Significance
The axon initial segment is a specialized domain responsible for the generation of action potential and maintenance of axonal polarity and is characterized by enrichment for voltage-gated ion channels and a scaffold protein, ankryin-G (AnkG). AnkG interacts with the dynein regulator Ndel1 to regulate cargo sorting at the AIS. Here we demonstrate that a specific and high-affinity interaction mediates AnkG/Ndel1 complex formation via biochemical and structural approaches. We further show that several somatodendritic cargoes are missorted at the AIS on disruption of the AnkG/Ndel1 complex. Our studies not only establish the mechanistic basis for AnkG–Ndel1 interaction and for the maintenance of the axonal polarity, but also provide insight into the AnkB/Ndel1 complex function potentially at distal axonal cargo transport.
Keywords: ankyrin-G/B, Ndel1/Nde1, axon initial segment, axonal polarity, selective transport
Abstract
Ankyrin-G (AnkG), a highly enriched scaffold protein in the axon initial segment (AIS) of neurons, functions to maintain axonal polarity and the integrity of the AIS. At the AIS, AnkG regulates selective intracellular cargo trafficking between soma and axons via interaction with the dynein regulator protein Ndel1, but the molecular mechanism underlying this binding remains elusive. Here we report that Ndel1’s C-terminal coiled-coil region (CT-CC) binds to giant neuron-specific insertion regions present in both AnkG and AnkB with 2:1 stoichiometry. The high-resolution crystal structure of AnkB in complex with Ndel1 CT-CC revealed the detailed molecular basis governing the AnkB/Ndel1 complex formation. Mechanistically, AnkB binds with Ndel1 by forming a stable 5-helix bundle dominated by hydrophobic interactions spread across 6 distinct interaction layers. Moreover, we found that AnkG is essential for Ndel1 accumulation at the AIS. Finally, we found that cargo sorting at the AIS can be disrupted by blocking the AnkG/Ndel1 complex formation using a peptide designed based on our structural data. Collectively, the atomic structure of the AnkB/Ndel1 complex together with studies of cargo sorting through the AIS establish the mechanistic basis for AnkG/Ndel1 complex formation and for the maintenance of axonal polarity. Our study will also be valuable for future studies of the interaction between AnkB and Ndel1 perhaps at distal axonal cargo transport.
Neurons are highly polarized cells with distinct compartments responsible for vital physiological functions. The axon initial segment (AIS) is a specialized region connecting the soma and axon characterized by enrichment for diverse proteins, including voltage-gated ion channels (e.g., sodium channels, potassium channels, calcium channels), cell adhesion molecules (Neurofascin-186 and NrCAM), cytoskeleton proteins (βIV-spectrin, MAP6, and MAP7D2), and a scaffold protein, ankyrin-G (AnkG) (1, 2). In addition to being responsible for the initial depolarization and generation of action potential, the AIS also serves as an axonal-somatodendritic filter, contributing to selective transport of specific molecules to somatodendrites or axons to maintain the neuronal polarity (1, 3). In this specialized membrane domain, AnkG is a master scaffold protein in organizing and stabilizing diverse complexes that it forms with Nav channels, Kv channels, L1 family CAMs, and βIV-spectrin (4, 5). Loss of AnkG, either in vitro or in vivo, leads to axonal polarity disruption and causes axons to acquire dendritic properties (6, 7). Given the known essential physiological roles of the AIS, it is not surprising that mutations in AIS-related proteins are closely related with diverse neurologic diseases, including epilepsy, bipolar disorder, schizophrenia, and autism spectrum disorder (8–12).
Previous studies have suggested that AnkG is the first protein that accumulates at the AIS during axonal development, and thus it has become a defining marker in studies investigating AIS function (13, 14). The position of the AIS at the interface of soma and axons in neurons supports its function in controlling and maintaining axonal polarity, although the underlying mechanisms of these AIS complex functions remain unclear. Previous studies have reported that an actin-based passive selective filter at the AIS allows entry of axonal, but not somatodendritic, vesicles into the distal axon (15–17). However, the existence of such a cytoplasmic filter has been challenged recently by several studies that used superresolution microscopy and reported finding only sparse actin filaments with mixed orientations, rather than the expected dense network (18–20). An alternative microtubule-based active sorting mechanism has been proposed in several studies (21–23). Microtubules are uniformly oriented with their plus-ends pointing to the distal axon, and the molecular motor protein dynein controls retrograde transport along microtubules in axons (24, 25). A recent study demonstrated that AnkG-dependent localization of the dynein regulator Ndel1 at the AIS is responsible for efficient local cargo reversal (22). Ndel1, or its homolog Nde1, together with Lis1 locally activates dynein for cargo transportation out of the axons (22, 26); however, the molecular mechanisms governing the Ndel1/AnkG complex formation remain elusive.
Here we biochemically and structurally characterized the interaction between AnkG and Ndel1/Nde1 in detail, and found that an 1,897- to 1,966-amino acid section of the AnkG giant insertion region strongly binds to the C-terminal coiled-coil domains of Ndel1/Nde1 and does so with a molar ratio of 1:2. We found that the 440-kDa giant AnkB (gAnkB), which is also specifically expressed in neurons, also binds to Ndel1/Nde1 in a similar manner. We then elucidated the precise molecular mechanisms of the interaction between neuronal-specific ankyrins and Ndel1/Nde1 by solving the crystal structure of the Ndel1-binding region of gAnkB bound to Ndel1. Further structural analysis not only revealed the molecular basis of the interaction, but also enabled us to design specific mutations to evaluate the functions of the AnkG/Ndel1 complex in cargo sorting at the AIS and a peptide to highly specifically block AnkG/Ndel1 complex formation at the AIS. Extensive cell biology analyses revealed that Ndel1 accumulation at the AIS requires specific interaction between AnkG and Ndel1. Finally, and supporting a gatekeeper function of AnkG at the AIS, we found that the sorting of 3 distinct somatodendritic cargoes—transferrin receptor (TfR), GluR1, and Coxsackie and adenovirus receptor (CAR)—is disrupted in cells lacking AIS-localized AnkG/Ndel1 complexes.
Results
AnkG Strongly Binds to Ndel1/Nde1 with 1:2 Stoichiometry.
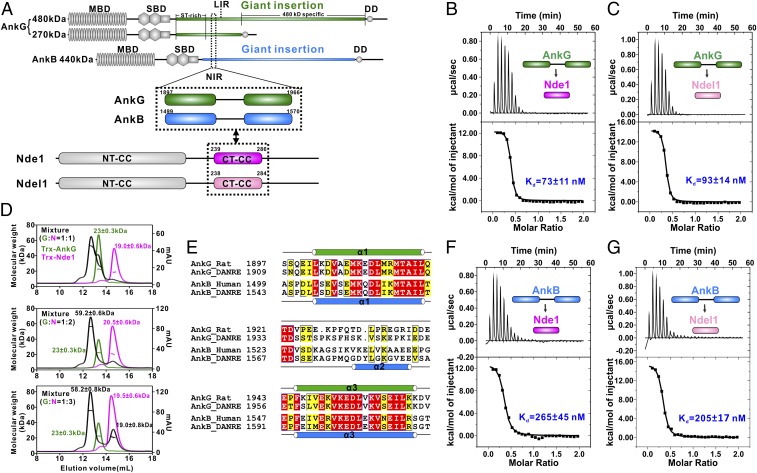
An earlier study established that interaction between AnkG and Ndel1/Nde1 is mediated by the 480/270-kDa AnkG giant insertion region and the C terminus of Ndel1/Nde1 (22). Seeking to characterize this interaction in considerable biochemical detail, we used isothermal titration calorimetry (ITC) assays to map the binding location of a highly purified recombinant C-terminal variant of Nde1 (amino acids 191 to 335) with AnkG exon 37 (which codes the giant insertion region). We found a predicted unstructured region in AnkG exon 37 (amino acids 1,897 to 1,966), which we term “AnkG-NIR” (Ndel1/Nde1-interacting region). AnkG-NIR is located immediately preceding AnkG-LIR (LC3 interacting region; amino acids 1,985 to 2,010) (27) and is both necessary and sufficient for binding to Nde1 with a dissociation constant (Kd) ∼75 nM (Fig. 1A and SI Appendix, Table S1). Further ITC-based mapping showed that the Nde1 C-terminal coiled-coil region (amino acids 239 to 286, which we term “Nde1 CT-CC”) binds to AnkG-NIR with essentially the same affinity (Kd ∼73 nM; Fig. 1 A–C and SI Appendix, Table S1), revealing that the Nde1 CT-CC alone is sufficient for binding to AnkG-NIR. Interestingly, the N values of the ITC curves obviously deviate from 1, suggesting that the binding stoichiometry of the AnkG-Nde1 might not be 1:1.
Fig. 1.
The giant insertions of AnkG and AnkB bind strongly to Ndel1/Nde1 at a 1:2 molar ratio. (A) Schematic diagram showing the domain organizations of 270/480-kDa AnkG and 440-kDa AnkB, Nde1, and Ndel1, as well as the location of the NIR sequences in AnkG/AnkB. In this drawing, the AnkG/AnkB and Nde1/Ndel1 interactions are further highlighted and indicated by 2-way arrows. MBD, membrane-binding domain; SBD, spectrin-binding domain; DD, death domain. (B and C) ITC-based measurements of the binding affinity of the AnkG-NIR with Nde1 CT-CC (B) or Ndel1 CT-CC (C). The Kd error is the fitting error obtained using a 1-site binding kinetics model to fit the ITC data. The ITC-measured N value, which is related to the binding stoichiometry, indicates that the binding stoichiometry of AnkG-NIR with Ndel CT-CC/Ndel1 CT-CC interaction is 1:2. (D) Analytical gel filtration chromatography analysis coupled with static light scattering analysis of Trx-AnkG-NIR (green line), Trx-Nde1 CT-CC (pink line), and the Trx-AnkG-NIR/Trx-Nde1 CT-CC complex (black line), indicating that AnkG-NIR and Nde1 CT-CC form a stable 1:2 complex in solution. (E) Sequence alignment of the NIRs from AnkG and AnkB. Residues that are identical and highly similar are indicated in red and yellow boxes, respectively. The secondary structure elements are labeled according to the crystal structure of the AnkB/Ndel1 complex and the NMR data of the AnkG/Nde1 complex. (F and G) ITC-based measurement of the binding affinity of AnkB-NIR with Ndel CT-CC (F) and with Ndel1CT-CC (G).
We next used analytical gel filtration chromatography coupled with a static light scattering assay to confirm the direct interaction and to determine the binding ratio between AnkG-NIR and Nde1 CT-CC. When AnkG-NIR and Nde1 CT-CC were mixed at a 1:1 molar ratio, they formed a heterogeneous complex (Fig. 1D). Subsequent SDS/PAGE analysis showed that a significant portion of AnkG-NIR remained in its free form. When mixed at a 1:2 molar ratio, a homogeneous complex peak was formed with the measured molecular mass of 59.2 kDa, matching well with the theoretical molecular weight of a complex comprising 1 copy of AnkG-NIR and 2 copies of Nde1 CT-CC (∼61.4 kDa). Further supporting the 1:2 binding stoichiometry, when we mixed the 2 components at a 1:3 molar ratio, the complex peak eluted in the same position with a similar molecular mass (∼58.2 kDa), but in addition to the complex peak, roughly one-third of the detected Nde1 CT-CC was in its free form (Fig. 1D). Collectively, these results demonstrate that 1 AnkG-NIR molecule is able to bind to 2 Nde1 CT-CC molecules.
Nde1 and Ndel1 are paralogous proteins sharing many redundant functions and sharing high sequence similarity, especially in the N-terminal coiled-coil (NT-CC) regions (amino acids 1 to 192) and CT-CC regions (∼68% and 72% sequence identities, respectively). We used similar methods to examine the interaction between AnkG-NIR and Ndel1 CT-CC and found that Ndel1 CT-CC can also bind to AnkG-NIR with high affinity (Kd ∼93 nM), and does so with 1:2 binding stoichiometry (Fig. 1C).
AnkB Also Contains an NIR and Binds to Ndel1/Nde1 with 1:2 Stoichiometry.
Both AnkG and AnkB have giant isoforms (270/480-kDa AnkG/gAnkG and 440-kDa AnkB/gAnkB) that are specifically expressed in the axons of neurons (28–30). gAnkG is enriched at the AIS and the node of Ranvier, whereas gAnkB is localized in distal axonal regions (28, 31). Evolutionary analysis suggests that the insertion of the vertebrate giant exon occurred before the divergence of AnkG and AnkB (32). As the AnkG-NIR is positioned immediately before the AnkG-LIR, and given that both gAnkG and gAnkB contain a similar LIR in the giant insertion (27), we anticipated that gAnkB might also have a similar NIR. Sequence analysis showed that there is indeed a region encompassing amino acids 1,499 to 1,570 preceding AnkB-LIR, and that this region aligns well with AnkG-NIR (and is thus denoted “AnkB-NIR”; Fig. 1 A and E).
We again used ITC and analytical gel filtration assays to test the binding between AnkB-NIR and Ndel1/Nde1. The ITC analysis showed that AnkB-NIR can also interact with both Nde1 and Ndel1 CT-CCs with high affinity (Kd ∼265 nM and ∼205 nM, respectively; Fig. 1 F and G). Consistent with our expectation, analytical gel filtration results also showed that 1 AnkB-NIR molecule can bind to 2 Ndel1 molecules (SI Appendix, Fig. S1).
Structural Characterization of the AnkB–Ndel1 Interaction.
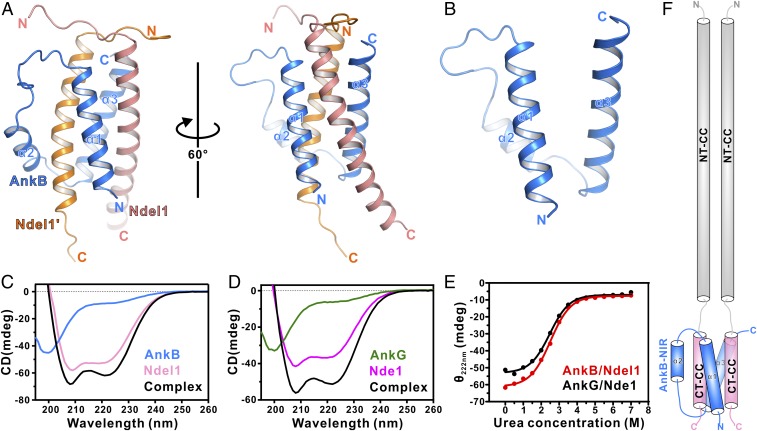
To help elucidate the molecular mechanisms governing complex formation between AnkG/AnkB and Ndel1/Nde1, we sought to determine the structures of AnkG/AnkB in complex with Ndel1/Nde1. Given that both AnkG and AnkB can interact strongly with either Ndel1 or Nde1, we performed crystal screenings on all 4 combinations. Although the AnkG/Ndel1 or AnkG/Nde1 complexes formed crystals, the crystals diffracted poorly (only 5 to 6 Å after extensive optimization). Fortunately, we successfully obtained the crystals of AnkB-NIR in complex with Ndel1 CT-CC that diffracted to 1.5 Å. We used the ab initio phasing program ARCIMBOLDO to solve a high-quality complex crystal structure (33) (SI Appendix, Table S2 and Fig. S2). In line with our biochemical findings, the AnkB/Ndel1 CT-CC complex structure consists of 1 AnkB molecule and 2 Ndel1 CT-CC molecules, which pack together to form a 5-helix bundle architecture (Fig. 2A).
Fig. 2.
Overall structure of AnkB-NIR in complex with Ndel1 CT-CC. (A) Ribbon diagram showing the overall structure of AnkB/Ndel1 complex. In this drawing, AnkB-NIR is shown in marine blue, and 2 copies of Ndel1 CT-CC are shown in orange and deep salmon, respectively. (B) Ribbon diagram showing that apo AnkB-NIR is composed of 3 α-helices widely separated from one another, indicating that AnkB-NIR alone lacks a well-formed folding core. (C) CD spectra showing that Ndel1 CT-CC alone is well folded, with an α-helical conformation, whereas AnkB-NIR alone predominantly forms a random coil. The AnkB/Ndel1 complex forms a robust α-helical structure. (D) CD spectra showing that Nde1 CT-CC alone is well folded, with an α-helical conformation, whereas AnkG-NIR alone predominantly forms a random coil. The AnkG/Nde1 complex forms a robust α-helical structure. (E) CD spectra-based urea-induced denaturation curves showing denaturation profiles the AnkB/Ndel1 complex and the AnkG/Nde1 complex. (F) Cartoon depicting full-length Ndel1 forming a homodimer via NT-CC regions and interacting with AnkB via 2 CT-CCs.
In the complex structure, the 2 Ndel1 CT-CC molecules each form an α-helix with an N-terminal extended loop, which was also observed in a solution structure of Ndel1 in complex with DISC1 (34). Interestingly, these 2 helices, arranged in parallel, are wrapped by 3 α-helices of AnkB-NIR (Fig. 2A). The 3 α-helices of AnkB-NIR in the complex are widely separated from one another, lacking any obvious contact (Fig. 2B). The absence of a folding core by itself suggests that AnkB-NIR alone is highly unlikely to fold, and thus the observed folding of AnkB-NIR may be induced by Ndel1 binding. Consistent with this structural observation, the circular dichroism (CD) spectrum of AnkB-NIR alone is typical of a random coil conformation (Fig. 2C). When Ndel1 CT-CC was added, the appearance of the characteristic minima at 208 nm and 222 nm indicated robust formation of α-helical structures (Fig. 2C).
Similar results were obtained for AnkG-NIR alone and for the AnkG-NIR/Nde1 CT-CC complex, revealing that AnkG-NIR also undergoes Ndel1/Nde1 binding-induced folding (Fig. 2D). Moreover, CD spectra-based urea denaturation results demonstrated that the formed AnkB/Ndel1 and AnkG/Nde1 complexes are quite stable (Fig. 2E). Since the full-length Ndel1 forms a parallel homodimer through its NT-CC region (35), the parallel configurations of the 2 Ndel1 CT-CC molecules in our solved complex suggest that the full-length Ndel1 dimer binds to 1 molecule of AnkB, forming a highly stable Ndel1/AnkB complex (Fig. 2F).
Detailed Structure of the AnkB/Ndel1 Complex.
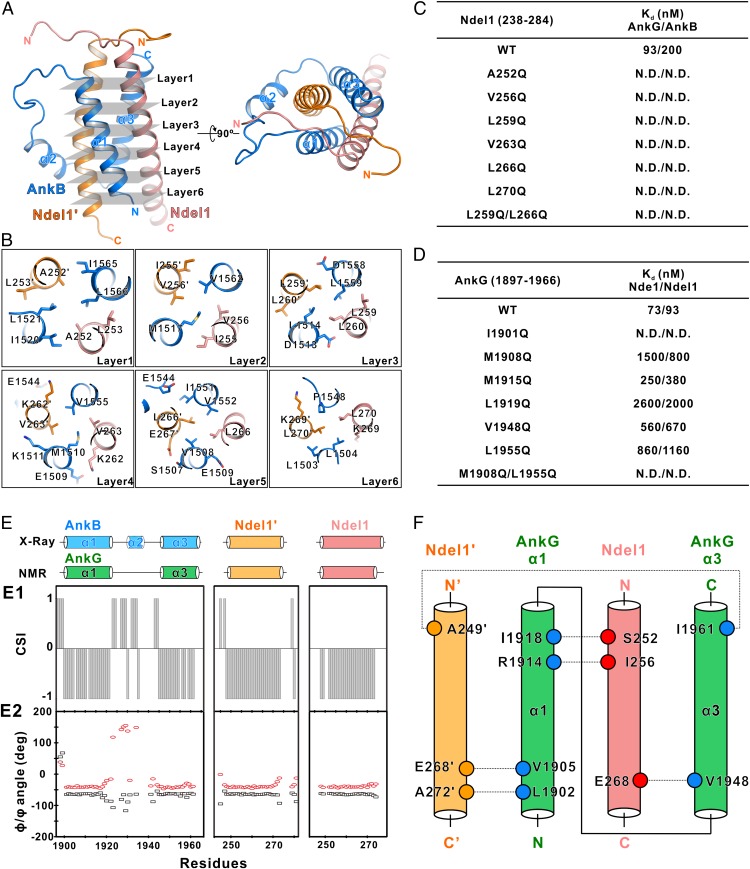
The binding interface of the AnkB/Ndel1 complex involves the α1/α3 of AnkB and the 2 Ndel1 CT-CCs; together, these form the core within the helical bundle (Fig. 3A). The interior of the core is formed by 6 layers of interacting amino acid side chains. The assemblies of the 6 layers are predominately mediated by hydrophobic interactions (Fig. 3B). Mutating any of the corresponding hydrophobic residues in Ndel1 to the polar residue glutamine completely abolished the binding to AnkB (Fig. 3C). Notably, charge–charge interactions also contribute to the binding. K262 from 1 Ndel1 molecule forms a salt bridge with the negatively charged E1544 from AnkB (Fig. 3B, layer 4). Simultaneously, the K262 residue from the other Ndel1 molecule is neutralized by 2 nearby negatively charged AnkB residues: E1506 and E1509 (Fig. 3B, layers 4 and 5).
Fig. 3.
Detailed molecular interface of the AnkB/Ndel1 complex. (A) Ribbon diagram showing the interior interactions of the AnkB/Ndel1 complex formed by 6 layers of interacting amino acid side chains, and the positions of all 6 layers. (B) Ribbon-stick diagram showing that the assemblies of the 6 layers are mediated mainly by hydrophobic interactions. (C) The measured binding affinities between various mutations of Ndel1 CT-CC and AnkG or AnkB from ITC-based binding assays. N.D. indicates that no binding was detected. (D) The measured binding affinities between various mutations of AnkG-NIR and Nde1 or Ndel1 from ITC-based binding assays. (E) Secondary structural analysis of the AnkG/Ndel1 complex based on NMR assignments. (E1) Chemical shift index analysis of the AnkG/Ndel1 complex using backbone assignments NH, N, CA, and CB. (E2) A TALOS dihedral angle prediction showing the α-helix distribution of AnkG/Ndel1. The secondary structures derived from X-ray crystallography structure and NMR analysis are compared at the top. (F) Schematic model showing the selected side chain NOEs of the AnkG/Ndel1 complex, suggesting direct contact of the key residues. The NOEs are highlighted by dashed lines. NOE analysis indicates that the complex topology observed in the solution NMR analysis is consistent with the AnkB/Ndel1 complex crystal structure.
Given the high similarity between AnkG and AnkB-NIRs, we next wondered whether the structure of AnkG resembles that of AnkB when binding to Ndel1 or Nde1. As we were unable to obtain well-diffracted crystals, we performed NMR spectroscopy as an alternative approach. The 1H-15N heteronuclear single quantum coherence spectrum of the AnkG/Ndel1 complex was homogeneous and well- dispersed, indicating that it formed a folded and stable structure. Furthermore, chemical shift index analysis revealed that the AnkG/Ndel1 also formed an α-helical structure, with the exception that the linker between α1 and α3 in AnkG was a loop, instead of a short α-helix (α2) as in AnkB (Fig. 3E). However, the 3D 15N- and 13C-separated nuclear Overhauser effect (NOE) spectra of the AnkG/Ndel1 complex severely overlapped in many regions, presumably due to the highly helical structure and the lack of aromatic residues in the core of the complex. This prevented us from determining the de novo high-resolution structure using NMR at high resolution. Nonetheless, a number of NOE signals assigned are highly consistent with the topology observed in our crystal structure of the AnkB/Ndel1 complex (Fig. 3F).
To further confirm this binding mode, we evaluated how the interaction between AnkG/Nde1 or AnkG/Ndel1 was impacted on mutation of hydrophobic residues positioned at the 6-layer interface. ITC results showed that individually changing each of the corresponding hydrophobic residues in AnkG to glutamine led to significant weakening or total loss of binding to Nde1 or Ndel1 (Fig. 3D). Moreover, mutations of corresponding hydrophobic residues in Ndel1 to glutamine also eliminated the binding (Fig. 3C). CD-based urea denaturation analysis also showed that the AnkG M1908Q mutant, which has significantly weakened binding ability, led to destabilization of the complex (SI Appendix, Fig. S3). Taken together, these results demonstrate that AnkG and AnkB-NIRs share a similar mode in binding to Ndel1 or Nde1; both form a stable helical bundle via an extensive 6-layer network of hydrophobic interactions supported by additional several charge–charge interactions.
AnkG-Mediated Accumulation of Ndel1/Nde1 at the AIS.
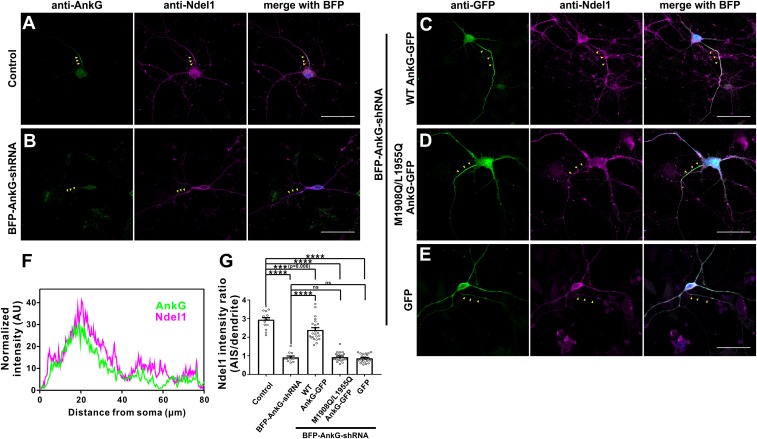
AnkG, the predominant scaffold protein at the AIS, is known to specifically localize at the AIS region in neurons (28, 30, 36). This unique localization pattern allowed us to investigate the potential role(s) of the AnkG-Ndel1/Nde1 interaction by examining their AIS-specific accumulation in cultured hippocampal neurons. Consistent with previously reported data (22), we found that Ndel1 and Nde1 showed a clear AIS enrichment in hippocampal neurons and colocalized well with AnkG (Figs. 4 A and F and SI Appendix, Fig. S4). Furthermore, the intensity of the AnkG signal was substantially reduced after BFP-AnkG-shRNA transfection, and nearly no AIS enrichment for Ndel1 was observed in AnkG knockdown neurons (Fig. 4 B and G), clearly indicating that the AIS localization of Ndel1 depends on AnkG binding. Overexpression of 480-kDa wild-type (WT) AnkG-GFP, but not GFP-tag alone, after BFP-AnkG-shRNA knockdown rescued the AIS localization of both AnkG and Ndel1 (Fig. 4 C, E, and G). Consistent with our biochemical results showing that the M1908Q/L1955Q AnkG variant cannot bind to Ndel1 (Fig. 3D), overexpression of this mutant could rescue AnkG but not Ndel1 AIS localization (Fig. 4 D and G).
Fig. 4.
The specific interaction between AnkG-NIR and Ndel1 is required for Ndel1 accumulation at the AIS. (A) Neurons transfected with control BFP-shRNA and stained for AnkG (green) and Ndel1 (magenta). Yellow arrowheads indicate the AIS. (Scale bar: 50 μm.) (B) Neurons transfected with BFP-AnkG-shRNA and stained for AnkG (green) and Ndel1 (magenta). (Scale bar: 50 μm.) (C) shRNA-resistant WT 480-kDa AnkG-GFP effectively rescues AnkG self-clustering and clustering of Ndel1 at the AIS. BFP marks the AnkG-shRNA transfected neurons. (Scale bar: 50 μm.) (D) The M1908Q/L1955Q AnkG-GFP is clustered at the AIS but fails to recruit Ndel1 to the AIS. BFP marks the AnkG-shRNA–transfected neurons. (Scale bar: 50 μm.) (E) Control GFP alone did not accumulate at the AIS and failed to recruit Ndel1 to the AIS. BFP marks the AnkG-shRNA–transfected neurons. (Scale bar: 50 μm.) (F) Fluorescence intensity plots of A provide a comparison of the immunosignal strength of AnkG (AIS, green) and Ndel1 (magenta). (G) Quantification of the antiendogenous Ndel1 fluorescence intensity ratio of the AIS to dendrites in control neurons (A; n = 15), in neurons depleted of endogenous 270/480-kDa AnkG (B; n = 12), and in neurons rescued with WT AnkG-GFP (C; n = 23), M1908Q/L1955Q AnkG-GFP (D; n = 20), and GFP alone (E; n = 22). Error bars indicate SE. ****P < 0.0001; ns, not significant.
We also tested whether various binding-deficient Ndel1/Nde1 mutants could localize at the AIS and found that L259Q, L266Q, and double-mutant L259Q/L266Q variants did not accumulate at the AIS (SI Appendix, Fig. S5 A and B). The same disrupted accumulation phenotypes were observed for the corresponding mutant variants of Nde1 (SI Appendix, Fig. S5C), again supporting that binding to AnkG is required for the recruitment of Ndel1 and Nde1 to the AIS. We also observed that shRNA-mediated knockdown of NDEL1 did not affect AnkG’s AIS enrichment, revealing that accumulation of AnkG at the AIS does not rely on Ndel1 or Nde1 (Fig. 4D and SI Appendix, Fig. S6).
The AnkG/Ndel1 Complex Regulates Selective Cargo Sorting at the AIS.
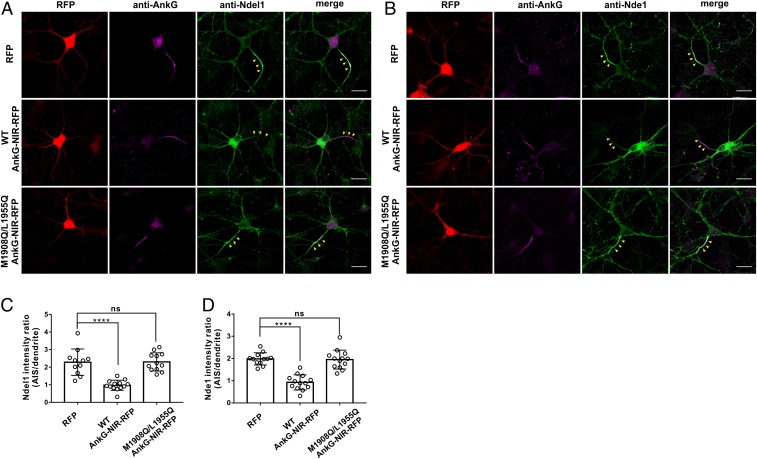
To investigate the physiological roles of the complexes formed between AnkG and Ndel1/Nde1 at the AIS, we next attempted to disrupt the complex. However, considering the high sequence similarity and functional redundancy of Ndel1 and Nde1, we anticipated that knockdown of either of the 2 proteins alone would potentially still allow for normal or near-normal function of the complex in question. Thus, guided by our biochemical and structural data, we adopted a strategy based on overexpression of an AnkG-NIR peptide (1,897 to 1,966; WT AnkG-NIR-red fluorescent protein [RFP]) in cultured hippocampal neurons to saturate the endogenous Ndel1 and Nde1. Entirely consistent with our biochemical data, overexpression of this peptide totally abolished AIS enrichment of Ndel1 and Nde1 (Fig. 5 A and B, Middle and Fig. 5 C and D). In contrast, overexpression of the RFP tag or an M1908Q/L1955Q AnkG-NIR-RFP mutant variant peptide had no effect on Ndel1 or Nde1 enrichment at the AIS (Fig. 5 A and B, Upper and Lower and Fig. 5 C and D). Note that localization of AnkG was unaffected by the expression of any of these peptides (Fig. 5 A and B).
Fig. 5.
Overexpression of an AnkG-NIR peptide abolished AIS enrichment of Ndel1 and Nde1. (A and B) Neurons transfected with RFP control, WT AnkG-NIR-RFP, or M1908Q/L1955Q AnkG-NIR-RFP and stained for AnkG (magenta) or endogenous Ndel1 (A) or Nde1 (B). Yellow arrowheads indicate the AIS. (Scale bars: 20 μm.) (C) Quantification of the Ndel1 fluorescence intensity ratio of the AIS to dendrites in neurons transfected with RFP (n = 11), WT AnkG-NIR-RFP (n = 13), or M1908Q/L1955Q AnkG-NIR-RFP (n = 12). Error bars indicate SE. ****P < 0.0001. (D) Quantification of the Nde1 fluorescence intensity ratio of AIS to dendrites in neurons transfected with RFP (n = 12), WT AnkG-NIR-RFP (n = 13), or M1908Q/L1955Q AnkG-NIR-RFP (n = 13). Error bars indicate SE. ****P < 0.0001; ns, not significant.
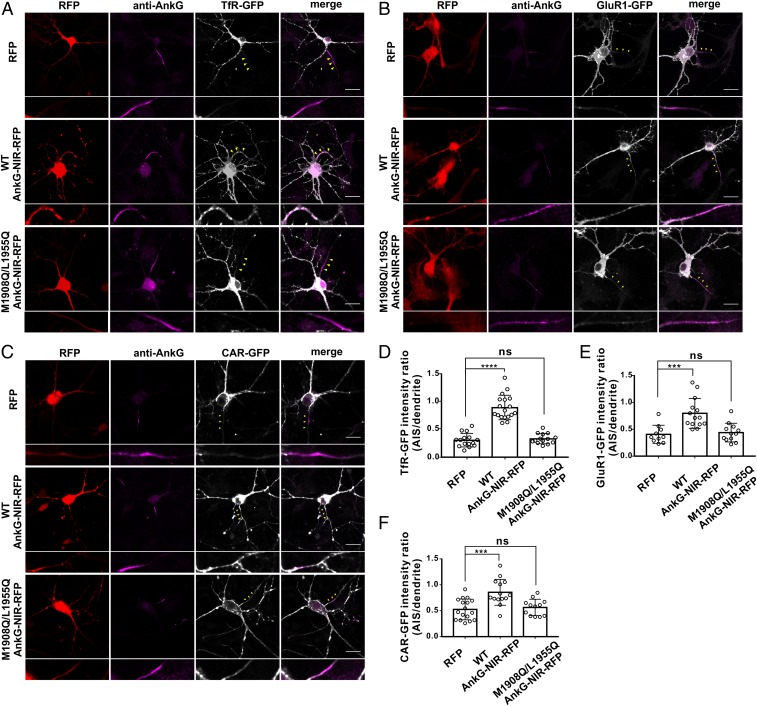
We next tested the neuronal functional consequences of eliminating formation of the AnkG/Ndel1 complex at the AIS by examining somatodendritic cargo sorting into axons. Given Ndel1/Nde1 enrichment at the AIS, we speculated that disruption of AnkG/Ndel1 complex formation would result in axonal-somatodendritic cargo sorting defects due to deregulation of the dynein motor activity at the AIS. To pursue this, we conducted experiments in cultured hippocampal neurons using a GFP reporter fusion variant of the specific somatodendritic distribution protein TfR, which is not sorted into axons under normal conditions (Fig. 6 A, Upper and Fig. 6D). On disruption of the AnkG/Ndel1(Nde1) complex at the AIS by overexpression of the WT AnkG-NIR-RFP peptide, a significant amount of TfR-GFP was sorted into axons, with pronounced accumulation at the AIS region (Fig. 6 A, Middle and Fig. 6D). Consistently, no TfR-GFP signal was detected in axons when the aforementioned Ndel1/Nde1-binding-deficient AnkG-NIR-RFP peptide was overexpressed (Fig. 6 A, Lower and Fig. 6D).
Fig. 6.
The AnkG/Ndel1 complex regulates selective cargo sorting at the AIS. (A–C) Neurons cotransfected with TfR-GFP (A), GulR1-GFP (B), or CAR-GFP (C) and RFP control, WT AnkG-NIR-RFP, or M1908Q/L1955Q AnkG-NIR-RFP and stained for AnkG (magenta). Yellow arrowheads indicate the AIS, which is shown at a higher magnification below each image. (Scale bars: 20 μm.) (D–F) Quantification of the TfR-GFP (D), GluR1-GFP (E), and CAR-GFP (F) fluorescence intensity ratios of the AIS to dendrites in neurons transfected with RFP, WT AnkG-NIR-RFP, or M1908Q/L1955Q AnkG-NIR-RFP; n = 10 to 18 cells. Error bars indicate SE. ****P < 0.0001 (D); ***P = 0.0007 (E); ***P = 0.0003 (F). ns, not significant.
We used a similar experimental approach to test other somatodendritic cargos, including the AMPA receptor subunit GluR1 and CAR, in polarized sorting into axons. As with TfR-GFP, significant amounts of GluR1-GFP and CAR-GFP fusion reporters were sorted into axons on WT AnkG-NIR-RFP peptide-mediated disruption of the AnkG/Ndel1 complex formation at the AIS (Fig. 6 B, C, E, and F). These results are in agreement with a previous study that for the first time showed mistargeting of dendritic cargoes, including TfR, CAR, GluR1, and GluR2, into axons by knockdown of Ndel1 (22). Taken together, our results demonstrate that direct and high-affinity interaction between AnkG and Ndel1 at the AIS is critical for the AIS to function as a gatekeeper controlling axonal-somatodendritic polarity, likely via selective sorting of axonal cargoes.
Discussion
In this study, we have characterized the binding interaction between the neuronal-specific gAnkG and the dynein regulator Ndel1 by solving the structure of AnkB, a close homolog of AnkG, in complex with Ndel1. AnkG is a master scaffold protein that functions to establish and maintain axonal polarity at the AIS; it tethers diverse membrane proteins to the underlying spectrin-based cytoskeleton (37). Canonical ankyrins (including all 3 family members, AnkG, AnkB, and AnkR) contain an N-terminal membrane-binding domain, which is composed of 24 ankyrin repeats, responsible for binding to diverse membrane targets; a middle spectrin binding domain that bridges the membrane proteins to cytoskeletons; and a C-terminal regulatory domain (36, 38, 39). Interestingly, the 480-kDa gAnkG, with a single exon (exon 37) encoding more than 2,600 amino acids, is the predominate isoform in the nervous system (28). AnkG cannot accumulate at the AIS without exon 37, although little is known about the functions of this requisite giant insertion.
In our previous study, we identified a super-potent LIR motif within a short segment of the exon 37-coded region of AnkB or AnkG that binds to the GABARAP family members with very high affinity (27). Interestingly, the LIR motif immediately follows the NIR in both AnkB and AnkG. The finding of shared binding motifs in both AnkB and AnkG giant insertions supports the evolutionary analysis that the divergence of AnkB and AnkG happens after the emergence of the giant exon (28). We also hypothesize that the giant insertion of AnkG may contain additional protein–protein interaction motifs, and thus the gAnkG can specifically assembly a large and specific list of protein complexes specifically at the AIS. In line with this hypothesis, the giant insertion of AnkG also functions in recruitment of spectrin to the AIS (28, 40), and mutations within the giant insertion have been reported to be closely associated with human diseases (11, 40).
Both giant AnkG and AnkB are specifically expressed in the nervous system; however, the distinct expression patterns of AnkG and AnkB enable the 2 proteins to play different functional roles in axons. AnkG serves as a master scaffold protein in organizing the AIS micromembrane domain, while AnkB facilities cargo transportation in the distal axon (41–43). We have demonstrated that the AnkG/Ndel1 complex is required for sorting of cargoes at the AIS by reversal transport of somatodendritic cargoes out of axon. Although the physiological role of the AnkB/Ndel1 complex is not fully understood at present, our study suggests that the stable complex may also play roles in axonal trafficking or microtubule network stabilization by linking the scaffold to the Ndel1-dynein complex in the distal axon. Further studies are needed to characterize the in vivo function of AnkB/Ndel1 in either scenario. Although the giant insertion of AnkG and AnkB share some sequence similarities, taking Ndel1 or GABARAP binding as an example, there are very distinct sequences in the giant insertions of AnkG and AnkB, providing a possible explanation for the functional divergence of the 2 proteins in axons. A recent study reported that autism mutations in the large insertion region of ANK2 (the gene encoding AnkB) lead to elevated axon branching and aberrant structural connectivity, shedding light on the critical functions of giant AnkB in vivo (44).
Ndel1 was first characterized as a binder with Lis1 and dynein, serving as a dynein regulator that functions in the development of the adult nervous system (45–47). Subsequently, the Ndel1-Lis1-dynein complex was demonstrated to function in cell cycle processes, including mitosis and mitotic spindle organization (48–50). The N-terminal region of Ndel1/Nde1 forms a stable dimer and binds to Lis1 (35). However, the C-terminal of Ndel1/Nde1 is less well characterized, although it is believed to bind to other targets (51). A recent study showed that Ndel1 can directly bind to DISC1 through the C-terminal coiled-coil domain, forming a stable 3-helix bundle complex structure, and demonstrated that this bundle functions in DISC1’s regulation of neurogenesis during mitosis (34). In the present study, we found that this proposed target-binding module can indeed bind to other proteins with distinct binding mechanisms. Our finding of the distinct 1:2 binding stoichiometry of the AnkG/Ndel1 complex fits well with the architecture of Ndel1; the long dimeric NT-CC of Ndel1 is capable of binding with Lis1 and with dynein (35, 52). Importantly, several severe microcephaly and profound mental retardation causing mutations of NDE1 result in loss of the Nde1 region containing the CT-CC identified in this study (53, 54), indicating indispensable roles for the CT-CC–mediated protein interactions in normal neuronal development.
In summary, we have characterized the detailed interaction between the neuronal scaffold AnkG/AnkB and the dynein regulator Ndel1/Nde1 through combined biochemical and structural approaches, and have revealed the essential functions of their binding in regulating selective cargo sorting at the AIS. Our study thus establishes the molecular and structural basis for understanding the functions of the giant insertions of both AnkG and AnkB, with implications for the maintenance of neuronal polarity and insights into the mechanisms for neuronal disorders.
Materials and Methods
All proteins used in this study was expressed in Escherichia coli BL21 (DE3) and purified by Ni2+-NTA affinity chromatography followed by size-exclusion chromatography. Crystals were obtained by the sitting-drop or hanging-drop vapor diffusion method at 16 °C. An extended method describing protein preparation, biochemical assays, CD measurements, NMR spectroscopy, crystallization, structure determination, hippocampal neuronal culture, immunostaining, and microscopy and data analysis can be found in SI Appendix, Materials and Methods. The structure factors and coordinates of the structure have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 6KZJ).
Supplementary Material
Acknowledgments
We thank the Shanghai Synchrotron Radiation Facility beamline 17U1 and National Facility for Protein Science beam line 19U1 for X-ray beam time. We also thank Dr. Zhongliang Zhu (University of Science and Technology of China) for help with the crystal diffraction in-house test. This work was supported by grants from the Ministry of Science and Technology of the People’s Republic of China (2019YFA0508402, to C.W. and M.Z.), the National Natural Science Foundation of China (31670734), the Fundamental Research Funds for the Central Universities (WK2070000092), the Innovative Program of the Development Foundation of Hefei Center for Physical Science and Technology (2018CXFX008, to C.W.), Research Grants Council of Hong Kong (16100517, C6004-17G, and AoE-M09-12, to M.Z.), and the National Natural Science Foundation of China (NSFC) Fund for Distinguished Young Scholars and NSFC major research grants (81425009, 31630028, and 91632305, to Y.Z.). C.W. is supported by the Chinese Academy of Sciences Pioneer Hundred Talents Program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The structure factors and coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 6KZJ).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916987117/-/DCSupplemental.
References
- 1.Rasband M. N., The axon initial segment and the maintenance of neuronal polarity. Nat. Rev. Neurosci. 11, 552–562 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Kevenaar J. T., Hoogenraad C. C., The axonal cytoskeleton: From organization to function. Front. Mol. Neurosci. 8, 44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nirschl J. J., Ghiretti A. E., Holzbaur E. L. F., The impact of cytoskeletal organization on the local regulation of neuronal transport. Nat. Rev. Neurosci. 18, 585–597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett V., Lorenzo D. N., Spectrin- and ankyrin-based membrane domains and the evolution of vertebrates. Curr. Top. Membr. 72, 1–37 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Bennett V., Healy J., Membrane domains based on ankyrin and spectrin associated with cell-cell interactions. Cold Spring Harb. Perspect. Biol. 1, a003012 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobotzik J. M., et al. , AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc. Natl. Acad. Sci. U.S.A. 106, 17564–17569 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedstrom K. L., Ogawa Y., Rasband M. N., AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J. Cell Biol. 183, 635–640 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C. Y., Rasband M. N., Axon initial segments: Structure, function, and disease. Ann. N. Y. Acad. Sci. 1420, 46–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X., et al. , Selective filtering defect at the axon initial segment in Alzheimer’s disease mouse models. Proc. Natl. Acad. Sci. U.S.A. 111, 14271–14276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez A. Y., et al. , Ankyrin-G isoform imbalance and interneuronopathy link epilepsy and bipolar disorder. Mol. Psychiatry 22, 1464–1472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson A. D., et al. , Ankyrin-G regulates forebrain connectivity and network synchronization via interaction with GABARAP. Mol. Psychiatry, 10.1038/s41380-018-0308-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iqbal Z., et al. , Homozygous and heterozygous disruptions of ANK3: At the crossroads of neurodevelopmental and psychiatric disorders. Hum. Mol. Genet. 22, 1960–1970 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Leterrier C., et al. , Ankyrin G membrane partners drive the establishment and maintenance of the axon initial segment. Front. Cell. Neurosci. 11, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Bras B., et al. , In vivo assembly of the axon initial segment in motor neurons. Brain Struct. Funct. 219, 1433–1450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song A. H., et al. , A selective filter for cytoplasmic transport at the axon initial segment. Cell 136, 1148–1160 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Nakada C., et al. , Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 5, 626–632 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Winckler B., Forscher P., Mellman I., A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature 397, 698–701 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Jones S. L., Korobova F., Svitkina T., Axon initial segment cytoskeleton comprises a multiprotein submembranous coat containing sparse actin filaments. J. Cell Biol. 205, 67–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Este E., Kamin D., Göttfert F., El-Hady A., Hell S. W., STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep. 10, 1246–1251 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Watanabe K., et al. , Networks of polarized actin filaments in the axon initial segment provide a mechanism for sorting axonal and dendritic proteins. Cell Rep. 2, 1546–1553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leterrier C., et al. , Nanoscale architecture of the axon initial segment reveals an organized and robust scaffold. Cell Rep. 13, 2781–2793 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Kuijpers M., et al. , Dynein regulator NDEL1 controls polarized cargo transport at the axon initial segment. Neuron 89, 461–471 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Petersen J. D., Kaech S., Banker G., Selective microtubule-based transport of dendritic membrane proteins arises in concert with axon specification. J. Neurosci. 34, 4135–4147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maday S., Twelvetrees A. E., Moughamian A. J., Holzbaur E. L., Axonal transport: Cargo-specific mechanisms of motility and regulation. Neuron 84, 292–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y., et al. , Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat. Cell Biol. 10, 1172–1180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klinman E., Tokito M., Holzbaur E. L. F., CDK5-dependent activation of dynein in the axon initial segment regulates polarized cargo transport in neurons. Traffic 18, 808–824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., et al. , Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins. Nat. Chem. Biol. 14, 778–787 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Jenkins P. M., et al. , Giant ankyrin-G: A critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 957–964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Bennett V., Restriction of 480/270-kD ankyrin G to axon proximal segments requires multiple ankyrin G-specific domains. J. Cell Biol. 142, 1571–1581 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett V., Lorenzo D. N., An adaptable spectrin/ankyrin-based mechanism for long-range organization of plasma membranes in vertebrate tissues. Curr. Top. Membr. 77, 143–184 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Galiano M. R., et al. , A distal axonal cytoskeleton forms an intra-axonal boundary that controls axon initial segment assembly. Cell 149, 1125–1139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jegla T., et al. , Bilaterian giant ankyrins have a common evolutionary origin and play a conserved role in patterning the axon initial segment. PLoS Genet. 12, e1006457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodríguez D. D., et al. , Crystallographic ab initio protein structure solution below atomic resolution. Nat. Methods 6, 651–653 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Ye F., et al. , DISC1 regulates neurogenesis via modulating kinetochore attachment of Ndel1/Nde1 during mitosis. Neuron 96, 1041–1054 e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derewenda U., et al. , The structure of the coiled-coil domain of Ndel1 and the basis of its interaction with Lis1, the causal protein of Miller-Dieker lissencephaly. Structure 15, 1467–1481 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Wang C., et al. , Structural basis of diverse membrane target recognitions by ankyrins. eLife 3, e04353 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett V., Baines A. J., Spectrin and ankyrin-based pathways: Metazoan inventions for integrating cells into tissues. Physiol. Rev. 81, 1353–1392 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Wang C., Yu C., Ye F., Wei Z., Zhang M., Structure of the ZU5-ZU5-UPA-DD tandem of ankyrin-B reveals interaction surfaces necessary for ankyrin function. Proc. Natl. Acad. Sci. U.S.A. 109, 4822–4827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K., Li J., Wang C., Wei Z., Zhang M., Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions. eLife 6, e29150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang R., et al. , Neurodevelopmental mutation of giant ankyrin-G disrupts a core mechanism for axon initial segment assembly. Proc. Natl. Acad. Sci. U.S.A. 116, 19717–19726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kline C. F., Scott J., Curran J., Hund T. J., Mohler P. J., Ankyrin-B regulates Cav2.1 and Cav2.2 channel expression and targeting. J. Biol. Chem. 289, 5285–5295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lorenzo D. N., et al. , βII-spectrin promotes mouse brain connectivity through stabilizing axonal plasma membranes and enabling axonal organelle transport. Proc. Natl. Acad. Sci. U.S.A. 116, 15686–15695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorenzo D. N., et al. , A PIK3C3-ankyrin-B-dynactin pathway promotes axonal growth and multiorganelle transport. J. Cell Biol. 207, 735–752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang R., et al. , ANK2 autism mutation targeting giant ankyrin-B promotes axon branching and ectopic connectivity. Proc. Natl. Acad. Sci. U.S.A. 116, 15262–15271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasaki S., et al. , A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681–696 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Niethammer M., et al. , NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697–711 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Feng Y., et al. , LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 28, 665–679 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Moon H. M., et al. , LIS1 controls mitosis and mitotic spindle organization via the LIS1-NDEL1-dynein complex. Hum. Mol. Genet. 23, 449–466 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shim S. Y., et al. , Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J. Biol. Chem. 283, 12232–12240 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Vergnolle M. A., Taylor S. S., Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 17, 1173–1179 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Bradshaw N. J., Hayashi M. A., NDE1 and NDEL1 from genes to (mal)functions: Parallel but distinct roles impacting on neurodevelopmental disorders and psychiatric illness. Cell. Mol. Life Sci. 74, 1191–1210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zyłkiewicz E., et al. , The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J. Cell Biol. 192, 433–445 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alkuraya F. S., et al. , Human mutations in NDE1 cause extreme microcephaly with lissencephaly [corrected]. Am. J. Hum. Genet. 88, 536–547 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bakircioglu M., et al. , The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am. J. Hum. Genet. 88, 523–535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.