Abstract
Objectives
Low genetic diversity can lead to reduced average fitness in a population or even extinction. Preserving genetic connectivity across fragmented landscapes is therefore vital to counteract the negative consequences of genetic drift and inbreeding. This study aimed to assess the genetic composition and consequently the conservation status of a nationwide sample of European hedgehogs (Erinaceus europaeus) in Denmark.
Methods
We applied an adaptation of the genotyping by sequencing (GBS) technique to 178 individuals from six geographically distinct populations. We used a Bayesian clustering method to subdivide individuals into genetically distinct populations. We estimated individual observed (iHO), observed (HO), and unbiased expected (uHE) heterozygosity, inbreeding coefficient (FIS), percentage of polymorphic loci (P%) and tested for deviations from Hardy-Weinberg equilibrium (HWE). We used linear models to test for potential anthropogenic effects on the genetic variability of hedgehogs with iHO, uHE, P% and FIS as response variables, and assessed the demographic history of the population.
Results
The Danish hedgehog population is composed of three genetic clusters. We found a mean P% of 54.44–94.71, a mean uHE of 0.126–0.318 and a mean HO of 0.124–0.293 in the six populations. The FIS was found to be significantly positive for three of the six populations. We detected a large heterogeneity of iHO values within populations, which can be due to inbreeding and/or fragmentation. FIS values decreased with increasing farmland density, but there was no significant association with human population or road density.
Conclusions
We found a low level of genetic variability and evidence for genetic substructure and low effective population size, which are all consequences of habitat fragmentation. We failed to detect signs of a recent population bottleneck or population increase or decline. However, because the test only identifies recent changes in population size, we cannot reject the possibility of a longer-term decline in the Danish hedgehog population.
Introduction
The western European hedgehog (Erinaceus europaeus, hereafter referred to as “hedgehog”), is a widely-distributed nocturnal generalist predator species found in a range of habitats in the British Isles, New Zealand and continental Europe, from Iberia and Italy northwards into Scandinavia [1, 2]. Despite their wide geographic distribution, the species is feared to be in decline, based on research conducted at collected at national and local scale from several western European countries including the UK, Belgium, the Netherlands, Sweden and Germany [3–12]. It is suggested that the decline is more severe in the rural than urban areas [5, 7]. Although we lack population trend data for Denmark, it is likely that the situation is similar because of comparable habitat fragmentation, landscape structure, farm management practices and climate across north-western European countries.
Potential drivers of this decline include habitat loss and fragmentation, which are a great concern in terrestrial ecosystems in general [13, 14]. In Europe, intensified agricultural practices often include the removal of hedgerows to create large, homogenous, and intensively managed fields [15]. These practices have particular relevance for hedgehogs because hedgerows and field margins are important habitats for rural hedgehogs [16, 17]. In addition, these areas function as corridors connecting suitable habitats [18].
Habitat fragmentation by roads is famously a source of hedgehog mortality via roadkill [10, 19–21], but also has more subtle effects. Fragmentation reduces habitat connectivity, which is important for movement and dispersal and therefore has a direct effect on gene flow with consequences for fitness, adaptability and survival of local populations [22, 23]. Although habitat fragmentation is considered most pronounced in cities [24], where the dwindling green spaces can form crucial habitat networks [25], rural fragmentation remains a concern.
The small home range sizes and short nightly travel distances of hedgehogs mean that they are likely to be vulnerable to even small amounts of fragmentation. Although they can travel 2–3 km a night, adult hedgehog home ranges are ~20–30 ha for males and ~10 ha for females, expanding temporarily during the mating season [2]. Juveniles do not disperse far when reaching independence and leaving their natal nests [26] and adults appear to remain in the same area throughout their lives [1]. In addition, studies on relocated hedgehogs indicate that they do not disperse far even when released into a foreign [27] and unfavourable habitat [28].
In addition to habitat fragmentation, poisons, pollution and competition can affect hedgehog populations. The widespread application of molluscicides, insecticides and rodenticides can negatively influence hedgehog populations via secondary poisoning and elimination of prey items [29–32] and predation by, and inter-specific competition with, badgers (Meles meles) can be an important mortality source [9, 33–36]. Hedgehogs are increasingly associated with residential areas, possibly due to greater food availability around humans (including natural prey and anthropogenic sources), more suitable nest sites and a lower risk of predation by badgers [9, 28, 33–37].
Genetic diversity of hedgehogs
Maintaining gene flow across fragmented landscapes is necessary to prevent the negative consequences of genetic drift, reduced genetic diversity, and inbreeding depression, which can result in less viable and less fertile offspring [38, 39]. The tools of conservation genetics, including genetic rescues, are useful for evaluating, conserving and managing genetically vulnerable, fragmented populations [40]. Research into the population genetics of hedgehogs is therefore useful for assessing conservation status.
Of particular concern in this context are the degrees of heterozygosity and inbreeding. Hedgehogs are promiscuous and have hetero-paternal superfecundation [41], which may reduce inbreeding, since a litter can consist of several half-siblings instead of full siblings [41]. This could be beneficial if mating does takes place between siblings in isolated populations. However, it is currently unknown whether hedgehogs are able to actively differentiate between kin and non-kin e.g. during the mating season. Nevertheless, it is clear that if a small, isolated population is severely inbred, and there is no distinction between kin and non-kin when choosing mates, the population will most likely become even more vulnerable due to the increased degree of inbreeding.
Previous genetic research on hedgehogs has been conducted using microsatellites [23, 41–53]. Bolfikova and Hulva (2012)[48] used population and landscape genetic approaches to describe the population structure and patterns of gene flow of E. europaeus and E. roumanicus in the central European contact zone between the two species. They found that a homogenous population of E. europaeus, had been divided by two large rivers in the Czech Republic (Vlatava and Elbe), into a subpopulation in the western part, and two subpopulations with a mosaic location pattern in the eastern part of the country. They found a significantly lower observed heterozygosity (HO) than expected heterozygosity (HE) in five of the nine microsatellite loci studied (n = 131), with a mean HO of 0.695 and a mean HE of 0.687, using the mitochondrial control region and nuclear microsatellites. In the UK, a study of 42 individuals in an isolated population of hedgehogs in the Regent’s Park, London (166 ha), showed a low genetic diversity, with a mean HE of 0.197, and a mean HO of 0.198 (nloci = 6) [52]. Becher and Griffiths (1998)[43] detected a restriction of gene flow between eight small populations of hedgehogs in a 15 km2 fragmented landscape in Oxfordshire, UK and found a statistically significant genetic differentiation among the studied populations (n = 160, nloci = 6) and a mean HO of 0.7. According to the authors, these results indicated that the hedgehogs of Oxfordshire had a restricted dispersal which may have been caused by human-mediated barriers such as roads and train tracks in the landscape [43]. Braaker et al. (2017)[23] studied the habitat connectivity and spatial genetic structure of 147 hedgehogs residing in Zurich, Switzerland, with an area of 88 km2 (nloci = 10). The population of hedgehogs in Zurich were divided into three genetic clusters, separated by two rivers and the major transportation axes. Genetic diversity measures were similar between the three clusters, and the F coefficients (FIS) were low. Mean HE ranged between 0.569–0.627 for the three clusters, and the mean HO ranged between 0.523–0.631 [23]. In summary, previous genetic studies on hedgehogs in Europe based on microsatellite techniques, have found a mean HE ranging between 0.197 and 0.687.
The Danish context
Hedgehogs have a long history in Denmark. Archaeological evidence from Mesolithic sites (Maglemosian cultures) confirms their presence in 7500 BC and other evidence suggests that they immigrated in the early Preboreal around 9550 BC [54]. Research into the glacial refugia and interglacial expansion of the European hedgehog showed three monophyletic clades [46]. The first from Italy northwards through Austria, Switzerland, Germany, the Netherlands, Scandinavia and Estonia. The second was only found in western Europe, from Spain northwards through France, the Netherlands and into the UK and Ireland. The third clade was restricted to Sicily. A single clade dominated in the Scandinavian hedgehogs [46].
Denmark consists of the large peninsula Jutland and several islands of varying sizes. The larger islands are connected by long bridges (0.75–17 km), which hedgehogs are unlikely to cross, isolating the local hedgehog populations in the different areas of Denmark. The total area of Denmark is 43,000 km2 [55], and 62% of this is arable land [56]. Altogether, Denmark has 74,728 km of roads and 1,737,000 cars for 5,800,000 people [57–59]. The number of cars in Denmark has increased with 22% since 2008 [58], and an increase in road traffic could likely influence the number of hedgehogs being killed by cars [60].
For this study we adapted and optimised the use of a second-generation genotyping technique, genotyping by sequencing (GBS) for the genetic analysis of European hedgehogs.
The main aims of this study were: i) To develop and test a set of SNPs which can be used for investigating the genetic structure and variability of the European hedgehog on a broader scale ii) to evaluate the patterns of the genetic diversity distribution in the Danish hedgehog populations, iii) to investigate potential anthropogenic effects on the genetic variability of the hedgehogs and iiii) to estimate the historical changes in their effective population size (Ne) through genetic signatures.
Materials and methods
The genetic samples were obtained as part of a nationwide citizen science project with the general aim of describing hedgehog ecology in Denmark. By use of local and national media and a project website, volunteers were encouraged to collect dead hedgehogs from May to December 2016. A total of 697 dead hedgehogs originating from all parts of Denmark were collected. The volunteers were instructed to record the date and location of the find and deliver the dead hedgehog to the nearest of 26 collection stations, distributed nationally. The hedgehog carcasses were stored locally at -20°C. Members of the research staff regularly transported the collected, dead hedgehogs to university laboratories, where they were thawed and necropsied from August 2016 to May 2018. During the necropsies, tissue samples from skin and muscle were obtained for the genetical analyses. The DNA samples were stored at -20°C.
Genetic samples from 178 individuals dispersed throughout Denmark were used in the study (Table 1, Supporting Information S1 Table and S2 Table [61]). The selection of samples was based on the quality of the extracted DNA from approximately 330 samples originating from all parts of Denmark. We chose 214 samples for further analyses. The later filtering of samples, described in the section “Filtering Raw Sequence Data, Mapping and SNP Calling”, reduced the sample size to 178 individuals.
Table 1. Distribution of genetic samples in the study.
| Location | Abbreviation | Area in km2 | Number of individuals |
|---|---|---|---|
| Jutland north of the Limfjord | JNL | 5340 | 9 |
| Jutland south of the Limfjord | JSL | 23873 | 71 |
| Funen | FN | 3479 | 15 |
| Zealand | Z | 7031 | 51 |
| Lolland and Falster | LFA | 1797 | 18 |
| Bornholm | BH | 588 | 14 |
| Total: | 178 |
An overview of the distribution of the genetic samples in the study.
Sample preparation
DNA was extracted from muscle and skin tissues (1–2 mg) using the DNeasy Blood and Tissue kit (QIAGEN, Germany) and subsequently digested with Sau96I (NEB) and ligated to adapters [62]. The ligated samples (50μL containing approximately 50 ng DNA pooled from 4 separate samples) were purified with AMPure XP beads (Beckman Coulter, USA) and amplified using the Phusion High-Fidelity PCR kit (Thermo Scientific, USA). The following PCR conditions were applied: 72°C in 5 min, 98°C in 30 sec, followed by 20 cycles of 98°C in 10 sec, 66°C in 30 sec and 72°C in 30 sec, with a final extension at 72°C for 5 min. Finally, the amplified barcoded DNA was purified with AMPure XP beads (Beckman Coulter, USA) and the DNA concentrations were determined by Qubit (Thermo Scientific, USA) and visualized on the TapeStation 2200 using a D1000 ScreenTape (Agilent, USA). Paired-end (2x151 bp) sequencing was performed on an Illumina HiSeq X platform by Admera Health (USA). The described method was previously tested in a pilot study using a few individuals from South Jutland by Rasmussen et al. (2019)[63].
Barcoding analysis
The i7 barcodes of the dual-barcoded sequenced reads were demultiplexed using bcl2fastq2 version 1.0.0 (Illumina, USA) allowing zero mismatch. The i5 barcodes were demultiplexed using Fastq-multx version 1.02.772 (https://github.com/brwnj/fastq-multx) allowing one mismatch in order to remove the barcode sequences from the sequenced reads.
Filtering raw sequence data, mapping and SNP calling
Adaptor quality trimming were accomplished by Trim Galore using default parameters (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore). Burrows-Wheeler Aligner (BWA) was used to align the reads against the hedgehog (Erinaceus europaeus) using the EriEur2.26 reference genome. Only reads with a mapping quality of at least 30 were used for the further analysis. Variants were called using The Genome Analysis Tool Kit’s HaplotypeCaller, and joint genotyping was performed using GenotypeGVCFs. Initial filtering was performed using SelectVariants and filtered for SNPs, bi-allelic sites, and mapping quality > 30 [64]. Minor allele frequency (MAF) was estimated from the read coverage, and SNPs were filtered using a minimum of 1% MAF (average variant allele frequency < 0.99 and > 0.01). Finally, SNPs were filtered with a read coverage between 20 and 100 and a maximum number of missing data of 25%. Individuals with more than 25% missing data were not included in the analysis.
Genetic variability and population structure
We assessed genetic diversity using four quantities: The percentage of polymorphic loci (P%), the FIS coefficient (FIS), unbiased expected (uHE) and observed (HO) heterozygosity. These were all estimated using GENALEX v. 6.5 [65]. In addition, the individual level of heterozygosity (iHO) was calculated for every individual, plotted in a box plot, and tested for significant differences among locations with a one-way ANOVA. The pairwise comparisons among locations were conducted with a Tukey’s HSD test. The mean iHO´s within every population were ranked from the lowest to the highest value and plotted together in order to show the populations with the lowest and the highest levels of iHO.
Furthermore, the degree of genetic differentiation among the six populations was quantified with fixation index (FST), and, for each population, a test for departure from Hardy-Weinberg equilibrium (HWE) was performed using GENALEX v. 6.5.
In addition to measuring genetic differentiation and variability, population genetic structure of the sampled individuals (n = 178) was assessed using a Bayesian clustering method, implemented with STRUCTURE v. 2.3, which clusters individuals into genetically distinct populations/groups (K) based on their allelic frequency at multiple loci and minimise deviations from HWE within groups [66]. To illustrate the population genetic structure of hedgehogs, 10 independent runs of K = 1–5 were carried out on the individuals with 106 Markov chain Monte Carlo (MCMC) iterations and 105 burn-in period on the basis of independent allele frequencies and admixture ancestry model. Additionally, we ran STRUCTURE of K = 1–7 separately for every population to test if further substructuring could be detected within the particular populations. The accurate number of populations (K) was determined according to the ΔK formula [66] using the program STRUCTURE HARVESTER [67]. In addition to this Bayesian clustering approach, a multivariate ordination of individual genotypes was obtained by principal component analysis (PCA) using the software GENODIVE [68] and plotted with the software PAST version 1.90 [69].
Assessment of the demographic history
Tests for recent population declines or expansions in the population size were performed for every population with the program BOTTLENECK v. 1.2, after 15,000 iterations assuming an infinite allele model (IAM) according to a deficiency of the rare alleles and an excess of the heterozygosity. The model assumes that a population (large size) is in mutation-drift equilibrium and that as a consequence of a severe restriction in size the loss of alleles (especially the rare ones) occurs at a higher pace that the loss of the genic heterozygosity. Consequently, a deficiency in the total number of alleles is detected when compared to the number of expected alleles in a large population with the same observed heterozygosity. The deficiency in allele number will only be apparent until the population reaches the mutation-drift equilibrium again [70, 71].
Estimation of potential anthropogenic effects on genetic variability of hedgehogs
To investigate whether the differences in genetic variability found between the hedgehog populations were associated with human population density in their area, road density and farmland density of the areas from which the studied hedgehogs derived, we fitted linear models in R [72]. The response variables were iHO, uHE, FIS and % polymorphic loci (%P) and the explanatory variables were human population density per km2 [73, 74], farmland per km2 [75], and kilometres of roads per km2 [58] (The dataset used is available in Supporting Information S3 Table [61]).
For each of the four response variables (iHO, uHE, FIS and %P) we first fitted models with a single explanatory variable at a time (i.e. three models for each of the three response variables), followed by linear models with two explanatory variables at a time. Then we fitted models that included all three explanatory variables as main effects. Lastly, we prepared models that included two explanatory variables at a time (e.g. road density and farmland density) and the interactions between them. We tested the significance of the interaction term by comparing models that included the interaction term to those that did not include the interaction term, using an ANOVA test.
Research ethics
Ethical approval was not required for this research, because the hedgehogs used in the study had already died of natural causes either in the wild or in care at a hedgehog rehabilitation centre. Additionally, the volunteers deciding to assist in the collection of dead hedgehogs did so of their own free will and were instructed on how to keep a high hygienic standard and prioritise traffic safety when collecting the dead hedgehogs through our project website.
Results
Genotypes
Using the Sau96I restriction enzyme we were able to recover a total of 2.4 million high-quality SNPs. Filtering for MAF > 1% estimated from the read coverage, a maximum of 25% missing data and a read coverage ranging from 20 to 100, resulted in 2902 applicable SNPs. Individuals containing less than 75% of the selected SNPs were not included in the analysis and this resulted in 178 individuals used for studying the genetic variability (the GENEPOP dataset is available in Supporting Information S2 Table [61]).
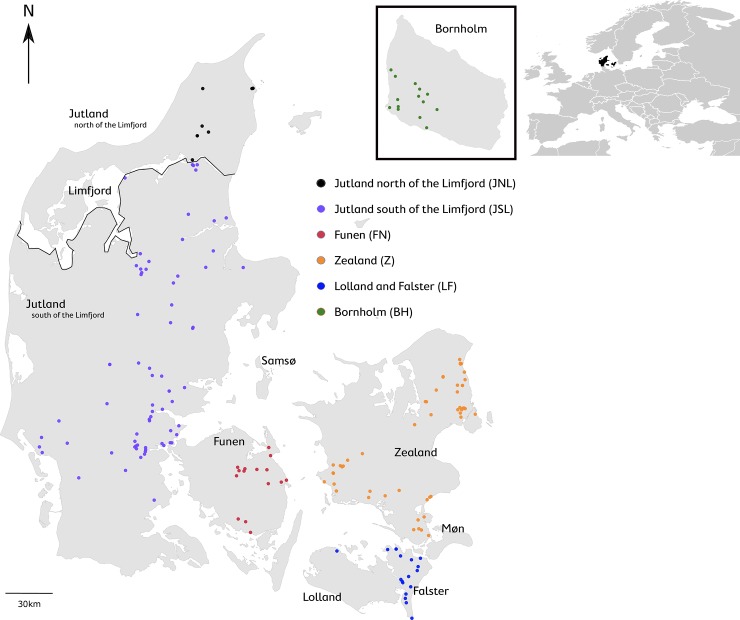
The geographical sampling locations reported by the volunteers are shown in Fig 1, representing Jutland north of the Limfjord (JNL), Jutland south of the Limfjord (JSL), Funen (FN), Zealand (Z), Lolland and Falster (LFA) and Bornholm (BH). The location data may have varying levels of accuracy, because volunteers provided this data without verification.
Fig 1. Geographical overview of the samples.
Map of Denmark indicating the locations of the 178 hedgehogs used in the study.
Genetic variability and structure
We found that the unbiased expected heterozygosity (uHE), mean individual heterozygosity iHO and the F coefficient (FIS), varied with location (Fig 2). uHE and iHO showed marked variation among regions, with the lowest value reported on Bornholm (BH) and the highest in Jutland south of the Limfjord (JSL). The measure of uHE was very similar in Funen (FN) and Jutland north of the Limfjord (JNL). Mean iHO values from Lolland and Falster (LFA), Jutland north of the Limfjord (JNL) and Zealand (Z) were almost identical. Of the three statistically significant and positive FIS values (FN, JSL, Z) the levels were similar between Jutland south of the Limfjord (JSL) and Zealand (Z), but markedly lower on Funen (Fig 2, Table 2).
Fig 2. Plots of genetic heterozygosity and location.
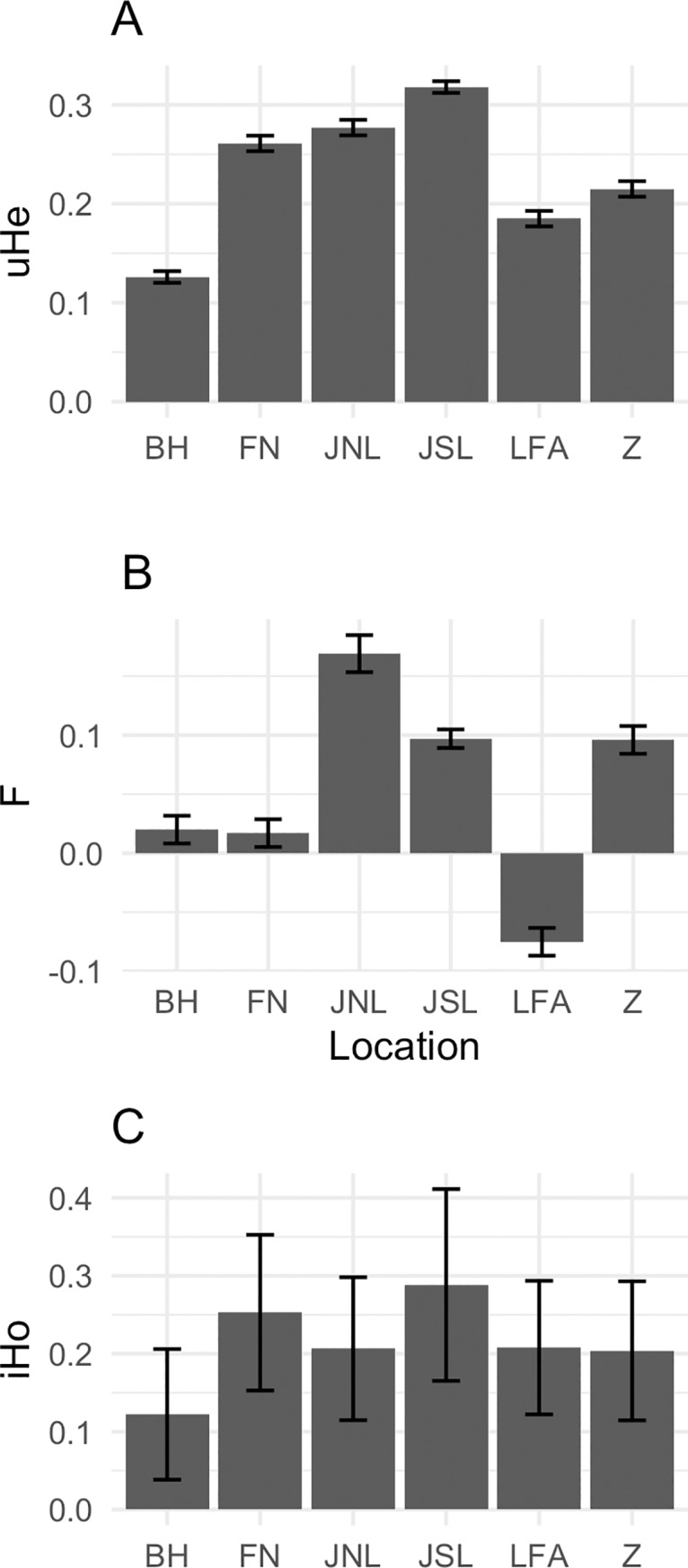
Measures of (A) unbiased expected heterozygosity (uHE) and (B) F coefficient (FIS) and (C) individual observed heterozygosity (iHO), vary with location. Height of the bars indicate the estimated value and error bars represent the 95% confidence intervals of these estimates. Locations are as follows: BH = Bornholm, FN = Funen, JNL = Jutland north of the Limfjord, JSL = Jutland south of the Limfjord, LFA = Lolland and Falster and Z = Zealand.
Table 2. Table of genetic heterozygosity.
| Locations | Abbreviations | Area km2 | uHE | FIS | HO | iHO | %P | HWE test | |
|---|---|---|---|---|---|---|---|---|---|
| Jutland north of the Limfjord | JNL | 5340 | Mean | 0.277 | 0.169 | 0.212 | 0.206 | 72.72 | NS |
| SE | 0.004 | 0.008 | 0.004 | 0.016 | |||||
| Jutland south of the Limfjord | JSL | 23873 | Mean | 0.318 | 0.097 | 0.293 | 0.288 | 94.79 | *** |
| SE | 0.003 | 0.004 | 0.004 | 0.008 | |||||
| Funen | FN | 3479 | Mean | 0.261 | 0.017 | 0.253 | 0.253 | 79.08 | *** |
| SE | 0.004 | 0.006 | 0.004 | 0.013 | |||||
| Zealand | Z | 7031 | Mean | 0.215 | 0.096 | 0.206 | 0.204 | 87.08 | *** |
| SE | 0.004 | 0.006 | 0.004 | 0.006 | |||||
| Lolland and Falster | LFA | 1797 | Mean | 0.185 | -0.075 | 0.208 | 0.208 | 67.73 | NS |
| SE | 0.004 | 0.006 | 0.005 | 0.010 | |||||
| Bornholm | BH | 588 | Mean | 0.126 | 0.02 | 0.124 | 0.122 | 54.44 | NS |
| SE | 0.003 | 0.006 | 0.004 | 0.011 |
Table presenting the percent of polymorphic loci (P%), F coefficient (FIS), unbiased expected (uHE), observed (HO) and individual observed (iHO) heterozygosity and a test for Hardy-Weinberg equilibrium (HWE) for the six hedgehog populations studied.
The genetic polymorphism increased according to the size of the regions, ranging from 54.44% in BH to 94.79% in JSL (Table 2).
The uHE ranged from uHE = 0.126 in BH to uHE = 0.318 in JSL whereas, the HO ranged from HO = 0.124 in BH to HO = 0.293 in JSL (see Table 2).
We detected a significant deviation for HWE in JSL, FN and Z and in all cases the deviations were due to a deficiency of heterozygotes which is also reflected by the positive FIS values which ranged from -0.075 in LFA to 0.17 in JNL (Table 2).
A box plot of the individual heterozygosity iHO for every individual is presented in the Supporting Information S1 Fig [61]. The one-way ANOVA conducted for testing significance of the mean iHO among the six populations investigated was highly significant: F 5,172 = 31.92, p < 0.0001). The Tukey’s test found several significant differences of the mean iHO between populations (Supporting Information S4 Table [61]). The JSL population had significantly higher iHO compared to all the remaining populations with the exception of FN population. In addition, BH had a significantly lower mean iHo than all the other populations. Lastly, FN has significantly higher iHO than Z. The plots of the iHO ranked in ascending order is showing that the populations with the lowest iHO are BH, followed by JSL which is however quickly increase in iHO (indicating that few individuals have low values), followed by Z, JNL, LFA and finally FN which showed the highest starting levels of iHO (Supporting Information S1 Fig and S2 Fig [61]).
We found that all the pairwise fixation index (FST) values between populations were highly significant (p < 0.001) with a FST range from 0.034 between JNL and JSL and FST = 0.321 between JNL and BH (see Table 3), indicating that the six populations of hedgehogs tested are genetically different.
Table 3. Pairwise FST values matrix.
| JNL | JSL | FN | Z | LFA | BH | |
|---|---|---|---|---|---|---|
| JNL | ||||||
| 0.034 | JSL | |||||
| 0.146 | 0.108 | FN | ||||
| 0.201 | 0.159 | 0.046 | Z | |||
| 0.237 | 0.191 | 0.063 | 0.030 | LFA | ||
| 0.321 | 0.268 | 0.192 | 0.182 | 0.204 | BH |
Pairwise FST values matrix. All the FST values were highly significant (p< 0.001). Locations: JNL = Jutland north of the Limfjord, JSL = Jutland south of the Limfjord, FN = Funen, Z = Zealand, LFA = Lolland and Falster and BH = Bornholm.
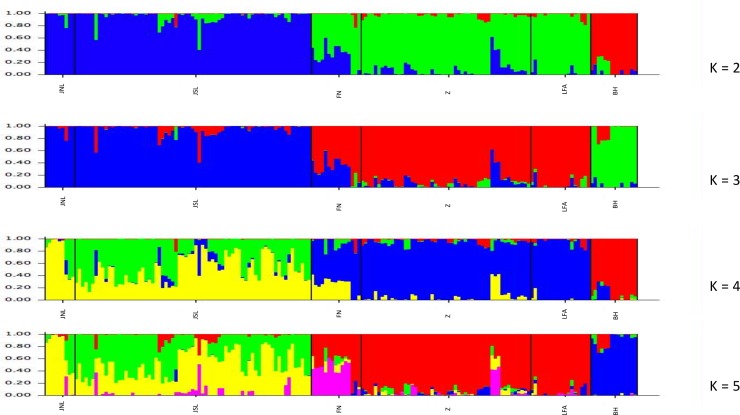
The Bayesian clustering of the genotyped data assigned the highest posterior probability: Estimated Ln Prob of Data = -144614.8, Variance of ln likelihood = 1958.7 for K = 3 (the plot of the Ln Prob of Data versus K is available in the Supporting Information S3 Fig [61]). We found that the three clusters included: 1) Jutland north of the Limfjord (JNL) and Jutland south of the Limfjord (JSL); 2) Funen (FN), Zealand (Z), Lolland and Falster (LFA); 3) Bornholm (BH). We did not observe further evidence of subtructuring within populations when testing for K = 4 and K = 5. These findings were also confirmed by the lack of further substructuring, when testing for K = 1–7 for each population analysed separately (Supporting Information S4 Fig [61]).
The PCA (Supporting Information S5 Fig [61]) was concordant with the clustering of STRUCTURE (Fig 3), the PC1 (Eigenvalues 19.54; variance explained; 79.24%) and PC2 (Eigenvalues 5.12; variance explained; 20.76%) and the convex hulls kept the three clusters well separated (Supporting Information S5 Fig [61]).
Fig 3. STRUCTURE analysis plots.
Plots of the STRUCTURE analysis for K = 2–5 for all the six detected populations: Jutland north of the Limfjord (JNL), Jutland south of the Limfjord (JSL), Funen (FN), Zealand (Z), Lolland and Falster (LFA) and Bornholm (BH).
Demographic changes
We found no signs of recent increase or decline in the population in the six populations tested by use of the infinite allele model (IAM).
Estimation of potential anthropogenic effects on genetic variability of hedgehogs
Testing for effects of farmland density, population density and road density in the particular areas from where the hedgehogs derived on the genetic variability observed in the study, we found no statistically significant associations between iHO, uHE and % polymorphic loci (%P) for each of the six populations and farmland density, road density or population density, fitted as single main effects (Table 4, the dataset used is available in Supporting Information S3 Table [61]). Models representing two or three explanatory variables at a time as main effects, with iHO, uHE or %P as response variables, showed no significant associations. We furthermore tested for a significant interactive effect between the different explanatory variables by comparing models with and without the interactions, but again found no significant relationship. Additionally, the explanatory variables road density and population density had no statistically significant effect on FIS. However, we found a statistically significant association between FIS and farmland density (Table 4). FIS decreases with increased farmland density (with a factor 1.0315). Models fitted with farmland density and road density, as well as all three explanatory variables at a time, as main effects, also showed significant associations between farmland density and FIS.
Table 4. Statistical results from the estimation of potential anthropogenic effects on genetic variability of hedgehogs.
| Explanatory variables | iHO | uHE | FIS | %P |
|---|---|---|---|---|
| Road density | F[1,4] = 0.5045, p = 0.517 | F[1,4] = 0.935, p = 0.388 | F[1,4] = 0.019, p = 0.898 | F[1,4] = 0.004, p = 0.95 |
| Farmland density | F[1,4] = 0.00006, p = 0.994 | F[1,4] = 0.405, p = 0.559 | F[1,4] = 9.603, p = 0.036* | F[1,4] = 0.456, p = 0.537 |
| Population density | F[1,4] = 0.011, p = 0.921 | F[1,4] = 0.003, p = 0.958 | F[1,4] = 0.356, p = 0.583 | F[1,4] = 1.184, p = 0.338 |
| Road density + Farmland density | F = 0.381, p = 0.581 F = 0.019, p = 0.898 | F = 0.89, p = 0.415 F = 0.81, p = 0.434 | F = 0.063, p = 0.394 F = 10.491, p = 0.048* | F = 0.006, p = 0.944 F = 0.342, p = 0.600 |
| Population density + Road density | F = 0.014, p = 0.913 F = 2.161, p = 0.238 | F = 0.006, p = 0.943 F = 4.808, p = 0.116 | F = 0.375, p = 0.584 F = 1.212, p = 0.351 | F = 11.700, p = 0.283 F = 2.746, p = 0.196 |
| Farmland density + Population density | F = 0.001, p = 0.972 F = 0.008, p = 0.933 | F = 0.321, p = 0.610 F = 0.003, p = 0.963 | F = 6.375, p = 0.086 F = 0.835, p = 0.4283 | F = 0.414, p = 0.566 F = 0.633, p = 0.484 |
| Road density + Farmland density + Population density | F = 0.389, p = 0.597 F = 0.020, p = 0.901 F = 1.063, p = 0.411 | F = 1.209, p = 0.386 F = 1.100, p = 0.404 F = 2.073, p = 0.287 | F = 0.124, p = 0.758 F = 20.851, p = 0.045* F = 3.962, p = 0.185 | F = 1.922, p = 0.300 F = 0.100, p = 0.782 F = 1.190, p = 0.389 |
| Road density: Farmland density | ANOVA, F = 0.305, p = 0.636 | ANOVA, F = 0.348, p = 0.615 | ANOVA, F = 0.030, p = 0.878 | ANOVA, F = 0.106, p = 0.927 |
| Road density: Population density | ANOVA, F = 1.416, p = 0.356 | ANOVA, F = 10.756, p = 0.082 | ANOVA, F = 0.377, p = 0.602 | ANOVA, F = 0.842, p = 0.456 |
| Farmland density: Population density | ANOVA, F = 1.187, p = 0.708 | ANOVA, F = 0.151, p = 0.735 | ANOVA, F = 0.018, p = 0.906 | ANOVA, F = 0.017, p = 0.909 |
Results from the analyses of potential anthropogenic effects on the genetic variability of hedgehogs, based on linear models including one or more of the three explanatory variables (road density, farmland density or population density) tested against the four response variables iHO, uHE, FIS and %P. The colon (:) indicates models with an interaction term between the explanatory variables, which were then compared to models that did not include the interaction term, using an ANOVA test. Significant p-values are marked with *.
Discussion
Genetic variability and structure
Previous studies have compared the applicability of microsatellite and SNP as linkage mapping markers [76, 77], mentioning that the advantage of microsatellite markers are their highly polymorphic characteristics compared to the biallelic SNPs. However, the high polymorphism in the microsatellite markers may cause relatively high genotyping errors. The advantage of SNPs is their greater density in the sample, providing a higher information content, even though they provide less information per locus. Hence, their lower variability causes a need for an increased number of markers compared to the microsatellite approach [76, 77].
Preceding genetic studies on hedgehogs in Europe based on microsatellite techniques, have found a mean HE ranging between 0.197 and 0.6872. A calculated mean HE of 0.197 and a mean HO of 0.198 (n = 42) was reported in an isolated population of hedgehogs in Regent’s Park, London [52], while a mean HO of 0.7 (n = 160) was determined in a study from Oxfordshire, UK [43]. A mean HE of 0.6872, a mean HO of 0.695 and a mean FIS of 0.0686 was determined in hedgehogs from the Czech and Slovak Republics (n = 131) [48]. Mean HE ranged between 0.569–0.627 (n = 147) and the mean FIS ranged between -0.006–0.070 for three genetic clusters in Zurich, Switzerland [23]. In our research based on the GBS technique, we found a mean uHE varying between 0.126–0.318 and a mean HO varying between 0.124–0.293 for the Danish hedgehogs studied (Table 2). As Denmark is a relatively small area compared to e.g. continental Europe, with an isolated island structure, and thereby has a reduced gene flow, a lower heterozygosity would be expected for the Danish hedgehogs. The peninsula of Jutland south of the Limfjord is connected to Germany, which could potentially have caused a higher gene flow, but we still found a positive FIS coefficient and low heterozygosity in this population (HO = 0.293, uHE = 0.318, FIS = 0.097). However, individuals from Jutland south of the Limfjord did in fact have the highest measures of heterozygosity (HO and uHE) of all the areas tested in the present study (Fig 2, Table 2). Furthermore, in comparison to using SNPs with only two possible alleles, the microsatellite approach in the previous studies may also overestimate heterozygosity due to the polymorphic nature of the microsatellites, causing a direct comparison between the results of the two approaches to be scientifically unsound [78].
Both the STRUCTURE analysis (Fig 3 and Supporting Information S4 Fig [61]) and PCA plot (Supporting Information S5 Fig [61]) in the present study were concordant. It is quite evident that the population in Jutland (JNL, JSL) forms one single cluster whereas all the remaining islands tested, apart from BH, form the second cluster. Lastly, the STRUCTURE analysis plots (Fig 3) are clearly indicating an admixed structure on the island of FN, as also supported by findings in the PCA plot (Supporting Information S5 Fig [61]). The plot of the iHO values for each of the six populations ranked from the lowest to the highest values within each population (Supporting Information S2 Fig [61]) are also showing that the iHO for FN has the steepest cumulative curve, indicating a higher heterogeneity among individuals for the iHO values, which can be due to further substructuring or the presence of inbreed individuals. Despite the evidence for an admixed structure on the island of FN, the log-likelihood plot for K > 1 for the island of FN failed to find significantly higher log-likelihood for K = 2 compared to the log-likelihood of K = 1, rejecting the evidence for genetic substructuring. One reason could be the small sample size of the hedgehogs analysed on the FN island. It is noteworthy that the iHO values for the other populations show heterogeneity among individuals both in terms of lowest iHO values and in terms of slopes, which indicate the presence of further substructuring and/or inbreeding (Supporting Information S2 Fig [61]).
Lastly, the island of BH is separated from the second cluster (FN, Z, LFA) even if partially overlapping with it, and is clearly separated from the first cluster (JSL, JNL). The island of Bornholm, which was previously connected to the continent around northern Germany, became an island in late Preboreal approximately 8000 BC [79]. The rest of Denmark was part of a large continent connected with current areas such as UK and southern Sweden [79], but was transformed into islands and the peninsula of Jutland around 6000–6500 BC [54], when the North American ice shield melted and made the oceans rise. At that point in time, Denmark was surrounded by the Littorina Sea, Lolland and Falster was still connected, and the area south and north of the Limfjord and Djursland as well as northern Zealand consisted of archipelagos [79]. It was not until around 6000 BC that the current geography of Denmark was shaped. Previous research indicates that Lolland and Falster was divided by the sound Guldborgsund around 4000 BC [80], but may have been periodically connected up until 1000 AD [81], which could explain the non-significant difference of FST found between the two islands before we decided to merge them into one population. Jutland north and south of the Limfjord have regularly been connected by different isthmuses closing off the western entrance to the Limfjord from the North Sea from around 1200 AD. In 1863 it was decided to artificially maintain an opening between the Limfjord and the North Sea [82]. The periodical connection between Jutland north and south of the Limfjord could have influenced the genetic cluster found between those two populations of hedgehogs.
The FST values were all highly significant, however, because several of the populations investigated are not in HWE the FST values should be interpreted with caution, as one of the assumptions for a correct estimate test is that the populations which are compared, are panmictic.
JNL, LFA and BH had a low genetic variability (Table 2). However, these three populations did have considerably smaller sample sizes than JSL and Z, which could have affected the results (as the small sample size increase the possibility of committing an error type II; false negative). As an example, JNL had an F coefficient (FIS) of 16.9%, but the HWE test was still negative. This could be due to the small sample size or the large standard errors of uHE and HO.
JSL, FN and Z showed a low genetic variability (Table 2), and as can be seen from the STRUCTURE plot for K = 3 (Fig 3), there is evidence for further substructuring and/or non- panmictic populations. The STRUCTURE analysis failed to find further substructuring. The significant deviation from HWE observed in three of the populations investigated could reinforce the hypothesis of barriers to gene-flow such as habitat fragmentation, as the deviation from HWE could be due to further substructuring of the populations investigated (Wahlund effect) which produces a heterozygosity deficiency due to the lack of panmixia, as seen in hedgehog populations, where competition for the favor of females often occur. Additionally, female hedgehogs are selective of their mates, which often results in courtship without mating [83].
An additional possible factor, which could have contributed to the deviations from HWE, is the effect of humans relocating hedgehogs into foreign habitats. During the past 20–30 years, the rehabilitation of sick, orphaned and injured wild hedgehogs has become an established practice in many western European countries. Denmark has a number of working hedgehog rehabilitation centres, where volunteers care for the hedgehogs with the purpose of releasing the surviving individuals back into the wild. In 2013 the Danish Nature Agency prepared guidelines for the care of wildlife, instructing rehabilitators to refrain from moving mammals over water, and ensuring that rehabilitated wildlife would be released back into their original habitat [84]. Injured wildlife from e.g. Jutland north of the Limfjord, should therefore only be admitted to a wildlife rehabilitation centre north of the Limfjord and be released into the original habitat. Only recently, in 2019, have the Danish authorities established legal frameworks and monitoring programs for the practice of wildlife rehabilitation. The practice before 2013 was to transport wildlife in need of care to the nearest rehabilitation centre, which could be situated far away from the original habitat. Often the animals would be released near the rehabilitation centre. There are previous examples of hedgehogs from e.g. the small island of Ærø being cared for in the northern part of Zealand 200 km away, crossing the seas of the South Funen Archipelago and the Great Belt, most likely because people had brought the sick hedgehog with them when returning from vacation. Furthermore, Kristiansson (1981) [85] showed that intentional introductions of hedgehogs could have influenced the distribution of hedgehogs in Sweden, Norway and Finland. The anthropogenic effects on hedgehog genetics in Denmark could largely be explained by the translocation of hedgehogs between different parts of Denmark.
One drawback to our citizen science sample collection methods was that we did not have precise and reliable records of the geographical location of the samples. This had consequences for the analyses we could carry out because analyses such as Isolation by Distance (IBD) and the Mantel test are very sensitive to even small deviations from the precise location point. We therefore refrain from interpreting the conducted Mantel and IBD tests. Future collection of genetic samples should endeavor to collect precise locations for the samples.
Effective population size and population bottleneck and expansion
The low level of genetic variability can be explained by inbreeding, genetic substructure and extremely low Ne or a combination between these factors which could be caused by habitat fragmentation and/or the large amount of farmland in Denmark. As intensified agricultural practises increase, arable land is gradually becoming a less suitable habitat for hedgehogs. The decline in the hedgehog population of the UK has even been found to be more severe in rural areas than urban [5]. Two-thirds of the area of Denmark is arable land [56]. In comparison, the share of total area by type and land cover in percentage of the EU countries show that the amount of cropland is particularly high in Denmark (50.2%) compared to e.g. Austria (15.3%), United Kingdom (19.7%), Slovakia (26.6%) and the Czech Republic (32%) [86], where previous studies on hedgehog genetics found remarkably high genetic variability using microsatellites [23, 43, 48]. However, as the approach to determine the genetic heterozygosity in the present study (the GBS technique using SNPs) differs from the previous research, the variation of genetic variability between the studies should be interpreted with caution and the results should not be compared directly.
We found an effect of farmland density on the F coefficients (FIS) in our study, but the effect was surprisingly that FIS decreased with increasing farmland density. This may be due to the tendency for a lower degree of road-associated fragmentation in farmland areas and less traffic in general. Or perhaps because the hedgehogs, which are able to survive in this habitat type, can move more freely in their search for mates, because the limitations of movement primarily present in urban areas such as buildings, fences and traffic-laden roads, are less pronounced in farmland areas. Further research is needed to understand this effect.
Habitat fragmentation can cause founder effects [87] and, because hedgehogs have relatively small home ranges and are not dispersing far from their birthplace, they are vulnerable to habitat fragmentation and barriers of movement in general. Consequently, a conservation campaign has been established in the UK, where citizens are encouraged to make holes in their fences to increase garden connectivity for the hedgehogs. Roads as barriers causing habitat fragmentation are also a challenge for hedgehogs, as they are often killed in traffic when crossing roads, especially during the mating season, where males increase their home ranges in search for mates [83]. We tested the possible effects of road density, as a measure of habitat fragmentation in the area from which the hedgehogs derived, on the genetic variability found. We failed to find an association between road density and iHO, FIS, %P and uHE.
We also tested whether the human population density per area for each population of hedgehogs studied, had an effect on iHO, FIS, %P and uHE. JSL, FN and Z, where we found inbreeding and/or subpopulations, had the highest number of citizens per area. This measure may indicate that hedgehogs in these areas are under stronger influence of anthropogenic effects caused by factors such as more cars and traffic and more construction sites replacing hedgehog habitats. However, we failed to find an association between population density and iHO, FIS, %P and uHE in the hedgehogs studied. However, the tests for effects of population density, farmland density and road density were based on a small sample size (n = 6 populations), which has likely influenced the statistical power of the tests.
The software BOTTLENECK 1.2. failed to detect signs of population bottlenecks or increase in population size. However, the software only detects decreases or increases in population size, which has occurred recently (within 0.2 Ne to 0.4 Ne generations). Therefore, we cannot reject the possibility that the population is declining and or have declined drastically before the scope of 0.2 to 0.4 Ne generations.
Conclusions
We adapted the GBS technique with the application of 2902 SNPs per individual to investigate the genetic structure and variability of the European hedgehog on a broader scale. By applying the technique to samples from 178 Danish hedgehogs, we found a low genetic variability (HO = 0.124–0.293). We detected differences between the mean iHO in the populations, which indicate some degree of inbreeding and fragmentation. The 178 Danish hedgehogs tested could be divided into six geographically distinct populations based on the Danish island structure and hence isolation by water. This division was furthermore confirmed by the pairwise fixation index (FST). The STRUCTURE analysis determined that the six populations were distributed inside three genetic clusters. Investigating the potential anthropogenic effects on the genetic variability of the hedgehogs, we discovered that the inbreeding coefficient (FIS) decreased with increasing farmland density, but we found no evidence for an effect of human population or road density. We found evidence for genetic substructure and low effective population size for all populations, which are all consequences of habitat fragmentation. Furthermore, no signs of a recent population bottleneck or population increase or decline were detected.
There is a valuable potential for further analyses such as individual-based landscape genomic studies testing for the effect of landscape attributes on the genetic diversity and connectivity, if precise location data and environmental parameters are provided, enabling the correlation of genetic parameters like uHE, HO, %P and FIS. Given the lack of knowledge on the population status of Danish hedgehogs, we believe that future research on hedgehog genetics should focus on the effects of low individual genetic heterozygosity to determine the impact of inbreeding on individual fitness including indicators such as dental health, parasitic load, microbiomes, toxicology and prevalence of cancer.
Supporting information
Overview of the individuals used in the genetic sampling.
(TIFF)
A presentation of the dataset from GENEPOP.
(TIFF)
The data applied in the analyses with linear models to investigate anthropogenic effects on hedgehog heterozygosity. Area km2 describes the area of the regions measured in km2 [74]. Population/Area is a measure of the human population density per km2 in the regions [73, 74]. Farm/Area is a farmland per km2 in the regions [75], and Roads/Area indicates km of roads per km2 in the regions [58].
(TIFF)
Tukey’s test matrix for testing pairwise significant differences of the mean iHO between the six populations: Jutland north of the Limfjord (JNL), Jutland south of the Limfjord (JSL), Funen (FN), Zealand (Z), Lolland and Falster (LFA) and Bornholm (BH).
(TIFF)
Box plot of the individual heterozygosity (iHO) estimated for the six populations: Jutland north of the Limfjord (JNL), Jutland south of the Limfjord (JSL), Funen (FN), Zealand (Z), Lolland and Falster (LFA) and Bornholm (BH).
(TIFF)
Plot of the iHO values for each of the six populations ranked from the lowest to the highest values within each population.
(TIFF)
Likelihood plot of STRUCTURE results. Ln P(D) is the mean likelihood of K, the number of simulated clusters. The most likely K is that where ln P(D) is maximized.
(TIFF)
The figures are representing the Likelihood plots of results for each of the populations run separately in STRUCTURE (JNL, JSL, FN, Z, LFA, BH). Ln P(D) is the mean likelihood of K, the number of simulated clusters. The most likely K is that where ln P(D) is maximized. Jutland north of the Limfjord (JNL), Jutland south of the Limfjord (JSL), Funen (FN), Zealand (Z), Lolland and Falster (LFA) and Bornholm (BH).
(TIFF)
Principal Component Analysis of the PC1 (Eigenvalues 19.54; variance explained; 79.24%) and PC2 (Eigenvalues 5.12; variance explained; 20.76%) and the convex hulls. The following colours are indicating the six discovered populations: JNL: Jutland north of the Limfjord (Black); JSL: Jutland south of the Limfjord (Purple); FN: Funen (Red); Z: Zealand (Orange); LFA: Lolland and Falster (Blue); BH: Bornholm (Green).
(TIFF)
Acknowledgments
We thank Dr Rien van Wijk for preparing the map for Fig 1. We thank Senior Researcher Ole Bennike, Senior Researcher Merete Binderup and Professor Emeritus Johannes Krüger for useful information on the formation of the Danish landscape. We thank research assistant Camilla Birch and Dr Rien van Wijk for helping with the necropsies, and the Natural History Museum of Denmark for offering to share their facilities for the necropsies.
Data Availability
The supplementary material is available from https://doi.org/10.5281/zenodo.3571408.
Funding Statement
SLR received grants from Beckett Fonden (grant number: 44200), (http://www.beckett-fonden.dk); Svalens Fond; Fonden til Værn for Værgeløse Dyr; Iwan Kliem Larsens Mindelegat (grant number: 108060); Ingeniør K. A. Rohde og hustrus legat; Bodil Pedersen Fonden (grant number: 16-2015), (http://bodilpedersenfonden.dk); and Aalborg Zoo Conservation Foundation (AZCF; grant number: 5-2017), (https://aalborgzoo.dk/aalborg-zoos-naturbevaringsfond.aspx). SLR and TBB both received grants 15. Juni Fonden (grant number: 2015-B-134) (http://www.15junifonden.dk) and Naturama (naturama.dk).
References
- 1.Reeve N. Hedgehogs London: Poyser; 1994. [Google Scholar]
- 2.Morris P. Hedgehogs Stansted, Essex: Whittet Books Ltd.; 2014. [Google Scholar]
- 3.SoBH. The state of Britain's Hedgehogs 2011. British Trust for Ornithology (BTO) commissioned by People's Trust for Endangered Species (PTES) and the British Hedgehog Preservation Society (BHPS): 2011. [Google Scholar]
- 4.SoBH. The State of Britain’s Hedgehogs 2015. British Hedgehog Preservation Society and People’s Trust for Endangered Species, 2015. [Google Scholar]
- 5.SoBH. The State of Britain’s Hedgehogs 2018. British Hedgehog Preservation Society and People’s Trust for Endangered Species: 2018. [Google Scholar]
- 6.Hof AR, Bright PW. Quantifying the long-term decline of the West European hedgehog in England by subsampling citizen-science datasets. Eur J Wildl Res. 2016;62(4):407–13. 10.1007/s10344-016-1013-1 WOS:000380127300003. [DOI] [Google Scholar]
- 7.Williams BM, Baker PJ, Thomas E, Wilson G, Judge J, Yarnell RW. Reduced occupancy of hedgehogs (Erinaceus europaeus) in rural England and Wales: The influence of habitat and an asymmetric intra-guild predator. Sci Rep. 2018;8(1):12156 Epub 2018/09/08. 10.1038/s41598-018-30130-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holsbeek L, Rodts J, Muyldermans S. Hedgehog and other animal traffic victims in Belgium: Results of a countrywide survey. Lutra. 1999;42(1):111–9. BIOSIS:PREV200000071214. [Google Scholar]
- 9.van de Poel JL, Dekker J, van Langevelde F. Dutch hedgehogs Erinaceus europaeus are nowadays mainly found in urban areas, possibly due to the negative effects of badgers Meles meles. Wildlife Biol. 2015;21(1):51–5. 10.2981/wlb.00072 WOS:000347398500006. [DOI] [Google Scholar]
- 10.Huijser MP, Bergers PJM. The effect of roads and traffic on hedgehog (Erinaceus europaeus) populations. Biological Conservation. 2000;95(1):111–6. BIOSIS:PREV200000306409. [Google Scholar]
- 11.Krange M. Change in the occurrence of the West European Hedgehog (Erinaceus europaeus) in western Sweden during 1950–2010. [MSc thesis]. Sweden: Karlstad University; 2015.
- 12.Müller F. Langzeit-Monitoring der Strassenverkehrsopfer beim Igel (Erinaceus europaeus L.) zur Indikation von Populationsdichteveränderungen entlang zweier Teststrecken im Landkreis Fulda. Beiträge zur Naturkunde in Osthessen. 2018;54:21–6. [Google Scholar]
- 13.Brooks TM, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Rylands AB, Konstant WR, et al. Habitat loss and extinction in the hotspots of biodiversity. Conservation Biology. 2002;16(4):909–23. 10.1046/j.1523-1739.2002.00530.x WOS:000177199500009. [DOI] [Google Scholar]
- 14.Crooks KR, Burdett CL, Theobald DM, King SRB, Di Marco M, Rondinini C, et al. Quantification of habitat fragmentation reveals extinction risk in terrestrial mammals. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(29):7635–40. 10.1073/pnas.1705769114 WOS:000405662300064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoate C, Boatman ND, Borralho RJ, Carvalho CR, de Snoo GR, Eden P. Ecological impacts of arable intensification in Europe. Journal of Environmental Management. 2001;63(4):337–65. 10.1006/jema.2001.0473 WOS:000173317500001. [DOI] [PubMed] [Google Scholar]
- 16.Hof AR, Bright PW. The value of agri-environment schemes for macro-invertebrate feeders: hedgehogs on arable farms in Britain. Animal Conservation. 2010;13(5):467–73. 10.1111/j.1469-1795.2010.00359.x WOS:000282370500008. [DOI] [Google Scholar]
- 17.Riber AB. Habitat use and behaviour of European hedgehog Erinaceus europaeus in a Danish rural area. Acta Theriologica. 2006;51(4):363–71. 10.1007/bf03195183 WOS:000241390700004. [DOI] [Google Scholar]
- 18.Michel N, Burel F, Butet A. How does landscape use influence small mammal diversity, abundance and biomass in hedgerow networks of farming landscapes? Acta Oecologica-International Journal of Ecology. 2006;30(1):11–20. 10.1016/j.actao.2005.12.006 WOS:000239342400002. [DOI] [Google Scholar]
- 19.Dowding CV, Harris S, Poulton S, Baker PJ. Nocturnal ranging behaviour of urban hedgehogs, Erinaceus europaeus, in relation to risk and reward. Animal Behaviour. 2010;80(1):13–21. 10.1016/j.anbehav.2010.04.007 WOS:000279153100004. [DOI] [Google Scholar]
- 20.Rondinini C, Doncaster CP. Roads as barriers to movement for hedgehogs. Functional Ecology. 2002;16(4):504–9. 10.1046/j.1365-2435.2002.00651.x WOS:000177256800009. [DOI] [Google Scholar]
- 21.Rautio A, Isomursu M, Valtonen A, Hirvela-Koski V, Kunnasranta M. Mortality, diseases and diet of European hedgehogs (Erinaceus europaeus) in an urban environment in Finland. Mammal Research. 2016;61(2):161–9. 10.1007/s13364-015-0256-7 WOS:000372880400008. [DOI] [Google Scholar]
- 22.Reed DH. Extinction risk in fragmented habitats. Animal Conservation. 2004;7:181–91. 10.1017/s1367943004001313 WOS:000221970100009. [DOI] [Google Scholar]
- 23.Braaker S, Kormann U, Bontadina F, Obrist M K,. Prediction of genetic connectivity in urban ecosystems by combining detailed movement data, genetic data and multi-path modelling. Landscape and Urban Planning. 2017;160:107–14. 10.1016/j.landurbplan.2016.12.011 WOS:000394403300011. [DOI] [Google Scholar]
- 24.Grimm NB, Faeth SH, Golubiewski NE, Redman CL, Wu J, Bai X, et al. Global change and the ecology of cities. Science. 2008;319(5864):756–60. 10.1126/science.1150195 WOS:000252963000045. [DOI] [PubMed] [Google Scholar]
- 25.Hof AR, Bright PW. The value of green-spaces in built-up areas for western hedgehogs. Lutra. 2009;52(2):69–82. BIOSIS:PREV201000094991. [Google Scholar]
- 26.Sæther HM. Overlevelse og tidlig spredning hos juvenile piggsvin (Erinaceus europaeus). [MSc thesis]. Norway: Norges Teknisk-Naturvitenskapelige Universitet; 1997.
- 27.Rasmussen SL. Personality, survivability and spatial behaviour of juvenile hedgehogs (Erinaceus europaeus): a comparison between rehabilitated and wild individuals. [MSc thesis]. Denmark: University of Copenhagen; 2013.
- 28.Doncaster CP, Rondinini C, Johnson PCD. Field test for environmental correlates of dispersal in hedgehogs Erinaceus europaeus. Journal of Animal Ecology. 2001;70(1):33–46. WOS:000167677400005. [Google Scholar]
- 29.Dowding CV, Shore RF, Worgan A, Baker PJ, Harris S. Accumulation of anticoagulant rodenticides in a non-target insectivore, the European hedgehog (Erinaceus europaeus). Environmental Pollution. 2010;158(1):161–6. 10.1016/j.envpol.2009.07.017 WOS:000273245000022. [DOI] [PubMed] [Google Scholar]
- 30.Robinson RA, Sutherland WJ. Post-war changes in arable farming and biodiversity in Great Britain. Journal of Applied Ecology. 2002;39(1):157–76. 10.1046/j.1365-2664.2002.00695.x WOS:000174307300014. [DOI] [Google Scholar]
- 31.Brakes CR, Smith RH. Exposure of non-target small mammals to rodenticides: short-term effects, recovery and implications for secondary poisoning. Journal of Applied Ecology. 2005;42(1):118–28. 10.1111/j.1365-2664.2005.00997.x WOS:000227175200013. [DOI] [Google Scholar]
- 32.Haigh A, Butler F, O'Riordan RM. Intra- and interhabitat differences in hedgehog distribution and potential prey availability. Mammalia. 2012;76(3):261–8. 10.1515/mammalia-2011-0110 WOS:000308551400004. [DOI] [Google Scholar]
- 33.Micol T, Doncaster CP, Mackinlay LA. Correlates of local variation in the abundance of hedgehogs Erinaceus europeaus. Journal of Animal Ecology. 1994;63(4):851–60. 10.2307/5262 WOS:A1994PM01600011. [DOI] [Google Scholar]
- 34.Young RP, Davison J, Trewby ID, Wilson GJ, Delahay RJ, Doncaster CP. Abundance of hedgehogs (Erinaceus europaeus) in relation to the density and distribution of badgers (Meles meles). Journal of Zoology. 2006;269(3):349–56. 10.1111/j.1469-7998.2006.00078.x WOS:000238186400009. [DOI] [Google Scholar]
- 35.Hubert P, Julliard R, Biagianti S, Poulle M-L. Ecological factors driving the higher hedgehog (Erinaceus europeaus) density in an urban area compared to the adjacent rural area. Landscape and Urban Planning. 2011;103(1):34–43. 10.1016/j.landurbplan.2011.05.010 WOS:000295771500004. [DOI] [Google Scholar]
- 36.Pettett CE, Moorhouse TP, Johnson PJ, Macdonald DW. Factors affecting hedgehog (Erinaceus europaeus) attraction to rural villages in arable landscapes. Eur J Wildl Res. 2017;63(54). 10.1007/s10344-017-1113-6 WOS:000403084900013. [DOI] [Google Scholar]
- 37.Morris PA. The effects of supplementary feeding on the movements of hedgehogs (Erinaceus europaeus) Mammal Rev. 1985;15(1):23–33. 10.1111/j.1365-2907.1985.tb00383.x WOS:A1985ADU2800006. [DOI] [Google Scholar]
- 38.Keller LF, Waller DM. Inbreeding effects in wild populations. Trends in Ecology & Evolution. 2002;17(5):230–41. 10.1016/s0169-5347(02)02489-8 WOS:000175024300013. [DOI] [Google Scholar]
- 39.Lacy RC. Importance of genetic variation to the viability of mammalian populations. Journal of Mammalogy. 1997;78(2):320–35. 10.2307/1382885 WOS:A1997XB31600005. [DOI] [Google Scholar]
- 40.Ralls K, Ballou JD, Dudash MR, Eldridge MDB, Fenster CB, Lacy RC, et al. Call for a Paradigm Shift in the Genetic Management of Fragmented Populations. Conservation Letters. 2018;11(2). 10.1111/conl.12381 WOS:000430118100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran S, Turner PD, O'Reilly C. Multiple paternity in the European hedgehog. Journal of Zoology. 2009;278(4):349–53. 10.1111/j.1469-7998.2009.00583.x WOS:000268222100011. [DOI] [Google Scholar]
- 42.Becher SA, Griffiths R. Isolation and characterization of six polymorphic microsatellite loci in the European hedgehog Erinaceus europaeus. Molecular Ecology. 1997;6(1):89–90. 10.1046/j.1365-294x.1997.00159.x WOS:A1997WH49800010. [DOI] [PubMed] [Google Scholar]
- 43.Becher SA, Griffiths R. Genetic differentiation among local populations of the European hedgehog (Erinaceus europaeus) in mosaic habitats. Molecular Ecology. 1998;7(11):1599–604. 10.1046/j.1365-294x.1998.00457.x WOS:000076933000018. [DOI] [PubMed] [Google Scholar]
- 44.Santucci F, Emerson BC, Hewitt GM. Mitochondrial DNA phylogeography of European hedgehogs. Molecular Ecology. 1998;7(9):1163–72. 10.1046/j.1365-294x.1998.00436.x WOS:000075779100007. [DOI] [PubMed] [Google Scholar]
- 45.Henderson M, Becher SA, Doncaster CP, Maclean N. Five new polymorphic microsatellite loci in the European hedgehog Erinaceus europaeus. Molecular Ecology. 2000;9(11):1949–51. 10.1046/j.1365-294x.2000.01098-18.x WOS:000165404800041. [DOI] [PubMed] [Google Scholar]
- 46.Seddon JM, Santucci F, Reeve NJ, Hewitt GM. DNA footprints of European hedgehogs, Erinaceus europaeus and E-concolor. Pleistocene refugia, postglacial expansion and colonization routes. Molecular Ecology. 2001;10(9):2187–98. 10.1046/j.0962-1083.2001.01357.x WOS:000171127700007. [DOI] [PubMed] [Google Scholar]
- 47.Berggren KT, Ellegren H, Hewitt GM, Seddon JM. Understanding the phylogeographic patterns of European hedgehogs, Erinaceus concolor and E-europaeus using the MHC. Heredity. 2005;95(1):84–90. 10.1038/sj.hdy.6800694 WOS:000230110700013. [DOI] [PubMed] [Google Scholar]
- 48.Bolfikova B, Hulva P. Microevolution of sympatry: landscape genetics of hedgehogs Erinaceus europaeus and E. roumanicus in Central Europe. Heredity. 2012;108(3):248–55. 10.1038/hdy.2011.67 WOS:000300596100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraser M, Sten S, Gotherstrom A. Neolithic Hedgehogs (Erinaceus europaeus) from the Island of Gotland show early contacts with the Swedish mainland. Journal of Archaeological Science. 2012;39(2):229–33. 10.1016/j.jas.2011.08.006 WOS:000298464300003. [DOI] [Google Scholar]
- 50.Bolfikova B, Konecny A, Pfaeffle M, Skuballa J, Hulva P. Population biology of establishment in New Zealand hedgehogs inferred from genetic and historical data: conflict or compromise? Molecular Ecology. 2013;22(14):3709–20. 10.1111/mec.12331 WOS:000321552800007. [DOI] [PubMed] [Google Scholar]
- 51.Bolfikova BC, Eliasova K, Loudova M, Krystufek B, Lymberakis P, Sandor AD, et al. Glacial allopatry vs. postglacial parapatry and peripatry: the case of hedgehogs. Peerj. 2017;5 10.7717/peerj.3163 WOS:000400303800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Reilly C. Genetic Analysis of hedgehogs in Regent’s Park. Unpublished report.: The Royal Parks, 2016.
- 53.Barthel L. Population structure and behaviour of the European hedgehog in an urbanized world. [PhD thesis]. Berlin: Freie Universität Berlin; 2019.
- 54.Aaris-Sorensen K. Diversity and dynamics of the mammalian fauna in Denmark throughout the last glacial-interglacial cycle, 115–0 kyr BP. Fossils and Strata. 2009;57:i–iv, 1–59. ZOOREC:ZOOR14603022625. [Google Scholar]
- 55.Statistics Denmark. Areal 2019 [cited 2019 May]. Available from: https://www.dst.dk/da/Statistik/emner/geografi-miljoe-og-energi/areal/areal.
- 56.Statistics Denmark. Afgrøder i dansk landbrug 2017 [cited 2019 May]. Available from: https://www.dst.dk/Site/Dst/Udgivelser/nyt/GetPdf.aspx?cid=24323.
- 57.Statistics Denmark. Population in Denmark 2019 [cited 2019 May]. Available from: https://www.dst.dk/en/Statistik/emner/befolkning-og-valg/befolkning-og-befolkningsfremskrivning/folketal.
- 58.The Danish Road Directorate. Længden af offentlige veje i kommuner (xls) 2019 [cited 2019 October]. Available from: https://www.vejdirektoratet.dk/side/trafikkens-udvikling-i-tal#2.
- 59.Statistics Denmark. Vejnet 2018 [cited 2019 May]. Available from: https://www.dst.dk/da/Statistik/emner/geografi-miljoe-og-energi/infrastruktur/vejnet.
- 60.Elmeros M, Andersen, P.N., Sunde, P. Haugaard, L., Skov, F. & Madsen, A.B. Påkørte større vilde dyr i Danmark 2003–2012. Aarhus Universitet, DCE–Nationalt Center for Miljø og Energi, 2014.
- 61.Rasmussen SL, Nielsen JL, Jones OR, Berg TB, Pertoldi C. Supplementary material for the manuscript: Genetic structure of the European hedgehog (Erinaceus europaeus) in Denmark. December 12 2019. ed. 10.5281/zenodo.3571408: Zenodo; 2019. [DOI] [Google Scholar]
- 62.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. Plos One. 2011;6(5). 10.1371/journal.pone.0019379 WOS:000290224800026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rasmussen SL, Yashiro E, Sverrisdóttir E, Nielsen KL, Lukassen MB, Nielsen JL, et al. Applying the GBS Technique for the Genomic Characterization of a Danish Population of European Hedgehogs (Erinaceus europaeus). Genetics and Biodiversity (GABJ). 2019;3(2):78–86. [Google Scholar]
- 64.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43(5):491–501. 10.1038/ng.806 WOS:000289972600023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28(19):2537–9. 10.1093/bioinformatics/bts460 WOS:000309687500024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. WOS:000087475100039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14(8):2611–20. 10.1111/j.1365-294X.2005.02553.x WOS:000229961500029. [DOI] [PubMed] [Google Scholar]
- 68.Meirmans PG, Van Tienderen PH. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Notes. 2004;4(4):792–4. 10.1111/j.1471-8286.2004.00770.x WOS:000225496600081. [DOI] [Google Scholar]
- 69.Hammer O, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4(1). ZOOREC:ZOOR13700068172. [Google Scholar]
- 70.Luikart G, Cornuet JM. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology. 1998;12(1):228–37. 10.1046/j.1523-1739.1998.96388.x WOS:000072094300025. [DOI] [Google Scholar]
- 71.Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW. Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conservation Genetics. 2010;11(2):355–73. 10.1007/s10592-010-0050-7 WOS:000275455700003. [DOI] [Google Scholar]
- 72.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 73.Statistics Denmark. By2: Population 1. January by municipality, size of the city, age and sex 2019 [cited 2019 August]. Available from: www.statistikbanken.dk/BY2.
- 74.Statistics Denmark. ARE207: Area 1. January by region 2019 [cited 2019 August]. Available from: www.statistikbanken.dk/ARE207.
- 75.Statistics Denmark. AFG07: CULTIVATED AREA BY REGION, UNIT AND CROP 2019 [cited 2019 October]. Available from: https://www.statistikbanken.dk/AFG07.
- 76.Schaid DJ, Guenther JC, Christensen GB, Hebbring S, Rosenow C, Hilker CA, et al. Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility loci. American Journal of Human Genetics. 2004;75(6):948–65. 10.1086/425870 WOS:000224866400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ball AD, Stapley J, Dawson DA, Birkhead TR, Burke T, Slate J. A comparison of SNPs and microsatellites as linkage mapping markers: lessons from the zebra finch (Taeniopygia guttata). Bmc Genomics. 2010;11 10.1186/1471-2164-11-218 WOS:000277271000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Putman AI, Carbone I. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecology and Evolution. 2014;4(22):4399–428. 10.1002/ece3.1305 WOS:000345316200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aaris-Sørensen K. Danmarks forhistoriske dyreverden 3rd ed. Denmark: Gyldendal; 1998. [Google Scholar]
- 80.Bennike O, Jensen JB. Postglacial, relative shore-level changes in Lillebaelt, Denmark. Geological Survey of Denmark and Greenland Bulletin. 2011;(23):37–40. WOS:000294389700009. [Google Scholar]
- 81.Jakobsen EM. Fribrødre Å: Geologi i vikingetid. VARV. 1987;4:104–15. [Google Scholar]
- 82.Bennike O, Nørgaard-Pedersen N, Jensen JB, Andresen KJ, Seidenkrantz M-S. Development of the western Limfjord, Denmark after the last deglaciation: a review with new data. Bulletin of the Geological Society of Denmark. 2019;in press. [Google Scholar]
- 83.Morris P. Hedgehog. Hedgehog [New Naturalist Vol 137]. 2018:i-ix, 1–404. ZOOREC:ZOOR15502009962.
- 84.Danish Nature Agency. Vejledning for vildtplejestationer 2013 [cited 2019 May]. Available from: https://naturstyrelsen.dk/media/178541/vejledning-for-vildtplejestationer-endelig-udgave.pdf.
- 85.Kristiansson H. Distribution of the European hedgehog (Erinaceau europaeus L.) in Sweden and Finland. Ann Zool Fenn. 1981;18(2):115–9. WOS:A1981MK35200003. [Google Scholar]
- 86.Eurostat. Land cover, 2015: European Union; 2015 [cited 2019 May]. Available from: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=File:Land_cover,_2015.png. [Google Scholar]
- 87.Provine WB. Ernst Mayr: genetics and speciation. Genetics. 2004;167(3):1041–6. WOS:000223109300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the individuals used in the genetic sampling.
(TIFF)
A presentation of the dataset from GENEPOP.
(TIFF)
The data applied in the analyses with linear models to investigate anthropogenic effects on hedgehog heterozygosity. Area km2 describes the area of the regions measured in km2 [74]. Population/Area is a measure of the human population density per km2 in the regions [73, 74]. Farm/Area is a farmland per km2 in the regions [75], and Roads/Area indicates km of roads per km2 in the regions [58].
(TIFF)
Tukey’s test matrix for testing pairwise significant differences of the mean iHO between the six populations: Jutland north of the Limfjord (JNL), Jutland south of the Limfjord (JSL), Funen (FN), Zealand (Z), Lolland and Falster (LFA) and Bornholm (BH).
(TIFF)
Box plot of the individual heterozygosity (iHO) estimated for the six populations: Jutland north of the Limfjord (JNL), Jutland south of the Limfjord (JSL), Funen (FN), Zealand (Z), Lolland and Falster (LFA) and Bornholm (BH).
(TIFF)
Plot of the iHO values for each of the six populations ranked from the lowest to the highest values within each population.
(TIFF)
Likelihood plot of STRUCTURE results. Ln P(D) is the mean likelihood of K, the number of simulated clusters. The most likely K is that where ln P(D) is maximized.
(TIFF)
The figures are representing the Likelihood plots of results for each of the populations run separately in STRUCTURE (JNL, JSL, FN, Z, LFA, BH). Ln P(D) is the mean likelihood of K, the number of simulated clusters. The most likely K is that where ln P(D) is maximized. Jutland north of the Limfjord (JNL), Jutland south of the Limfjord (JSL), Funen (FN), Zealand (Z), Lolland and Falster (LFA) and Bornholm (BH).
(TIFF)
Principal Component Analysis of the PC1 (Eigenvalues 19.54; variance explained; 79.24%) and PC2 (Eigenvalues 5.12; variance explained; 20.76%) and the convex hulls. The following colours are indicating the six discovered populations: JNL: Jutland north of the Limfjord (Black); JSL: Jutland south of the Limfjord (Purple); FN: Funen (Red); Z: Zealand (Orange); LFA: Lolland and Falster (Blue); BH: Bornholm (Green).
(TIFF)
Data Availability Statement
The supplementary material is available from https://doi.org/10.5281/zenodo.3571408.