Significance
Epithelial cell transforming 2 (Ect2) is an oncoprotein. It activates RhoA and functions as a signaling node for cell division, cell growth, and many other cellular processes. Its dysregulation is associated with cancers. However, the regulation of Ect2 activity is poorly understood. We determined the crystal structure of human Ect2 and illustrate the mechanisms of Ect2 regulation, including autoinhibition and allosteric activation, which suggests a positive-feedback loop in RhoA signaling. Together, our findings provide mechanistic insights into the central signaling axis that controls cell division, and the basis for future Ect2-targeting drug development as well.
Keywords: cytokinesis, Ect2, RhoA, cancer, biophysics
Abstract
Epithelial cell transforming 2 (Ect2) protein activates Rho GTPases and controls cytokinesis and many other cellular processes. Dysregulation of Ect2 is associated with various cancers. Here, we report the crystal structure of human Ect2 and complementary mechanistic analyses. The data show the C-terminal PH domain of Ect2 folds back and blocks the canonical RhoA-binding site at the catalytic center of the DH domain, providing a mechanism of Ect2 autoinhibition. Ect2 is activated by binding of GTP-bound RhoA to the PH domain, which suggests an allosteric mechanism of Ect2 activation and a positive-feedback loop reinforcing RhoA signaling. This bimodal RhoA binding of Ect2 is unusual and was confirmed with Förster resonance energy transfer (FRET) and hydrogen–deuterium exchange mass spectrometry (HDX-MS) analyses. Several recurrent cancer-associated mutations map to the catalytic and regulatory interfaces, and dysregulate Ect2 in vitro and in vivo. Together, our findings provide mechanistic insights into Ect2 regulation in normal cells and under disease conditions.
Epithelial cell transforming 2 (Ect2) is a guanine nucleotide exchange factor (GEF) that activates Rho family GTPases (1). It is involved in a myriad of cellular processes, including cell division, growth, polarity, adhesion, migration, and centromere maintenance (2–7). Mounting evidence indicates that dysregulation of Ect2 is associated with a wide spectrum of cancers, and Ect2 inhibition is reported to suppress tumor growth, suggesting that Ect2 is an oncoprotein and important in cancer development (8–14).
The best-known function of Ect2 is its central role in cytokinesis. In many cells, Ect2 is recruited by the centralspindlin complex at the onset of anaphase to the division plane, where it promotes RhoA activation, driving the assembly and contraction of the actomyosin contractile ring (15–20). A positive-feedback loop in RhoA signaling has been proposed (21, 22), in which Ect2 is activated at the cell equator and the positive feedback to Rho autoactivation allows the cortex to respond to spindle microtubules via autoamplification. Ect2 depletion leads to cytokinesis failure even though there are many other GEFs in cells (23). This spindle–centralspindlin–Ect2–RhoA–actomyosin signaling axis is well known to control the position of the division plane (24–26), and may be related to the positioning signal from the spindle that Rappaport (27) described decades ago, but the detailed molecule mechanisms remain not completely clear.
In particular, how Ect2 is regulated is poorly understood. Ect2 contains tandem N-terminal BRCT domains and a C-terminal catalytic DH–PH domain. Ect2 by itself is autoinhibited (28–30). Truncation of an N-terminal fragment abrogates the Ect2 autoinhibition, leading to a malignant transforming activity, which suggests that Ect2 is autoinhibited by its N-terminal domains (1, 28). Plk1 phosphorylates MagRacGAP/Cyk4 (Cyk4 hereafter) subunit of the centralspindlin complex, which then binds to the BRCT domains of Ect2 and is implicated in Ect2 activation (31–33). Whereas Cyk4 is required for Ect2 activation (15, 22, 34), its binding to the BRCT domains plays a role in recruitment rather than direct activation (15).
In this study, we report the crystal structure of human Ect2, which reveals the mechanism of Ect2 autoinhibition. We demonstrate that Ect2 is activated by GTP-bound RhoA, which is essential for faithful cytokinesis. We show that several recurring cancer-associated mutations cause dysregulation of the enzyme in vitro and in vivo, clarifying the mechanism of Ect2 regulation under disease conditions.
Results
Structural Determination of Ect2.
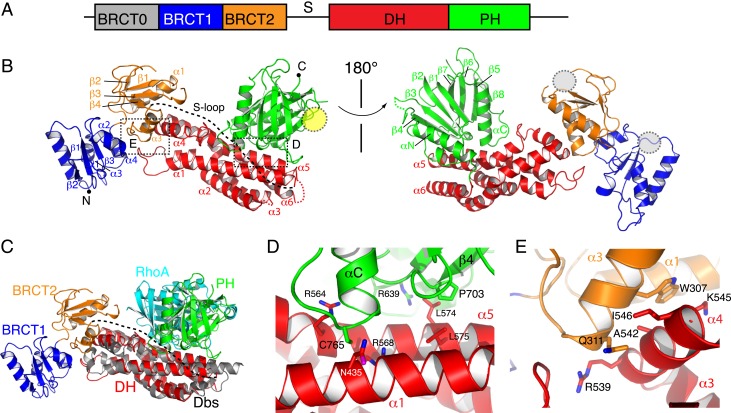
Human wild-type (WT) Ect2 contains 3 BRCT domains (33) and a DH–PH domain (Fig. 1A). To facilitate crystallization, we removed the first BRCT domain (BRCT0), and determined the crystal structure of Ect2 (ΔB0) that contains 2 BRCT domains (BRCT1 and BRCT2) and the DH–PH domain at a resolution of 2.8 Å (Fig. 1B). The structure was solved with molecular replacement using models of the BRCT domains (Protein Data Bank [PDB] ID code 4N40), the DH domain and the PH domain (PDB ID code 1lb1) as the initial templates (33, 35).
Fig. 1.
Structure of Ect2. (A) Domain organization of Ect2. (B) Two different views of the overall structure. The S-loop (dotted line) is disordered. The boxed regions are enlarged in D and E. The putative Cyk4 binding sites of the BRCT domains and the lipid-binding site of the PH domain are illustrated as gray and yellow ovals, respectively. (C) Structural alignment of the DH domains of Ect2 (color coded) and Dbs (in gray) bound with RhoA (in cyan) (PDB ID code 1lb1) (35). (D) DH–PH interactions. (E) DH–BRCT2 interactions.
There is one molecule of Ect2 in an asymmetric unit (Table 1). Because the loop connecting the BRCT domains and the DH–PH domain (the S-loop; SI Appendix, Fig. S1) is largely disordered in the crystals, there are potentially 2 alternative ways to organize these domains into one copy of Ect2 (SI Appendix, Fig. S2A). In one possibility, the BRCT2 domain packs against the tip of DH domain, and is distal to the PH domain (2BRCT_a); in the other possibility, the BRCT1 domain packs against the center of the DH domain and is close to the PH domain (2BRCT_b). The packing interface of 2BRCT_a, but not that of 2BRCT_b, is highly conserved, suggesting the model of 2BRCT_a is biological relevant. To distinguish these 2 possibilities, we conducted small-angle X-ray scattering (SAXS) analyses of the molecule in solution (SI Appendix, Table S1). Whereas the backcalculated profile from 2BRCT_b fits poorly (χ2 = 19.69), the backcalculated scattering profile from 2BRCT_a fits well to the experimental data (χ2 = 0.80), indicating that the solution structure of ΔB0 Ect2 has a similar conformation to 2BRCT_a (SI Appendix, Fig. S2 B and C).
Table 1.
Data collection and refinement statistics (molecular replacement)
| Data collection | |
| Space group | I222 |
| Cell dimensions | |
| a, b, c, Å | 106.97 130.60 131.79 |
| α, β, γ, ° | 90.00 90.00 90.00 |
| Resolution, Å | 30–2.80 (2.91–2.80)* |
| Rsym or Rmerge | 0.07 (0.99) |
| I/σI | 21.42 (1.93) |
| Completeness, % | 98 (100) |
| Redundancy | 6.1 (6.2) |
| CC1/2 | 0.72 |
| Refinement | |
| Resolution, Å | 32.85–2.80 (2.93–2.80) |
| No. reflections | 23,040 (2,836) |
| Rwork/Rfree | 0.24/0.27 |
| No. atoms in an AU | 4,178 |
| Protein | 4,178 |
| Ligand/ion | 0 |
| Water | 0 |
| B factors | |
| Protein | 107.9 |
| Ligand/ion | |
| Water | |
| R.m.s. deviations | |
| Bond lengths, Å | 0.004 |
| Bond angles, ° | 0.685 |
Values in parentheses are for highest-resolution shell.
Superposition of the tandem BRCT domains determined in this study with the ones containing 3 BRCT domains (33) indicates that the BRCT1–BRCT2 domain adopts very similar conformations (SI Appendix, Fig. S3A), with BRCT0 projecting away from the catalytic DH domain. To consolidate this idea, we performed SAXS analyses of the WT Ect2. The molecular weights calculated from volume of correlation (Vc) indicated that both WT and ΔB0 Ect2 are monomeric in solution (SI Appendix, Fig. S2D) (36). The proposed structure of WT Ect2, which is modeled using the structures of ΔB0 Ect2 and the triple BRCT domains, docked well to the reconstructed molecular envelope based on the SAXS measurements, with the different electron density between WT and ΔB0 Ect2 matching the proposed position of BRCT0 (SI Appendix, Fig. S2C). Together, the data indicate that the structure we determined is not from crystal packing artifacts, but represents the intrinsic folding of Ect2 in solution.
Mechanism of Ect2 Autoinhibition.
The catalytic DH domain is flanked by the BRCT domains, S-loop, and the PH domain, all of which play a role in Ect2 autoinhibition. A striking feature of the Ect2 structure is the interactions between the DH and PH domains. The DH domain of Ect2 adopts a typical structure as that found in the GEF proteins Dbs (35) and SOS (37), but with the PH domain positioned very differently (SI Appendix, Fig. S3B). Compared with the structure of Dbs, the last helix of the DH domain of Ect2 (α6) is about 2 helical turns shorter. The PH domain of Ect2 folds back and binds to a highly conserved surface of the DH domain (SI Appendix, Fig. S3C). Remarkably, comparison of Ect2 with the structure of Dbs bound to RhoA indicates that this conserved surface is the catalytic center, which is nevertheless blocked by the PH domain (Fig. 1C). The sequestration of the catalytic center by the PH domain is unexpected, which provides a mechanism of Ect2 autoinhibition. This mechanism of Ect2 autoinhibition shows some similarity to that of SOS, in which the PH domain partially occlude the Rho GTPase binding site of the DH domain (37).
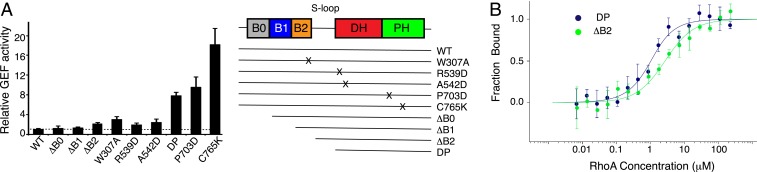
The PH domain of Ect2 binds to the DH domain mainly through hydrophobic interactions (Fig. 1D). Specifically, the side chain of Pro703 at the tip of β4 of the PH domain packs against Leu574 and Leu575 of the DH domain. Similarly, the side chain of Cys765 of the PH domain inserts into a pocket formed by Asn435, Arg564, and Arg568 of the DH domain. To validate the importance of the autoinhibited structure, we introduced mutations to disrupt the DH–PH interaction and measured the GEF activity. Compared with the WT protein, mutations of the DH–PH interface (P703D and C765K) dramatically increased the GEF activity over 10-fold (Fig. 2A and SI Appendix, Fig. S4A and Table S2). Consistent with the hyperactivity in vitro, overexpression of the P703D or C765K mutant Ect2 in HeLa cells caused noticeable changes in interphase cell morphology (SI Appendix, Fig. S5), such as cell rounding and formation of stress fibers, suggesting ectopic RhoA activation. These findings indicate that the PH domain is important for Ect2 autoinhibition.
Fig. 2.
Mechanism of Ect2 inhibition. (A) Relative GEF activity of Ect2 with WT interface (WT) and various mutants. (A, Right) Schematic of the Ect2 constructs. Error bars indicate SD (n = 3 independent experiments). (B) Microscale thermophoresis (MST) analyses of Ect2 binding to RhoA. To enhance the binding, the assays were done with RhoA in the apo form in the presence of EDTA. Dissociation constants: Kd (DP) = 0.6 ± 0.2 μM; Kd (ΔB2) = 2.5 ± 0.6 μM. Error bars indicate SD (n = 3 independent experiments).
The tandem N-terminal BRCT domains, BRCT2 in particular, interact with the periphery of the DH domain. Multiple residues of α4 in the DH domain, including Arg539, Ala542, Lys545, and Ile546, bind to BRCT2, but with few interactions with the BRCT0 and BRCT1 domains (Fig. 1E). In support of the structure, truncation analyses showed that deletion of BRCT0 (ΔB0) or BRCT0+BRCT1 (ΔB1) did not greatly change the GEF activity, whereas further removal of BRCT2 (ΔB2) increased the activity by ∼2-fold (Fig. 2A and SI Appendix, Fig. S4B), supporting the idea that BRCT2 is directly involved in DH binding and Ect2 inhibition. Likewise, disruption of the BRCT2–DΗ interface by point mutations on BRCT2 (W307A) or the DH domain (R539D or A542D) increased the Ect2 activity by ∼2-fold (Fig. 2A and SI Appendix, Fig. S4A). In sharp contrast to the marked effect induced by the PH domain mutations, the release of Ect2 inhibition by BRCT deletion modestly increased the GEF activity. This is consistent with the notion that BRCT2 binds to the DH domain at a distal site, playing a peripheral role in Ect2 regulation.
Previous studies indicated that truncation of an N-terminal fragment of Ect2, which includes the BRCT domains and the S-loop, resulted in Ect2 activation (7, 16, 28, 30). Interestingly, truncation of the BRCT domains led to a low/no cellular transformation activity (28), suggesting that the S-loop is important for Ect2 inhibition. Consistent with these studies, removal of the BRCTs and S-loop (DP Ect2), which is also monomeric in solution (SI Appendix, Fig. S2E), notably increased the GEF activity, ∼4-fold more active than the BRCT truncation mutant (ΔB2), Fig. 2A. The S-loop is unstructured, connected to the catalytic α1, and adjacent to the putative RhoA-binding site (Fig. 1C). Truncation of the S-loop increased the RhoA-binding affinity by ∼4-fold (Fig. 2B), suggesting that the S-loop inhibits Ect2 by perturbing RhoA binding.
Together, our structure and mechanistic analyses indicate that Ect2 is autoinhibited by 3 elements, the BRCT domains, the S-loop, and the PH domain. The PH domain directly blocks the catalytic center, playing an essential role in Ect2 inhibition, whereas the BRCT2 domain and the S-loop are in close proximity to the RhoA-binding site, which may impede RhoA binding, providing a rationale for their inhibitory functions.
Allosteric Activation of Ect2 by RhoA.
During cytokinesis of many cells, Ect2 is recruited to the division plane and activated by yet-unknown mechanisms, which is a major question in cytokinesis. Phosphorylation of the S-loop is known to regulate the Ect2 activity in cells (18, 38–40). The S-loop is largely disordered, suggesting that phosphorylation may regulate Ect2 through nonspecific and/or indirect manners. It remains controversial whether Ect2 is activated by the binding of phosphorylated Cyk4 to the BRCT domains (15, 31–33). The Cyk4 binding mode of Ect2 is currently unclear. Given the highly similar structures of the tandem BRCT domains of Ect2 and Rad4 (41), the substrate-binding mode of Rad4 suggests that the Cyk4 binding sites of the BRCT domains in Ect2 are distal to the inhibition center (Fig. 1B and SI Appendix, Fig. S3 D and E), consistent with the notion that BRCT mutations that prevent centralspindlin binding has a minor effect on Ect2 activation in human cells (15).
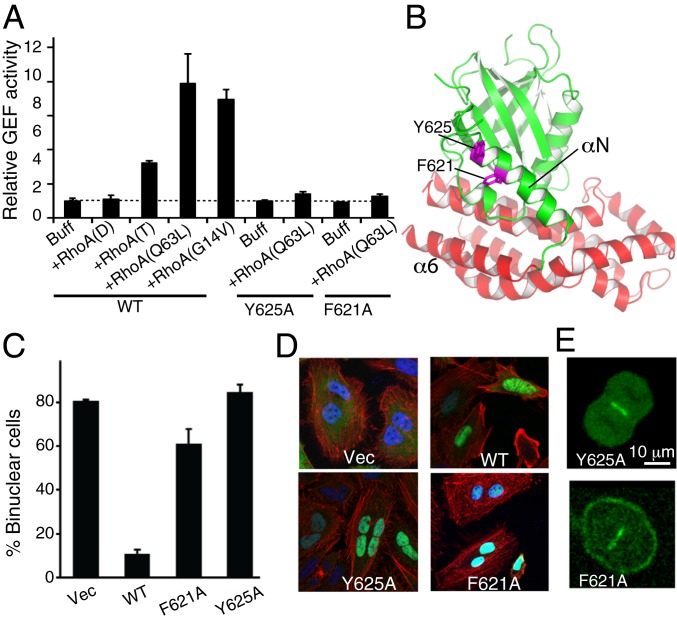
To obtain insights into the mechanism of Ect2 regulation, we searched for different Ect2-activating ligands. Addition of Anillin (42) or a phosphorylated Cyk4 peptide negligibly activated Ect2 (SI Appendix, Fig. S6). Considering small GTPases allosterically activate the GEF activities of SOS/Ras and p63RhoGEF (43, 44), and a positive-feedback loop is proposed to regulate ECT-2 in Caenorhabditis elegans (22), we reason that RhoA is a potential activating ligand. Remarkably, we found that GTP-bound, but not GDP-bound, RhoA promoted the Ect2 activity (Fig. 3A). The constitutively activated RhoA (Q63L) mutant stimulated Ect2 even more obviously. The hyperactivation of Ect2 by RhoA (Q63L) is not an artifact of the mutation, because marked activation was also observed with another constitutively activated mutant RhoA (G14V). Consistent with the model of BRCT and RhoA binding both individually and together, the presence of RhoA (Q63L) further increased the activity of the partially activated DP mutant (SI Appendix, Fig. S4D). These results suggest that activated RhoA binds to Ect2, promoting the release of its autoinhibition.
Fig. 3.
Ect2 activation by GTP-bound RhoA. (A) Relative GEF activities of WT, 2 Ect2 mutants in the presence and absence of activated RhoA. Buff, buffer without addition of RhoA; RhoA (D), GDP-bound RhoA; RhoA(T), GTP-bound RhoA. Error bars indicate SD (n = 3 independent experiments). (B) Mapping the activated RhoA binding sites to αN of the PH domain. (C) Quantification of the cytokinesis failure of HeLa cells with endogenous Ect2 knocked down and rescued with WT and various mutant Ect2. Error bars indicate SD (>200 cells). (D) Y625A and F621A mutant Ect2 failed to support cytokinesis. F-actin was stained with phalloidin (red) and nuclei with DAPI (blue). (E) Cellular localizations of Y625A and F621A mutant Ect2. Endogenous Ect2 was knocked down in HeLa cells, which was rescued with GFP-tagged WT or mutant Ect2.
We then looked for the cryptic activated-RhoA binding site by examining the exposed residues. We tested a series of Ect2 mutants for their ability to selectively compromise RhoA activation without perturbation of the basal GEF activity (SI Appendix, Fig. S7), and identified helix αN in the PH domain (Fig. 3B). Mutation of an exposed residue of αN (Y625A), which is distal to the catalytic surface of the DH domain, did not perturb the basal GEF activity, but severely diminished Ect2 activation by RhoA (Q63L) (Fig. 3A and SI Appendix, Fig. S4E). Similarly, mutation of a nearby residue (F621A) also selectively blocked Ect2 activation by RhoA. Together, these data suggest an allosteric activation mechanism, in which activated RhoA binds to αN, stimulates the release of PH inhibition and promotes the binding of substrate RhoA to the catalytic center. This regulation of Ect2 appears analogous to that of p63RhoGEF (SI Appendix, Fig. S3G), in which the GEF activity of p63RhoGEF is inhibited by the PH domain (45), and activated by binding Gαq to the DH and PH domains (43).
To evaluate the importance of allosteric activation of Ect2 by RhoA, we introduced the allosteric mutants into HeLa cells and examined their cytokinesis. As reported before (16), knocking down endogenous Ect2 by small interfering RNA (siRNA) resulted in cytokinesis failure and formation of multinucleate cells (Fig. 3 C and D and Movie S1). This was rescued by the expression of the siRNA-resistant WT Ect2 (Movie S2). In contrast, the Ect2 mutant (F621A), which was expressed at a level comparable to the siRNA-resistant WT protein (SI Appendix, Fig. S8A), was localized correctly inside the cells (Fig. 3E) but failed to rescue the cytokinesis failure (Fig. 3C and Movie S3). Similarly, the Ect2 mutant (Y625A) did not support cytokinesis either (Movie S4). Together, these data support the critical role of allosteric Ect2 activation by RhoA in cell division.
Förster Resonance Energy Transfer, Pulldown, and Hydrogen–Deuterium Exchange Mass Spectrometry Analyses Support That Two RhoA Molecules Bind to Ect2.
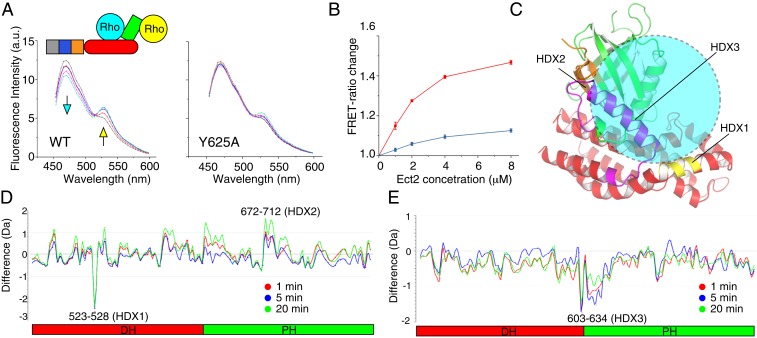
Our findings suggest bimodal RhoA binding, with one molecule of RhoA functioning as an activator and binding to the PH domain (allosteric site), and the other acting as the substrate, binding to the DH domain (catalytic site) and exchanging the bound nucleotide. To further confirm this model, we performed Förster resonance energy transfer (FRET)-based assays (46), in which one RhoA molecule was labeled with the donor probe (cyan fluorescent protein [CFP]) and the other RhoA molecule (Q63L) with the acceptor probe (yellow fluorescent protein [YFP]) (Fig. 4A). Binding of these 2 RhoA molecules to Ect2 is expected to bring the fluorescent probes close together, enhancing the FRET efficiency. In the absence of Ect2, the fluorescence intensity from the acceptor was barely detected, indicating that the 2 RhoA molecules did not associate by themselves in solution. Titration with increasing concentrations of Ect2 enhanced the acceptor fluorescence and reduced the donor fluorescence in a dose-dependent manner. The increased FRET ratio supports the notion that 2 RhoA molecules bind to Ect2 (Fig. 4B). Consistent with the enzymatic measurements, the Ect2 mutation Y625A clearly reduced the FRET ratio, supporting the model of αN as the allosteric RhoA binding site.
Fig. 4.
Binding of 2 RhoA molecules to Ect2. (A) FRET assays of the binding of 2 RhoA molecules by WT or Y625A Ect2. Schematic diagram of the assay is shown at the Top. RhoA (WT) and RhoA (Q63L) were labeled with CFP and YFP, respectively. Cyan and yellow arrows indicate the reductions and increases in the signals from the donor and acceptor fluorescent probes, respectively. Three independent experiments were performed, and a representative one is shown. (B) Quantification of the FRET ratio change in the presence of WT (red) or Y625A (blue) Ect2. Error bars indicate SD (n = 3 independent experiments). (C) Summary of the HDX-MS results shown on the structure of Ect2 in the inhibited state. Only the DH and PH domains are shown. The HDX1, HDX2, and HDX3 regions are colored yellow, orange, and magenta, respectively. Substrate RhoA binding to the canonical catalytic site is schematically illustrated as a cyan circle. (D) Difference plot of the HDX data from Ect2 (DP) in the presence and absence of RhoA (G17A). Differently colored lines indicate different labeling times. Negative values indicate less exchange and more protection upon RhoA binding. Two independent experiments were conducted, and one representative is shown. (E) Difference plot of the HDX data from the Ect2 (DP)–RhoA (G17A) complex in the presence and absence of RhoA (Q63L).
We also performed a pulldown assay to validate the model of 2 RhoA binding using the dominant-negative mutant RhoA (G17A), RhoA (Q63L), and DP Ect2. We found that RhoA (G17A), which binds strongly to Ect2 (30), pulled down RhoA (Q63L) in an Ect2-dependent manner, whereas the Y625A mutation abrogated this interaction (SI Appendix, Fig. S9). The affinity between Ect2 and the activator RhoA seems to be low, so other components (22) and/or clustering of the molecules at the cleavage furrow may promote Ect2 activation by RhoA in cells.
To directly investigate the positions of the 2 RhoA-binding sites, we conducted hydrogen–deuterium exchange mass spectrometry (HDX-MS) analyses. In order to maximize the efficiency of complex formation, we used RhoA (G17A) and the partially activated DP Ect2. We first compared the HDX behavior of DP Ect2 in the presence and absence of RhoA (G17A). Peptide segments resulting from pepsin proteolysis of Ect2 collectively covered ∼89.2% of the protein (SI Appendix, Fig. S10A) and were used to map the substrate RhoA-binding site. Upon incubation with RhoA (G17A), the solvent accessibility of DP Ect2 exhibited dramatic changes, particularly at 2 regions spanning residues 523 to 528 (HDX1) and residues 672 to 712 (HDX2), respectively (Fig. 4D and SI Appendix, Fig. S10 B and C). The first region encompasses an exposed surface of the DH domain that is close to the canonical RhoA-binding site (Fig. 4C), and that became less accessible with the binding of RhoA (G17A), consistent with substrate RhoA binding to the canonical catalytic site. The second region comprises residues of the PH domain that are buried in the inhibited state and that became more accessible upon RhoA binding, consistent with a displacement of the PH domain from the inhibited conformation. These data support the notion that substrate RhoA binds to the catalytic surface of the DH domain, and the PH domain inhibition is released in the activated state.
Next, we compared the HDX behavior of the DP Ect2–RhoA (G17A) complex in the presence and absence of RhoA (Q63L). In binding to the RhoA (Q63L), the most notable changes were detected at the region spanning several peptides (residues 603 to 622, 605 to 622, 623 to 634, HDX3), which displayed reduced HDX (Fig. 4E and SI Appendix, Fig. S10 D–F). The HDX3 region maps approximately to the αN of the PH domain (Fig. 4C), supporting the notion that activated RhoA (Q63L) binds to the PH domain. Given the alternation of HDX occurred predominately at HDX3, the region around αN of the PH domain seems to the major site for RhoA (Q63L) binding, although we cannot formally exclude the possibility that RhoA (Q63L) may also bind somewhere else. Taking together, our findings support the model of 2 RhoA binding, in which the activator RhoA binds to the regulatory PH domain and the substrate RhoA binds to the catalytic DH domain.
Implications of Ect2 Dysregulation in Cancers.
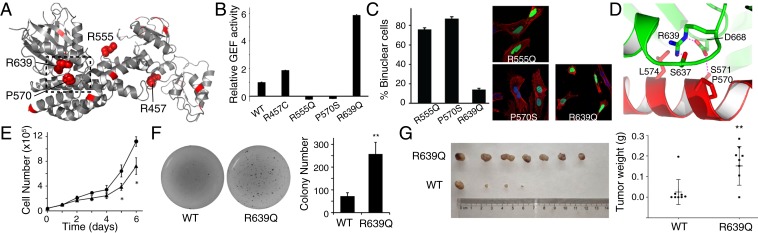
Up-regulation of Ect2 occurs in a variety of tumor types, and is associated with poor prognoses in cancer patients (23, 47, 48). The structure of Ect2 allows assessment of the impact of cancer-associated mutations. About 400 cancer-associated missense mutations of Ect2 were detected (in The Cancer Genome Atlas and cBioPortal Databases) and 42 were found in 2 or more cases (49). Of these recurring mutational residues, 21 map to the structurally resolved region (Fig. 5A), and several, including P570S/L, R555Q/L/W, R457C/H, and R639Q/L, are located at the catalytic DH–PH domain (50–57). The P570S/L mutations are at the catalytic center involved in substrate binding (SI Appendix, Fig. S3F). As a result, the P570S mutant completely lost the GEF activity (Fig. 5B) and failed to support cytokinesis (Fig. 5C). The R555Q/L/W mutations are proximal to the catalytic center. The equivalent residue in Dbs (Lys758) interacts with an acidic surface of RhoA (SI Appendix, Fig. S3F) and is important for the GEF activity (35). As expected, the R555Q mutation abrogated the GEF activity of Ect2 (Fig. 5B), and the mutant failed to support cytokinesis (Fig. 5C). Thus, the loss of function of Ect2 caused by the P570L and R555Q mutations results in cytokinesis failure, which may jeopardize the genomic integrity and serve as a starting point for aneuploidy and tumorigenesis in patients (47). The loss of function of Ect2 may lead to other defects because it is involved in many other biological processes.
Fig. 5.
Dysregulation of Ect2 by recurring cancer-associated mutations. (A) Recurring cancer-associated mutations (in red) mapped to the structure of Ect2. The residues tested in this study are shown as spheres. The boxed region is enlarged for further analysis in D. (B) Relative GEF activity of WT and the cancer-associated mutants of Ect2. Error bars indicate SD (n = 3 independent experiments). (C) Cytokinesis defects caused by 3 cancer-associated mutant Ect2. (D) Structure of Ect2 around Arg639. (E) In vitro cell proliferation assays with A375 cells expressing WT (triangles) or R639Q (circles) mutant Ect2. Error bars indicate SD (n = 3 independent experiments). *P < 0.05. (F) Soft agar colony formation assays of A375 cells expressing WT or R639Q mutant Ect2. Error bars indicate SD (n = 3 independent experiments). **P < 0.01. (G) Xenograft tumor models with A375 cells expressing WT or R639Q mutant Ect2. Tumor weights were analyzed after 4-wk inoculation. **P < 0.01.
In contrast, the R457C/H mutations map to the DH–BRCT binding interface, and the R457C mutant partially released the Ect2 inhibition (Fig. 5B). Moreover, the R639Q/L mutations map to the DH–PH interface that is directly involved in blocking the catalytic center (Fig. 5D and SI Appendix, Fig. S3H). Consistent with the structure, the R639Q mutation markedly increased the GEF activity (Fig. 5B). Therefore, the R639Q/L mutations found in cancer patients affect the central element in Ect2 regulation, releasing the PH inhibition and endowing the enzyme with a constitutive GEF activity.
Because Ect2 up-regulation activates multiple signaling pathways and is associated with poor survival in cancer patients (9, 12, 13), we also tested the biological function of the R639Q mutant. This mutant largely rescued the cytokinesis defect caused by depletion of endogenous Ect2, suggesting it is competent in supporting cell division (Fig. 5C). We tested whether the hyperactive R639Q mutation promotes tumor cell growth. We generated a stable cancer cell line (A375) expressing WT or the R639Q mutant at similar levels (SI Appendix, Fig. S8B). Consistent with previous studies (12), higher levels of phosphorylation of Akt, FAK, and ERK were detected in the R639Q-expressing cells (SI Appendix, Fig. S8B). Notably, the cells expressing the R639Q mutant proliferated faster in vitro than those expressing the WT protein (Fig. 5E). We used a soft agar colony formation assay to evaluate the oncogenic potential of the mutant. Relative to the WT protein, the R639Q-expressing clones displayed significant more colonies on soft agar (Fig. 5F). A subcutaneous (s.c.) xenograft model was used to study the tumorigenicity of the mutant. The R639Q-expressing tumors were found significantly larger than the ones expressing the WT protein (Fig. 5G). Together, the data support the idea that the R639Q mutant gains a stronger potency in promoting tumor cell growth.
Discussion
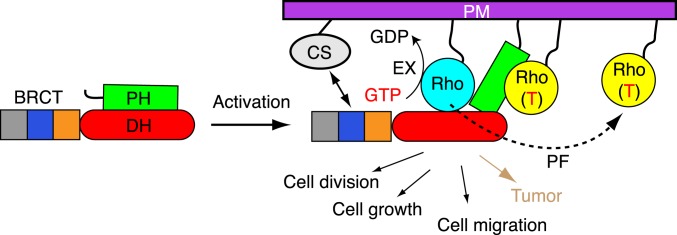
The most notable finding of our study is Ect2 inhibition by the PH domain, which is ultimately linked to the allosteric activation by RhoA (Fig. 6). These findings provide insights into Ect2 signaling, particularly in control of cell division.
Fig. 6.
Model of Ect2 regulation. CS, the centralspindlin complex; PF, positive feedback; PM, plasma membrane; RhoA (T), GTP-bound RhoA; double arrow, centralspindlin–Ect2 interaction. Ect2 is autoinhibited in the ground state. The activator RhoA (GTP-bound, yellow) binds to the PH domain, allosterically releasing the Ect2 inhibition, and the substrate RhoA (cyan) binds to the catalytic center, ready for GTP–GDP exchange (EX).
It is well known that Ect2 is inhibited by the N-terminal domains (28, 29). We show here that the N-terminal inhibition is composed of BRCT inhibition and S-loop inhibition. Overexpression of the N terminus of Ect2 was shown to inhibit cytokinesis, functioning as a dominant-negative mutant (7, 29). This probably results from sequestration of important Ect2 binding proteins, such as Cyk4, and may not be due to transinhibition of the GEF activity of Ect2, which is nevertheless autoinhibited intramolecularly. In C. elegans, an E129K substitution in BRCT1 activates the biological activity of ECT-2 in vivo (22). E129 of C. elegans ECT-2 is not conserved in HsEct2 (SI Appendix, Fig. S1). Based on the overall sequence homology, E129 is located at the β1–α1 loop, which is 2 residues shorter than that in HsEct2. The reason of ECT-2 activation by the E129K mutation is unclear, which may disrupt interaction with other regulators in cells, or perturb the tight packing between β1 and α1 of the BRCT1 domain, leading to partial release of the ECT-2 inhibition. Further work is required to make rigorous conclusions of the positioning of the BRCT domains relative to the GEF domain.
Compared with the well-established N-terminal inhibition, we found that the enzyme is even more potently inhibited by the PH domain. Thus, our findings add an unexpected level of Ect2 regulation by PH inhibition. The model that Ect2 is strongly inhibited by the PH domain provides the rationale for the findings that binding of the centralspindlin complex to the N-terminal BRCT domains recruits Ect2 but does not play a major role in Ect2 activation in human cells (15). In addition, Ect2 is allosterically activated by RhoA. Interestingly, the G707D mutation in the PH domain of ECT-2 in C. elegans activated the enzyme (22). G707 is located at a nonconserved β5–β6 loop of the PH domain (SI Appendix, Fig. S1). The reason for ECT-2 activation by the G707D mutation is unknown, which may destabilize the inhibitory PH–DH interaction.
The finding that Ect2 is activated by GTP-bound RhoA reveals the molecular mechanism underlying the positive-feedback loop of RhoA signaling at the division plane (15, 21, 22). During cytokinesis, Ect2 is recruited to the plasma membrane at the division plane, where its autoinhibition is released by GTP-bound RhoA, which may result from the basal activity of Ect2 and other GEFs under the plasma membrane. The activated Ect2 in turn leads to more GTP-bound RhoA, achieving RhoA autoamplification. This model is consistent with the previous genetic studies implicating GTP-bound RHO-1 activates RhoGEF during cytokinesis in C. elegans embryos (22). This positive-feedback loop provides the mechanism that accelerates the formation of the active RhoA zone and confers its spatial restriction during cytokinesis, underpinning the essentiality of Ect2 in cell division. Analogous feedback activation of the nucleotide exchange factor SOS/Ras and Lbc family of RhoGEFs are also found (44, 58).
Our structure also sheds mechanistic light on the malfunctions of Ect2 mutants in cancers and offers a framework for future biochemical and cellular analyses. In this study, we demonstrate that both gain of function and loss of function caused by Ect2 mutation are indeed targeted in cancers. It is interesting that some of the cancer-associated mutations are loss-of-function mutations, raising the question of how those cells are dividing. In line with these findings, Ect2 knockdown induced cytokinesis defects in nontransformed cells but not in non–small-cell lung cancer (NSCLC) cells (9), which may cripple the genomic integrity and signaling networks to induce an Ect2-independent cytokinesis mechanism. Although Ect2 is not essential for cytokinesis in NSCLC cells, Ect2 knockdown blocked transformed growth and tumorigenicity (9), in which Ect2 probably serves as a Rac GEF (13). It is widely observed that Ect2 knockdown inhibits cancer growth and induces cell senescence (8–14), suggesting that Ect2 is a potential drug target for cancer treatments. The discovery of the unique structure of Ect2, the inhibitory DH–PH interaction in particular, paves the way for developing new, highly specific drugs for the treatment of cancers. It is not known that Rho-mediated feedback is directly relevant to Rac activation, and more studies are needed in the future.
Methods
Protein Expression and Purification.
The full-length HsEct2(NP_001245245.1) was cloned from human cDNA and was ligated to a pMal-p2(GE Healthcare) vector with a His tag added to the C terminus of the Ect2 gene. RhoA was subcloned and ligated to a pGEX4T2 vector. The various truncation and mutations of HsEct2 and RhoA used in this study were obtained from the full-length WT plasmid by quick change or other cloning methods.
WT and various mutants Ect2 were overexpressed in Escherichia coli strain BL21 (DE3) containing chaperone GroES-GroEL-tig. Chaperones were induced with 10 ng/mL tetracycline, and proteins were induced with 0.2 mM isopropyl β-d-thiogalactoside after chaperone expression at 18 °C. Cells were collected at speed 4,000 rpm (Beckman; rotor JA4.2) for 20 min, and lysed in 20 mM Tris⋅HCl, pH 8.0, 500 mM NaCl, 10% (vol/vol) glycerol, and 1 mM PMSF, pH 8.0, using a nano homogenize machine (ATS) at 4 °C. The supernatant was loaded onto Ni-NTA resins and eluted with 250 mM imidazole after wash. Four millimolar EDTA and 10 mM DTT were added after elution. The sample was then reloaded onto MBP beads and washed with 20 column volumes of lysis buffer, after which 5% (wt/wt) TEV was added to perform on-column cutting overnight. Ect2 was further purified by ion exchange (Source S or Source Q; GE Healthcare; according to the isoelectric points of the constructs), and gel filtration (Superdex200; GE Healthcare) chromatography through AKTA system. The purified proteins were concentrated in 10 mM Hepes, pH 7.0, 300 mM NaCl, 2% glycerol, and 10 mM DTT and stocked at −80 °C.
RhoA was purified as described (59). GDP/GTP/N-methylanthraniloyl (Mant)-GDP–state RhoA were charged with GDP/GTP/Mant-GDP for 3 h after treated with EDTA, and MgCl2 was added to stop the reaction. The proteins were then desalted to different buffers as required.
Crystallization, Data Collection, and Structure Solution.
Crystals of HsEct2 (residues: 142 to 834) and HsEct2 (residues: 142 to 790) were grown at 18 °C by hanging-drop vapor diffusion methods. The HsEct2 (residues: 142 to 834) crystals grew from 0.1 M CHES, pH 9.5, 0.5 M NaCl, 4% sucrose, 2 mM EDTA, 1 to 1.25 M AmSO4, and 1 mM TCEP with protein-to-reservoir volume ratio of 1:1. The HsEct2 (residues: 142 to 790) crystals grew from 0.1 M CHES, pH 9.5, 0.95 to 1.05 M Na·K tartrate, 0.2 M Li2SO4, and 1 mM TCEP. Crystals were harvested in cryoprotectant containing 10 to 25% glycerol and then flash-frozen in liquid nitrogen.
Diffraction data from the crystals were collected at BL17U1 of the beamline of Shanghai Synchrotron Radiation Facility. All of the data were processed with the HKL2000. The model was manually completed with Coot and refined with Phenix (60). The HsEct2 (residues: 142 to 790) crystals diffracted to a higher resolution and were used for model building. The final structure was refined to 2.8 Å, with Rwork/Rfree = 0.24/0.27, Ramachandran outlier 0.20%, allowed 2.77%, and favored 97.04%.
GEF Assays.
The GEF assays were performed as described (61). Release of the bound Mant-GDP from RhoA was monitored using VARIOSKAN FLASH (Thermo Scientific). The reaction was in 10 mM Hepes, pH 7.0, 150 mM NaCl, 2 mM MgCl2, 5% glycerol, 0.02 mg/mL BSA, 1 mM DTT, 100 μM GDP, with 250 nM RhoA and 2.5 μM Ect2. When used as the Ect2 activator, WT RhoA (2.5 μM) was precharged with GTP or GDP as indicated, whereas the RhoAQ63L and RhoAG14V mutants were directly used after purification. The fluorescence time courses were fit to a single-exponential decay with Eq. 1:
| [1] |
ρF is total fluorescence decrease, R is decay rate constant, and F0 is equilibrium fluorescence intensity. Normalized GEF activities of different Ect2 constructs were calculated by Eq. 3:
| [2] |
| [3] |
Rcontrol is the decay constant of RhoA(Mant-GDP) in the buffer conditions without Ect2; Ropen is the decay constant of the DP protein, which is referred as to the open state of Ect2; Ri is the decay constants of different Ect2 constructs.
Microscale Thermophoresis.
ΔB2 (residues 408 to 790) and DP (residues 326 to 790) Ect2 mutants were labeled with the fluorescent dye NT-647 (following the instructions of the kit MO-L001; NanoTemper), and desalted to the reaction buffer (10 mM Tris, 8.0, 150 mM NaCl, 4 mM EDTA, 0.005% Tween 20, 5 mM DTT). RhoA (1 to 189) was dialyzed to the reaction buffer for 3 h and was quantified with BCA protein assay kit (Macgene) immediately after dialysis. Ect2 was titrated with different concentrations of RhoA before loading onto capillaries. The difference of the thermophoretic properties of Ect2 was measured with Monolith NT.115 (NanoTemper Technologies) with the laser power 40%. The dissociation constant was measured with the MO. Affinity Analysis software suit. Each experiment was repeated 3 times.
FRET Assays.
FRET assays were performed in 10 mM Hepes, 60 mM NaCl, 1mM MgCl2, 10 mM DTT, 10% glycerol, 0.01% Tween 20, 1 mg/mL BSA, pH 7.0, with 1 μM RhoA(WT)–CFP and 1 μM RhoA(Q63L)–YFP under various concentrations of Ect2 DP or Ect2 DP Y625A (0, 0.5, 1, 2, 4, and 8 μM). After incubation on ice for 2 h, the measurements were performed in a Microplate Reader (VARIOSKAN FLASH; Thermo Scientific) with 433-nm excitation. Emissions between 450 and 600 nm were scanned. FRET ratio change was calculated as follows:
where I is the fluorescence intensity, D is the donor molecule RhoA(WT)–CFP, and A is the acceptor molecule RhoA(Q63L)–YFP. ON and OFF are the FRET-on and FRET-off states with or without Ect2, respectively.
HDX-MS.
For HDX labeling, the following samples were prepared before the labeling reactions: 1) Ect2 (DP) with and without equal molar quantity of RhoA G17A; 2) Ect2 (DP) and RhoA G17A (1:1 molar ratio) with and without 3-fold molar excess of RhoA Q63L. To initiate deuterium labeling, 5 μL of each protein solution was diluted with 45 μL of labeling buffer (10 mM Hepes, 7.6, 80 mM NaCl, 99.8% D2O) at 10 °C, resulting in 10 μM Ect2 during labeling. The labeling reactions were stopped at various time points (1, 5, and 20 min) by adding 50 μL of chilled quench buffer (4 M guanidine hydrochloride, 200 mM citric acid, and 500 mM TCEP, pH 1.8). After pepsin solution (5 μL; 1 μM) was added for digestion for 3 min, samples were placed into Thermo-Dionex Ultimate 3000 HPLC system autosampler. The peptides were eluted by a 40-min gradient of acetonitrile (7 to 45%) in 0.1% formic acid at 100 µL/min with Thermo-Dionex Ultimate 3000 HPLC system, which is connected to a Thermo Scientific Q Exactive mass spectrometer. Peptides were separated on a reverse-phase column (Acquity UPLC BEH C18 column, 1.7 µm, 2.1 × 50 mm; Waters). The Q Exactive mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 2.0.0.0 software, acquiring over a m/z of 350 to 2,000. Peptides were identified using an in-house Proteome Discoverer (version PD1.4; Thermo Fisher Scientific), and HDX-MS data were processed by HD Examiner from Thermo Fisher Scientific.
SAXS Data Collection and Processing.
WT (residues 45 to 834) and ΔB0 (residues 142 to 834) Ect2 at ∼2.5 mg⋅mL−1 in 10 mM Hepes, 300 mM NaCl, 5% glycerol, 10 mM DTT, pH 7.0 were used. SAXS analyses were conducted similarly as described before (62). Basically, scattering profiles of the proteins were calculated using the program PRIMU (63) and standard procedures (64). The radius of gyration (Rg) and the forward scattering intensity I(0) were calculated from the data of infinite dilution, and also estimated using the program GNOM (65). The molecular weights were calculated using the Rg/Vc power law (36). The theoretical scattering intensity was obtained by fitting to the experimental data using the program CRYSOL (66). Missing residuals of 2BRCT_a, 2BRCT_b and 3BRCT were added by the SWISS model server (67).
Multiangle Light Scattering.
Protein at 2 mg/mL was injected onto a Superdex200 column equilibrated in the buffer: 20 mM Tris, 8.0, 200 mM NaCl, 4 mM EDTA, 2 mM DTT, and 5% Gly. The chromatography system was coupled to an 18-angle light scattering detector (DAWN HELEOS; Wyatt Technology). Data were collected at every 0.5 s at a flow rate of 0.5 mL/min. BSA (68 kDa) was used as a standard to calibrate the system. Data analysis was carried out using the program ASTRA6.1 and yields the molar mass and mass distribution of the sample.
GST–RhoA Pulldown Assays.
GST pulldown experiments were performed using 8 μM GST–RhoA (G17A), 8 μM Ect2, 8 μM RhoA (Q63L), and 50-μL GST resins in 200 μL of binding buffer (10 mM Hepes, pH 7.6, 60 mM NaCl, 1 mM MgCl2, 10 mM DTT, 10% glycerol, and 0.01% Tween 20). GST-tagged RhoA (G17A) was first incubated with GST resins, then mixed with DP Ect2 or the Y625A mutant, and then with RhoA (Q63L). The samples were incubated for 1 h at each binding step, washed 4 times with 1 mL of binding buffer and eluted with 30 mM glutathione. The whole process was performed on a rotating platform at 4 °C. The samples were analyzed with SDS–PAGE containing 6 M urea and visualized with Coomassie blue staining.
Ect2 RNA Interference and Rescue Assay.
siRNA of HsEct2 was generated to target the following sequence: UAUCACUCUUGUUUCAAUCUGAGGC/CAAGAGUGAUAUUGGUUCA. HsEct2 RNA interference (RNAi)-resistant form was generated by creating synonymous mutations in the sequence: 141 5′-AcaAatCgaGacCagGgtAatCttAgtC-3′ 168, and related Ect2 mutant derivations were obtained by quick-change PCR method from this RNAi-resistant template. Ect2 and its mutants were fused to EGFP(pEGFP-C1). During RNAi and rescue assay, totally 40 fmol of siRNA and 600 ng of DNA were cotransfected into HeLa cell using Lipofectamine 3000 (Invitrogen) with 0.8 × 105 cells for each well of 24-well plate.
Cells were fixed with 4% paraformaldehyde (PFA) 48 h after transfection and were viewed by immunofluorescence. Mouse monoclonal anti-GFP (Roche; 1:200) was used for GFP targeting. Secondary, polyclonal goat anti-mouse antibody conjugated to Alexa Fluor 488 (Roche; 1:250) was used for IF detection. DNA was stained with 1 μg/mL DAPI (Molecular Probes). F-Actin was stained with Alexa Fluor 647 Phalloidin (Molecular Probes; 1:40).
Samples were observed by confocal microscope (Zeiss; LSM 780) with 63×/1.4 N.A. oil-immersion lens. Binuclear cells and multinuclear cells were counted for each species. Every single experiment was repeated 3 times.
Western Blotting.
Transfected HeLa cells or A375 stable cell lines were prepared in RIPA buffer contained protease inhibitor mixture and additional phosphatase inhibitor mixture. Protein concentrations were measured with a BCA protein assay kit. Western blotting was performed as standard method. The following antibodies were used: rabbit anti-Ect2 C20 (Santa Cruz; 1:1,000), anti-GAPDH (EasyBio; 1:5,000), anti–p-ERK1/2 (Thr202/Tyr204), anti–total-ERK1/2, anti–p-FAK (Tyr397), anti–total-FAK, anti–p-AKT (Ser473), anti–total-AKT (Cell Signaling, 1:1,000; anti–total-ERK1/2, 1:2,500). HRP-conjugated secondary anti-rabbit antibody (EasyBio, 1:5,000) was used in detecting primary antibodies.
Calculation of Surface Conservation.
Ect2 sequences from 24 homologs were used to calculate the surface conservation using Consurf Sever (68), which scored the residues from 1 to 9 according to increasing conservation. Residues scored 7 to 9, whose conservation were above 80% among the selected sequences, were taken into consideration. The sequences of Ect2 orthologs used in the calculation are as follows: XP_005155777.1, XP_009445134.1, XP_012173507.1, XP_012245924.1, XP_015493690.1, XP_016063740.1, XP_016767892.1, XP_017683284.1, XP_020735256.1, XP_020836134.1, XP_021251947.1, XP_021459787.1, XP_022427755.1, XP_022525285.1, ADY41526.1, CDW58828.1, CEF69107.1, KRZ08246.1, NP_496319.1, NP_729306.1, NP_001084407.1, NP_001106617.1, NP001171096.1, and NP_001245245.1.
Cancer-Associated Mutations.
Somatic mutations in human cancers associated with Ect2 were from the database COSMIC (Catalogue of Somatic Mutations in Cancer) and cBioPortal.
Live-Cell Imaging.
Ect2 and its mutants were fused to EGFP and transfected into HeLa cells using Lipofectamine 3000 (Invitrogen). Totally 2.5 × 105 cells were plated on 35-mm glass-bottom dish and kept at 37 °C with 5% CO2. The EGFP signals were visualized at 24 to 48 h after transfection using a Perkin-Elmer Spinning Disk confocal microscope system equipped with Olympus IX83 microscope, a UPLSAPO 40 × 2 air lens (Olympus; N.A. 0.95), a CSUX1 spinning disk scanner (Yokogawa), and a C9100-23B EMCCD (Hamamatsu), and controlled by Volocity 6.3.1 software (Perkin-Elmer). Typically, few fractions of the cells on the dish are in cytokinesis, and we record at least 5 movies for each construct.
Stable Cell Line Generation.
Three plasmids packing system was used for lentivirus packing. The 3 plasmids are pLVX-EGFP-N1 inserted with the target genes, ps-PAX2 and pCMV-VSV-G. The packing cell line is HEK293T, and the lentivirus was harvested 48 h after plasmid transfection. Fresh lentivirus containing media were used to infect A375 cells for 3 times until we can see the expression of EGFP. Positive cells were selected by puromycin at 2 μg/mL for 1 wk.
Cell Proliferation Assay and Soft Agar Colony Formation Assay.
Cells were seeded in 6-well cell culture dishes in triplicate at a density of 40,000 cells as indicated per well in 2-mL medium without FBS. The medium was changed every day. The cell numbers at the indicated time points were determined by counting using a hemocytometer. For the soft agar assay, 3,000 cells were seeded per well in 6-well plates with 10% FBS DMEM containing 0.3% agarose on top of a layer of 0.6% agarose. Cells were then cultured in a 37 °C incubator for 2 wk. The colonies were fixed with 4% PFA, stained with 0.005% (wt/vol) crystal violet, and counted.
Tumor Xenograft Model.
A375 cells stably expressing WT and the R639Q mutant Ect2 were grown in cell culture as described above. The cell concentration was adjusted to 4 × 106 cells per mL. Mice were fixed and skin in armpit was cleaned with ethanol before tumor cell injection. A suspension of 4 × 105 cells in 0.1 mL of DMEM media were injected s.c. into the 2 flanks of male 5-wk-old athymic BALB/c nu/nu nude mice. Tumor growth was evaluated at 4 wk postinjection. All animal experiments were performed in accordance with relevant guidelines and regulations and were approved by the Animal Care and Use Committee of Peking University. The tumor weights were recorded as zero when undetected in some of the mice. Student’s t test was used to evaluate difference in tumor weight between WT and the mutant.
Supplementary Material
Acknowledgments
We thank Shilong Fan at the Center of Structure Biology (Tsinghua University) and the staff at beamline BL17U of Shanghai Synchrotron Radiation Facility for help with diffraction data collection; Xiaolin Tian at the Center of Biomedical Analysis (Tsinghua University) for the HDX-MS sample analysis; and the Tsinghua University Branch of China National Center for Protein Sciences Beijing for providing facility support. We thank the reviewers for their constructive suggestions. This work was supported by the grants of National Key Research and Development Program (2019YFA0508900 and 2017YFA0102900 to Z.C.); the National Natural Science Foundation of China (31570731, 31270762, and 31630046 to Z.C.); and Advanced Innovation Center for Structural Biology, Tsinghua–Peking Joint Center for Life Sciences.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors for Ect2 have been deposited in the Protein Data Bank (PDB), https://www.rcsb.org/ (ID code 6L30).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913054117/-/DCSupplemental.
References
- 1.Miki T., Smith C. L., Long J. E., Eva A., Fleming T. P., Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature 362, 462–465 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Ratheesh A., et al. , Centralspindlin and α-catenin regulate Rho signalling at the epithelial zonula adherens. Nat. Cell Biol. 14, 818–828 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagana A., et al. , A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 12, 1186–1193 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Liu X. F., Ishida H., Raziuddin R., Miki T., Nucleotide exchange factor ECT2 interacts with the polarity protein complex Par6/Par3/protein kinase Czeta (PKCzeta) and regulates PKCzeta activity. Mol. Cell. Biol. 24, 6665–6675 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motegi F., Sugimoto A., Sequential functioning of the ECT-2 RhoGEF, RHO-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. Nat. Cell Biol. 8, 978–985 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Morita K., Hirono K., Han M., The Caenorhabditis elegans ect-2 RhoGEF gene regulates cytokinesis and migration of epidermal P cells. EMBO Rep. 6, 1163–1168 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatsumoto T., Xie X., Blumenthal R., Okamoto I., Miki T., Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell Biol. 147, 921–928 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sano M., et al. , Expression level of ECT2 proto-oncogene correlates with prognosis in glioma patients. Oncol. Rep. 16, 1093–1098 (2006). [PubMed] [Google Scholar]
- 9.Justilien V., Fields A. P., Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene 28, 3597–3607 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirata D., et al. , Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal cancer progression. Clin. Cancer Res. 15, 256–266 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Huff L. P., et al. , The role of Ect2 nuclear RhoGEF activity in ovarian cancer cell transformation. Genes Cancer 4, 460–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., et al. , ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J. Hepatol. 62, 1287–1295 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Justilien V., et al. , Ect2-dependent rRNA synthesis is required for KRAS-TRP53-driven lung adenocarcinoma. Cancer Cell 31, 256–269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cairney C. J., et al. , A “synthetic-sickness” screen for senescence re-engagement targets in mutant cancer backgrounds. PLoS Genet. 13, e1006942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotýnková K., Su K. C., West S. C., Petronczki M., Plasma membrane association but not midzone recruitment of RhoGEF ECT2 is essential for cytokinesis. Cell Rep. 17, 2672–2686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su K. C., Takaki T., Petronczki M., Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev. Cell 21, 1104–1115 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Rosa A., Vlassaks E., Pichaud F., Baum B., Ect2/Pbl acts via Rho and polarity proteins to direct the assembly of an isotropic actomyosin cortex upon mitotic entry. Dev. Cell 32, 604–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yüce O., Piekny A., Glotzer M., An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170, 571–582 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piekny A. J., Glotzer M., Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr. Biol. 18, 30–36 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Petronczki M., Glotzer M., Kraut N., Peters J. M., Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell 12, 713–725 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Bement W. M., et al. , Activator-inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium. Nat. Cell Biol. 17, 1471–1483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D., Glotzer M., The RhoGAP activity of CYK-4/MgcRacGAP functions non-canonically by promoting RhoA activation during cytokinesis. eLife 4, e08898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook D. R., Rossman K. L., Der C. J., Rho guanine nucleotide exchange factors: Regulators of Rho GTPase activity in development and disease. Oncogene 33, 4021–4035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishima M., Centralspindlin in Rappaport’s cleavage signaling. Semin. Cell Dev. Biol. 53, 45–56 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Basant A., Glotzer M., Spatiotemporal regulation of RhoA during cytokinesis. Curr. Biol. 28, R570–R580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollard T. D., O’Shaughnessy B., Molecular mechanism of cytokinesis. Annu. Rev. Biochem. 88, 661–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappaport R., Experiments concerning the cleavage stimulus in sand dollar eggs. J. Exp. Zool. 148, 81–89 (1961). [DOI] [PubMed] [Google Scholar]
- 28.Saito S., et al. , Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J. Biol. Chem. 279, 7169–7179 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Kim J. E., Billadeau D. D., Chen J., The tandem BRCT domains of Ect2 are required for both negative and positive regulation of Ect2 in cytokinesis. J. Biol. Chem. 280, 5733–5739 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Solski P. A., et al. , Requirement for C-terminal sequences in regulation of Ect2 guanine nucleotide exchange specificity and transformation. J. Biol. Chem. 279, 25226–25233 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Burkard M. E., et al. , Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 7, e1000111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfe B. A., Takaki T., Petronczki M., Glotzer M., Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7, e1000110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou Y., et al. , Crystal structure of triple-BRCT-domain of ECT2 and insights into the binding characteristics to CYK-4. FEBS Lett. 588, 2911–2920 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Tse Y. C., et al. , RhoA activation during polarization and cytokinesis of the early Caenorhabditis elegans embryo is differentially dependent on NOP-1 and CYK-4. Mol. Biol. Cell 23, 4020–4031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snyder J. T., et al. , Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat. Struct. Biol. 9, 468–475 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Rambo R. P., Tainer J. A., Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soisson S. M., Nimnual A. S., Uy M., Bar-Sagi D., Kuriyan J., Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell 95, 259–268 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Hara T., et al. , Cytokinesis regulator ECT2 changes its conformation through phosphorylation at Thr-341 in G2/M phase. Oncogene 25, 566–578 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Niiya F., Tatsumoto T., Lee K. S., Miki T., Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene 25, 827–837 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Justilien V., Jameison L., Der C. J., Rossman K. L., Fields A. P., Oncogenic activity of Ect2 is regulated through protein kinase C iota-mediated phosphorylation. J. Biol. Chem. 286, 8149–8157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu M., et al. , Phosphorylation-dependent assembly and coordination of the DNA damage checkpoint apparatus by Rad4(TopBP1). Mol. Cell 51, 723–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frenette P., et al. , An anillin-Ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PLoS One 7, e34888 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutz S., et al. , Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science 318, 1923–1927 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Margarit S. M., et al. , Structural evidence for feedback activation by Ras⋅GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Rojas R. J., et al. , Galphaq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J. Biol. Chem. 282, 29201–29210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen A. W., Daugherty P. S., Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23, 355–360 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Lens S. M. A., Medema R. H., Cytokinesis defects and cancer. Nat. Rev. Cancer 19, 32–45 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Fields A. P., Justilien V., The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv. Enzyme Regul. 50, 190–200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crompton B. D., et al. , The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 4, 1326–1341 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Giannakis M., et al. , RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 46, 1264–1266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Research Network et al. , Integrated genomic characterization of oesophageal carcinoma. Nature 541, 169–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson G., et al. , Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C., et al. , Whole exome sequencing identifies frequent somatic mutations in cell-cell adhesion genes in Chinese patients with lung squamous cell carcinoma. Sci. Rep. 5, 14237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.South A. P., et al. , NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J. Invest. Dermatol. 134, 2630–2638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen K., et al. , Mutational landscape of gastric adenocarcinoma in Chinese: Implications for prognosis and therapy. Proc. Natl. Acad. Sci. U.S.A. 112, 1107–1112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cancer Genome Atlas Network , Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medina F., et al. , Activated RhoA is a positive feedback regulator of the Lbc family of Rho guanine nucleotide exchange factor proteins. J. Biol. Chem. 288, 11325–11333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun L., et al. , Mechanistic insights into the anchorage of the contractile ring by anillin and Mid1. Dev. Cell 33, 413–426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li P., Martins I. R., Amarasinghe G. K., Rosen M. K., Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat. Struct. Mol. Biol. 15, 613–618 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong Y., et al. , Structural basis of ubiquitin modification by the Legionella effector SdeA. Nature 557, 674–678 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I., PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 (2003). [Google Scholar]
- 64.Wang J., et al. , A method for helical RNA global structure determination in solution using small-angle X-ray scattering and NMR measurements. J. Mol. Biol. 393, 717–734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svergun D. I., Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 (1992). [Google Scholar]
- 66.Svergun D., Barberato C., Koch M. H. J., CRYSOL a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28, 768–773 (1995). [Google Scholar]
- 67.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashkenazy H., et al. , ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.