Significance
Transcription factors bound to enhancers that are distant from target genes control gene transcription to regulate many biological processes. LDB1/SSBP complexes function as a common core of diverse important protein complexes that are known to be critical for development and can connect enhancers and genes. SSBPs stabilize these complexes by preventing LDB1 degradation by the 26S proteasome. Here we provide a glimpse of the structure of the human core LDB1/SSBP2 protein complex and how it may be regulated.
Keywords: LIM domain binding protein 1 (LDB1), single-stranded DNA binding protein 2 (SSBP2), dimerization, crystal structure, Wnt enhanceosome
Abstract
The Lim domain binding proteins (LDB1 and LDB2 in human and Chip in Drosophila) play critical roles in cell fate decisions through partnership with multiple Lim-homeobox and Lim-only proteins in diverse developmental systems including cardiogenesis, neurogenesis, and hematopoiesis. In mammalian erythroid cells, LDB1 dimerization supports long-range connections between enhancers and genes involved in erythropoiesis, including the β-globin genes. Single-stranded DNA binding proteins (SSBPs) interact specifically with the LDB/Chip conserved domain (LCCD) of LDB proteins and stabilize LDBs by preventing their proteasomal degradation, thus promoting their functions in gene regulation. The structural basis for LDB1 self-interaction and interface with SSBPs is unclear. Here we report a crystal structure of the human LDB1/SSBP2 complex at 2.8-Å resolution. The LDB1 dimerization domain (DD) contains an N-terminal nuclear transport factor 2 (NTF2)-like subdomain and a small helix 4–helix 5 subdomain, which together form the LDB1 dimerization interface. The 2 LCCDs in the symmetric LDB1 dimer flank the core DDs, with each LCCD forming extensive interactions with an SSBP2 dimer. The conserved linker between LDB1 DD and LCCD covers a potential ligand-binding pocket of the LDB1 NTF2-like subdomain and may serve as a regulatory site for LDB1 structure and function. Our structural and biochemical data provide a much-anticipated structural basis for understanding how LDB1 and the LDB1/SSBP interactions form the structural core of diverse complexes mediating cell choice decisions and long-range enhancer–promoter interactions.
The LDBs are nuclear proteins that are highly conserved from worms to man and have no known enzymatic or DNA-binding activity (1, 2). LDB1 (also known as NLI and CLIM2) functions together with Lim-homeodomain (LIM-HD) and Lim-only (LMO) proteins to regulate cell fate choice in numerous developmental systems (3–5). In accord, Ldb1-null mouse embryos display severe patterning defects as early as E8.5 and embryonic lethality (6). LDB1 is required for both primitive and definitive hematopoiesis (7, 8). Mechanistic studies show that, in erythroid cells, LDB1 and the Lim-only protein LMO2 are part of an erythroid DNA-binding complex that occupies enhancers and genes and that LDB1 dimerization brings these regulatory regions into proximity for gene activation (9–11).
LDB proteins contain 4 well-conserved domains (Fig. 1A and SI Appendix, Fig. S1): an N-terminal dimerization domain (DD), an LDB1/Chip conserved domain (LCCD), a nuclear localization signal (NLS), and a C-terminal LIM interaction domain (LID) (12–14). LDB1-DD mediates self-interaction in vitro (12, 13) and is required for establishing the long-range β-globin/locus control region (LCR) interaction and transcriptional activation of the β-globin gene (11). The highly conserved LCCD is necessary and largely responsible for binding single-stranded DNA binding proteins (SSBP2/3/4, also known as SSDP in Drosophila) (14, 15). The LDB1-LID interacts with tandem LIM domains in LIM-HD and LMO proteins, which play critical roles in cell-fate determination, tissue development, and cytoskeletal organization (2, 16). Structures of the LMO LIM domains in complex with LDB-LID have been previously reported (16–20). In contrast, 3D structures of the DD and LCCD remain unresolved.
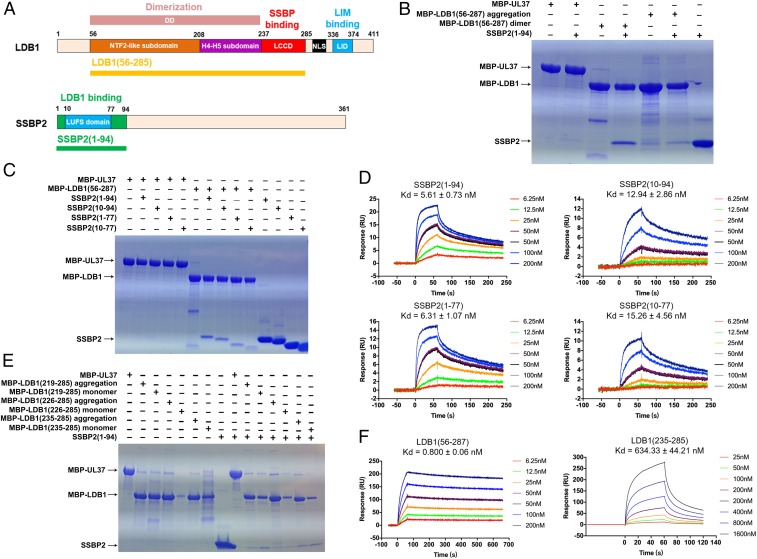
Fig. 1.
Biochemical characterization of the LDB1/SSBP2 interaction. (A) Schematic diagram of human LDB1 and SSBP2 domain organization. The dimerization domain (DD), LDB1/Chip conserved domain (LCCD), nuclear localization sequence (NLS), and LIM interaction domain (LID) are indicated for human LDB1. The LUFS domain (residues 10 to 77) is indicated for human SSBP2. We determined the crystal structure of the LDB1(56–285)/SSBP2(1–94) complex. (B) LDB1 dimer binds to SSBP2 tightly, as shown by MBP pull-down assays. MBP-UL37 was used as a negative control. (C) SSBP2 LUFS domain is sufficient for its interaction with LDB1 as shown by MBP pull-down assays. (D) Binding affinities of 4 SSBP2 fragments with LDB1 were measured by SPR assays. Values are presented ± SD. (E) LDB1 LCCD is sufficient for its binding with SSBP2 as shown by MBP pull-down assays. (F) Binding-affinities of SSBP2 with 2 LDB1 fragments were measured by SPR assays.
The N-terminal regions of SSBP proteins are highly conserved and contain a LUFS (LUG/LUH, Flo8 and SSBP/SSDP) domain (residues 10 to 77 in SSBP2), which promotes homotetramerization and is also found in a number of proteins, including some transcriptional corepressors (21–23). The N-terminal 94 residues of SSBP2 can specifically bind the LCCD of LDB/Chip proteins (14). SSBP proteins may functionally depend on Ldb1 in multiple developmental processes, including axis formation in Xenopus, wing development in Drosophila, and head morphogenesis in mice (14, 15, 24, 25). For example, SSBP requires its LUFS domain and LDB1/Chip for correct localization to the nucleus, suggesting that SSBP is brought to the nucleus via its interaction with LDB1/Chip to play a role in transcriptional regulation (14). SSBP2 appeared to promote the SSBP/LDB1/LMO2 complex assembly, and its concentration may be limiting for complex formation (26).
One of the critical biochemical functions of SSBP2 is to protect LDB1 from proteasomal degradation by inhibiting LDB1 interaction with RING finger protein 12 (RNF12/RLIM), an E3 ubiquitin ligase that promotes rapid turnover of LDB1 (26, 27). The stoichiometry of LDB1, LIM-HD, and LMO proteins is tightly controlled in the cell and is likely a critical determinant of their joint biological functions (28–30). The LDB1/SSBP interaction may be the key for cellular stability of LDB1, which in turn acts as a core for forming multiple transcriptional regulatory complexes in addition to the SSBP/LDB1/LMO2 complex, including the nuclear enhanceosome of the Wnt/β-catenin signaling pathway (31–34). Despite functional significance and high conservation, 3D structures of LDB1 DD and LCCD remain unknown. In addition, it is unclear how SSBP2/3/4 proteins interact with LDB1 and exert their biochemical function. Here, we report a crystal structure of LDB1 DD and LCCD domains (hLDB1 DD-LCCD) in complex with the conserved SSBP2 N-terminal domain. Our work lays a solid foundation for understanding LDB1 structure, LDB1/SSBP interaction, and functional regulation of this complex.
Results
Biochemical Characterization of LDB1/SSBP2 Interaction.
Size-exclusion chromatography-coupled multiangle light scattering (SEC-MALS) analysis showed that the LDB1/SSDP complex contains an LDB1 dimer and an SSDP tetramer in solution (32). To map the critical region for the LDB1/SSBP2 interaction, we overexpressed and purified a number of LDB1 fragments that contain both the DD and LCCD domains, including N-terminal maltose-binding protein (MBP)-tagged human LDB1(56–287). MBP-LDB1(56–287) tends to form large aggregates as judged from its SEC profile, with only a small fraction forming stable soluble dimers (SI Appendix, Fig. S2A). Our MBP pull-down data showed that the soluble dimeric form of LDB1 can interact with SSBP2(1–94) more tightly than the aggregated form (Fig. 1B). On the SSBP2 side, the entire N-terminal conserved region SSBP2(1–94) interacts with LDB1(56–287) slightly more avidly than the conserved LUFS domain (residues 10 to 77), with a Kd of 5.6 nM vs. 15.3 nM (Fig. 1 C and D). Therefore, we used the purified dimeric form of LDB1(56–287) and SSBP2(1–94) for most of our structural and biochemical studies.
It was previously shown that the LCCD domain per se is sufficient for SSBP2 interaction with LDB1 (14). We generated a number of MBP-tagged LDB1 fragments that contain the LCCD but not the DD domain. These MBP-LDB1 fragments also have a strong tendency to form aggregates, and only a small fraction appear in monomeric form. We found that monomeric forms of LCCD fragments bind significantly tighter than corresponding aggregated forms (Fig. 1E). Importantly, our surface plasmon resonance (SPR) data demonstrate that the dimerization domain of LDB1 contributes to the SSBP2 interaction, since SSBP2(1–94) binds to LDB1 DD-LCCD (residues 56 to 287; with a Kd of ∼0.8 nM) dramatically more tightly than LDB1-LCCD (residues 235 to 285; Kd of ∼634 nM; Fig. 1F).
Overall Structure of the LDB1/SSBP2 Complex.
To provide a structural basis for understanding the molecular architecture of the LDB/SSBP complexes that play important roles in various biological systems, we have determined the crystal structure of the human LDB1(56–285)/SSBP2(1–94) complex and refined the structure at 2.8-Å resolution (SI Appendix, Table S1). In our crystal lattice, there are 1 LDB1 and 2 SSBP2 molecules in each asymmetric unit (Fig. 2 A and B). With the analysis of crystallographic packing patterns, combined with mutagenesis analysis (as detailed later), we can clearly define the symmetric LDB1 dimerization interface in the 2:4 molar ratio LDB1/SSBP2 complex, which is formed by 2 neighboring asymmetric units related by a 2-fold crystallographic axis (Fig. 2C). In the LDB1/SSBP2 complex, 2 LDB1 dimerization domains form the core of the complex, which is flanked by 2 LCCD domains. SSBP2 dimers mainly interact with the flanking LCCD domains, with additional contacts with the dimerization domain (as detailed later).
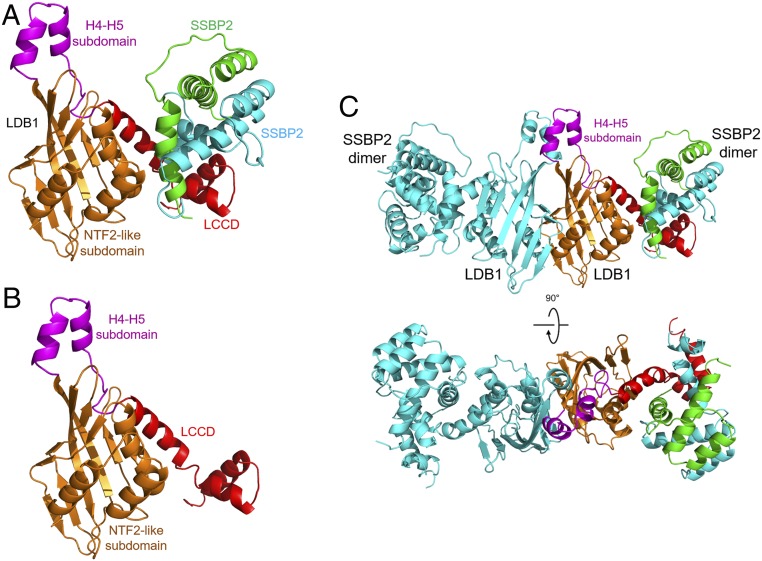
Fig. 2.
Overall structure of the human LDB1/SSBP2 complex. (A) Structure of 1 LDB1 in complex with 1 SSBP2 dimer. One SSBP2 dimer binds to 1 LDB1 LCCD domain. LDB1 NTF2-like subdomain, H4-H5 subdomain, and LCCD are colored in orange, magenta, and red, respectively. SSBP2 dimers are colored in green and cyan. (B) The “LDB1-only” structure in the complex. There is an H4-H5 subdomain between the main dimerization domain (NTF2-like subdomain) and LCCD. (C) Two orthogonal views of the LDB1/SSBP2 complex structure, which contains a symmetric LDB1 dimer, with each LDB1 monomer interacting with 1 SSBP dimer.
The structure of LDB1-DD can be divided into 2 parts: a subdomain containing a β-sheet with 3 α-helices on one side, which is defined by the Dali server to be a nuclear transport factor 2 (NTF2)-like fold, despite the lack of sequence homology between LDB1 and any protein with the NTF2-like fold (SI Appendix, Table S2 and Fig. S3) (35), and a small subdomain formed by helices H4 and H5 that is also part of the LDB1 dimerization interface (as detailed later). The versatile “α + β conical barrel” NTF2 fold, originally observed in NTF2 protein (36), is adopted by a variety of quite different sequences and found in proteins with many different functions (37, 38).
C-terminal to the H4-H5 subdomain is a short, conserved linker, followed by the highly conserved LCCD domain that contains 3 helices (H6 to H8) with the helix H6 packing on the surface of the NTF2-like subdomain. Helices H6 and H7 are otherwise one continuous helix except for a kink due to a conserved proline residue (Pro251) between them. Helices H7 and H8 form extensive interactions with SSBP2 (as detailed later).
The LDB1 Dimerization Interface and Mutagenesis Analysis.
The LDB1 homodimerization surface involves both the NTF2-like and the H4-H5 subdomains. On the NTF2-like subdomain, there are 2 hydrophobic interfaces formed by hydrophobic residues. The first hydrophobic interface mainly involves residues Tyr69, Phe72 of helix 1 (H1) and their symmetric partners, as well as a hydrogen bond pair between Asp68 and Lys76, whereas the second hydrophobic interface involves LDB1 Tyr140 and Val142 (Fig. 3A). In total, the LDB1 dimerization interface buries an area of interface of 1,585 Å2, while the NTF2-like subdomain alone buries an area of interface of 901 Å2.
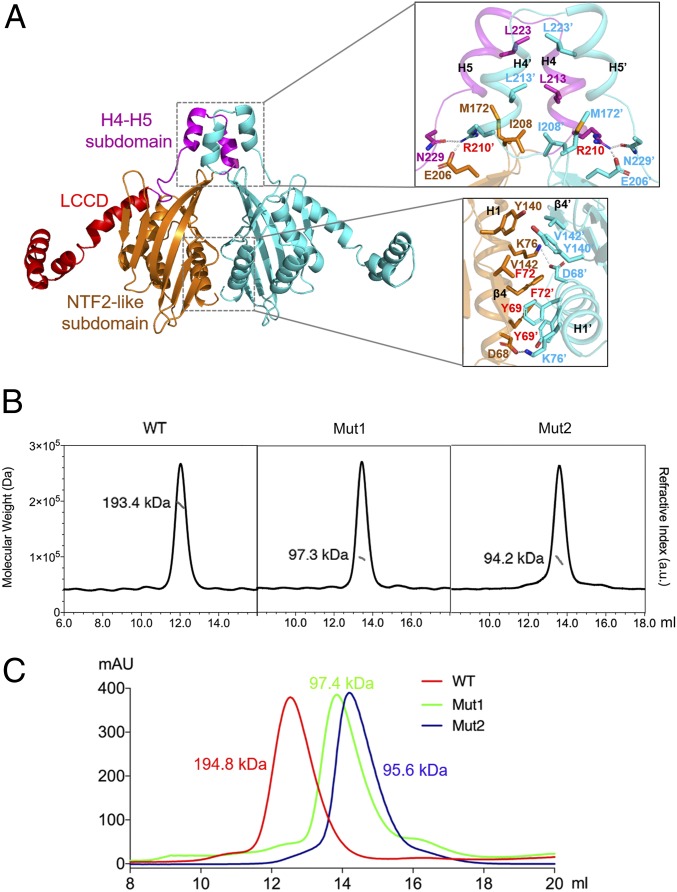
Fig. 3.
Structure and mutagenesis analysis of the LDB1 dimerization interface. (A) Close-up view of the LDB1 dimerization interface of the NTF2-like subdomain and the H4-H5 subdomain. Residues examined in mutagenesis analysis are labeled in red. (B) SEC-MALS analysis of MBP-tagged wild-type LDB1(56–287) (WT), LDB1(56–287) mut1 (Y69D/F72D/R210A), and LDB1(56–287) mut2 (replacing H4-H5 with a short linker) in complex with SSBP2. Molecular weights measured by SEC-MALS are shown. (C) Alignment of SEC profiles of MBP-tagged LDB1 WT (in red), mut1 (in green), and mut2 (in blue) in complexes with SSBP2. Calculated molecular weights for WT MBP-tagged LDB1 dimer in complex with 2 SSBP2 dimers and MBP-tagged LDB1 mut1 and mut2 monomers in complex with 1 SSBP2 dimer are labeled.
The 2 neighboring H4-H5 subdomains in an LDB1 dimer form a small H4-H5/H4′-H5′ 4-helix bundle. The H4-H5/H4′-H5′ interface is stabilized by hydrophobic interactions involving Leu213, Leu223, and buttressing hydrophobic residues from the “top” side of the NTF2-like subdomain, including Ile208, Met172. Interestingly, Arg210 of H4 uses its long sidechain to penetrate through the hydrophobic core between the partnering NTF2-like and H4′-H5′ subdomains and forms a hydrogen bond and salt bridge network with Glu206′ and Asn229′ (Fig. 3A). These interactions may stabilize the homodimeric H4-H5/H4′-H5′ subdomains on “top” of the NTF2-like homodimeric subdomains.
We next generated 2 LDB1 mutants on its dimerization interface. LDB1(56–287) mutant 1 (Y69D/F72D/R210A) replaces key dimer-interface residues in both the NTF2-like and H4-H5 subdomains. In comparison with the corresponding wild-type LDB1 protein that is dimeric, purified MBP-tagged LDB1(56–287; mut1) appeared predominantly as a monomer in SEC-MALS, SEC, and AUC experiments (Fig. 3 B and C and SI Appendix, Fig. S4), confirming our observed LDB1 dimer interface. To test the role of H4-H5 subdomain in LDB1 dimer, we also generated mut2 that replaces the H4-H5 subdomain (208IPRSILAMHAQDPQMLDQLS227) with a short 6-residue flexible linker (ASGGAS). The purified MBP-LDB1(56–287; mut2) is also predominantly monomeric, demonstrating a role for the H4-H5 subdomain in dimerization in vitro.
The LDB1/SSBP2 Interface and Mutagenesis Analysis.
For SSBP2(1–94), the LDB1 binding surface largely overlaps with its tetramerization (dimer-onto-dimer) interface (23) (SI Appendix, Fig. S5). Thus, each subunit of the LDB1 homodimer interacts with one SSBP2 dimer. On the LDB1 side, the interactions mostly involve the LCCD motif H6 and H7, while there is also significant interaction with the NTF2-like subdomain (Fig. 4 A and B). It should be noted that, consistent with our biochemical data (Fig. 1), the conserved SSBP2 region C-terminal to the LUFS domain (residues 77 to 94) is also visible in one copy of our asymmetric SSBP2 dimer structure and makes direct contacts with the helix H8 of the LDB1-LCCD (Fig. 4C).
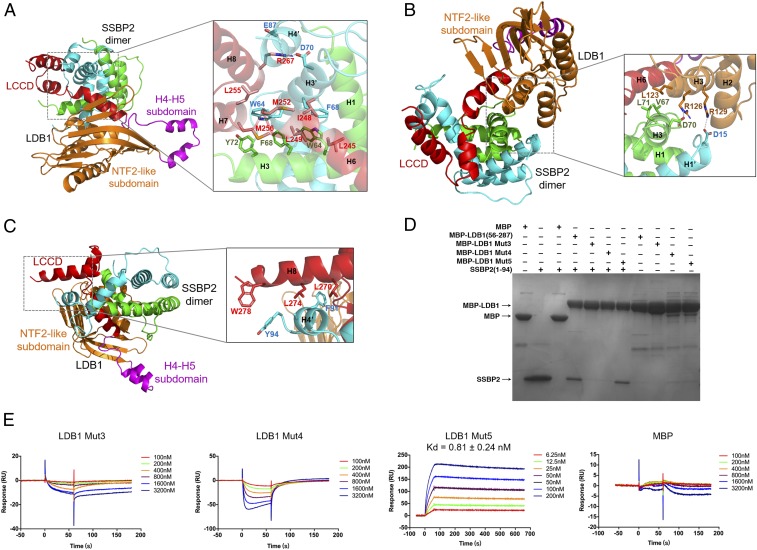
Fig. 4.
The LDB1/SSBP2 interface and mutagenesis analysis of the LDB1/SSBP2 interface. (A) Close-up view of the LDB1 LCCD/SSBP2 interface. Residues examined in mutagenesis analysis are labeled in red. (B) Close-up view of the LDB1-DD/SSBP2 interface. (C) Close-up view of the interface between LDB1-LCCD and SSBP2 residues 77 to 94 that is C-terminal to the SSBP2 LUFS domain. (D) Mutagenesis analysis of LDB1/SSBP2 interface, as shown by MBP pull-down assays. (E) Binding affinities of 3 MBP-tagged LDB1 mutants with SSBP2(1–94) were measured by SPR assays. MBP is a negative control, and the values are presented ± SD.
The first of 2 SSBP2 molecules in the SSBP2 dimer mostly uses its helix H3 (last helix in the LUFS domain consisting of residues 10 to 77) to interact with the N-terminal half of LCCD by forming tight hydrophobic interactions with Leu245, Ile-248, and Leu249 of LDB1 H6 (Fig. 4A). The second SSBP2 molecule uses the helix H3 and conserved C terminus (residues 78 to 94) to interact with the surface hydrophobic patch formed by both helices H7 and H8 of LDB1, including LDB1 residues Met252, Leu255, Met256, Leu270, Leu274, and Try278 (Fig. 4 A and C). In addition to extensive hydrophobic interactions, there are also a number of salt bridges, including binding of LDB1 Arg267 to a negatively charged cluster formed by Asp70 and Glu87 of the second SSBP2 (Fig. 4A). There are also additional interactions between SSBP2 dimer and the LDB1 NTF2-like subdomain. These interactions mainly involve hydrophobic interface formed by residues Val67, Leu71 of the first SSBP2 helix H3 and Leu123 of LDB1 helix H3, salt bridges formed by Asp70 of the first SSBP2 helix H3 and Arg126 of LDB1 helix H3, as well as Asp15 of the second SSBP2 helix H1 and Arg129 of LDB1 helix H3.
We have generated 3 mutants (mut3, mut4, and mut5) in the LDB1-LCCD to examine the importance of the aforementioned LDB1/SSBP2 interactions. The purified LDB1(56–287; mut3) protein, with the L245A/I248A/L249A mutation, apparently loses its ability to interact with SSBP2(1–94) in both MBP pull-down and SPR assays (Fig. 4 D and E). The purified LDB1(56–287; mut4) protein, with the M252A/L255A/M256A mutation, also apparently loses its ability to interact with SSBP2(1–94) in both assays. In contrast, we did not observe any significant change of SSBP2 binding to LDB1(56–287; mut5) carrying the R267A mutation. Of note, complete deletion of the DD also resulted in an unstable LDB1 protein in cells (11), suggesting that both the DD and LCCD contacts are important for SSBP2 interactions with LDB1.
Functional Test of LDB1 Dimerization and LDB1/SSBP2 Interaction.
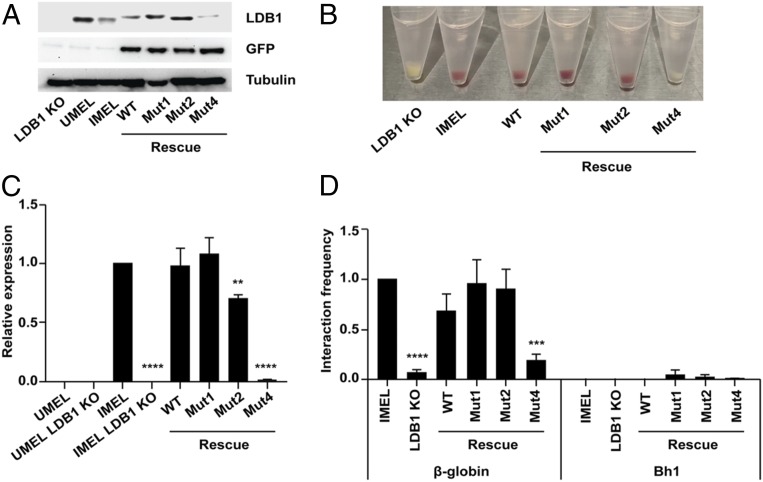
We examined β-globin transcriptional activity and chromatin looping through loss- and gain-of-function studies for 3 LDB1 mutants: LDB1 mut1 (Y69D/F72D/R210A) that disrupts the LDB1 dimer interface, LDB1 mut2 that replaces the H4-H5 helices with a 6-residue flexible linker, and LDB1 mut4 (M252A/L255A/M256A) that disrupts the LDB1/SSBP2 interaction. The ability of full-length LDB1 (wt, mut1, mut2, or mut4) to rescue β-globin expression and LCR looping was tested in the background of LDB1-knockout (KO) mouse erythroid leukemia (MEL) cells (Fig. 5A). Surprisingly, the changes in LDB1 mut1 did not have a significant effect on the ability of LDB1 to rescue β-globin expression or chromatin looping (Fig. 5 B–D), suggesting that, in the context of full-length LDB1, in vivo, disruption of these sites of LDB1 self-interaction is not sufficient to disrupt dimerization. LDB1 mut2 was able to rescue LCR chromatin looping but had a significantly reduced β-globin expression level. This is consistent with our previous published results (11), in which LDB1-delta4/5 with straight deletion of helices H4 and H5 allowed LCR looping but with reduced β-globin expression. The protein level of LDB1 mut4, which disrupts the SSBP2/LCCD interaction, is significantly reduced, consistent with the previous conclusion that SSBP2 is critical for LDB1 stability (26, 27). Furthermore, with reduced LDB1 level, β-globin expression was not rescued, and the looping interactions were lost for LDB1 mut4. This result provides further support for the critical role of SSBP binding in LDB1 stabilization and function.
Fig. 5.
β-Globin transcriptional activity and chromatin looping analysis of LDB1 mutants. (A) Western blots with LDB1 and GFP antibodies. MEL cell differentiation was induced with 1.5% vol/vol dimethyl sulfoxide (DMSO) for 4 d. UMEL, uninduced MEL cells; IMEL, induced MEL cells. (B) Induction of LDB1 KO MEL, MEL, and cells rescued by expression of LDB1 mutants from lentiviruses. Red-colored pellets are indicative of β-globin (hemoglobin) production in induced cells. (C) β-Globin expression after induction. LDB1 KO MEL cells are rescued with wild-type LDB1, mut1, mut2, or mut4. UMEL, uninduced MEL; IMEL, induced MEL. (D) Long-range interactions in the β-globin locus. A chromosome conformation capture (3C) analysis was performed for cells expressing LDB1 and LDB1 mutants. Relative interaction frequency between the HS2 as anchor fragment and β-globin. Bh1 locus served as a control. Error bars in the figures represent standard error (SEM; ***P < 0.001, ****P < 0.0001).
Discussion
LDB proteins contain 4 conserved structural domains: a dimerization domain (DD), an LDB/Chip conserved domain (LCCD), a nuclear localization signal (NLS), and a C-terminal LIM interaction domain (LID). Protein folding propensity analyses indicate that the DD-LCCD region forms a continuously folded structure, while the rest of LDB1 protein may be largely flexible, with the LID forming defined structures when complexed with tandem LIM domains (SI Appendix, Fig. S6). Indeed, part of LCCD (helix H6) docks on the surface of LDB1-DD, and the overall DD-LCCD 3D structure may be relatively rigid. Due to structural flexibility outside of the LDB1 DD-LCCD region, SSBP2 binding is not expected to interfere with the interaction between LDB1-LID and LMO2 LIM domains. Consistent with this, LDB1, SSBP2, and LMO can coexist in the same complex (26).
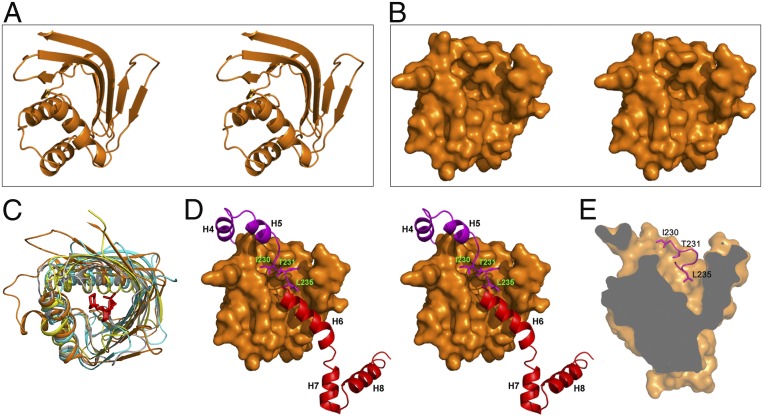
The bulk of LDB1-DD forms an NTF2-like subdomain, despite the lack of apparent sequence homology with all known NTF2-like domains (Fig. 6 A and B and SI Appendix, Table S2). It should be noted that most of the known proteins with an NTF2-like fold contain a central pocket between the β-sheet and the layer of helices, forming a convenient location for creating an enzyme active site or a small molecule binding site (37) (Fig. 6C and SI Appendix, Table S2 and Fig. S3). In our LDB1-DD structure, there is also a pocket in the NTF2-like subdomain that is largely covered by the linker between LDB1-DD H5 and LDB1-LCCD H6 (Fig. 6). Three conserved sidechains in this linker (Ile230, Thr231, and Leu235) point into the pocket and fill the majority of the pocket space (Fig. 6 D and E). It is tempting to speculate that an unknown small-molecule ligand (or a protein ligand) may bind in this LDB1 NTF2-like subdomain pocket and dislodge the DD-LCCD linker from the pocket, which may in turn destabilize the docking of LCCD H6 from the DD surface and lead to an open conformation of LDB1. This potential LDB1 open-closed conformational change may allow for ligand-induced selection of different binding partners of LDB1, such as CTCF, estrogen receptor, and the E3 ubiquitin-ligase RLIM, which bind to the LDB1 DD-LCCD region (27, 31, 33). In this regard, it is particularly interesting that SSBP2 interacts with the LDB1-LCCD fragment much more weakly (∼800 fold) than the LDB1 DD-LCCD fragment (Fig. 1F), suggesting that a potential open conformation may promote SSBP2 dissociation from LDB1. Future studies will be needed to test this hypothesis. It should also be noted that this pocket may also be a potential anticancer drug target, since compounds occupying this pocket may dislodge SSBP2 from the complex and lead to LDB1 destabilization and subsequent Wnt pathway inhibition (26, 39).
Fig. 6.
The LDB1 NTF2-like subdomain contains a potential small-molecule binding pocket. (A and B) Cartoon and surface representation of LDB1 NTF2-like subdomain in stereo view. (C) Structural superposition of different NTF2-like folds: those of LDB1 (in orange); LinA (in yellow; PDB code 3A76); CDL2.2, a computationally designed Vitamin-D3 binder (in gray; PDB code 5IEN); and NTF2 (in cyan; PDB code 1U5O). The ligand from 5IEN is colored in red. (D) This pocket is covered by the LDB1 229NITRCGL235 motif between the H5 in LDB1 DD and the LCCD H6. A, B, and D have the same orientation. (E) A cutaway illustration of the potential ligand-binding pocket of the LDB1 NTF2-like subdomain. Sidechains of I230, T231, and L235 cover this pocket in the LDB1/SSBP2 complex structure.
The importance of the LDB1 dimerization domain to its biological function has long been appreciated (12, 13). Mechanistic studies using the β-globin locus provided the insight that LDB1 dimerization is necessary and sufficient to establish proximity between the β-globin LCR and genes, which activates their expression (11). The present studies provide in-depth understanding of the self-interaction surfaces of LDB1. It is surprising that the LDB1 mut1 missense mutations that disrupted the dimerization interface in vitro were unable to prevent rescue of LDB1/β-globin proximity and transcription activation in cells (Fig. 5). It is possible that, in vivo, other structural components in the LDB1/SSBP-seeded large complex may promote oligomerization of LDB1, but we cannot rule out that elevated levels of mut1 in transfected cells contributed to interactions. On the contrary, the finding that LDB1 mut2, in which a flexible linker replaces the H4-H5 subdomain, can rescue LCR looping but is defective for transcription activation is congruent with the in vivo studies (11). Deletion of H4-H5 did not compromise dimerization/looping in erythroid cells, but RNA pol II recruitment was reduced, as was β-globin transcriptional output (11). Together, these results suggest that the NTF2-like subdomain dimerization interface may suffice for LDB1 dimerization in cells rescued by LDB1 missing the H4-H5 subdomain. Furthermore, these observations support the idea that the H4-H5 subdomain may have other functions beyond participating in long-range looping, for example, in transcription activation. It is also possible, and not mutually exclusive, that H4-H5 may play a role in modulating potential ligand binding in the neighboring pocket of the NTF2-like subdomain. During the review process of this manuscript, Renko et al. (39) reported crystal structure of an LDB1 dimerization domain, which appeared to have a dimerization interface very similar to that in our LDB1/SSBP2 complex structure. Our complex structure is also consistent with all biochemical data reported in that work (39).
It is intriguing that the DD and LCCD domains mediating the LDB1/SSBP2 interaction are conserved even in plants. For example, the Arabidopsis LUFS domain-containing protein LEUNIG cooperates with SEUSS, a protein that shares similarity with LDB/Chip proteins, to regulate flower development (14, 21, 40). We notice that key residues in the LDB1/SSBP2 interface are also conserved in SEUSS and LEUNIG (SI Appendix, Fig. S7), suggesting that SEUSS and LEUNIG may also interact in a manner similar to that of LDB1/SSBP2 we describe here. Therefore, our structure reveals an ancient and fundamentally important structural framework used by many organisms and diverse biological processes to enable precise control of transcription.
Materials and Methods
Protein Purification.
Proteins were overexpressed in BL21(DE3) cells and purified with Ni-NTA resin followed by anion-exchange chromatography and gel filtration.
Crystallization and Structure Determination.
The LDB1/SSBP2 complex was crystallized using hanging-drop vapor diffusion. The structure was determined by the single-wavelength anomalous dispersion (SAD) method and refined to 2.8-Å resolution.
In Vitro Protein–Protein Interaction.
LDB1/SSBP2 interactions and oligomerization states of the LDB1/SSBP2 complex and their mutants were analyzed by SEC-MALS, MBP pull-down, and SPR analysis.
Functional Analysis of LDB1 Dimerization and LDB1/SSBP2 Interaction.
Potential functions of LDB1 dimerization and LDB1/SSBP2 interaction were analyzed through β-globin transcriptional activity and chromatin looping assays. More detailed information on structural, biochemical, and functional experiments is provided in SI Appendix, Supplementary Materials and Methods.
Data Availability.
The atomic coordinates and structure factors of the LDB1/SSBP2 complex have been deposited in the Protein Data Bank, https://www.rcsb.org/ under the accession code 6TYD.
Supplementary Material
Acknowledgments
We appreciate assistance in data collection from staff of beamlines 8.2.1 and 8.2.2, Advanced Light Source. We also thank Y. Chen, Z. Yang, and X. Yu for SPR or SEC-MALS technical support at the protein science research platform of the Institute of Biophysics. This work was supported by the National Laboratory of Biomacromolecules and by National Natural Science Foundation of China Grants 31570794 and 31629002; and Chinese Academy of Sciences Grant XDB08010303 to X.-X.Y. and W.X., respectively. J.K. and A.D. were supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors of the LDB1/SSBP2 complex have been deposited in the Protein Data Bank, https://www.rcsb.org/ under the accession code 6TYD.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914181117/-/DCSupplemental.
References
- 1.Matthews J. M., Visvader J. E., LIM-domain-binding protein 1: A multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 4, 1132–1137 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu G., Dean A., Enhancer long-range contacts: The multi-adaptor protein LDB1 is the tie that binds. Biochim. Biophys. Acta. Gene Regul. Mech. 1862, 625–633 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Agulnick A. D., et al. , Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384, 270–272 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Jurata L. W., Kenny D. A., Gill G. N., Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc. Natl. Acad. Sci. U.S.A. 93, 11693–11698 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach I., Carrière C., Ostendorff H. P., Andersen B., Rosenfeld M. G., A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 11, 1370–1380 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay M., et al. , Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development 130, 495–505 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Li L., et al. , A requirement for Lim domain binding protein 1 in erythropoiesis. J. Exp. Med. 207, 2543–2550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L., et al. , Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood 121, 4575–4585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song S. H., Hou C., Dean A., A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell 28, 810–822 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng W., et al. , Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krivega I., Dale R. K., Dean A., Role of LDB1 in the transition from chromatin looping to transcription activation. Genes Dev. 28, 1278–1290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breen J. J., Agulnick A. D., Westphal H., Dawid I. B., Interactions between LIM domains and the LIM domain-binding protein Ldb1. J. Biol. Chem. 273, 4712–4717 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Jurata L. W., Pfaff S. L., Gill G. N., The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J. Biol. Chem. 273, 3152–3157 (1998). [DOI] [PubMed] [Google Scholar]
- 14.van Meyel D. J., Thomas J. B., Agulnick A. D., Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development 130, 1915–1925 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Chen L., et al. , Ssdp proteins interact with the LIM-domain-binding protein Ldb1 to regulate development. Proc. Natl. Acad. Sci. U.S.A. 99, 14320–14325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deane J. E., et al. , Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4. EMBO J. 22, 2224–2233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deane J. E., et al. , Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex. EMBO J. 23, 3589–3598 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffries C. M., et al. , Stabilization of a binary protein complex by intein-mediated cyclization. Protein Sci. 15, 2612–2618 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Omari K., et al. , Structure of the leukemia oncogene LMO2: Implications for the assembly of a hematopoietic transcription factor complex. Blood 117, 2146–2156 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Gadd M. S., et al. , A structural basis for the regulation of the LIM-homeodomain protein islet 1 (Isl1) by intra- and intermolecular interactions. J. Biol. Chem. 288, 21924–21935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conner J., Liu Z., LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. U.S.A. 97, 12902–12907 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sridhar V. V., Surendrarao A., Gonzalez D., Conlan R. S., Liu Z., Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc. Natl. Acad. Sci. U.S.A. 101, 11494–11499 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., Wang Z., Tang Q., Yan X. X., Xu W., Crystal structure of the LUFS domain of human single-stranded DNA binding protein 2 (SSBP2). Protein Sci. 28, 788–793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishioka N., et al. , Ssdp1 regulates head morphogenesis of mouse embryos by activating the Lim1-Ldb1 complex. Development 132, 2535–2546 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Enkhmandakh B., Makeyev A. V., Bayarsaihan D., The role of the proline-rich domain of Ssdp1 in the modular architecture of the vertebrate head organizer. Proc. Natl. Acad. Sci. U.S.A. 103, 11631–11636 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Z., et al. , Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 21, 942–955 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostendorff H. P., et al. , Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature 416, 99–103 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Fernández-Fúnez P., Lu C. H., Rincón-Limas D. E., García-Bellido A., Botas J., The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J. 17, 6846–6853 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milán M., Cohen S. M., Regulation of LIM homeodomain activity in vivo: A tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol. Cell 4, 267–273 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Rincón-Limas D. E., Lu C. H., Canal I., Botas J., The level of DLDB/CHIP controls the activity of the LIM homeodomain protein apterous: Evidence for a functional tetramer complex in vivo. EMBO J. 19, 2602–2614 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnsen S. A., et al. , Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 69, 128–136 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiedler M., et al. , An ancient Pygo-dependent Wnt enhanceosome integrated by Chip/LDB-SSDP. eLife 4, e09073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J., Krivega I., Dale R. K., Dean A., The LDB1 complex co-opts CTCF for erythroid lineage-specific long-range enhancer interactions. Cell Rep. 19, 2490–2502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Tienen L. M., Mieszczanek J., Fiedler M., Rutherford T. J., Bienz M., Constitutive scaffolding of multiple Wnt enhanceosome components by Legless/BCL9. eLife 6, e20882 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullock T. L., Clarkson W. D., Kent H. M., Stewart M., The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2). J. Mol. Biol. 260, 422–431 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Eberhardt R. Y., et al. , Filling out the structural map of the NTF2-like superfamily. BMC Bioinf. 14, 327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.PDB-101. Structural Biology Highlights: New NTF2-like Domains. Available at https://pdb101.rcsb.org/learn/structural-biology-highlights/new-ntf2-like-domains. Accessed 2 November 2019.
- 39.Renko M., et al. , Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome. Proc. Natl. Acad. Sci. U.S.A. 116, 20977–20983 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franks R. G., Wang C., Levin J. Z., Liu Z., SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129, 253–263 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of the LDB1/SSBP2 complex have been deposited in the Protein Data Bank, https://www.rcsb.org/ under the accession code 6TYD.