Abstract
This review focuses on DNA–dependent protein kinase (DNA–PK), which is the key regulator of canonical non–homologous end–joining (NHEJ), the predominant mechanism of DNA double–strand break (DSB) repair in mammals. DNA–PK consists of the DNA–binding Ku70/80 heterodimer and the catalytic subunit DNA–PKcs. They assemble at DNA ends, forming the active DNA–PK complex, which initiates NHEJ–mediated DSB repair. Paradoxically, both Ku and DNA–PKcs are associated with telomeres, and they play crucial roles in protecting the telomere against fusions. Herein, we discuss possible mechanisms and contributions of Ku and DNA–PKcs in telomere regulation.
Keywords: DNA–PK, Shelterin, Telomerase, Telomere, hnRNP–A1
Introduction
DNA–dependent protein kinase (DNA–PK) consists of the DNA–binding Ku70/80 heterodimer and the catalytic subunit DNA–PKcs. It is the key regulator of the canonical non–homologous end–joining (HNEJ) mechanism for DNA double–strand break (DSB) repair. The Ku heterodimer, which is highly abundant in mammals, has an extremely high affinity for broken DNA ends, with its ring–shaped structure easily sliding into place. The loading of Ku at the DNA ends quickly recruits DNA–PKcs to form the active DNA–PK complex, which is essential for NHEJ–mediated end–joining activity (see references in [1]). The catalytic DNA–PKcs subunit is a member of the phosphatidylinositol–3 kinase–like kinase (PIKK) family, which includes ataxia–telangiectasia mutated (ATM) and ATM– and Rad3–related (ATR). Together, they are key upstream kinases in sensing DNA damage and promoting DNA damage repair to preserve genome integrity [2, 3].
Telomere maintenance is crucial to protect the integrity of linear chromosomes in eukaryotes. Mammalian telomeres, which have highly repetitive TTAGGG sequences with a single–stranded, G–rich extension (“overhang”) at the 3′ ends, are protected by the shelterin protein complexes [4, 5]. These contain six subunits, including the double–stranded telomeric DNA–binding factor TRF1/2 and the single–stranded telomeric DNA–binding factor POT1. They enable the formation of the t–loop structure where the single–stranded telomeric overhang hides inside the duplex part of the telomere to prevent the recognition of telomere ends by the DNA repair machinery [6]. The critical issues of telomere maintenance mostly occur during the transition between DNA replication and reestablishment of the t–loop telomeric capping structure to guard the G–rich 3′ overhangs. Furthermore, DNA replication cannot be completed at the very end of the telomere since the lagging strand replication requires upfront synthesis of Okazaki fragments. Telomere shortening, which is inevitable through each cell division, results in replicative senescence [7, 8]. Telomerase and the alternative lengthening of telomeres (ALT) mechanism evolved to extend telomere length and counterbalance telomere shortening during each cell cycle. Such telomere expansion strategies are crucial for continuous expansion of stem cell populations, although they also contribute to cancer development [7, 9]. For example, the ALT mechanism, which is dependent on homologous recombination (HR), is involved in roughly 10–15% of human cancers [10].
The NHEJ mechanism downstream from the DNA–PK complex is responsible for fusion of unprotected telomeres. Interestingly, Ku and DNA–PKcs are required for telomere protection at multiple steps. In this review, we focus on their participation and mechanism in this crucial process for chromosome integrity.
Association of DNA–PK with the shelterin complex
The repetitive TTAGGG sequences of mammalian telomeres are primarily protected by the specialized six–subunit shelterin complex, which comprises TRF1, TRF2, POT1, TPP1, TIN2 and RAP1 [4, 5]. This complex guarantees the stability of the t–loop structure to shelter the telomeres against a series of harmful situations [5]. TRF1 and TRF2 are connected by their association with TIN2, and they abundantly bind to the duplex part of telomeres with distinct roles in telomeric protection. TRF1 facilitates efficient telomeric replication and prevents replication fork stalling by recruiting and/or activating a class of helicases [11, 12]. TRF2 promotes the maintenance of the telomeric overhang by recruiting the Snm1b/Apollo nuclease to the newly replicated blunt–ended leading–strand telomeres and prevents telomeric overhang degradation by nucleases [13–15]. It also protects the telomere against fusions in part by counteracting ATM kinase activation and thus suppressing DDR signaling at telomeres [16–18]. Similarly, POT1, which binds specifically to the single–stranded telomeric DNA, represses ATR kinase activation at telomeres [19].
Ku and DNA–PKcs have been found to independently associate with different shelterin complex components. The DNA–binding ku70/80 heterodimer is able to interact physically with TRF1, TRF2 and RAP1 [20–22]. Ku is known to have a very high affinity to all DNA termini regardless of the sequences, although it appears that Ku does not bind to duplex telomeric DNA directly but tethers with TRF1 to bind indirectly [23]. Such indirect binding of Ku could contribute to the inhibition of NHEJ activity at telomeres [24, 25]. The catalytic DNA–PKcs is able to interact with TRF2 and RAP1 at telomeres, and their association prevents end–joining [17]. Additionally, the DNA–PKcs–interacting protein KIP/CIB is required to mediate DNA–PKcs recruitment to telomeres and bridge the association between DNA–PKcs and TRF2 for telomere protection [16]. These results suggest that the DNA–PK complex is recruited to the internal region of the telomeres rather than the very end, and that it participates in telomeric maintenance through TRF1 and TRF2.
DNA–PK on modulation of telomerase activity
Telomerase is a ribonucleoprotein complex that contains a catalytic telomerase reverse transcriptase (TERT) subunit and an integral telomerase RNA component (Terc, also referred to as TR, telomerase RNA) subunit for telomere maintenance and elongation [26–29]. TR is required to serve as a template for the synthesis and extension of the G–rich 3′ telomeric overhang by TERT [26]. The regulation of telomerase activity at telomeres is complex and involves several accessory factors associated with TERT, including Ku [30]. It was reported that telomerase was co–immunoprecipitated with antibodies against Ku in human cells, and that Ku physically interacts with in vitro translated human TERT in the absence of human TR (hTR) and telomeric DNA [31]. Studies from yeast Ku indicate that it interacts with telomerase–associated TLC1 RNA (yeast TR) and Cdc13, which recognizes single–strain telomeric DNA [32–35]. In fact, yeast Ku is capable of binding to the stem–loop structure of TLC1 RNA [33, 34], and facilitates nuclear retention of TLC1 critical for telomere homeostasis [35]. This is consistent with the studies that human Ku could bind directly to hTR and elicit DNA–PK kinase activity [36, 37]. Mutation analysis revealed that the same DNA–binding surface of yeast Ku80 is required for interactions with TLC1 and that it facilitates telomerase recruitment to telomeres [38]. Furthermore, the expression of a Cdc13–Ku70 fusion protein leads to telomeric extension [32]. Additional protein–protein associations among yeast Ku70/80, telomeric transcriptional silencing protein Sir4 and Rap1 likely also contribute to telomerase recruitment [39–41]. These results show that Ku plays a crucial role in promoting or stabilizing telomerase to the telomeric DNA in yeast for telomere maintenance.
The involvement of DNA–PKcs in telomerase regulation is less clear. It is able to form a protein complex with TERT through its interacting protein KIP, and overexpression of KIP improves telomerase activity in human cells [42]. Conversely, it was reported that hTR interacts with the Ku heterodimer and stimulates DNA–PK kinase activity on heterogeneous nuclear ribonucleoprotein A1 (hnRNP–A1), which binds to single–strand telomeric DNA and plays a critical role in telomere biogenesis [36, 37, 43]. Mouse genetics analyses reveal that in telomerase deficient background (Terc−/−), disruption of Ku or DNA–PKcs genes result in progressively shorter telomeres [44, 45], suggesting that the DNA–PK complex coordinates with telomerase to preserve normal telomeres.
Implication of DNA–PK for telomere length regulation
The function of telomeric capping for the DNA–PK complex is superficially paradoxical in light of its role in promoting the NHEJ pathway. This probably reflects its distinct roles at telomeric versus broken ends. Multiple studies from different groups suggested that all three subunits of the DNA–PK complex contribute to telomeric capping protection, since deficiency in either subunit results in increased incidents of telomere fusion in mouse and human cells (see further discussion below). It is less clear whether the DNA–PK complex contributes to the maintenance of telomere length. While loss of Ku results in telomere shortening in most eukaryotes, telomeric expansion was found in Drosophila and Arabidopsis in the absence of Ku [46]. It is possible that Ku is required to restrict telomere lengthening through telomerase or HR–mediated ALT mechanisms, particularly in those eukaryotic species where HR is the predominant type of DSB repair.
Knockout of the mouse Ku86 gene causes the accumulation of telomere fusions but there are conflicting reports regarding telomere length regulation, with either shortening or lengthening being shown [23, 44, 47, 48]. In the absence of telomerase activity (in a Terc−/− background), knockout of Ku86 results in progressively shorter telomeres in later generations of Terc−/−/Ku86−/− mice [44].
Loss of DNA–PKcs in mice also lead to mixed reports on telomere length regulation [45, 49]. In the absence of telomerase activity, DNA–PKcs deficiency accelerates telomere shortening even in the first generation of Terc−/−/DNA–PKcs−/− double knockout mice. This is accompanied by decreased proliferation of germ cells, contrasting to the development of these phenotypical defects in later generations in Terc−/− mice [45, 50]. These results demonstrate an accelerated rate of telomeric shortening in the absence of telomerase and the DNA–PK complex. They suggest that the DNA–PK complex in association with telomerase does play a role in telomere length maintenance.
Implication of DNA–PK on telomeric capping
The role of the Ku heterodimer on telomeric capping
It is apparent that Ku is involved in telomere length modulation in all eukaryotic species [46]. The evidence for its role in telomeric capping and prevention of fusion came from studies in mouse and human cells. It is speculative that its participation in telomeric capping protection is restricted in higher organisms or only found in vertebrates. Ku is clearly crucial in protecting telomeres from end–to–end fusions in mouse cells since Ku knockout increases the frequency in telomeric fusions [47, 51]. However, Ku also appears to promote telomere fusions when telomeres are critically shortened in telomerase–deficient mouse cells [44]. These results indicate that critical telomere length and telomeric interacting proteins are necessary to modulate Ku activity in telomere protection or end–to–end fusions via the NHEJ mechanism. Mutation analyses reveal that the helix 5 (α5) of yeast Ku70 has a selective impact on NHEJ, whereas mutations in the α5 of yeast Ku80 have a selective impact on telomeric maintenance. A spatially organized ‘two–face’ model of the Ku heterodimer was proposed with an outward Ku70 NHEJ–specific α–helix surface dealing with DSB repair and an inward Ku80 telomeric silencing α–helix dealing with telomeric regulation [24].
Notably, TRF2 was reported to interact with Ku70 in a way that involved α5, suggesting a mechanism by which TRF2 can impedes the NHEJ function of Ku on synapsing telomere ends [25]. This is consistent with the report that TRF2 can remodel telomeric DNA into t–loop configurations to block the end–loading of the Ku heterodimer, in turn preventing telomeres from engaging in Ku–dependent NHEJ [52, 53]. The distinctive features of yeast Ku70 and Ku80 are conserved in mouse and human Ku proteins. The same mechanism is likely to apply to mammalian Ku proteins in telomeric capping and protection, although further validation is needed. Additionally, mouse Ku cooperates with TRF2 and POT1 to prevent sister telomere exchanges mediated by HR–dependent recombination between sister telomeres [53, 54].
Knockout of mouse Ku70 or Ku86 is associated with retarded growth, dwarfism and premature aging characteristics, but it does not cause developmental lethality [55–58]. On the contrary, partial deletion of Ku by siRNA or inactivation of a single allele of Ku in human cells leads to increased apoptosis and severe loss of telomere integrity, including telomere fusions and length shortening [59–61]. Furthermore, complete elimination of both copies of the Ku86 gene results in somatic lethality and massive telomere loss in the form of open circular telomeric DNA [62]. These studies demonstrate that the Ku heterodimer is essential for telomere maintenance and cell viability in humans.
Involvement of DNA–PKcs kinase activity and its Thr2609 cluster on telomeric capping
Although Ku proteins are evolutionarily conserved and required for telomere protection in all eukaryotic species, DNA–PKcs homologs are primarily found in vertebrates. Information on the role of DNA–PKcs in telomere protection was mostly generated in mouse or human cells. DNA–PKcs deficiency occurs naturally in mice, dogs and horses and results in the severe combined immunodeficiency (SCID) phenotype [63–67]. Investigation of SCID mouse cells revealed an increase in spontaneous chromosome aberrations including both chromosome– and chromatid–type telomere fusions, suggesting that DNA–PKcs plays an important role in telomeric capping [51]. Similar conclusions were subsequently validated in genetically engineered DNA–PKcs null or mutant mouse models [68–72]. Furthermore, specific leading–to–leading chromatid–type telomere fusions were reported in DNA–PKcs−/− mouse cells [45, 70] and in DNA–PKcs3A/3A mouse cells defective in DNA–PKcs Thr2609 cluster phosphorylation [71] (see below for further discussion).
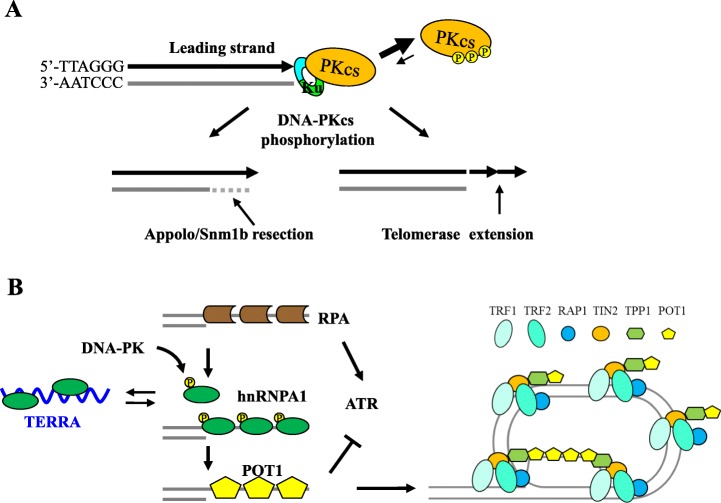
Thus, DNA–PKcs could play an important role in processing the blunt–ended leading–strand telomeres after DNA synthesis to produce the single–stranded G–overhangs (Fig. 1a), whereas the overhangs occur naturally at the lagging strand and shield themselves following replication [73, 74]. Such leading–to–leading telomere fusions were not reported in Ku70 or Ku80 knockout mouse cells. It is possible that Ku and DNA–PKcs contribute to the telomeric capping and maturation process in distinct ways. For example, DNA–PKcs is required to resolve the stalled replication fork in telomeres and participates in replication stress signaling independently of Ku [75, 76].
Fig. 1.
DNA–PK facilitates G–overhang production and telomeric capping. a DNA–PKcs phosphorylation at the Thr2609 cluster promotes dissociation of the DNA–PK complex at newly replicated blunt–ended leading telomeres. This enables G–overhang production through i) telomerase–mediated telomere extension, or ii) Snm1b/Apollo exonuclease–mediated end–resection. b DNA–PK–dependent hnRNP–A1 phosphorylation improves hnRNP–A1’s ability to displace RPA and favor POT1 loading at single–stranded telomeric DNA. This supports T–loop formation. TERRA negatively regulates T–loop formation by trapping hnRNP–A1 away from telomere overhangs. The RPA–to–POT1 displacement also prevents ATR signaling activation at single–stranded telomeric DNA
Significant loss of telomere protection, but not telomere shortening, was found in post–replicated leading telomeres in DNA–PKcs3A/3A mutant mouse cells lacking a functional DNA–PKcs Thr2609 phosphorylation cluster [71, 77]. The Thr2609 cluster is crucial for DNA–PKcs activity in DSB repair and radiation resistance [78–80]. Although it was initially identified as an autophosphorylation event, subsequent analyses revealed that the Thr2609 cluster is respectively targeted by ATM and ATR kinases in response to DSBs and replication stress [79, 81]. Consequently, DNA–PKcs phosphorylation at the Thr2609 cluster triggers a series of conformational changes and modulates the dynamic association and dissociation of the DNA–PK complex at DNA termini [82, 83].
The importance of the DNA–PKcs Thr2609 cluster was further demonstrated using DNA–PKcs3A mutant mice harboring three alanine substitutions to eliminate DNA–PKcs phosphorylation at the Thr2609 cluster. Homologous DNA–PKcs3A/3A mice all die prematurely after birth due to the loss of hematopoietic stem cells (HSCs) and congenital bone marrow failure, which are not found in DNA–PKcs null or SCID mice [77]. Furthermore, DNA–PKcs3A/3A cells displayed significant telomere fusions without apparent telomere shortening. Approximately 46% of DNA–PKcs3A/3A metaphase spreads display telomere abnormalities compared to 20% in DNA–PKcs−/− and 1% in DNA–PKcs+/+ metaphase spreads [71]. Similarly, high incidents of telomere fusions were also found in kinase dead DNA–PKcsKD/KD mouse cells [72], or upon treatment with DNA–PKcs kinase inhibitors [84, 85].
These results suggest that DNA–PK kinase activity and Thr2609 cluster phosphorylation are crucial for telomere protection. Whether DNA–PK kinase inhibition impairs Thr2609 cluster phosphorylation and telomere deprotection is not clear since ATM and ATR also contribute to DNA–PKcs Thr2609 cluster regulation in vivo. Nonetheless, DNA–PK kinase activity is elicited during G2/M phases and necessary for Thr2609 cluster phosphorylation [86]. DNA–PK kinase activation is probably required to modulate additional telomere regulators such as hnRNP–A1 (see below for further discussion) and Werner (Wrn) syndrome protein to assist in telomere capping development. It was reported that DNA–PKcs stimulates Wrn helicase activity (but not its exonuclease activity) to unwind and release the D–loop substrate, and that overexpression of Wrn reversed telomeric G–overhang shortening in DNA–PKcs knockdown cells [87].
Significant and spontaneous γH2AX signals were observed specifically in mitotic DNA–PKcs3A/3A cells from cell culture and tissue analyses. Furthermore, these mitotic γH2AX signals predominantly occur at leading–strand telomeres [71]. The newly synthetized leading–strand telomeres are nearly blunt–ended or carry a few nucleotides in overhang, and require a G2/M phase processing event for final maturation of the G–overhang [74, 88]. The leading G–overhang can be produced through telomerase–dependent telomere elongation [73, 74]. Alternatively, it can be generated through an end–resection by Snm1b/Apollo, a TRF2–interacting exonuclease involved in production of leading overhangs after replication and protection of leading telomeres from engagement with NHEJ–mediated repair [14, 15].
By contrast, lagging–strand telomeres form the G–rich overhangs automatically due to the removal of the RNA primer in the terminal Okazaki fragment and failure to position the fragment at the chromosome terminus. Leading strand–specific γH2AX signals caused by DNA–PKcs3A mutant protein suggest that DNA–PKcs and the Ku heterodimer are present leading telomeric ends immediately after replication. Subsequent DNA–PKcs phosphorylation and conformational change triggers DNA–PKcs dissociation from the very end of leading telomeres [82, 83]. This dynamics allows an orderly processing of G–overhang at the leading daughter telomeres, otherwise the prolonged occupancy by the mutant DNA–PK3A protein complex interferes with overhang production at leading telomeres, elicits DDR signaling, and results in loss of HSCs and presaging phenotypes [71, 77]. This hypothesis predicts that, upon removal of the Ku heterodimer, DNA–PKcs3A mutant protein alone cannot disrupt overhang synthesis at the leading–strand telomeres. Indeed, the Ku86−/−/DNA–PKcs3A/3A double mutant mice survive much longer than DNA–PKcs3A/3A mice (BC unpublished result). It is also possible that DNA–PKcs phosphorylation influences its interaction with TRF2 or the ability of TRF2 to recruit the Snm1b/Apollo exonuclease to leading telomeres [14–17].
The short lifespan and HSC loss characters of DNA–PKcs3A/3A mice can be rescued by bone marrow transplantation (BMT) although the BMT–rescued DNA–PKcs3A/3A mice are prone to cancer at multiple sites including high incidence of skin squamous cell carcinoma (SCC) and lymphoma [71, 77]. This indicates that a functional DNA–PKcs T2609 cluster is required for proper maintenance of telomeres to prevent genomic instability and cancinogenesis.
In support of this, a DNA–PKcs Thr2609Pro mutation was previously identified from a breast cancer biopsy [89]. Expresion of Thr2609Pro mutant DNA–PKcs protein resulted in leading–strand telomeric deprotection as shown in DNA–PKcs3A/3A mouse cells [71]. The DNA–PKcs3A mouse model resembles dyskeratosis congenita (DC), a rare bone marrow failure syndrome that is characterized by defects in telomere maintenance [90, 91]. DC patients are known to be at high risk of developing head and neck SCC and hematologic malignancies [92]. Considering the phenotypical similarity, it is reasonable to speculate that mutations in the DNA–PKcs PRKDC gene could be found in DC patients.
DNA–PK–dependent hnRNP–A1 phosphorylation facilitates telomeric capping
The main challenges in telomere maintenance occur during the transition between DNA replication and reestablishment of telomeric capping protection. The newly synthesized G–overhangs are protected by the replication protein A (RPA) complex, which is the predominant single–stranded DNA–binding (ssDNA–binding) protein and is essential for both DNA replication and damage repair [93]. An extended ssDNA–RPA filament at stalled replication forks will trigger the ATR–Chk1 S–phase checkpoint pathway and promote DNA repair [94, 95]. Thus, it is critical that POT1 rapidly displaces RPA at newly synthetized telomeric overhangs to prevent unnecessary DDR. POT1 is the main single–stranded telomeric DNA–binding factor of the shelterin complex, but it cannot out–compete RPA on its own: it requires additional support from hnRNP–A1 [96]. HnRNP–A1 is versatile factor involved in multiple processes during RNA biogenesis and a critical regulator of telomere homeostasis [97, 98]. It is capable of binding to single–stranded telomeric DNA and the RNA component of telomerase. It also promotes telomerase activation and telomere length extension [99, 100].
Since hnRNP–A1 is the direct substrate of DNA–PK kinase, its role in telomere protection has been linked to DNA–PK [37, 101]. HnRNP–A1 phosphorylation by DNA–PK in vivo coincides with telomeric overhang synthesis during G2/M phases. Consequently, hnRNP–A1 phosphorylation promotes its ability to bind to single–stranded telomeric DNA and facilitates the RPA–to–POT1 switch [43]. Conversely, cells lacking hnRNP–A1 or expressing the phospho–dead mutant hnRNP–A1 display an elevated γH2AX signal at telomeres and higher incidents of telomere aberrations, including sister telomere fusions [43]. How hnRNP–A1 phosphorylation improves its ability to bind to telomeric DNA is not clear. It is notable that hnRNP–A1 Ser95, one of the two key phosphorylation residues, is located between the RNA– and DNA–binding RRM1 and RRM2 motifs, suggesting that phosphorylation induces a conformational change to improve their access to RNA and DNA. Alternatively, DNA–PKcs–dependent hnRNP–A1 phosphorylation could modulate the intermolecular dimerization of hnRNP–A1 and affect its RNA– and DNA–binding ability [102]. These results demonstrate that DNA–PK kinase activity promotes the RPA–to–POT1 switch through hnRNP–A1 phosphorylation to facilitate telomeric capping protection (Fig. 1b).
The ability of hnRNP–A1 to bind to single–stranded telomeric DNA is modulated by telomeric repeat–containing RNA (TERRA), the non–coding RNA species produced from the sub–telomeric region by RNA Pol–II–mediated transcription [103]. The direct interaction between hnRNP–A1 and TERRA could trap hnRNP–A1 away from telomeric overhangs to promote the RPA–to–POT1 switch. Nonetheless, the abundance of TERRA peaks during G1 and decreases gradually from S phase to mitosis, thus releasing the TERRA–bound hnRNP–A1 to compete with RPA and promote POT1 loading to telomeric overhangs [96, 104]. These findings suggest that the balance between hnRNP–A1 and TERRA is crucial for telomere homeostasis and telomerase activity, since excessive TERRA molecules prevent telomere extension by telomerase and the RPA–to–POT1 switch, whereas excessive hnRNP–A1 proteins could overload telomeric overhangs and prevent their access to telomerase or POT1 [105]. It is interesting to note that DNA–PKcs has been identified among TERRA RNA–binding proteins [106]. Considering its weak DNA affinity [107], it is unlikely that DNA–PKcs binds to TERRA directly but is rather tethered to it through other TERRA–binding proteins. In addition, its ability to phosphorylate hnRNP–A1 and to regulate RNA Pol–II transcription could potentially influence TERRA production and regulation in telomere maintenance [43, 108].
DNA–PK coordinates with topoisomerase–II to resolve stalled replication fork at telomeres
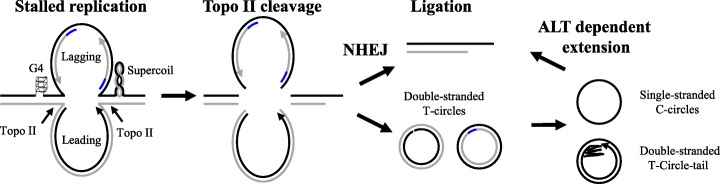
The G–rich and repetitive nature of telomere is prone to G–quadruplex secondary structure formation, which blocks the progression of DNA replication fork and induces replication stress [109]. For example, treatment with TMPyP4, a stabilizer of G–quadruplex, induces severe replication fork stalling at the telomere and results in significant production of fragile telomeres [110]. Resolution of replication stress at telomeres leads to the formation of circular telomeric DNA with a single–stranded tail or the “t–circle–tail” structure, mediated by topoisomerase II (Topo–II) and DNA–PK–mediated NHEJ activities. The t–circle–tail structure resembles cyclized leading or lagging replication intermediates after excision by Topo–II from the genome. Inhibition of Topo–II cleavage activity by ICRF–187 decreases production of this extrachromosomal t–circle–tail. Similarly, inhibition of DNA–PK kinase or Lig4 activities decrease production of extrachromosomal t–circle–tail [76]. These results support a “looping–out” mechanism through ordination between topoisomerase II and NHEJ to resolve stalled replication fork at the telomeres (Fig. 2). This is consistent with our findings that DNA–PKcs is critically involved in the cellular response to replication stress, and it coordinates with the ATR signaling pathway for optimal replication checkpoint and fork recovery [75, 81, 111].
Fig. 2.
A “looping–out” mechanism to resolve a stalled replication fork at telomeres via the topoisomerase II (Topo–II) and NHEJ mechanism. Unresolved G–quadruplex (G4) structures hinder completion of DNA replication at telomeric regions. Topo–II cleavages on both sides release the stalled replication fork and generate both leading and lagging daughter DNA. Repair and ligation via the NHEJ, HR or other repair mechanisms support replication, which resumes at newly ligated telomeres, causing the production of T–circles from the released leading and lagging DNA. The T–circles progress into T–circle–tail or single–stranded C–circles, which could participate in telomere extension through the ALT mechanism. This figure is modified from Zeng et al., EMBO Rep 18: 1412–1428
The circular types of extrachromosomal telomeric DNA, including double–stranded T–circles and the single–stranded C–circles, are commonly identified in cells with long telomeres through telomerase–mediated elongation. Cells with this characteristic include cancer and stem cell populations. These circular DNA types are associated with replication stress and the ALT mechanism [10, 112]. Excessive elongation of telomeres compromises telomere stability and is counterbalanced by a telomerase–trimming mechanism that involves a HR mechanism and production of extrachromosomal telomeric circles to maintain telomere homeostasis [113–115]. It is likely that the extensive telomeres are prone to replication stalling due to increased incidence of G–quadruplexes. The looping–out mechanism provide some explanations to the trimming of large telomeric segments and T–circle production in cells with long telomeres [76]. It is interesting to note that knockout of the Ku80 gene in human cells results in massive telomere loss due to HR–mediated t–circles and rapid deletion of the telomere, suggesting that the DNA–PK complex is essential for telomere homeostasis and cellular viability in human cells [62]. It is possible that DNA–PKcs–dependent NHEJ counterbalances and restricts the ability of the HR machinery to resolve stalled replication fork or DSB repair at telomeres. It is unclear whether the extrachromosomal telomeric circles are merely byproducts in resolving stalled replication forks or are actively involved in telomere maintenance. Telomeric circles have been suggested as the template for telomere extension by a rolling circle mechanism, under which the single–stranded C–circle serves as a template for the extension of the G–rich telomeric overhang [10].
DNA–PK could play additional roles in regulating the stability of telomeric G–quadruplex structure. For example, POT1 and hnRNP–A1 are capable of disrupting telomeric G–quadruplex [116, 117]. It is likely that DNA–PK kinase activity facilitates the removal of G–quadruplex through these telomeric DNA binding proteins during telomere replication. Alternatively, DNA–PK could influence G–quadruplex stability through RecQ helicases such as Wrn for telomere maintenance [118]. Notably, telomeric G–quadruplex also functions as a scaffold and is recognized by TLS/FUS (translocated in liposarcoma/fused in sarcoma) proto–oncoprotein through its C′ terminal RGG–rich domain, and that overexpression of TLS/FUS results in heterochromatin and telomere shortening in vivo [119]. It is possible that TLS/FUS binding stabilizes G–quadruplex structure and leads to progressive telomere shortening through hindering the completion of telomere replication. An independent study reported that TLS/FUS is a downstream phosphorylation target of DNA–PK [120], although it is not clear whether TLS/FUS phosphorylation by DNA–PK plays a role in telomere homeostasis regulation. Further investigation is needed to unveil DNA–PK’s impact on TLS/FUS regulation.
Conclusions and future perspectives
The DNA–PK complex is crucial for telomere homeostasis regulation, particularly in human cells since depletion of the Ku heterodimer leads to severe telomere erosion and loss of cell viability. It is likely that the Ku heterodimer and catalytic DNA–PKcs subunit contribute to both overlapping and distinctive regulations to foster the integrity of telomeres, especially during telomere replication where they are involved in reestablishment of telomeric capping protection. The detailed mechanisms underlying DNA–PK promotion of telomere stability through protein–protein interactions and targeted phosphorylation remain to be elucidated.
Loss of DDR regulators is commonly associated with genomic instability and cancer development [121, 122]. On the contrary, overexpression of DNA–PKcs seemingly occurs in many cancer types [123]. Whether DNA–PKcs overexpression contributes to telomere homeostasis during carcinogenesis requires further study. Nonetheless, a combination of anti–DNA–PKcs and anti–telomere strategies have proposed and might offer additional tools in combatting aggressive and radioresistant tumors [124–127]. Further investigation will help to determine the benefit of these combined modality approaches to cancer patients.
Acknowledgements
Not applicable.
Abbreviations
- ATM
Ataxia–telangiectasia mutated
- ATR
Ataxia–telangiectasia and Rad3–related
- Chk1
Checkpoint kinase 1
- DNA–PK
DNA–dependent protein kinase
- DNA–PKcs
DNA–dependent protein kinase catalytic subunit
- DSBs
DNA double–strand breaks
- HR
Homologous recombination
- Ku
Ku70/80
- NHEJ
Non–homologous end–joining
- PI3K
Phosphatidylinositol–3 kinase
- PIKK
Phosphatidylinositol–3 kinase–like kinase
- POT1
Protection of telomeres 1
- RAP1
Repressor activator protein 1
- Terc
Telomerase RNA component
- Tert
Telomerase reverse transcriptase
- TIN2
TRF1–interacting nuclear protein 2
- TLS/FUS
Translocated in liposarcoma/fused in sarcoma
- TPP1
Telomere protection protein 1
- TRF1
Telomeric repeat–binding factor 1
- TRF2
Telomeric repeat0binding factor 2
Authors’ contributions
All the authors contributed to the manuscript as well as reading and approving the final manuscript.
Funding
This work is supported by grants from the National Natural Science Foundation of China (No. 81802740 to JS), Natural Science Foundation of Chongqing City (No. cstc2018jcyjAX0741 to JS), Cancer Prevention Research Institute of Texas (RP160268 to BPC), and National Institutes of Health (CA233594 to BPC).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiangdong Sui, Email: Jiangdong.Sui@hotmail.com.
Benjamin P. C. Chen, Email: benjamin.chen@utsouthwestern.edu
References
- 1.Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair (Amst) 2009;8(9):1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66(6):801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279(49):51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 5.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19(18):2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 6.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–514. doi: 10.1016/S0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 7.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26(5):867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 8.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12(10):1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 9.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5(7):577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 10.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11(5):319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 11.Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, et al. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23(17):2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12(6):1489–1498. doi: 10.1016/S1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 14.Wu P, van Overbeek M, Rooney S, de Lange T. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell. 2010;39(4):606–617. doi: 10.1016/j.molcel.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, et al. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J. 2010;29(13):2230–2241. doi: 10.1038/emboj.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khadka P, Lee JH, Baek SH, Oh SY, Chung IK. DNA-PKcs-interacting protein KIP binding to TRF2 is required for the maintenance of functional telomeres. Biochem J. 2014;463(1):19–30. doi: 10.1042/BJ20131395. [DOI] [PubMed] [Google Scholar]
- 17.Bombarde O, Boby C, Gomez D, Frit P, Giraud-Panis MJ, Gilson E, et al. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 2010;29(9):1573–1584. doi: 10.1038/emboj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, et al. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2(8):E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448(7157):1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 20.Hsu HL, Gilley D, Blackburn EH, Chen DJ. Ku is associated with the telomere in mammals. Proc Natl Acad Sci U S A. 1999;96(22):12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song K, Jung D, Jung Y, Lee SG, Lee I. Interaction of human Ku70 with TRF2. FEBS Lett. 2000;481(1):81–85. doi: 10.1016/S0014-5793(00)01958-X. [DOI] [PubMed] [Google Scholar]
- 22.O'Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279(27):28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 23.Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, et al. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14(22):2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat Struct Mol Biol. 2007;14(4):301–307. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- 25.Ribes-Zamora A, Indiviglio SM, Mihalek I, Williams CL, Bertuch AA. TRF2 interaction with Ku heterotetramerization interface gives insight into c-NHEJ prevention at human telomeres. Cell Rep. 2013;5(1):194–206. doi: 10.1016/j.celrep.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, et al. The RNA component of human telomerase. Science. 1995;269(5228):1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277(5328):955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 28.Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90(4):785–795. doi: 10.1016/S0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 29.Jaiswal RK, Kumar P, Yadava PK. Telomerase and its extracurricular activities. Cell Mol Biol Lett. 2013;18(4):538–554. doi: 10.2478/s11658-013-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29(11):1095–1105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chai W, Ford LP, Lenertz L, Wright WE, Shay JW. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J Biol Chem. 2002;277(49):47242–47247. doi: 10.1074/jbc.M208542200. [DOI] [PubMed] [Google Scholar]
- 32.Grandin N, Damon C, Charbonneau M. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol. 2000;20(22):8397–8408. doi: 10.1128/MCB.20.22.8397-8408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, et al. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27(1):64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 34.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17(19):2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008;27(5):748–757. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ting NS, Yu Y, Pohorelic B, Lees-Miller SP, Beattie TL. Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res. 2005;33(7):2090–2098. doi: 10.1093/nar/gki342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ting NS, Pohorelic B, Yu Y, Lees-Miller SP, Beattie TL. The human telomerase RNA component, hTR, activates the DNA-dependent protein kinase to phosphorylate heterogeneous nuclear ribonucleoprotein A1. Nucleic Acids Res. 2009;37(18):6105–6115. doi: 10.1093/nar/gkp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfingsten JS, Goodrich KJ, Taabazuing C, Ouenzar F, Chartrand P, Cech TR. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell. 2012;148(5):922–932. doi: 10.1016/j.cell.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16(12):1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy R, Meier B, McAinsh AD, Feldmann HM, Jackson SP. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J Biol Chem. 2004;279(1):86–94. doi: 10.1074/jbc.M306841200. [DOI] [PubMed] [Google Scholar]
- 41.Hass EP, Zappulla DC. The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. Elife. 2015;4:e07750. doi: 10.7554/eLife.07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee GE, Yu EY, Cho CH, Lee J, Muller MT, Chung IK. DNA-protein kinase catalytic subunit-interacting protein KIP binds telomerase by interacting with human telomerase reverse transcriptase. J Biol Chem. 2004;279(33):34750–34755. doi: 10.1074/jbc.M401843200. [DOI] [PubMed] [Google Scholar]
- 43.Sui J, Lin YF, Xu K, Lee KJ, Wang D, Chen BP. DNA-PKcs phosphorylates hnRNP-A1 to facilitate the RPA-to-POT1 switch and telomere capping after replication. Nucleic Acids Res. 2015;43(12):5971–5983. doi: 10.1093/nar/gkv539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Espejel S, Franco S, Rodriguez-Perales S, Bouffler SD, Cigudosa JC, Blasco MA. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 2002;21(9):2207–2219. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Espejel S, Franco S, Sgura A, Gae D, Bailey SM, Taccioli GE, et al. Functional interaction between DNA-PKcs and telomerase in telomere length maintenance. EMBO J. 2002;21(22):6275–6287. doi: 10.1093/emboj/cdf593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher TS, Zakian VA. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair. 2005;4(11):1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 47.Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1(3):244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.d'Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, et al. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol. 2001;11(15):1192–1196. doi: 10.1016/S0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 49.Hande P, Slijepcevic P, Silver A, Bouffler S, van Buul P, Bryant P, et al. Elongated telomeres in scid mice. Genomics. 1999;56(2):221–223. doi: 10.1006/geno.1998.5668. [DOI] [PubMed] [Google Scholar]
- 50.Herrera E, Samper E, Martin-Caballero J, Flores JM, Lee HW, Blasco MA. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18(11):2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, et al. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci U S A. 1999;96(26):14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155(2):345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8(8):885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 54.Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29(2):471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, et al. Requirement for Ku80 in growth and immunoglobulin V(D) J recombination. Nature. 1996;382(6591):551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 56.Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D) J recombination intermediates. Cell. 1996;86(3):379–389. doi: 10.1016/S0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 57.Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7(5):653–665. doi: 10.1016/S1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 58.Ouyang H, Nussenzweig A, Kurimasa A, Soares VC, Li X, Cordon-Cardo C, et al. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J Exp Med. 1997;186(6):921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaco I, Munoz P, Blasco MA. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 2004;64(20):7271–7278. doi: 10.1158/0008-5472.CAN-04-1381. [DOI] [PubMed] [Google Scholar]
- 60.Myung K, Ghosh G, Fattah FJ, Li G, Kim H, Dutia A, et al. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol Cell Biol. 2004;24(11):5050–5059. doi: 10.1128/MCB.24.11.5050-5059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fattah KR, Ruis BL, Hendrickson EA. Mutations to Ku reveal differences in human somatic cell lines. DNA Repair. 2008;7(5):762–774. doi: 10.1016/j.dnarep.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106(30):12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirchgessner CU, Patil CK, Evans JW, Cuomo CA, Fried LM, Carter T, et al. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267(5201):1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 64.Blunt T, Gell D, Fox M, Taccioli GE, Lehmann AR, Jackson SP, et al. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc Natl Acad Sci U S A. 1996;93(19):10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danska JS, Holland DP, Mariathasan S, Williams KM, Guidos CJ. Biochemical and genetic defects in the DNA-dependent protein kinase in murine scid lymphocytes. Mol Cell Biol. 1996;16(10):5507–5517. doi: 10.1128/MCB.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Araki R, Fujimori A, Hamatani K, Mita K, Saito T, Mori M, et al. Nonsense mutation at Tyr-4046 in the DNA-dependent protein kinase catalytic subunit of severe combined immune deficiency mice. Proc Natl Acad Sci U S A. 1997;94(6):2438–2443. doi: 10.1073/pnas.94.6.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding Q, Bramble L, Yuzbasiyan-Gurkan V, Bell T, Meek K. DNA-PKcs mutations in dogs and horses: allele frequency and association with neoplasia. Gene. 2002;283(1–2):263–269. doi: 10.1016/S0378-1119(01)00880-0. [DOI] [PubMed] [Google Scholar]
- 68.Gilley D, Tanaka H, Hande MP, Kurimasa A, Li GC, Oshimura M, et al. DNA-PKcs is critical for telomere capping. Proc Natl Acad Sci U S A. 2001;98(26):15084–15088. doi: 10.1073/pnas.261574698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goytisolo FA, Samper E, Edmonson S, Taccioli GE, Blasco MA. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol Cell Biol. 2001;21(11):3642–3651. doi: 10.1128/MCB.21.11.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293(5539):2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- 71.Zhang S, Matsunaga S, Lin YF, Sishc B, Shang Z, Sui J, et al. Spontaneous tumor development in bone marrow-rescued DNA-PKcs(3A/3A) mice due to dysfunction of telomere leading strand deprotection. Oncogene. 2016;35(30):3909–3918. doi: 10.1038/onc.2015.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang W, Crowe JL, Liu X, Nakajima S, Wang Y, Li C, et al. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol Cell. 2015;58(1):172–185. doi: 10.1016/j.molcel.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell. 2006;21(3):427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 74.Chow TT, Zhao Y, Mak SS, Shay JW, Wright WE. Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev. 2012;26(11):1167–1178. doi: 10.1101/gad.187211.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin YF, Shih HY, Shang ZF, Kuo CT, Guo J, Du C, et al. PIDD mediates the association of DNA-PKcs and ATR at stalled replication forks to facilitate the ATR signaling pathway. Nucleic Acids Res. 2018;46(4):1847–1859. doi: 10.1093/nar/gkx1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang T, Zhang Z, Li F, Hu Q, Liu H, Tang M, et al. Looping-out mechanism for resolution of replicative stress at telomeres. EMBO Rep. 2017;18(8):1412–1428. doi: 10.15252/embr.201643866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol. 2011;193(2):295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16(18):2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, et al. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282(9):6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 80.Reddy YV, Ding Q, Lees-Miller SP, Meek K, Ramsden DA. Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J Biol Chem. 2004;279(38):39408–39413. doi: 10.1074/jbc.M406432200. [DOI] [PubMed] [Google Scholar]
- 81.Yajima H, Lee KJ, Chen BP. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol. 2006;26(20):7520–7528. doi: 10.1128/MCB.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285(2):1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jette N, Lees-Miller SP. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117(2–3):194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bailey SM, Brenneman MA, Halbrook J, Nickoloff JA, Ullrich RL, Goodwin EH. The kinase activity of DNA-PK is required to protect mammalian telomeres. DNA Repair. 2004;3(3):225–233. doi: 10.1016/j.dnarep.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 85.Rybanska-Spaeder I, Ghosh R, Franco S. 53BP1 mediates the fusion of mammalian telomeres rendered dysfunctional by DNA-PKcs loss or inhibition. PLoS One. 2014;9(9):e108731. doi: 10.1371/journal.pone.0108731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee KJ, Lin YF, Chou HY, Yajima H, Fattah KR, Lee SC, et al. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J Biol Chem. 2011;286(14):12796–12802. doi: 10.1074/jbc.M110.212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kusumoto-Matsuo R, Opresko PL, Ramsden D, Tahara H, Bohr VA. Cooperation of DNA-PKcs and WRN helicase in the maintenance of telomeric D-loops. Aging. 2010;2(5):274–284. doi: 10.18632/aging.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, et al. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138(3):463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang X, Szabo C, Qian C, Amadio PG, Thibodeau SN, Cerhan JR, et al. Mutational analysis of thirty-two double-strand DNA break repair genes in breast and pancreatic cancers. Cancer Res. 2008;68(4):971–975. doi: 10.1158/0008-5472.CAN-07-6272. [DOI] [PubMed] [Google Scholar]
- 90.Nishio N, Kojima S. Recent progress in dyskeratosis congenita. Int J Hematol. 2010;92(3):419–424. doi: 10.1007/s12185-010-0695-5. [DOI] [PubMed] [Google Scholar]
- 91.Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73(2):103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 92.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 94.Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21(8):879–885. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 95.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 96.Flynn RL, Centore RC, O'Sullivan RJ, Rai R, Tse A, Songyang Z, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471(7339):532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14(9):18999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roy Rajat, Huang Yueyang, Seckl Michael J., Pardo Olivier E. Emerging roles of hnRNPA1 in modulating malignant transformation. Wiley Interdisciplinary Reviews: RNA. 2017;8(6):e1431. doi: 10.1002/wrna.1431. [DOI] [PubMed] [Google Scholar]
- 99.Fiset S, Chabot B. hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res. 2001;29(11):2268–2275. doi: 10.1093/nar/29.11.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang QS, Manche L, Xu RM, Krainer AR. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA. 2006;12(6):1116–1128. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Le PN, Maranon DG, Altina NH, Battaglia CL, Bailey SM. TERRA, hnRNP A1, and DNA-PKcs interactions at human telomeres. Front Oncol. 2013;3:91. doi: 10.3389/fonc.2013.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13(9):1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 104.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol. 2010;30(20):4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Redon S, Zemp I, Lingner J. A three-state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res. 2013;41(19):9117–9128. doi: 10.1093/nar/gkt695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35(4):403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci U S A. 1998;95(2):525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bunch H, Lawney BP, Lin YF, Asaithamby A, Murshid A, Wang YE, et al. Transcriptional elongation requires DNA break-induced signalling. Nat Commun. 2015;6:10191. doi: 10.1038/ncomms10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8(10):825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 110.Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, et al. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342(6155):239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 111.Lin YF, Shih HY, Shang Z, Matsunaga S, Chen BP. DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response. Nucleic Acids Res. 2014;42(7):4463–4473. doi: 10.1093/nar/gku116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27(12):1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 113.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28(7):799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pickett HA, Henson JD, Au AYM, Neumann AA, Reddel RR. Normal mammalian cells negatively regulate telomere length by telomere trimming. Hum Mol Genet. 2011;20(23):4684–4692. doi: 10.1093/hmg/ddr402. [DOI] [PubMed] [Google Scholar]
- 115.Rivera T, Haggblom C, Cosconati S, Karlseder J. A balance between elongation and trimming regulates telomere stability in stem cells. Nat Struct Mol Biol. 2017;24(1):30–39. doi: 10.1038/nsmb.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci U S A. 2005;102(31):10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kruger AC, Raarup MK, Nielsen MM, Kristensen M, Besenbacher F, Kjems J, et al. Interaction of hnRNP A1 with telomere DNA G-quadruplex structures studied at the single molecule level. Eur Biophys J. 2010;39(9):1343–1350. doi: 10.1007/s00249-010-0587-x. [DOI] [PubMed] [Google Scholar]
- 118.Singh DK, Ghosh AK, Croteau DL, Bohr VA. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat Res. 2012;736(1–2):15–24. doi: 10.1016/j.mrfmmm.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Takahama K, Takada A, Tada S, Shimizu M, Sayama K, Kurokawa R, et al. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem Biol. 2013;20(3):341–350. doi: 10.1016/j.chembiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 120.Deng Q, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, et al. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J Neurosci. 2014;34(23):7802–7813. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 122.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 123.Hsu FM, Zhang S, Chen BP. Role of DNA-dependent protein kinase catalytic subunit in cancer development and treatment. Transl Cancer Res. 2012;1(1):22–34. doi: 10.3978/j.issn.2218-676X.2012.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yasaei H, Gozaly-Chianea Y, Slijepcevic P. Analysis of telomere length and function in radiosensitive mouse and human cells in response to DNA-PKcs inhibition. Genome Integr. 2013;4(1):2. doi: 10.1186/2041-9414-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou X, Zhang X, Xie Y, Tanaka K, Wang B, Zhang H. DNA-PKcs inhibition sensitizes cancer cells to carbon-ion irradiation via telomere capping disruption. PLoS One. 2013;8(8):e72641. doi: 10.1371/journal.pone.0072641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gurung RL, Lim HK, Venkatesan S, Lee PS, Hande MP. Targeting DNA-PKcs and telomerase in brain tumour cells. Mol Cancer. 2014;13:232. doi: 10.1186/1476-4598-13-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang M, Guo X, Gao Y, Lu D, Li W. Tumor cell-accelerated senescence is associated with DNA-PKcs status and telomere dysfunction induced by radiation. Dose Response. 2018;16(2):1559325818771527. doi: 10.1177/1559325818771527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.