Significance
Although >100 genes have been implicated in the etiology of autism spectrum disorder (ASD), we need to better understand their functions to reveal mechanisms underlying ASD and develop treatments. We developed a pipeline to discover functions of ASD-associated genes by inactivating each gene in the model organism Caenorhabditis elegans and observing the phenotypic consequences using machine vision. We quantified 26 phenotypes spanning morphology, locomotion, tactile sensitivity, and learning in >27,000 animals representing 135 genotypes, allowing us to identify disruptions in habituation (a neural circuit’s plastic ability to decrease responding to repeated sensory stimuli) as a shared impairment. We then demonstrated how this database can facilitate experiments that determine the functional consequences of missense variants and whether phenotypic alterations are reversible.
Keywords: autism spectrum disorder, neurodevelopmental disorders, Caenorhabditis elegans, habituation learning, variants of uncertain significance
Abstract
A major challenge facing the genetics of autism spectrum disorders (ASDs) is the large and growing number of candidate risk genes and gene variants of unknown functional significance. Here, we used Caenorhabditis elegans to systematically functionally characterize ASD-associated genes in vivo. Using our custom machine vision system, we quantified 26 phenotypes spanning morphology, locomotion, tactile sensitivity, and habituation learning in 135 strains each carrying a mutation in an ortholog of an ASD-associated gene. We identified hundreds of genotype–phenotype relationships ranging from severe developmental delays and uncoordinated movement to subtle deficits in sensory and learning behaviors. We clustered genes by similarity in phenomic profiles and used epistasis analysis to discover parallel networks centered on CHD8•chd-7 and NLGN3•nlg-1 that underlie mechanosensory hyperresponsivity and impaired habituation learning. We then leveraged our data for in vivo functional assays to gauge missense variant effect. Expression of wild-type NLG-1 in nlg-1 mutant C. elegans rescued their sensory and learning impairments. Testing the rescuing ability of conserved ASD-associated neuroligin variants revealed varied partial loss of function despite proper subcellular localization. Finally, we used CRISPR-Cas9 auxin-inducible degradation to determine that phenotypic abnormalities caused by developmental loss of NLG-1 can be reversed by adult expression. This work charts the phenotypic landscape of ASD-associated genes, offers in vivo variant functional assays, and potential therapeutic targets for ASD.
Autism spectrum disorders (ASDs) encompass a clinically and genetically heterogeneous group of neurodevelopmental disorders characterized by deficits in social communication and interaction, restrictive repetitive behaviors, and profound sensory processing abnormalities (1–4). The fifth edition of the Diagnostic and Statistical Manual of Mental disorders combines autistic disorder, Asperger disorder, childhood disintegrative disorder, and pervasive developmental disorder not otherwise specified into the single grouping of autism spectrum disorder (1). Despite extensive study, there is currently no unanimously agreed upon structural or functional neuropathology common to all individuals with ASD, and there is little understanding of the biological mechanisms that cause ASD (3). The most promising avenue for research into ASDs has stemmed from the observation that they have a strong genetic component, with monozygotic concordance estimates of ∼70 to 90% and several distinct highly penetrant genetic syndromes (3, 4).
Rapid advances in copy number variation association, whole-exome, and more recently, whole-genome sequencing technology and the establishment of large sequencing consortia, have dramatically increased the pace of gene discovery in ASD (5–9). There are now >100 diverse genes with established ties to ASD, many of which are being used in diagnosis. Importantly, each gene accounts for <1% of cases and none have shown complete specificity for ASD, with many implicated in multiple neurodevelopmental disorders (3, 4, 8). Some of these genes have fallen into an encouragingly small set of broadly defined biological processes such as gene expression regulation (e.g., chromatin modification) and synaptic neuronal communication (3, 6–8, 10). Seminal studies using mouse models, genetically stratified populations of individuals with ASD, human induced pluripotent stem cells (iPSCs), and, more recently, high-throughput genetic model organisms such as Drosophila and zebrafish have investigated the molecular, circuit, and behavioral phenotypic disruptions that result from mutations in diverse ASD-associated genes. These systems have offered valuable insights into the biological mechanisms underlying this heterogeneous group of disorders (11–24). However, thousands of additional mutations in these and many other genes have been identified in individuals with ASD, and their roles as causative agents, or their pathogenicity, remain ambiguous. Thus, there are 2 major challenges facing ASD genetics: (1) the large, growing number of candidate risk genes with poorly characterized biological functions and (2) the inability to predict the functional consequences of the large number of rare missense variants. Difficulties in rare missense variant interpretation stem in part from constraints on computational variant effect prediction and a paucity of in vivo experimental variant functional assays (25, 26). This lag between gene discovery and functional characterization is even more pronounced when assessing the role of putative ASD risk genes and variants in complex sensory and learning behaviors. As such, there is a great need to rapidly determine the functions of ASD-associated genes and the functional consequences of variants of uncertain significance and to delineate complex functional genetic networks among ASD-associated genes in vivo.
The genetic model organism Caenorhabditis elegans is a powerful system for the functional analysis of disease-associated genetic variation, particularly for high-throughput in vivo characterization of risk genes identified through genomics (27). C. elegans’s fully sequenced and thoroughly annotated genome and complete connectome have fueled numerous disease discoveries, including the identification of presenilins as part of the gamma secretase complex and the role of the insulin signaling pathway in normal and pathological aging (27–30). There are clear C. elegans orthologs for >50% of human genes, and human genes have repeatedly been shown to be so structurally and functionally conserved that they can directly replace their C. elegans ortholog (27, 31–33). There are well-annotated libraries of C. elegans strains with deletion alleles available for >16,000 of the ∼20,000 protein coding genes (34). C. elegans’s small size and rapid hermaphroditic mode of reproduction (3 d from egg to egg-laying adult) allows for the routine cultivation of large numbers of isogenic animals. Further, CRISPR-Cas9 genome engineering is reliable and efficient in C. elegans, and, unlike most organisms analyzed to date, rigorous whole-genome sequencing studies have revealed no significant off-target effects due to CRISPR-Cas9 genome editing in this organism (34, 35). Finally, we developed the Multi-Worm Tracker (MWT), a machine vision system that allows for comprehensive phenotypic analysis of large populations of freely behaving animals while they perform complex sensory and learning behaviors (36, 37). Multiplexing by running several trackers in parallel allows for analysis of multiple measures of morphology, locomotion, mechanosensory sensitivity, and several forms of learning in hundreds of animals simultaneously.
Here, we developed a scalable phenomic characterization pipeline to discover functions of ASD-associated genes by systematically inactivating each gene in a model organism and observing the phenotypic consequences using machine vision (SI Appendix, Fig. S1A). We present data summarizing scores on 26 quantitative phenotypes spanning morphology, baseline locomotion, tactile sensitivity, and habituation learning in 135 strains of C. elegans each carrying a mutation in an ortholog of an ASD-associated gene, revealing hundreds of shared and unique genotype–phenotype relationships. We clustered strains based on phenotypic similarity to discover functional interactions among ASD-associated genes, which we validate with epistasis analysis. Further, we use the phenotypic profiles for in vivo functional assays to assess missense variant effect, and to determine whether phenotypic alterations are reversible using targeted protein degradation methods based on degrons.
Results
ASD-Associated Genes Are Highly Conserved to C. elegans.
We identified C. elegans orthologs of the 102 genes associated with increased risk for ASD (FDR ≤ 0.1) according to Satterstrom et al. (8), the largest exome sequencing study of ASD to date (35,584 individuals, 11,986 ASD cases). We also identified numerous orthologs from SFARI Gene (https://gene.sfari.org/) “syndromic” and “high confidence” categories and the top 50 ASD-associated genes by variant count from our ongoing meta-analysis of rare variants, VariCarta (https://varicarta.msl.ubc.ca/index) (38). Of note, 72% (18/25) of genes in the SFARI high confidence category and 78% (80/102) of genes associated with ASD by Satterstrom et al. (8) have a clear C. elegans ortholog according to OrthoList2 and the Alliance of Genome Resources, compendiums of human–C. elegans orthology prediction algorithm results. This is substantially higher than the 53% estimated genome-wide orthology between humans and C. elegans (10,678/23,010) (32), suggesting an exceptionally high conservation of biological processes core to ASD pathology (SI Appendix, Fig. S1 B and C).
Rapid advances in gene discovery and orthology prediction have altered the gene lists used during the course of this project. Despite this challenge, our list covers 92% (58/63) of the most strongly associated genes from Satterstrom et al. (8) and 100% (15/15) of the SFARI Gene high confidence category genes (Datasets S1 and S2) that had an assay-suitable ortholog deletion or severe missense allele available. This mix of currently defined high- and mid-confidence ASD-associated genes offered a unique opportunity to study putative ASD-associated genes of unknown function alongside genes with known roles in neurodevelopment and sensory processing (Datasets S1 and S2 provide a complete listing of characterized strains, orthology relationships, and ASD-association confidence).
We then used our MWT (SI Appendix, Fig. S1D) to systematically characterize the 135 C. elegans strains with a mutation in an ortholog of an ASD-associated gene (98 strains with mutations in different genes and 37 strains with additional alleles of a subset of these genes; Dataset S1 and SI Appendix). We developed software to measure a comprehensive range of parameters while animals were subjected to an automated short-term habituation learning behavioral paradigm (SI Appendix, Fig. S1 E–I; further details provided in Materials and Methods and SI Appendix). We measured habituation because it is a sensitive in vivo assay of synaptic function and behavioral plasticity. Habituation is also impaired in individuals with ASD, and abnormalities in tactile sensitivity are present in >95% of cases (2, 39, 40). The degree of habituation impairment in ASD correlates with the severity of social impairment, and recent studies in monogenic mouse models of ASD suggest peripheral tactile hypersensitivity and impaired habituation precede, and may even lead to, more complex cognitive and social impairments (2, 39, 40).
We wrote custom scripts to extract 26 quantitative phenotypic features that fall into 5 categories: morphology, baseline locomotion, initial sensitivity, habituation learning, and spontaneous recovery/short-term memory retention (SI Appendix, Fig. S2A and Table S1 provide complete feature descriptions). Features were designed to minimize redundancy while maintaining interpretability. For example, we quantified multiple features of habituation because we have previously shown these response components habituate to different extents and with different time courses (SI Appendix, Fig. S1 F–I) and because growing evidence indicates different components of a single habituating response are mediated by different molecular mechanisms (37, 41–43). Quantification of correlations between all possible phenotypic feature pairs revealed expected moderate correlations (e.g., between length and width) and clustering that reflected the feature categories we predefined (SI Appendix, Fig. S2B). Digital representations of all strains are freely available in their raw and processed forms (https://dataverse.scholarsportal.info/dataset.xhtml?persistentId=doi:10.5683/SP2/FJWIL8) (44), allowing the code to be modified to extract any simple or compound phenotypic feature of choice.
Quantitative Phenotypic Profiles Identify Shared Functions Among ASD-Associated Genes.
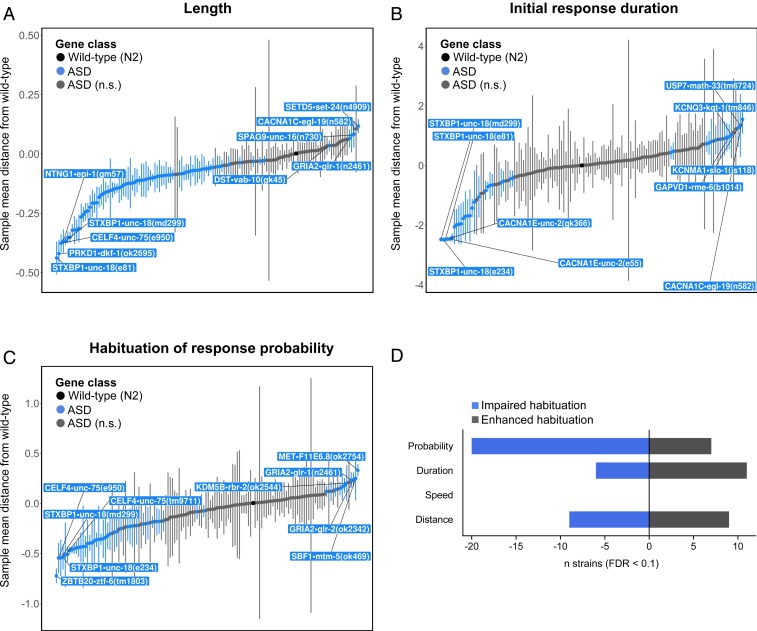
One way to visualize these data is as a series of 26 quantitative reverse genetic screens, where each strain is compared to wild-type on each phenotypic feature to determine if and how that gene plays a role in the biological process of interest (Fig. 1 and Dataset S3). Phenotypes shared by a large number of ASD risk genes were of particular interest, as they would suggest common biological functions among these seemingly diverse genes. We found that mutations in the vast majority of ASD-associated gene orthologs decrease multiple measures of size, most notably length (77/135, 57%), in age-synchronized populations (Fig. 1A). Thus, the majority of genes putatively associated with ASD/neurodevelopmental disorders delay development or impede growth when inactivated.
Fig. 1.
Phenotypic profiles enable rapid reverse genetic screens to identify shared functions among ASD-associated genes. Plots illustrate sample mean distance of each genotype group from wild-type. Strains outside the 95% confidence interval of the wild-type distribution are labeled and colored blue. A maximum of 5 strains are labeled in each direction to prevent overplotting. (A) Length by genotype. (B) Initial response duration by genotype. (C) Habituation of response probability by genotype. Error bars represent 95% confidence interval. (D) The number of strains with normal initial responses that either impair habituation (blue) or enhance habituation (gray) across the 4 response metrics quantified.
Analysis of baseline locomotion revealed that mutations in a large proportion of ASD-associated gene orthologs increase forward movement bias and path length (distance traveled; SI Appendix, Fig. S3A). This means many strains with mutations in ASD-associated gene orthologs spent more time moving forward before reversing. The frequency and duration of spontaneous reversals is modulated by the integration of multiple cross-modal sensory inputs (45), suggesting a widespread imbalance in the neural circuits that control spontaneous forward movement behavior toward increased activity in strains harboring mutations in orthologs of ASD-associated genes.
Assessing the duration, distance, and speed of reversal responses to the first mechanosensory stimulus indicated that mutations in orthologs of ASD-associated genes are approximately equally likely to result in initial hyper- or hyporesponsivity to tactile stimuli (SI Appendix, Fig. S3B; bidirectional analysis of initial sensitivity was precluded only for response probability due to a ceiling effect with >90% of animals responding to the initial stimulus). These results identify positive and negative roles for multiple ASD-associated genes in modulating mechanosensory processing.
Analysis of habituation learning across 135 strains revealed that mutations in many ASD-associated gene orthologs specifically impair habituation of response probability (Fig. 1C). Even after filtering out strains with abnormal initial response probability, ASD-associated gene orthologs were ∼3-fold more likely to impair habituation than enhance it (Fig. 1D and Dataset S3). This is a remarkably specific phenotype that causes the neural circuit to continue responding in an inflexible manner, versus merely impairing the ability to detect stimuli or respond. Importantly, we only observed this phenomenon for response probability; there was no such consistent pattern of habituation impairment for the duration, distance, or speed of the same measured responses (Fig. 1D and Dataset S3). Together, these results suggest that many ASD-associated genes normally mediate plasticity of the likelihood, but not vigor, of responding to mechanosensory stimuli.
Finally, we discovered that distinct sets of genes alter initial sensitivity, habituation, and retention of the same component of the same behavioral response (Fig. 1C and SI Appendix, Fig. S3, and Dataset S3). Adding to the complexity, we also found that different genes affect different components of the same behavior (e.g., some genes affect only response duration but not probability and vice versa). These results support the hypothesis that habituation is controlled by several dissociable genetic mechanisms (43) and underscore the need to assess multiple complex phenotypes to understand the functions of ASD-associated genes.
Phenotypic Profiles of Strains with Mutations in ASD-Associated Genes Define Shared and Unique Functions.
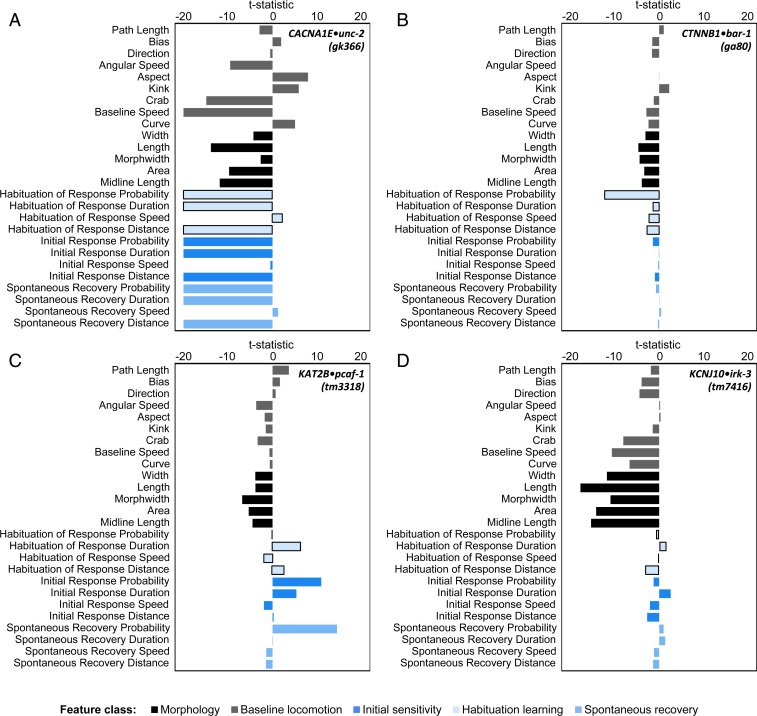
In addition to visualizing scores of all strains on each phenotype, one can also visualize the scores of each strain on all phenotypes, or the “phenotypic profile” for each strain (Fig. 2 and Dataset S4). Classical uncoordinated “unc” mutants, such as the calcium channel subunit CACNA1E•unc-2(gk366), displayed the most severe phenotypic profiles, consistent with their documented roles in neuronal development and function (Fig. 2A). Phenotypic profiles also revealed previously unknown phenotypes even in well-characterized mutants. For example, β-catenin has well-known roles in development, so it follows that CTNNB1•bar-1(ga80) mutants are smaller than wild-type animals. However, here we identify a previously unknown role for β-catenin in habituation learning, evidenced by a profound deficit in habituation of response probability in CTNNB1•bar-1(ga80) mutants (Fig. 2B). Animals carrying mutations in KAT2B•pcaf-1(tm3318), involved in CREB-related transcription coactivation and acetyltransferase activity, have relatively normal baseline locomotion but profound alterations in sensory and learning behaviors that are only revealed through stimulation (Fig. 2C). Conversely and more surprisingly, animals carrying mutations in genes such as KCNJ10•irk-3(tm7416), an inward rectifying potassium channel, have relatively normal sensory and learning behaviors despite profoundly abnormal morphology and baseline locomotion (Fig. 2D). Finally, several strains were not significantly different from wild-type on almost all phenotypic features, such as the membrane palmitoylated protein encoded by MPP6•C50F2.8(ok533) (SI Appendix, Fig. S4 and Dataset S4). Together, these results indicate a remarkable degree of phenotypic modularity and provide a catalog of the unique phenotypic profiles of ASD-associated genes.
Fig. 2.
Phenotypic profiles of strains with mutations in ASD-associated genes define shared and unique functions. (A) Phenotypic profile for CACNA1E•unc-2(gk366). (B) Phenotypic profile for CTNNB1•bar-1(ga80). (C) Phenotypic profile for KAT2B•pcaf-1(tm3318). (D) Phenotypic profile for KCNJ10•irk-3(tm7416).
A Phenomic Database of Strains with Mutations in ASD-Associated Genes.
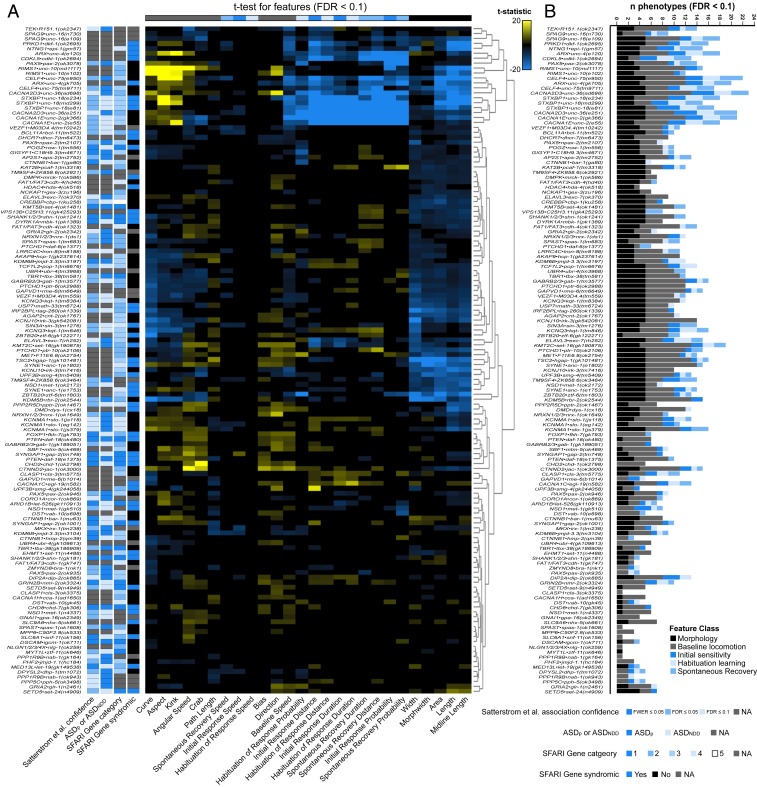
Fig. 3A summarizes the scores of ∼27,000 animals (∼200 animals per genotype) across 135 genotypes and 26 phenotypes for a total of 3,510 in vivo genotype-to-phenotype assessments (Fig. 3A and Datasets S5 and S6). We included several annotations on this heat map and reverse genetic screen plots to denote current estimates of ASD-association confidence and specificity (Fig. 3A and Datasets S7 and S8). For the phenotypes quantified and strains tested, no single phenotype is affected by all putative ASD-associated gene orthologs (Fig. 3A). However, all strains were significantly different from wild-type on at least 1 phenotypic metric (Fig. 3B). A larger number of strains displayed altered morphological and baseline locomotion phenotypes compared to sensory or learning phenotypes (Fig. 3B). Finally, our results indicate that distinct, partially overlapping sets of genes influence the different classes of phenotypes (e.g., naïve sensitivity and habituation learning are influenced by different sets of ASD-associated genes; Fig. 3 A and B).
Fig. 3.
A phenomic database of strains with mutations in ASD-associated genes. (A) Heat map summarizing the phenotypic profiles of 135 strains harboring a mutation in an ortholog of an ASD-associated gene. Cells represent directional t-statistics from comparisons to wild-type controls. t-statistics are clipped at 20, and only cells significant at FDR < 0.1 are colored for ease of interpretation. Interactive heat maps illustrating the full range of t-statistics are provided in Dataset S5. ASDp, ASD predominant; ASDNDD, ASD and neurodevelopmental disorders (8). (B) The number of significantly different features for each strain tested. Stacked bars are color-coded according to feature subclasses.
While there are many potential downstream uses for this database, we endeavored to illustrate a few of the most promising applications. First, we clustered the mutant strains based on their similarity in overall phenotypic profiles with the hypothesis, supported by recent large-scale model organism phenotyping efforts, that phenotypic similarity would enrich for functional genetic interactions among ASD-associated genes. However, while it has been successful for C. elegans morphology and baseline locomotion profiles in the past (46–52), a large-scale phenotypic clustering approach has not been attempted in combination with complex sensory and learning phenotypes. Before clustering, we confirmed the sensitivity and consistency of our phenotypic measures by examining the correlation of independently generated alleles targeting the same gene within the set of strains we tested. Our sample contained 33 genes represented by multiple independently generated alleles (29 genes with secondary alleles and 4 with both secondary and tertiary alleles targeting the same gene). Analysis of the distribution of the overall phenomic correlations between the same gene-independent allele pairs revealed that the average correlation was indeed higher than all other possible gene pairs (n = 9,004) and that the same gene allele pair distribution was skewed toward highly positive correlations (SI Appendix, Fig. S5A). We also included heat maps illustrating only genotype–phenotype relationships seen in all independent alleles of a given gene (Dataset S6).
We then used several clustering methods to group genes based on phenotypic similarity to predict genetic interactions. Hierarchical clustering accurately identified several well-known interactions, such as those between voltage-gated calcium channels CACNA1E•unc-2, Syntaxin binding proteins STXBP1•unc-18, and Rab3 binding proteins RIMS1•unc-10 (Fig. 3A and Datasets S5 and S9) (53, 54). Our analysis also confirmed recently discovered interactions, such as those between the dual specificity kinase DYRK1A•mbk-1 and the histone acetyltransferase CREBBP•cbp-1, and predicted several new interactions (Fig. 3A and Datasets S5 and S9) (55). We then investigated the overall phenotypic architecture of ASD-associated genes. t-distributed stochastic neighbor embedding (t-SNE) and multiple other clustering methods revealed that ASD-associated genes were largely continuously distributed in phenotypic space (SI Appendix, Fig. S5B). We found no evidence for a small number of highly discrete clusters that would suggest distinct “phenotypic classes” of ASD-associated genes (Fig. 3A and SI Appendix, Fig. S5B).
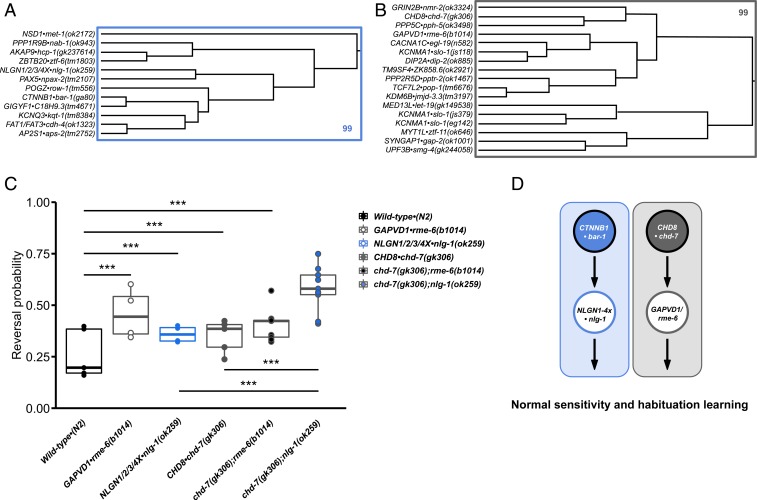
Next, we used epistasis analysis to test some of our interactions predicted by phenotypic proximity in vivo. Using sensory and habituation learning features for hierarchical clustering revealed 2 high-confidence clusters with members that display impaired habituation of response probability and hyperresponsivity to mechanosensory stimuli (increased initial reversal response duration; Fig. 4 A and B and SI Appendix, Fig. S6 A and B and Dataset S9). Genes within these clusters were selected for epistasis analysis based on confidence of ASD association and confirmation of genotype-to-phenotype relationships using analysis of a second allele or transformation rescue. Crossing strains within the same cluster revealed a functional interaction between the Chromodomain Helicase DNA Binding Protein CHD8•chd-7(gk306) and the GTPase-activating protein GAPVD1•rme-6(b1014); the impairment in habituation of response probability of double mutants was not significantly different from single mutants, suggesting that they function in the same genetic pathway to mediate short-term behavioral plasticity (Fig. 4C). A second allele of GAPVD1•rme-6(tm6649) and an additional allele of CHD8•chd-7(gk209) tested after the large-scale characterization also displayed the same phenotypic profile, confirming genotype-to-phenotype relationships (SI Appendix, Fig. S7). CHD8 is a high-confidence ASD-associated gene whereas GAPVD1 is relatively low confidence, yet, when they are inactivated in a model organism, they cause strikingly similar phenotypic profiles and function together to promote normal habituation learning.
Fig. 4.
Combining phenotypic clustering and epistasis analysis to map parallel genetic networks underlying hyperresponsivity and impaired habituation. (A and B) Hierarchical clustering based on sensory and learning features identified 2 sets of genes with members that display impairments in habituation of response probability and hyperresponsivity to mechanosensory stimuli (increased initial reversal response duration). Rectangles outline the largest clusters with AU P values >95%. (C) Final reversal probability across genotypes (average of the 28th to 30th responses). Dots represent individual plate means, and horizontal lines represent median of plate replicates. Binomial logistic regression followed by Tukey’s honestly significant difference (HSD) criterion was used to determine significance of the habituated level (proportion reversing at tap 30) for each pair of strains (***P < 0.001). (D) Parallel genetic pathways of ASD-associated genes underlie impaired habituation learning.
Crossing between clusters revealed that CHD8•chd-7(gk306) and the sole C. elegans ortholog of vertebrate neuroligins NLGN1/2/3/4X•nlg-1(ok259) function in parallel genetic pathways, with double mutants exhibiting additive impairments in habituation (Fig. 4C). Interestingly, we also discovered a synthetic lethal interaction between CHD8•chd-7(gk306) and CTNNB1•bar-1(ga80), suggesting CHD8 can function independently of its canonical role in mediating the Wnt/β-catenin signaling pathway (56). These results are consistent with the observation that Wnt/β-catenin targets are neither up-regulated nor the cause of lethality in CHD8 homozygous knockout mice (57), suggesting that Wnt-independent functions of CHD8 are present in multiple species (58). We combined our results with the recent observation that Wnt/β-catenin signaling increases expression and synaptic clustering of NLGN3 (59) to draw parallel pathways underlying impaired habituation (Fig. 4D). Together, these results demonstrate that systematic phenotypic clustering and epistasis analysis of complex sensory and learning phenotypes present an in vivo approach to map functional genetic network interactions among ASD-associated genes and prioritize candidates that would be missed by focusing on currently high-confidence genes.
Phenomic Profiles Can Be Leveraged for In Vivo Variant Functional Assays.
Another challenge facing ASD is the inability to interpret the functional consequences of the large number of rare variants of uncertain significance (25). Many variants found in ASD are so rare they preclude traditional genetic approaches to infer pathogenicity (25, 26). Experimental variant functional assays, where the variant is introduced into a model system, can be combined with computational approaches to guide clinical assessment, but there is a paucity of such assays for most ASD-associated genes due to a lack of understanding of their normal biological functions (25, 60, 61). Here, we show how the phenomic functional data we generated by studying inactivating mutations in ASD-associated gene orthologs can be combined with the genetic tractability of C. elegans to develop in vivo variant functional assays using complex sensory and learning behaviors as a readout.
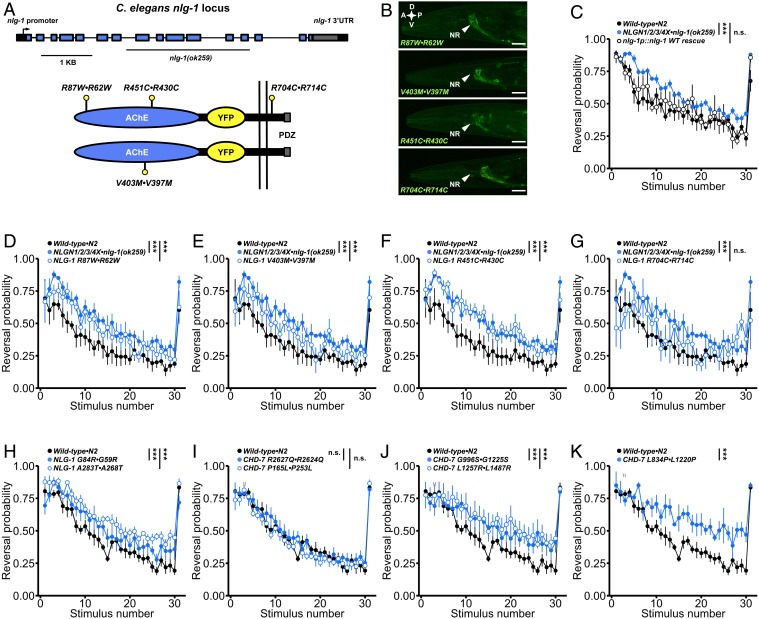
In our large-scale characterization, we discovered that mutations in the C. elegans ortholog of vertebrate neuroligins cause impairments in habituation of response probability. We generated a wild-type nlg-1::YFP fusion transgene and found that expression of this construct in nlg-1(ok259) deletion mutants was sufficient to restore normal habituation learning (Fig. 5 A–C and SI Appendix, Fig. S8A). We then used this transgenic rescue to assess the functional consequences of mutations equivalent to 4 conserved ASD-associated neuroligin missense variants by testing their ability to rescue, revealing varied partial loss of function (Fig. 5 D–G). We found similar results for these variants on 3 additional behavioral functional assays: octanol avoidance chemotaxis, thermotaxis, and sensory integration (SI Appendix, Fig. S9). Each of these functional assays involve distinct neural circuits, suggesting a general partial loss-of-function mechanism due to ASD-associated neuroligin variants in whole organism behavior. All ASD-associated variants were expressed at similar levels and properly localized to synapses in vivo, likely ruling out a simple pathogenic mechanism of improper trafficking or severely reduced cellular abundance (Fig. 5B). We also characterized 2 additional neuroligin variant lines generated with a precise CRISPR-Cas9 genome engineering method (61) and again observed varied partial loss of function (Fig. 5H). Finally, we assessed 5 additional missense variants in chd-7 (orthologous to CHD8 and CHD7) also generated with CRISPR-Cas9 and observed that P165L•P253L and R2627Q•R2624Q displayed wild-type function and habituated normally, while L834P•L1220P, G996S•G1225S, and L1257R•L1487R displayed impairments in habituation learning (Fig. 5 I–K). Importantly, all ASD-associated gene orthologs, including those with no prior functional annotation, were significantly different from wild-type animals on at least one phenotypic metric in our characterization (Fig. 3B), suggesting this will be a broadly applicable approach to decipher variants of uncertain significance.
Fig. 5.
Functional assessment of ASD-associated missense variants. (A) Schematic of the nlg-1(ok259) deletion allele and NLG-1::YFP transgene. Lollipops indicate the approximate locations of the equivalent ASD-associated variants assessed. Note the R430C variant in C. elegans NLG-1 corresponds to a mutation in human NLGN3 whereas the R62W, V397M, and R714C NLG-1 variants correspond to mutations in human NLGN4X. (B) All neuroligin variants were expressed at similar levels and localized properly to synapses in the nerve ring and nerve cords. A, anterior; P, posterior; D, dorsal; V, ventral; NR, nerve ring. (Scale bar: 0.02 mm.) (C) Expression of wild-type NLG-1::YFP rescued nlg-1(ok259) deletion mutant impaired habituation of response probability. (D–G) Each neuroligin variant was scored for its ability to rescue impaired habituation of response probability, revealing varied partial loss of function. (H) Missense variants in nlg-1 generated with CRISPR-Cas9 also displayed varied partial loss of function with corresponding impairments in habituation. Note the G59R and A268T variants in C. elegans NLG-1 correspond to variants in human NLGN4X. (I–K) P165L•P253L and R2627Q•R2624Q missense variants in C. elegans CHD-7 did not display habituation impairments, while L834P•L1220P, G996S•G1225S, and L1257R•L1487R displayed impairments in habituation. Note that the G1225S, L1487R, and R2624Q missense variants in C. elegans CHD-7 correspond to variants in human CHD7, while L1220P and P253L correspond to variants in human CHD8. (C–K) Data shown as mean ± SEM using plates as n. Binomial logistic regression followed by Tukey’s HSD criterion was used to determine significance of the habituated level (proportion reversing at tap 30) for each pair of strains (***P < 0.001). n.s., not significant.
CRISPR-Cas9 Auxin-Inducible Degradation Reveals That nlg-1 Phenotypes Are Reversible by Adult-Specific Reexpression.
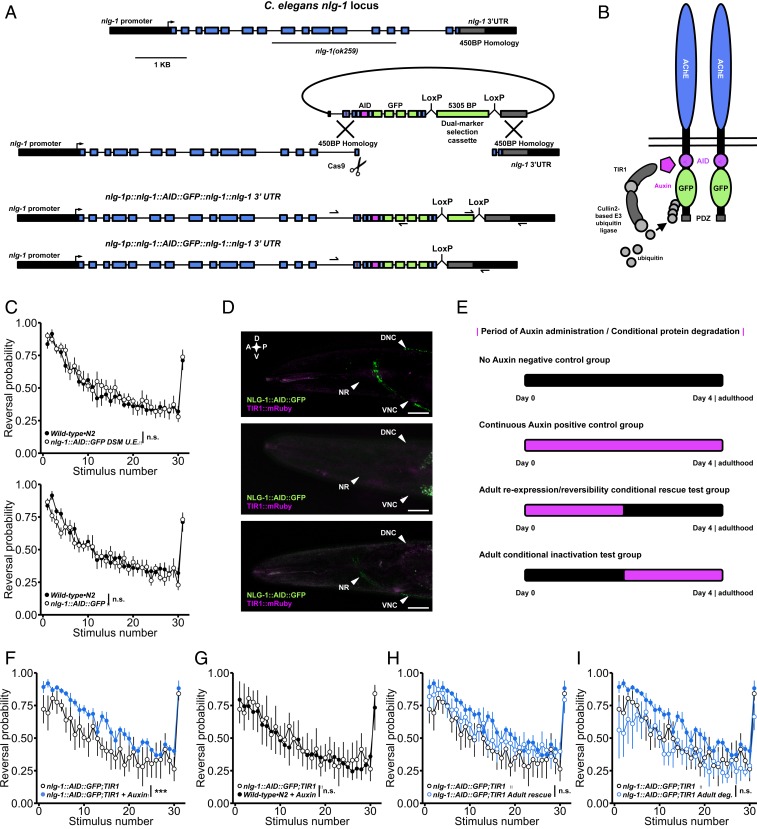
Historically, it was believed that the neurodevelopmental insults caused by monogenic risk factors for ASD were so severe that they would not be reversible in adulthood, and thus any treatment would have to be administered early to be effective. The seminal discoveries that phenotypes caused by mutations in MECP2 could be reversed by transgenic expression of the protein in adulthood, after a presumed critical developmental period had passed, offered tremendous hope for families suffering from Rett syndrome (62). Conversely, the observation that inactivation of MECP2, neuroligins, and several other ASD-associated genes in adulthood could cause severe electrophysiological and behavioral impairments demonstrated that they were not simply neurodevelopmental genes, and instead that they function throughout the lifespan to promote normal sensory and learning behaviors (63–65). Such conditional rescue and inactivation experiments are valuable to ASD research, but they are expensive, technically difficult, and time-consuming in mammalian model systems and are thus not yet practical for many genes. With C. elegans, however, conditional and reversible protein depletion is precise, rapid, and straightforward owing to their genetic tractability and the recent advent of CRISPR-Cas9 auxin-inducible degradation (AID) (66). AID relies on transgenic expression of TIR1, an inducible E3 ubiquitin ligase that targets any protein with an AID degron peptide tag for degradation by the proteasome only when in the presence of its activating hormone auxin (Fig. 6). Moreover, upon removal from auxin, TIR1 is inactivated, allowing rapid reconstitution of protein expression in large populations of freely behaving and intact animals without the need for surgery or vector delivery (66).
Fig. 6.
CRISPR-Cas9 auxin-inducible degradation reveals phenotypes caused by developmental loss of neuroligin can be rescued by adult reexpression. (A) The modified DMS cassette genome editing strategy used to insert GFP and a short degron tag into the endogenous neuroligin locus. The maroon line in the repair template indicates the location of an engineered silent mutation in the protospacer adjacent motif to prevent cleavage of exogenous DNA. (B) Schematic of the NLG-1::AID::GFP transgene. In the presence of auxin, TIR1 is activated targeting the fusion protein for degradation. Following auxin treatment, TIR1 is inactivated, allowing conditional degradation and reexpression of the fusion protein. (C) The fusion protein is fully functional; NLG-1::AID::GFP animals did not display habituation impairments before (Top) or after (Bottom) DMS cassette excision. (D) The fusion protein localizes properly to synapses in the nerve ring and nerve cords (Top). Treatment with 0.025 mM auxin is sufficient for complete degradation of the fusion protein (Middle) that is reversible 48 h after removal from auxin (Bottom). A, anterior; P, posterior; D, dorsal; V, ventral; NR, nerve ring. (Scale bar: 0.02 mm.) (E) Period of auxin administration for each group. (F) Continuous auxin administration recapitulated impairments in habituation of response probability. (G) Wild-type•N2 animals continuously treated with auxin exhibited normal habituation. (H) Adult-specific reexpression of neuroligin partially rescued impaired habituation. Solid blue circles represent continuous auxin administration group (as in F). (I) Adult-specific degradation of neuroligin did not induce habituation impairments (solid blue circles as in F). (C, F, and I) Binomial logistic regression followed by Tukey’s HSD criterion was used to determine significance of the habituated level (proportion reversing at tap 30) for each pair of strains (***P < 0.001). n.s., not significant. Data shown as mean ± SEM using plates as n.
Despite extensive study, adult transgenic rescue tests of behavioral phenotype reversibility have not been completed for any member of the vertebrate neuroligin family (53, 67). We used the CRISPR-Cas9 dual marker selection (DMS) cassette genome editing strategy to insert GFP and a short AID degron peptide tag into the endogenous C. elegans neuroligin locus 13 residues before the stop codon (Fig. 6A; further details provided in SI Appendix, SI Materials and Methods). This fusion protein localized properly to synapses and is fully functional; there were no habituation impairments or gross neuroanatomical abnormalities in genome-edited animals (Fig. 6 C and D and SI Appendix, Fig. S8B). We then crossed this strain into animals expressing TIR1 under a ubiquitous promoter and observed that treatment with 0.025 mM auxin for <10.0 h was sufficient for complete degradation of the functional NLG-1::AID::GFP fusion protein at multiple life stages (Fig. 6D). Moreover, NLG-1::AID::GFP was partially recovered 48 h after removal from auxin (Fig. 6D). We used this approach to test several conditional rescue and inactivation groups simultaneously (Fig. 6E). Animals reared on auxin for continuous degradation displayed habituation impairments equivalent to those of nlg-1(ok259) null mutants, confirming effective degradation (Fig. 6F). Importantly, wild-type animals continuously exposed to auxin displayed no morphological or behavioral abnormalities (Fig. 6G). Strikingly, we observed that, for animals reared on auxin to degrade NLG-1 throughout development, adult-specific expression of neuroligin was sufficient to partially rescue the habituation impairment phenotype (Fig. 6H). Surprisingly, adult-specific degradation of neuroligin did not lead to impaired habituation (Fig. 6I). These results indicate a critical role for neuroligin in generating a circuit properly tuned for normal mechanosensory processing and short-term behavioral plasticity. Further, they suggest behavioral disruptions caused by developmental loss of neuroligins might be at least partially reversible by adult expression. Given the speed of machine vision phenotyping and relative ease of CRISPR-Cas9 genome editing in C. elegans, AID represents a scalable approach that can be applied to diverse ASD-associated genes.
Discussion
We quantified 26 phenotypes for >27,000 animals across 135 genotypes and identified hundreds of genotype–phenotype relationships. When combined with a recent aggregation of all other machine vision behavioral datasets collected in this organism to date, our database more than doubles the number of tracked animals reported in the literature from ∼15,000 to >30,000 (68). By making the raw and processed data available online, we have created open and shareable phenotypic atlas of C. elegans strains carrying mutations in orthologs of ASD-associated genes. We have shown how this database can be used to identify shared functions, map genetic networks, gauge missense variant effect, and determine reversibility of phenotypic disruptions.
We found that the vast majority of strains with mutations in ASD-associated genes displayed delayed development or growth impediments, a finding consistent with a recent large-scale analysis of inactivating mutations in constrained genes in humans (69). We also found that a large number of ASD-associated genes impair habituation, a finding consistent with a recently reported large-scale analysis of ASD- and intellectual disability/developmental delay-associated genes in Drosophila and humans (70). Interestingly, our data show that ASD-associated genes specifically impair habituation of response probability. These results provide a genotype-to-phenotype list (Fig. 1C) that may hint at a common pathological mechanism that impairs plasticity of a neural circuit’s decision to respond without altering response vigor. Indeed, these Drosophila and C. elegans results may reflect a behavioral outcome of circuit-level hyperexcitability recently discovered in several human iPSC-derived neuronal culture models of a number of monogenic ASD risk factors (71, 72). The results also provide a potentially plausible explanation for inconsistent reports of impaired habituation in humans, which variably employ diverse response metrics most often without genetic stratification of patient populations (15, 39). While these shared phenotypes are exciting, our results and those of several other model systems (11, 15, 19, 22, 70, 71) reveal a remarkable diversity in phenotypic disruptions, suggesting that single phenotype functional validation and characterization efforts will be insufficient to capture the complex multifaceted phenotypic disruptions that stem from mutations in ASD-associated genes.
Phenomic Clustering and Epistasis to Map Genetic Networks Among ASD-Associated Genes In Vivo.
We combined phenotypic clustering with epistasis analysis to map functional genetic networks among ASD-associated genes and identified parallel genetic networks centered on CHD8•chd-7 and NLGN1/2/3/4X•nlg-1 that underlie hyperresponsivity and impaired habituation. These results provide in vivo functional support to the proposed broad categorization of ASD-associated genes into those involved in synaptic neuronal communication (neuroligin) and gene expression regulation (CHD8) (6, 8). An exciting question for future research will be to determine how well the phenomic functional interactions delineated here map onto biochemical interactions. Indeed, in vivo genetic networks will serve as an important benchmark from which to compare and extend existing networks of ASD-associated genes. Phenomic clustering will be particularly useful for capturing long-range functional interactions between proteins expressed in different cells or even different points in development, which cannot be detected by measures of direct protein–protein interactions or coexpression.
The strains we characterized were generally continuously distributed in phenotypic space, and we did not detect highly separated discrete phenotypic clusters. Even in this carefully controlled genetic background and environment, we found no evidence for distinct phenotypic classes of ASD-associated genes. This is in contrast to the molecular level, where multiple studies and our work suggest there are functionally distinct classes of ASD-associated genes (3, 8, 73). While phenotypic profiles were continuously distributed, certain genes were closer to each other than others in a manner that reflected underlying molecular interactions. The observation that ASDs are a group of etiologically distinct and variably phenotypically similar disorders provides further motivation for tailored treatments designed on the bases of shared molecular pathway disruptions (3, 73).
For the majority of genes studied in this work, there is a clear C. elegans ortholog with a loss-of-function mutation available from stock centers, oftentimes in an outcrossed strain to minimize the effects of background mutations. For some genes, orthology relationships are less clear, and there was a paucity of outcrossed deletion alleles, making the functional annotations added onto these genes provisional at this point. This limitation becomes less important when the same phenotypes were observed in 2 strains carrying independently generated alleles of the same gene, when the phenotypes were rescued by expression of the wild-type allele, or when the phenotypes were observed in a strain generated with CRISPR-Cas9 genome editing. As such, the reader should gauge the confidence of the genotype-to-phenotype relationships reported for individual strains accordingly when designing follow-up studies, an approach that has proven to be straightforward and extremely valuable when applied to previous large-scale phenotyping of baseline locomotion in similarly generated C. elegans knockout libraries (46, 47, 68, 74).
It is also important to consider whether the functions of ASD-associated genes discovered here will be conserved to their human orthologs, and, if they are conserved, whether they display specificity to ASD. Regardless of the qualitative diagnostic label applied, understanding the functions of these poorly characterized neurodevelopmental disorder-associated genes will be valuable for understanding pathology and designing treatments. Further, understanding gene functions that may not be specific to ASD pathology (e.g., seizures) can still be useful in diagnosis or treating comorbidities. Even in cases where a given gene may be implicated in multiple disorders, or where a deletion allele may not be the best model of genetic etiology, understanding the consequences of loss of function in that gene can offer valuable insights that aid in the interpretation of the functional impact of particular risk variants that may be specific to a given disease pathology. Regarding conservation of function, several of the shared functions for ASD-associated gene orthologs identified here, such as promoting normal development or habituation of response probability, have concurrently been discovered in higher model organisms and human iPSCs, suggesting many of the other functions reported here will also be conserved. Indeed, our analysis and previous work in other model systems suggest that ASD-associated genes are highly conserved throughout evolution (75). It is also worth reiterating that many human genes have been shown to functionally replace their C. elegans orthologs. For example, human NLGN1 and NLGN4 have both been shown to rescue multiple sensory abnormalities caused by loss of C. elegans nlg-1 (33, 76). We have also recently shown that directly replacing daf-18 with only a single copy of its human ASD-associated ortholog PTEN at the endogenous locus using CRISPR-Cas9 is able to rescue multiple sensory abnormalities caused by complete deletion of daf-18 (31). Indeed, it would not be surprising if a highly conserved human gene could replace its C. elegans ortholog. All model systems have their relative strengths and weaknesses, and the fastest and most generalizable insights in ASD research will undoubtedly come from synthesis of large amounts of information derived from diverse model systems and studies of individuals with ASD.
Phenomic Profiles Can Be Leveraged for In Vivo Assays of Missense Variant Effect.
We used neuroligin as a proof of principle to show how our phenomic profiles can be leveraged to establish in vivo variant functional assays. We found that all neuroligin variants tested displayed partial loss of function despite effective expression and proper subcellular localization. Further, we observed that variant functional results were consistent across multiple behavioral functional assays involving distinct neural circuits. While the neurobiological mechanisms underlying the behavioral effects of ASD-associated neuroligin variants currently remain controversial (different studies have implicated altered receptor trafficking, circuit-specific electrophysiological imbalances, etc.) (77–82), our results suggest that a treatment should be designed to increase neuroligin function while taking into account the presence of an existing dysfunctional protein. We also assessed several missense variants in chd-7 and observed habituation impairments in a subset, further prioritizing these candidates for future study (61). Importantly, all strains were significantly different from wild-type on at least one metric, allowing for many diverse in vivo variant functional assessments moving forward. Many genes were significantly different on multiple metrics, giving researchers with interests in particular biological processes the ability to choose a phenotypic functional assay most suited to their needs. A strength of C. elegans will continue to be the ability to rapidly assess multiple variants in complex sensory and learning behaviors in vivo.
Harnessing Phenotypic Profiles for Tests of Adult Reversibility.
We found that adult expression of neuroligin could partially reverse the impaired habituation phenotypes of animals that developed without neuroligin. Interestingly, we also found that adult-specific inactivation did not produce phenotypic disruptions. These results are surprising, as inactivation of NLGN1, NLGN2, and NLGN3 in mature vertebrates produces abnormalities in several complex behaviors (63, 64, 83). However, a plausible explanation could be that neuroligin is only necessary in adulthood for forms of learning and memory that require structural plasticity. Indeed, all studies reporting an adult requirement for neuroligin examined different forms of long-term memory, which, in contrast to the short-term memories studied here, require de novo activity-dependent synapse growth and maturation. Further, the phenotypes observed often did not manifest until weeks after neuroligins were inactivated in adult animals (64). These results led to a model in which neuroligin is sufficient to build a circuit capable of normal sensitivity and short-term habituation at any point throughout the lifespan, but, once that circuit has been built, its function is no longer required for short-term learning. Continued function of neuroligin would then remain necessary only for more complex forms of long-term learning and memory that require de novo activity-dependent growth and maturation of synaptic connections. The C. elegans nervous system is thought to have largely invariant connections across individuals, and there is no clear evidence yet of new synapse formation underlying long-term learning in C. elegans, making this model currently difficult to test. However, an adult form of experience/activity-dependent neural circuit remodeling where neural activity driving male mating behavior rewires synaptic connections has recently been discovered in C. elegans, and neuroligin was identified as a critical component in this process (84). These results suggest that some phenotypic alterations due to developmental loss of neuroligin may be reversible in adulthood. More broadly, they provide a rapid and inexpensive strategy where AID can be used to test reversibility of phenotypic disruptions caused by diverse ASD-associated genes and thereby prioritize candidates for further study in less evolutionarily distant model systems with greater translational potential.
Conclusions
We have completed a systematic phenomics analysis of ASD-associated genes and identified shared delays in development, hyperactivity, and impairments in habituation learning. Our data add to the rapidly expanding use of model organism phenomics to discover the functions of poorly characterized genes identified through genomic sequencing (9, 31, 85–87). We have shown how such data can be used to identify genetic interactions, establish variant functional assays, and develop tests of phenotypic reversibility. This work sets the stage for numerous future experiments using cell-specific and inducible genetic manipulations to delineate each of the specific mechanisms through which mutations in these genes produce their emergent phenotypic profiles. There is substantial evidence that an insufficient understanding of the biology of many disease-associated genes has prevented the successful development of therapies and that preclinical research is biased toward experimentally well-accessible genes (88). It is ideal to be systematic and unbiased whenever possible, an opportunity high-throughput model organisms such as C. elegans afford and thus one we should continue to exploit. As we continue to chart the phenotypic landscape of ASD-associated genes, the complicated paths to understanding mechanisms and developing personalized treatments become simpler to navigate.
Materials and Methods
Ortholog identification, machine vision phenotypic analyses, behavioral assays, statistical analyses, cloning, genome editing, auxin administration, and confocal imaging are described in SI Appendix, SI Materials and Methods. Briefly, Bristol N2 animals were used as the wild-type C. elegans strain. Strains were maintained on NGM (nematode growth medium) plates seeded with Escherichia coli strain OP50 according to standard experimental procedures. At 96 h posthatch, hermaphrodite animals grown and tested at 20 °C were used for all experiments in the large-scale characterization. All C. elegans strains used in this study are listed in the SI Appendix. Transgenic strains were generated by microinjection of transgene DNA or by genetic crosses.
Supplementary Material
Acknowledgments
We thank Dr. Evan Ardiel for discussions regarding the manuscript; Dr. Paul Sternberg, Dr. John Calarco, Dr. Erik Jorgensen, and Dr. Don Moerman and their labs for sharing constructs and protocols or making them available; the C. elegans knockout consortium for alleles and the National Bioresource Project and the C. elegans Genetics Center (National Institutes of Health Office of Research Infrastructure Programs, P40623 OD010440) for strains; Lexis Kepler, Aaron Reiss, and Anna Willms for assistance with experiments; and Christine Ackerley for discussions regarding figure design. This work was supported by a Canadian Institutes of Health Research (CIHR) Doctoral Research Award to T.A.M.; Simons Foundation Autism Research Initiative (205081) and Autism Speaks Grants (1975) to J.B.R., a SFARI award (573845) to K.H. (PI), P.P., and C.H.R. (co-PIs); a SFARI award (367560) to Paul Sternberg (PI) supporting W.-R.W.; and a CIHR project grant (CIHR MOP 130287) to C.H.R.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All data generated in this work are available free online in their raw and processed forms at https://dataverse.scholarsportal.info/citation?persistentId=doi:10.5683/SP2/FJWIL8. All analysis code is freely available at https://github.com/PavlidisLab/McDiarmid-etal-2019_Multi-Worm-Tracker-analysis. All strains, sequence confirmation files, and reagents are available from the corresponding author upon request.
See Commentary on page 26.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912049116/-/DCSupplemental.
References
- 1.American Psychiatric Association , Diagnostic and Statistical Manual of Mental Disorders (American Psychiatric Association Publishing, Washington, DC, ed. 5, 2013). [Google Scholar]
- 2.Sinclair D., Oranje B., Razak K. A., Siegel S. J., Schmid S., Sensory processing in autism spectrum disorders and Fragile X syndrome–From the clinic to animal models. Neurosci. Biobehav. Rev. 76, 235–253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Torre-Ubieta L., Won H., Stein J. L., Geschwind D. H., Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 22, 345–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez B. A., Scherer S. W., Syndromic autism spectrum disorders: Moving from a clinically defined to a molecularly defined approach. Dialogues Clin. Neurosci. 19, 353–371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I., et al. , The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders S. J., et al. ; Autism Sequencing Consortium , Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rubeis S., et al. ; DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satterstrom F. K., et al. , Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. bioRxiv:10.1101/484113 (24 April 2019). [DOI] [PMC free article] [PubMed]
- 9.Stessman H. A. F., et al. , Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 49, 515–526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Roak B. J., et al. , Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellegood J., et al. , Clustering autism: Using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol. Psychiatry 20, 118–125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grissom N. M., et al. , Male-specific deficits in natural reward learning in a mouse model of neurodevelopmental disorders. Mol. Psychiatry 23, 544–555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernier R., et al. , Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beighley J. S., et al. , Clinical phenotypes of carriers of mutations in CHD8 or its conserved target genes. Biol. Psychiatry, 10.1016/j.biopsych.2019.07.020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stessman H. A., Bernier R., Eichler E. E., A genotype-first approach to defining the subtypes of a complex disease. Cell 156, 872–877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Voet M., Nijhof B., Oortveld M. A. W., Schenck A., Drosophila models of early onset cognitive disorders and their clinical applications. Neurosci. Biobehav. Rev. 46, 326–342 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Silverman J. L., Yang M., Lord C., Crawley J. N., Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 11, 490–502 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peñagarikano O., et al. , Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Sci. Transl. Med. 7, 271ra8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoghbi H. Y., Bear M. F., Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb. Perspect. Biol. 4, a009886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon C.-H., et al. , Pten regulates neuronal arborization and social interaction in mice. Neuron 50, 377–388 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paşca S. P., et al. , Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 17, 1657–1662 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin N. D., Paşca S. P., Building models of brain disorders with three-dimensional organoids. Neuron 100, 389–405 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Zaslavsky K., et al. , SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons. Nat. Neurosci. 22, 556–564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coll-Tané M., Krebbers A., Castells-Nobau A., Zweier C., Schenck A., Intellectual disability and autism spectrum disorders ‘on the fly’: Insights from Drosophila. Dis. Model. Mech. 12, dmm039180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starita L. M., et al. , Variant interpretation: Functional assays to the rescue. Am. J. Hum. Genet. 101, 315–325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karczewski K. J., et al. , Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv:10.1101/531210 (30 January 2019).
- 27.Kaletta T., Hengartner M. O., Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 5, 387–398 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Levitan D., Greenwald I., Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer’s disease gene. Nature 377, 351–354 (1995). [DOI] [PubMed] [Google Scholar]
- 29.White J. G., Southgate E., Thomson J. N., Brenner S., The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 30.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R., A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 (1993). [DOI] [PubMed] [Google Scholar]
- 31.McDiarmid T. A., et al. , CRISPR-Cas9 human gene replacement and phenomic characterization in Caenorhabditis elegans to understand the functional conservation of human genes and decipher variants of uncertain significance. Dis. Model. Mech. 11, dmm036517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim W., Underwood R. S., Greenwald I., Shaye D. D., OrthoList 2: A new comparative genomic analysis of human and Caenorhabditis elegans genes. Genetics 210, 445–461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calahorro F., Ruiz-Rubio M., Functional phenotypic rescue of Caenorhabditis elegans neuroligin-deficient mutants by the human and rat NLGN1 genes. PLoS One 7, e39277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Au V., et al. , CRISPR/Cas9 methodology for the generation of knockout deletions in Caenorhabditis elegans. G3 9, 135–144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195, 1167–1171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swierczek N. A., Giles A. C., Rankin C. H., Kerr R. A., High-throughput behavioral analysis in C. elegans. Nat. Methods 8, 592–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDiarmid T. A., Yu A. J., Rankin C. H., Beyond the response-High throughput behavioral analyses to link genome to phenome in Caenorhabditis elegans. Genes Brain Behav. 17, e12437 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Belmadani M. et al., VariCarta: A comprehensive database of harmonized genomic variants found in autism spectrum disorder sequencing studies. Autism Res., 10.1002/aur.2236 (9 November 2019). [DOI] [PubMed]
- 39.McDiarmid T. A., Bernardos A. C., Rankin C. H., Habituation is altered in neuropsychiatric disorders-A comprehensive review with recommendations for experimental design and analysis. Neurosci. Biobehav. Rev. 80, 286–305 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Orefice L. L., et al. , Peripheral mechanosensory neuron dysfunction underlies tactile and behavioral deficits in mouse models of ASDs. Cell 166, 299–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ardiel E. L., et al. , Insights into the roles of CMK-1 and OGT-1 in interstimulus interval-dependent habituation in Caenorhabditis elegans. Proc. Biol. Sci. 285, 20182084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randlett O., et al. , Distributed plasticity drives visual habituation learning in larval zebrafish. Curr. Biol. 29, 1337–1345.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDiarmid T. A., Yu A. J., Rankin C. H., Habituation is more than learning to ignore: Multiple mechanisms serve to facilitate shifts in behavioral strategy. BioEssays 41, e1900077 (2019). [DOI] [PubMed] [Google Scholar]
- 44.McDiarmid T. A., et al. , Systematic phenomics analysis of autism-associated genes reveals parallel networks underlying reversible impairments in habituation. Scholars Portal Dataverse. 10.5683/SP2/FJWIL8. Deposited 26 June 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Bono M., Maricq A. V., Neuronal substrates of complex behaviors in C. elegans. Annu. Rev. Neurosci. 28, 451–501 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Yemini E., Jucikas T., Grundy L. J., Brown A. E. X., Schafer W. R., A database of Caenorhabditis elegans behavioral phenotypes. Nat. Methods 10, 877–879 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu H., et al. , Systematic profiling of Caenorhabditis elegans locomotive behaviors reveals additional components in G-protein Gαq signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 11940–11945 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sieburth D., et al. , Systematic analysis of genes required for synapse structure and function. Nature 436, 510–517 (2005). [DOI] [PubMed] [Google Scholar]
- 49.San-Miguel A., et al. , Deep phenotyping unveils hidden traits and genetic relations in subtle mutants. Nat. Commun. 7, 12990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geng W., Cosman P., Baek J.-H., Berry C. C., Schafer W. R., Quantitative classification and natural clustering of Caenorhabditis elegans behavioral phenotypes. Genetics 165, 1117–1126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Green R. A., et al. , A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145, 470–482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown A. E. X., de Bivort B., Ethology as a physical science. Nat. Phys. 14, 653–657 (2018). [Google Scholar]
- 53.Tong X.-J., et al. , Retrograde synaptic inhibition is mediated by α-neurexin binding to the α2δ subunits of N-type calcium channels. Neuron 95, 326–340.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaeser P. S., et al. , RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell 144, 282–295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S., et al. , DYRK1A interacts with histone acetyl transferase p300 and CBP and localizes to enhancers. Nucleic Acids Res. 46, 11202–11213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakamoto I., et al. , A novel beta-catenin-binding protein inhibits beta-catenin-dependent Tcf activation and axis formation. J. Biol. Chem. 275, 32871–32878 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Nishiyama M., et al. , CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat. Cell Biol. 11, 172–182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barnard R. A., Pomaville M. B., O’Roak B. J., Mutations and modeling of the chromatin remodeler CHD8 define an emerging autism etiology. Front. Neurosci. 9, 477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medina M. A., et al. , Wnt/β-catenin signaling stimulates the expression and synaptic clustering of the autism-associated Neuroligin 3 gene. Transl. Psychiatry 8, 45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun S., et al. , An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 26, 670–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong W.-R., et al. , Autism-associated missense genetic variants impact locomotion and neurodevelopment in Caenorhabditis elegans. Hum. Mol. Genet. 28, 2271–2281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guy J., Gan J., Selfridge J., Cobb S., Bird A., Reversal of neurological defects in a mouse model of Rett syndrome. Science 315, 1143–1147 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang M., et al. , Conditional ablation of neuroligin-1 in CA1 pyramidal neurons blocks LTP by a cell-autonomous NMDA receptor-independent mechanism. Mol. Psychiatry 22, 375–383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang J., et al. , Conditional neuroligin-2 knockout in adult medial prefrontal cortex links chronic changes in synaptic inhibition to cognitive impairments. Mol. Psychiatry 20, 850–859 (2015). [DOI] [PubMed] [Google Scholar]
- 65.McGraw C. M., Samaco R. C., Zoghbi H. Y., Adult neural function requires MeCP2. Science 333, 186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L., Ward J. D., Cheng Z., Dernburg A. F., The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baudouin S. J., et al. , Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science 338, 128–132 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Javer A., et al. , An open-source platform for analyzing and sharing worm-behavior data. Nat. Methods 15, 645–646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganna A., et al. ; GoT2D/T2D-GENES Consortium; SIGMA Consortium Helmsley IBD Exome Sequencing Project; FinMetSeq Consortium; iPSYCH-Broad Consortium , Quantifying the impact of rare and ultra-rare coding variation across the phenotypic spectrum. Am. J. Hum. Genet. 102, 1204–1211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fenckova M., et al. , Habituation learning is a widely affected mechanism in Drosophila models of intellectual disability and autism spectrum disorders. Biol. Psychiatry 86, 294–305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deneault E., et al. , Complete disruption of autism-susceptibility genes by gene editing predominantly reduces functional connectivity of isogenic human neurons. Stem Cell Reports 11, 1211–1225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deneault E., et al. , CNTN5- /+ or EHMT2- /+ human iPSC-derived neurons from individuals with autism develop hyperactive neuronal networks. eLife 8, e40092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winden K. D., Ebrahimi-Fakhari D., Sahin M., Abnormal mTOR activation in autism. Annu. Rev. Neurosci. 41, 1–23 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Brown A. E. X., Yemini E. I., Grundy L. J., Jucikas T., Schafer W. R., A dictionary of behavioral motifs reveals clusters of genes affecting Caenorhabditis elegans locomotion. Proc. Natl. Acad. Sci. U.S.A. 110, 791–796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shpigler H. Y., et al. , Deep evolutionary conservation of autism-related genes. Proc. Natl. Acad. Sci. U.S.A. 114, 9653–9658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hunter J. W., Of Autism and Worms: Neuroligin Mutants and Synaptic Function in C. elegans (University of Oklahoma, Oklahoma City, 2011). [Google Scholar]
- 77.Chanda S., Aoto J., Lee S.-J., Wernig M., Südhof T. C., Pathogenic mechanism of an autism-associated neuroligin mutation involves altered AMPA-receptor trafficking. Mol. Psychiatry 21, 169–177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bemben M. A., et al. , Autism-associated mutation inhibits protein kinase C-mediated neuroligin-4X enhancement of excitatory synapses. Proc. Natl. Acad. Sci. U.S.A. 112, 2551–2556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakanishi M., et al. , Functional significance of rare neuroligin 1 variants found in autism. PLoS Genet. 13, e1006940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chadman K. K., et al. , Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism Res. 1, 147–158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Etherton M., et al. , Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc. Natl. Acad. Sci. U.S.A. 108, 13764–13769 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tabuchi K., et al. , A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science 318, 71–76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bariselli S., et al. , Role of VTA dopamine neurons and neuroligin 3 in sociability traits related to nonfamiliar conspecific interaction. Nat. Commun. 9, 3173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hart M. P., Hobert O., Neurexin controls plasticity of a mature, sexually dimorphic neuron. Nature 553, 165–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thyme S. B., et al. , Phenotypic landscape of schizophrenia-associated genes defines candidates and their shared functions. Cell 177, 478–491.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kochinke K., et al. , Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am. J. Hum. Genet. 98, 149–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wangler M. F., et al. ; Members of the Undiagnosed Diseases Network (UDN) , Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 207, 9–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stoeger T., Gerlach M., Morimoto R. I., Nunes Amaral L. A., Large-scale investigation of the reasons why potentially important genes are ignored. PLoS Biol. 16, e2006643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.