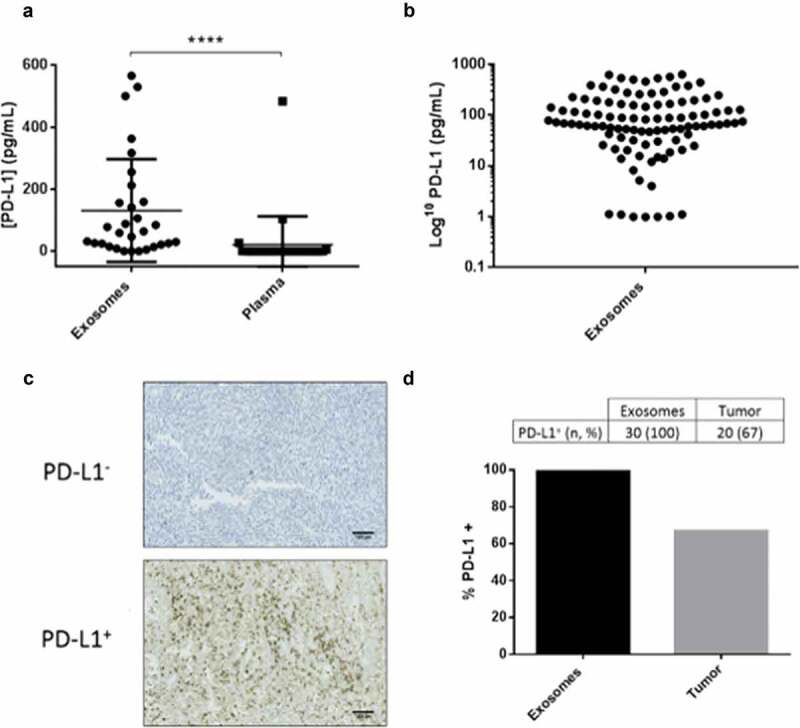
PD-L1 is easily detected in exosomes, when compared with soluble PD-L1 in plasma or in tumour biopsies. (a) Levels of PD-L1 in exosomes isolated from the plasma of melanoma patients compared with PD-L1 levels free in the plasma (n = 30) (****p < 0.0001), determined by ELISA. (b) Levels of ExoPD-L1 isolated from the 100 patient plasma samples of the EXOMEL cohort. (c) Representative IHC image of PD-L1 negative (PD-L1−) or positive (PD-L1+) tumours (22C3 antibody). Scale bars indicated 100 µm. (d) Percentage of patients positive for PD-L1 when measured in circulating exosomes versus tumour biopsies.
Association between baseline ExoPD-L1 expression and clinical variables
Baseline ExoPD-L1 levels (S1) were not associated with clinical/pathologic characteristics such as age, gender, primary melanoma histology subtype and location, tumour burden, prior therapy, or disease status, with the exception of lymphopenia that associated with a significantly higher ExoPD-L1 level (p = 0.048) (Supplementary Table 2).
Association between changes in exosomal-PD-L1 expression and tumour response
We evaluated whether a variation in ExoPD-L1 expression could be predictive of tumour response to therapy. Two blood samples, before and after treatment (S1+ S2), were obtained from 46 patients. Median interval between S1 and S2 was 4.5 months. Reasons for S2 sample unavailability were either patient death (n = 23) or the patient was lost to follow-up (n = 31). Among these 46 patients, 36 patients received anti-PD-1 therapy. The other patients received Ipilimumab (n = 2, including 1 in association with anti-PD-1 therapy), anti-BRAF and anti-MEK targeted therapies (n = 8, including 4 in association with anti-PD-1 therapy). The mean change in exosomal-PD-L1 (ΔExoPD-L1) between S1–S2 was 43.4 pg/mL. Association between ΔExoPD-L1 and disease status is shown in Figure 3(a–b). In patients experiencing complete and PR (n = 27), ΔExoPD-L1 decreased without statistical significance (mean S1: 121.06 ± 26.65 vs mean S2: 104.78 ± 17.11, p = 0.8607). In patients experiencing progression (n = 9), ΔExoPD-L1 increased significantly (mean S1 85.90 ± 24.46 vs mean S2 344.20 ± 70.30 p = 0.0002), (Figure 3(a)). Furthermore, ΔExoPD-L1 was significantly associated with disease status, i.e. responders (CR+PR+SD) vs non-responders (p = 0.001) (Figure 3(b–e)).
Figure 3.
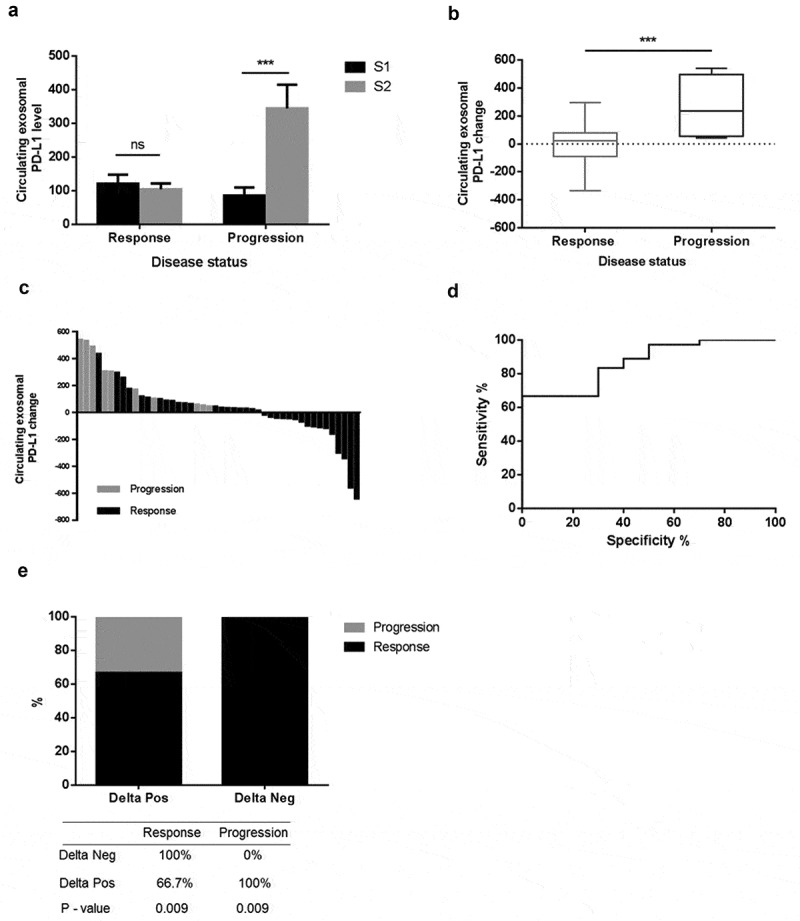
Changes in the level of ExoPD-L1 stratify melanoma patients according to disease status. (a) Mean value of circulating ExoPD-L1 evaluated at S1 and S2 in patients grouped according to disease status (response, n = 36) or progression, n = 10). Mann–Whitney (***p = 0.0002). (b) Comparison of changes in level of ExoPD-L1 in melanoma patients between S1/S2, according to disease status. Mann–Whitney (***p = 0.0002). (c) Waterfall plots showing changes in level of ExoPD-L1 between S1/S2, according to disease status. Black bars represent responder patients (n = 36) and grey bars represent progressive patients (n = 10). (d) ROC curve analysis of changes in level of ExoPD-L1 (S1/S2) in responder patients compared with non-responders (AUC = 0.867, SE 0.057, Cl95% 0.755–0.978; p < 0.001). (e) An increase in ExoPD-L1 is associated with disease progression p < 0.009. A decrease in ExoPD-L1 is associated with response p < 0.009. All patients with a decrease in level of ExoPD-L1 experienced a tumour response and all patients with an increase in level of ExoPD-L1 experienced progression.
Using ROC curve analysis, ΔExoPD-L1 showed good discrimination between patients experiencing disease response (n = 36) and those experiencing disease progression (n = 10) (AUC = 0.867, SE: 0.057, CI): 95% = 0.755–0.978; p < 0.001) (Figure 3(d)). Based on ROC curve analysis, we calculated an optimal cut-off. The ΔExoPD-L1 cut-off >100 pg/mL demonstrated an 83% sensitivity, a 70% specificity, a 91% positive predictive value and a 54% negative predictive value for disease progression.
Association between changes in ExoPD-L1 expression and clinical outcome
The median follow-up was at 9.9 months. No significant difference was found between the two groups of patients for either the total number of circulating exosomes or exosome size (Supplementary Figure 2A-B). In univariate analysis, ExoPD-L1 blood levels at S1 were not associated with OS and PFS. The small number of events regarding OS and PFS (5 events for OS and 15 events for PFS) precluded multivariable analysis. However, ΔExoPD-L1 was associated with PFS (HR: 1.003, CI 95% = 1.001 − 1.006; p = 0.006) and OS (HR: 1.004, CI: 95% = 1.001–1.008, p = 0.034) (Supplemental Table 3). The use of a ΔExoPD-L1 cut-off allowed stratification in two groups of patients statistically different in terms of OS (median not reached, p = 0.048) and PFS (ΔExoPD-L1 < 100, median PFS not reached vs. ΔExoPD-L1 > 100, median PFS = 9 months, CI: 95% = 4–14.2, p = 0.011) (Figure 4(a–b)). Altogether, these results suggest that monitoring changes in circulating ExoPD-L1 might be used to predict tumour response and clinical outcome.
Figure 4.
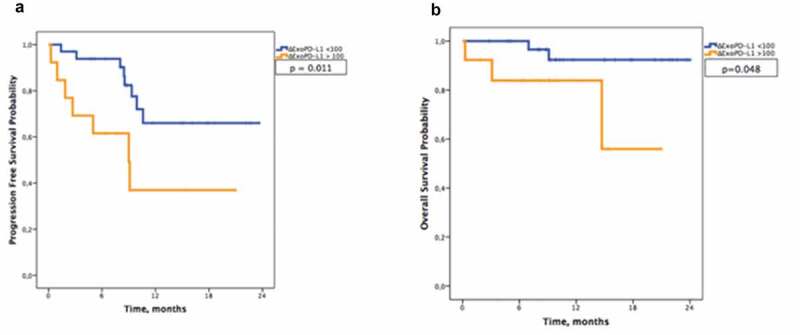
Melanoma-derived ExoPD-L1 can be used as a marker of survival. Kaplan–Meier estimates of (a) progression-free survival (p = 0.011) and (b) overall survival (p = 0.048) in patients according to ΔExoPD-L1 (n = 46).
ExoPD-L1 as a biomarker to detect early responders
Finally, we explored the possibility of following the evolution of the disease by measuring changes in ExoPD-L1 levels. For eight patients, we studied in detail whether PD-L1 levels in circulating exosomes could be used as a potential marker of response to therapy. At key points during different therapies, tumour progression/response was evaluated by medical imagery scanning and, in parallel, we quantified ExoPD-L1 in blood samples. As shown in Figure 5(a–d), tumour response detected in the scan was associated with a decrease in the level of circulating ExoPD-L1 and, inversely, tumour progression in the scan associated with an increase in circulating ExoPD-L1. Interestingly, this inverse association between ExoPD-L1 level and response to the treatment was observed independently of the therapy used (anti-PD-1 or targeted therapies). These encouraging results open the possibility of using exosomes expressing PD-L1, which can be easily isolated from blood samples during cancer patients’ follow-up assessments, as predictors of response to the treatment.
Figure 5.
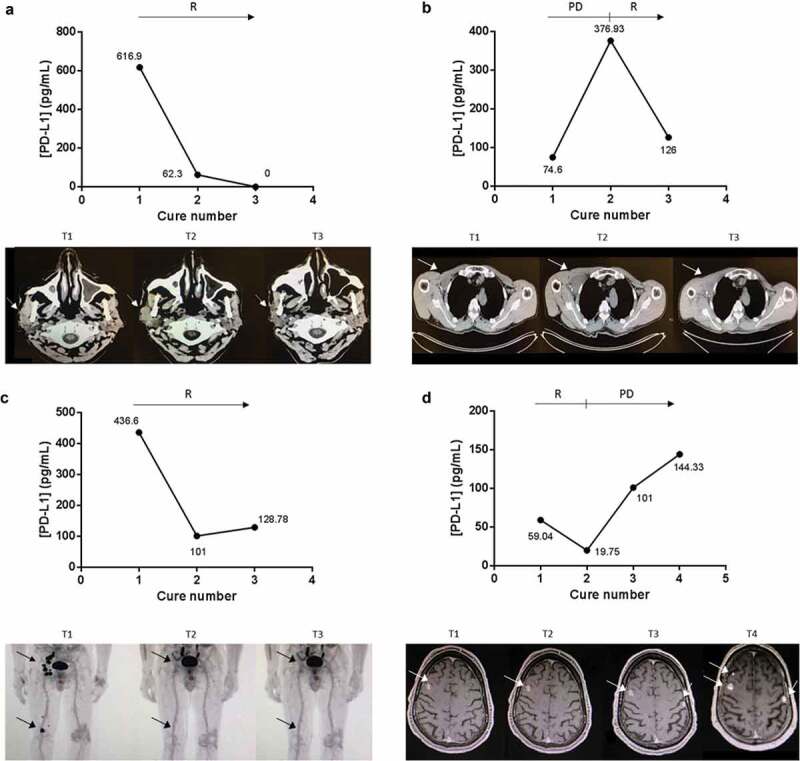
ExoPD-L1 can be used as follow-up markers in melanoma patients. Case study of the correlation between ExoPD-L1 levels in plasma samples from melanoma patients and response to PD-L1-based therapy. Concomitant imaging and ExoPD-L1 sampling in four patients experiencing (a) response as observed in the parotid gland and cervical lymph nodes on CT scan; (b) complete response in the subcutaneous tissue, popliteal and ilioinguinal lymph nodes on PET-CT; (c) disease response in the muscle and infraclavicular lymph nodes on CT scan; (d) initial response (at cure 1) then disease progression as detected in the brain on MRI. R: response, PD: progression of the disease. Tumour metastases in the scans are indicated by an arrow.
Discussion
In this study aiming to explore ExoPD-L1 as a potential tumour biomarker in melanoma cells and patients, we first confirmed previous reports showing that PD-L1 was found in exosomes including on their surface [14]. However, in contrast to what was reported by Liao X et al [23], we found that PD-L1 within the exosomes was detectable and measurable in all patients examined in our EXOMEL cohort but soluble PD-L1 could not be detected (or at very low levels). Soluble PD-L1 is probably not a reliable marker as compared with ExoPD-L1 and the most likely explanation is that in exosomes, PD-L1 is stabilized by the cholesterol-rich membrane [10]. We demonstrate that while PD-L1 within the circulating exosomes was detected in all 100 patients of the cohort (although at different levels, providing a rationale for a cut-off value), only 67% of tumour biopsies were positive for PD-L1. This lack of reliability on tumour biopsies can be explained by heterogeneity and dynamic PD-L1 expression changes within the tumour. Moreover, not only do circulating exosomes provide a reliable way to quantify PD-L1 levels, but also they allow for a non-invasive procedure with a blood sample. This is a clear advantage considering that biopsies have inherent risks. The abundance of circulating ExoPD-L1 can be explained by the fact that a single cancer cell can release thousands of exosomes, thereby resulting in a signal amplification [24]. In addition, other cells such as myeloid-derived suppressive cells, macrophages, or dendritic cells can also release exosomes with PD-L1 [22,25–29,30], thereby contributing to the overall number of circulating exosomes expressing PD-L1 observed in patients’ blood. The identification of each subpopulation of circulating exosomes expressing PD-L1 with cell-specific markers may help to improve the use of PD-L1 as a circulating predictor of tumour growth.
We also demonstrate here that PD-L1 is present at the exosome membrane in the supernatant of cancer cells (but at very low levels in normal cells) and in the blood of cancer patients (but not in healthy donors). The exosomes expressing PD-L1 are as efficient as the cancer cells from which they derive to render T-cells anergic, thereby blocking immune surveillance. These observations are consistent with the work of Chen et al [17]. We can hypothesize that, through exosomes, PD-L1 not only acts on tumours and close tumour microenvironments but also at distal sites. ExoPD-L1 can, therefore, play a key role in immunosuppression involving PD-1/PD-L1 by targeting T lymphocytes in secondary lymphoid organs. Although there are no data concerning the circulating ExoPD-L1 threshold beyond which immunosuppression is induced, this observation provides a rationale to combine PD-1/PD-L1 inhibitors with a targeted therapy.
In agreement with these results, analysis of the EXOMEL cohort indicates that the level of ExoPD-L1 inversely correlates with the response to the therapy: A high increase in ExoPD-L1 is associated with tumour progression while a decrease is associated with tumour regression, as determined by imaging scan analysis. The rate of circulating PD-L1 might reflect anti-tumour immunity involving CD8 + T-cells elicited by different therapies including, but not exclusively, checkpoint inhibitors. Since it has been reported that the tumour mutation burden (TMB) is positively associated with the response to therapy [31,32], it will be interesting to correlate ExoPD-L1 levels with patient TMB.
Conclusions
The present study offers the rationale for using ExoPD-L1 as a predictor of treatment response in melanoma patients, with the advantages of its non-invasive collection and real-time monitoring. It has already been suggested as a marker of PD-1/PD-L1 response [14,17]. In this work, we go further and demonstrate in a larger and prospective cohort that ExoPD-L1 may be a marker of different therapies (immuno- or targeted-therapy). We are currently conducting a multicenter clinical study with melanoma patients to establish precise cut-off values. The interest in ExoPD-L1 as a marker of patients’ response to therapy undoubtedly goes beyond melanoma and might be useful in other cancer types in which tumour evasion from T-cell surveillance plays a crucial role in patients’ survival and response to therapy, such as with lung cancer. The ability to capture Exo-PD-L1 directly from blood using a microchip requiring only a few microlitres of blood demands urgent investigation.
Supplementary Material
Acknowledgments
We would like to thank Isabel Grégoire and Kathryn Nason for editing and reviewing this manuscript for English language.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Trial registration
AC-2015-2496/DC-2014-2086
References
- [1].Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-pd-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ribas A, Hamid O, Daud A, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016. April 19;315(15):1600–1609. [DOI] [PubMed] [Google Scholar]
- [3].Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015. January 22;372(4):320–330. [DOI] [PubMed] [Google Scholar]
- [4].Kaunitz GJ, Cottrell TR, Lilo M, et al. Melanoma subtypes demonstrate distinct PD-L1 expression profiles. Lab Investig J Tech Methods Pathol. 2017. September;97(9):1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Madore J, Vilain RE, Menzies AM, et al. PD-L1 expression in melanoma shows marked heterogeneity within and between patients: implications for anti-PD-1/PD-L1 clinical trials. Pigment Cell Melanoma Res. 2015. May;28(3):245–253. [DOI] [PubMed] [Google Scholar]
- [6].Colombo M, Raposo G, Théry C.. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. [DOI] [PubMed] [Google Scholar]
- [7].Hessvik NP, Llorente A.. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci CMLS. 2018;75(2):193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. [DOI] [PubMed] [Google Scholar]
- [9].Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016. February 23;113(8):E968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cordonnier M, Chanteloup G, Isambert N, et al. Exosomes in cancer theranostic: diamonds in the rough. Cell Adhes Migr. 2017. March 4;11(2):151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016. April 1;126(4):1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006. April;Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- [13].Sheridan C. Exosome cancer diagnostic reaches market. Nat Biotechnol. 2016. April;34(4):359–360. [DOI] [PubMed] [Google Scholar]
- [14].Theodoraki M-N, Yerneni SS, Hoffmann TK, et al. Clinical significance of PD-L1+ exosomes in plasma of head and neck cancer patients. Clin Cancer Res Off J Am Assoc Cancer Res. 2018. February 15;24(4):896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ludwig S, Floros T, Theodoraki M-N, et al. Suppression of lymphocyte functions by plasma exosomes correlates with disease activity in patients with head and neck cancer. Clin Cancer Res. 2017. August 15;23(16):4843–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ricklefs FL, Alayo Q, Krenzlin H, et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv. 2018. March;4(3):eaar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Poggio M, Hu T, Pai -C-C, et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 2019. April 4;177(2):414–427.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nishino M, Giobbie-Hurder A, Gargano M, et al. Developing a common language for tumor response to immunotherapy: immune-related response criteria using unidimensional measurements. Clin Cancer Res Off J Am Assoc Cancer Res. 2013. July 15;19(14):3936–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Soluble PD-L1 as a biomarker in malignant melanoma and checkpoint blockade | Cancer immunology research [Internet] [cited 2019 February12]. Available from: http://cancerimmunolres.aacrjournals.org/content/early/2017/05/18/2326-6066.CIR-16-0329 [DOI] [PMC free article] [PubMed]
- [21].Gobbo J, Marcion G, Cordonnier M, et al. Restoring anticancer immune response by targeting tumor-derived exosomes with a HSP70 peptide aptamer. J Natl Cancer Inst. 2016 Mar;108(3), djv330. [DOI] [PubMed] [Google Scholar]
- [22].Sage PT, Schildberg FA, Sobel RA, et al. Dendritic cell PD-L1 limits autoimmunity and follicular T cell differentiation and function. J Immunol. 2018. April 15;200(8):2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gibbons Johnson RM, Dong H. Functional expression of programmed death-ligand 1 (B7-H1) by immune cells and tumor cells. Front Immunol [Internet]. 2017. August 10;8 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5554355/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alsaab HO, Sau S, Alzhrani R, et al. PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front Pharmacol [Internet]. 2017. August 23;8 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5572324/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lu C, Redd PS, Lee JR, et al. The expression profiles and regulation of PD-L1 in tumor-induced myeloid-derived suppressor cells. Oncoimmunology. 2016;5(12):e1247135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hartley GP, Chow L, Ammons DT, et al. Programmed cell death ligand 1 (PD-L1) signaling regulates macrophage proliferation and activation. Cancer Immunol Res. 2018. October;6(10):1260–1273. [DOI] [PubMed] [Google Scholar]
- [27].Differential regulation of PD-L1 expression by immune and tumor cells in NSCLC and the response to treatment with atezolizumab (anti–PD-L1) | PNAS [Internet] [cited 2019 February12]. Available from: https://www.pnas.org/content/115/43/E10119 [DOI] [PMC free article] [PubMed]
- [28].PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers | Annals of oncology | Oxford academic [Internet] [cited 2019 February12]. Available from: https://academic.oup.com/annonc/article-abstract/29/10/2085/5079837 [DOI] [PMC free article] [PubMed]
- [29].Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kowanetz M et al. Differential regulation of PD-L1 expression by immune and tumor cells in NSCLC and the response to treatment with atezolizumab (anti-PD-L1). Proc Natl Acad Sci U S A.. 2018;23;115(43):E10119–E10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sabari JK, Leonardi GC Shu CA, et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann Oncol. 2018;1;29(10):2085––2091.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16(11):25-98-2608.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Soluble PD-L1 as a biomarker in malignant melanoma and checkpoint blockade | Cancer immunology research [Internet] [cited 2019 February12]. Available from: http://cancerimmunolres.aacrjournals.org/content/early/2017/05/18/2326-6066.CIR-16-0329 [DOI] [PMC free article] [PubMed]
- Differential regulation of PD-L1 expression by immune and tumor cells in NSCLC and the response to treatment with atezolizumab (anti–PD-L1) | PNAS [Internet] [cited 2019 February12]. Available from: https://www.pnas.org/content/115/43/E10119 [DOI] [PMC free article] [PubMed]


