Significance
Expression of bacterial genes in response to various cues is predominantly regulated by 2- or multicomponent systems with autophosphorylation of a histidine kinase (HK), the first component, being controlled by an N-terminal sensor domain. This is followed by phosphotransfer to the receiver domain (Rec) of a cognate transcription factor. In about 20% of all cases, HK core and Rec are fused to form a hybrid HK (HHK). Here, we show the first full-length structure of an HHK and reveal how it gets activated by the second-messenger c-di-GMP that binds to a dedicated pseudo-Rec domain. The mechanism is fundamentally distinct from the canonical mechanism of HK regulation, but may be operational in many HHKs with a predicted pseudo-Rec domain.
Keywords: c-di-GMP, autophosphorylation, hybrid histidine kinase
Abstract
Cytosolic hybrid histidine kinases (HHKs) constitute major signaling nodes that control various biological processes, but their input signals and how these are processed are largely unknown. In Caulobacter crescentus, the HHK ShkA is essential for accurate timing of the G1-S cell cycle transition and is regulated by the corresponding increase in the level of the second messenger c-di-GMP. Here, we use a combination of X-ray crystallography, NMR spectroscopy, functional analyses, and kinetic modeling to reveal the regulatory mechanism of ShkA. In the absence of c-di-GMP, ShkA predominantly adopts a compact domain arrangement that is catalytically inactive. C-di-GMP binds to the dedicated pseudoreceiver domain Rec1, thereby liberating the canonical Rec2 domain from its central position where it obstructs the large-scale motions required for catalysis. Thus, c-di-GMP cannot only stabilize domain interactions, but also engage in domain dissociation to allosterically invoke a downstream effect. Enzyme kinetics data are consistent with conformational selection of the ensemble of active domain constellations by the ligand and show that autophosphorylation is a reversible process.
Two-component systems (TCSs) are widespread in microorganisms and mediate signal-dependent regulation of gene expression. They link diverse intracellular and extracellular stimuli to specific cellular responses, including development, cell division, and antibiotic resistance (1, 2). In their simplest form, TCSs are composed of a sensory histidine kinase (HK) and its cognate response regulator (for recent review, see refs. 3 and 4). The sensory HK is usually a homodimer that autophosphorylates a histidine in response to an input signal and then catalyzes transfer of the phosphoryl group to a conserved aspartate on the receiver domain (Rec) of its partner response regulator, thereby modifying its activity. As well as a Rec domain, response regulators usually harbor an output effector domain that often functions as a transcription factor. The HK catalytic core is composed of a dimerization and histidine transfer (DHp) domain for carrying the active histidine, and a catalytic ATP binding (CA) domain. The DHp domain contains 2 α-helices that mediate homodimerization by forming a 4-helix bundle. An exposed histidine residue in the DHp domain is autophosphorylated by the CA domain, which forms an α/β-sandwich that binds ATP. Phosphorelays, also called multicomponent systems, are more complex in that they include a histidine phosphotransferase (Hpt) domain to enable phosphate transfer between 2 Rec domains. In hybrid HKs (HHK) that function as part of phosphorelays, the first Rec domain is fused C terminally to the HK core. Although HHKs comprise almost 20% of bacterial HKs (5), no detailed structural or mechanistic studies on their regulation are available, apart from studies on the relaxed specificity between the HK core and Rec (6, 7).
Typically, HKs perceive the signal by dedicated periplasmic domains, but membrane spanning and cytosolic domains/proteins are known to regulate them as well (8). Input domains, such as PAS, GAF, and HAMP, are located N terminal to the HK core and affect the conformation and dynamics of the dimeric DHp helix bundle upon signal recognition, thereby controlling autophosphorylation of the conserved histidine on the first DHp α-helix (9–11) and initiating signal transduction. In contrast, ShkA, investigated here, is a noncanonical HHK lacking any N-terminal input domain, but exhibiting a pseudoreceiver domain (Rec1) between the kinase core and the C-terminal receiver domain (Fig. 1A). It was suggested recently (12) that such a domain organization may be common among HHKs and that the pseudoreceiver domain can act as signal input domain by binding to small signaling molecules.
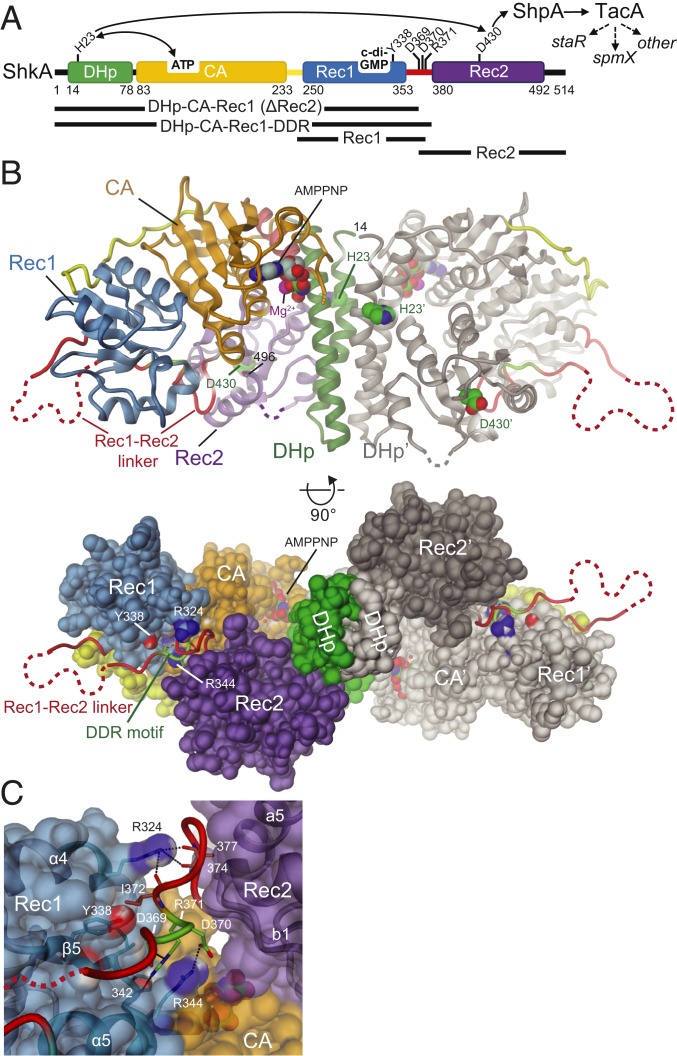
Fig. 1.
Crystal structure of full-length ShkA dimer shows compact, autoinhibited domain arrangement. (A) ShkA domain organization with functional residues indicated. The phospho-relay from ShkA via ShpA to transcription factor TacA, which controls the expression of staR, spmX, and other genes (broken arrows), is indicated by solid arrows. Constructs used in this study are shown at the bottom (see also SI Appendix, Table S4). (B) Side and bottom view of SkhA dimer in cartoon or surface representation with 1 protomer colored as in A and the other (primed labels) in gray. Disordered loops are indicated by dashed lines. His23, Asp430, and bound AMPPNP/Mg2+ are shown in CPK representation. The structure represents an inactive, autoinhibited conformation, since these potentially reacting groups are dispersed. For more structural details, see SI Appendix, Fig. S1. (C) Detailed view of Rec1-Rec2 linker (red, with DDR motif residues in light-green) bound to the α4-β5-α5 face of Rec1. Residues of the motif interact with Rec1 main-chain carbonyl 342 and R344 side-chain. Main-chain carbonyls 374 and 377 of the linker interact with Rec1 R324, and I372 forms an apolar contact (with I327). Despite the absence of direct Rec1/Rec2 contacts, these interactions tether tightly the N terminus of Rec2 to Rec1.
The second messenger, c-di-GMP, has been recognized as a major player in bacterial signal transduction, regulating diverse functions, including bacterial lifestyle, behavior, and development (13). c-di-GMP is produced by diguanylate cyclases, and diguanylate cyclase domains (GGDEF), together with phosphodiesterase (EAL) domains (14), comprise the second-most common group of TCS output domains, establishing a firm link between the 2 major systems in bacterial signaling. A connection in the opposite direction has been discovered only recently in Caulobacter crescentus with the cell cycle HK CckA being controlled by c-di-GMP (15). Increasing levels of c-di-GMP during the G1 phase cause CckA to switch from default kinase to phosphatase mode upon S-phase entry, thereby licensing cells for replication initiation. Based on structural and functional analyses, c-di-GMP–mediated cross-linking of the DHp and CA domain was proposed as the molecular mechanism of the switch (16). Most recently, it was found that ShkA, the other major kinase involved in the regulation of the C. crescentus cell cycle progression and morphogenesis (17), is also controlled by c-di-GMP (12). ShkA is a soluble HK that is activated during the C. crescentus cell cycle by increasing levels of the second messenger. Activation of ShkA initiates a multistep phosphorylation cascade (Fig. 1A), consisting of the phosphotransferase protein ShpA and the transcription factor TacA. Activation of TacA by phosphorylation initiates a G1/S-specific transcriptional program ultimately leading to cell morphogenesis and S-phase entry (12, 17). Direct targets of TacA include spmX and staR, encoding a critical morphogen and a transcriptional regulator responsible for stalk elongation, respectively. In contrast to CckA, which upon binding of c-di-GMP switches into phosphatase mode, the ligand is required for ShkA kinase activity, suggesting a distinct mechanism of allosteric control.
Here, we reveal the molecular mechanism of ShkA regulation, which is based on c-di-GMP–mediated mobilization of a locked, autoinhibited domain arrangement. We have determined crystal structures of full-length ShkA and of its isolated Rec1 domain in complex with c-di-GMP and have analyzed the dynamics of the full-length enzyme by NMR spectroscopy, employing isoleucine methyl group isotope labeling. Comprehensive enzymatic analyses revealed the underlying thermodynamics of the regulatory mechanism. Finally, we propose a general model for the structural transitions during the catalytic cycle of HHKs.
Results
Full-Length ShkA Crystal Structure Reveals an Autoinhibited State of ShkA.
ShkA is a HHK composed of a DHp-CA core domain, which catalyzes autophosphorylation of H23, and a Rec2 receiver domain carrying the phospho-acceptor D430 (Fig. 1A). A second, but degenerate, receiver domain Rec1 precedes Rec2. The crystal structure of full-length ShkA was determined by molecular replacement to 2.65-Å resolution in the presence of AMPPNP/Mg2+ (SI Appendix, Table S1). The structure shows a crystallographic dimer stabilized by contacts mediated by the DHp helices A1 and A2 (Fig. 1B and SI Appendix, Fig. S1A). The DHp domain is followed by a canonical CA domain with a tightly associated Rec1 domain (SI Appendix, Fig. S1B). Rec1 shows the canonical (βα)5 receiver domain fold (see also Fig. 3A), but with the fourth cross-over helix degenerated to a loop (β3-β4 loop). The acidic pocket is incomplete in that it has the 2 canonical acidic groups at the end of β1 replaced by S255, P256, explaining why the domain is not involved in phosphotransfer (17), despite the presence of an aspartate (D297) in the canonical phospho-acceptor position (SI Appendix, Fig. S2). Finally, the partly disordered Rec1-Rec2 linker leads to the canonical Rec2 domain that is found associated with the central DHp bundle (SI Appendix, Fig. S1C). Notably, the N-terminal end of the Rec2 domain appears firmly tethered to Rec1 not through a direct contact, but via multiple ionic interactions mediated mainly by a DDR motif on the Rec1-Rec2 linker (light green in Fig. 1 B and C). Although the precise position of the Rec2 may depend on its crystal contact with a neighboring CA domain, it is clear that dissociation of the DDR motif from the Rec1 domain would be required to allow autophosphorylation or phosphotransfer. The structure of the AMPPNP/Mg2+ ligand bound to the canonical CA site is well resolved, but 8 residues of the ATP lid are unstructured (SI Appendix, Fig. S1 D and E).
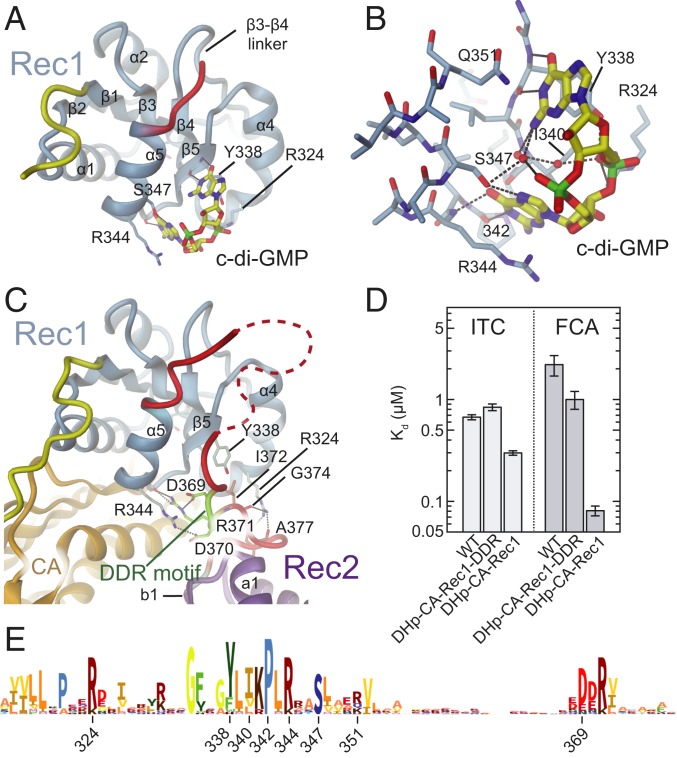
Fig. 3.
C-di-GMP and Rec1-Rec2 linker compete for Rec1 binding. (A and B) Crystal structure of ShkARec1 in complex with c-di-GMP with interacting residues shown in full. The Watson–Crick edges of both guanines form H-bonds with Rec1 main-chain atoms. The bases stack with Y338, Q351 and with R344. The hydroxyl of S347 is H-bonded to a guanine N7 group. Both Y338 and R324 interact with 1 of the phosphates. See also SI Appendix, Fig. S4. (C) Structure of Rec1 of full-length ShkA in same orientation as ShkARec1 in A. The comparison shows that the DDR motif (light-green) is bound to the same site on Rec1 as c-di-GMP (for a superposition see SI Appendix, Fig. S4B). Note that R324 and R344 of Rec1 are involved both in c-di-GMP and linker binding. (D) C-di-GMP binding to various ShkA constructs as measured by ITC (Upper) and FCA (Lower). Construct DHp-CA-Rec1 shows a significantly lower dissociation constant compared to the other variants, which can be attributed to the absence of the competing DDR segment. Full data are given in SI Appendix, Fig. S5. (E) Sequence logo encompassing the c-di-GMP binding site of the ShkARec1 domain.
The structure clearly represents an inactive, autoinhibited, state of ShkA. Large conformational changes would be required to allow access of bound ATP to H23 and of H23∼P to D430 for autophosphorylation and phosphotransfer, respectively. Similarly, DHp mediated dephosphorylation of D430∼P would require a substantial rotation of Rec2 toward the putative hydrolytic water thought to be bound to Thr27, 4 residues C terminal to the active histidine as in CpxA (18) or HK853 (19).
C-di-GMP Binding Induces Large Conformational and Dynamic Changes in ShkA.
C-di-GMP binds to the Rec1 domain as shown by NMR studies with the isolated domain (12). To investigate the mechanism of c-di-GMP–mediated relief of the autoinhibited, locked state (Fig. 1B), we next investigated the full-length enzyme by NMR. A 2D [15N,1H]-TROSY spectra of uniformly deuterated ShkA and ShkA/AMPPNP show only around 40 strong signals in the random-coil region of the spectrum (Fig. 2A and SI Appendix, Fig. S3A), indicating that they arise from residues located in locally flexible regions of the protein. The large majority of the 489 nonproline residues of ShkA are, however, not detected in this spectrum, due to the large molecular size of the rigid dimer and the associated slow molecular tumbling.
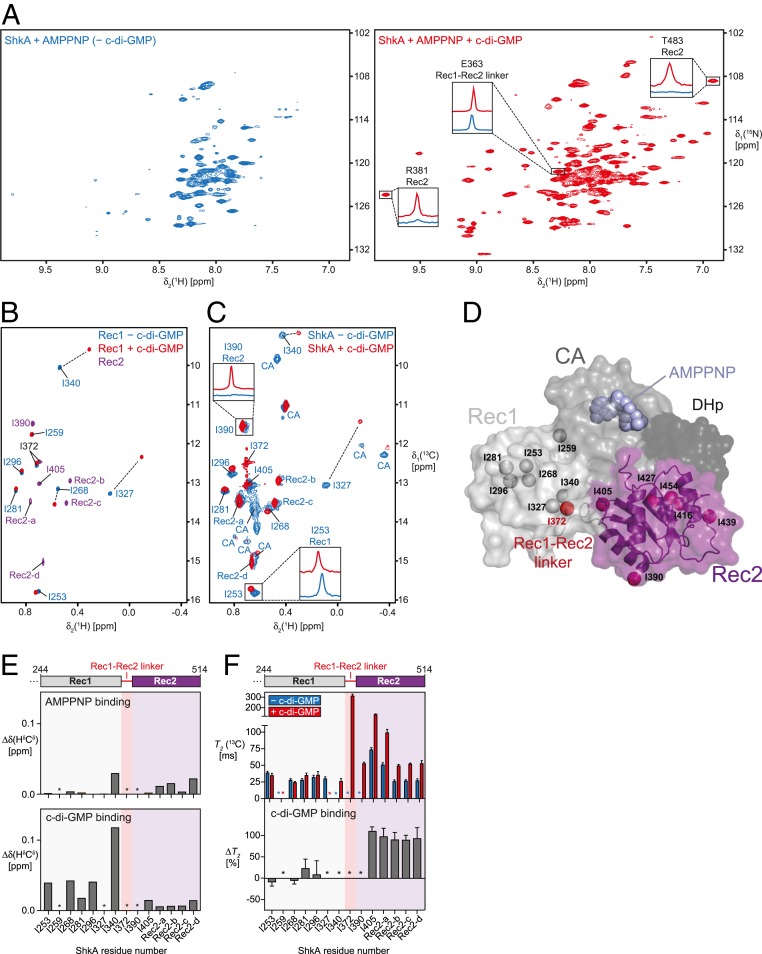
Fig. 2.
C-di-GMP binding to ShkA leads to release of the Rec2 domain. (A) Two-dimensional [15N,1H]-TROSY spectra of ShkA/AMPPNP (blue spectrum) and ShkA/AMPPNP/c-di-GMP (red spectrum). The Insets show 1D [1H]-cross sections for representative amide protons. (B) Overlay of 2D [13C,1H]-HMQC spectra of the isolated [13C/1Hδ1-Ile]Rec1 domain in apo state (blue) and c-di-GMP-bound state (red), and the isolated [13C/1Hδ1-Ile]Rec2 domain (purple). (C) Spectral overlay of 2D [13C,1H]-HMQC spectra of [13C/1Hδ1-Ile]ShkA in apo state (blue) and c-di-GMP-bound state (red). The 2 Insets show 1D [1H]-cross sections for representative methyl protons. Sequence-specific resonance assignments of the δ1 methyl groups are either indicated or assigned to the Rec2 or CA domain. (D) Crystal structure of ShkA/AMPPNP. Surface representation of the monomer composed of 4 domains: DHp (dark gray), CA with bound AMPPNP (gray), Rec1 (light gray), and Rec2 (purple). The δ1 methyl groups of isoleucine residues in the Rec1 and Rec2 domains are shown as spheres together with I372 (red sphere) that is part of the Rec1-Rec2 linker. (E) Chemical shift perturbation of isoleucine δ1 methyl groups upon addition of AMPPNP to ShkA (Upper) and upon addition of c-di-GMP to ShkA/AMPPNP (Lower). (F) Transverse relaxation times of isoleucine δ1 methyl carbons of ShkA/AMPPNP (blue bars) and ShkA/AMPPNP/c-di-GMP (red bars). The Lower panel shows the relative change of T2(13C) upon addition of c-di-GMP (ΔT2). (E and F) Asterisks indicate methyl groups that are either not assigned or line-broadened beyond detection in at least 1 of the 2 states.
Binding of AMPPNP to ShkA does not lead to major changes in the spectrum. In contrast, binding of c-di-GMP to either apo ShkA or ShkA/AMPPNP leads to dramatic spectral changes with around 120 well-resolved additional resonances being clearly detected, indicating a substantial change in the dynamics of ShkA for a large part of the protein (Fig. 2A and SI Appendix, Fig. S3A). This massive increase in signal intensity is exemplified for selected signals with their 1D 1H-cross sections (Fig. 2A and SI Appendix, Fig. S3D). A spectral overlay of ShkA/c-di-GMP with the isolated Rec2 domain shows that the new resonances perfectly overlap (SI Appendix, Fig. S3A), including Rec2 residues that are in contact with either the CA or DHp domain in the ShkA crystal structure (A421, G422, R446, A447, K448, A471, A472, G473). This indicates that upon binding of c-di-GMP to full-length ShkA, the Rec2 domain loses its contacts with the other domains and detaches completely from the otherwise rigid main architecture of the protein (SI Appendix, Fig. S3A). Since no significant chemical shift differences are detected between the 2 spectra, the detached Rec2 domain in ShkA has essentially the same 3D structure as the isolated Rec2 domain.
In order to obtain site-specific information of the effect of c-di-GMP on the structure and dynamics of ShkA, we increased the experimental sensitivity by employing methyl-NMR spectroscopy of specifically δ1-[13C/1H]-isoleucine–labeled full-length ShkA and isolated Rec1 and Rec2 domains (Fig. 2 B–D and SI Appendix, Fig. S3B). Sequence-specific resonance assignments for Rec1 were obtained by identifying unambiguous nuclear Overhauser effects in agreement with the crystal structure. Additional assignments for isoleucine methyl groups δ1-I340, δ1-I390, and δ1-I405 were obtained using selective mutagenesis of full-length ShkA (SI Appendix, Fig. S3C). The mutation I259V located within the Rec1 domain caused major spectral changes, indicating the importance of residue I259 in stabilizing the Rec1–CA domain interface (SI Appendix, Fig. S1B).
AMPPNP binding to the CA domain of full-length ShkA leads to only minimal chemical shift perturbation of the δ1-Ile methyl groups of the entire protein, implying that the structures of ligand-free and AMPPNP-bound ShkA are very similar (Fig. 2E and SI Appendix, Fig. S3B). In contrast, binding of c-di-GMP to either apo ShkA or to ShkA/AMPPNP causes substantial spectral changes. All δ1-Ile methyl group signals of the Rec2 domain strongly increase in intensity (SI Appendix, Fig. S3C), in full agreement with the spectral behavior of the backbone amides. Similar chemical shift changes were observed for c-di-GMP binding either to the isolated ShkA-Rec1 domain or to full-length ShkA, showing that c-di-GMP binds to the β5-α5 surface of prototypical Rec domains (Fig. 2 C and E and SI Appendix, Fig. S3B).
Overall, these data indicate that c-di-GMP binding to ShkA leads to liberation of the Rec2 domain from the protein core. To validate this hypothesis, we conducted NMR spin relaxation experiments in the presence and absence of c-di-GMP (Fig. 2F and SI Appendix, Fig. S3F). For the isolated receiver domains, average 13C transverse relaxation times for ILV methyl groups of Rec2 and Rec1 are quite similar (174 ms and 160 ms, respectively), in agreement with the similar molecular mass of both domains (16.3 kDa and 12.5 kDa, respectively). For full-length ShkA/AMPPNP in the absence of c-di-GMP, the transverse relaxation times of all δ1-Ile resonances is smaller (25 to 75 ms), as expected from the respective protein sizes (Fig. 2F). Addition of c-di-GMP strongly increases the 13C T2 times of Rec2 such that 13C T2 times are effectively doubled, but does not significantly affect those of δ1-Ile of Rec1 (Fig. 2F and SI Appendix, Fig. S3F). These experiments thus quantitatively confirm that c-di-GMP binding to ShkA leads to the specific detachment of the Rec2 domain from the protein core into a dynamic, multiconformational state, which enables the enzyme to undergo the large motions required for the phosphoryl transfer reactions.
C-di-GMP Competes with Rec1-Rec2 Linker for Binding to Rec1 Domain.
To reveal the molecular mechanism of c-di-GMP–induced Rec2 domain mobilization, we set out to determine the structure of the ShkA/c-di-GMP complex. Well-diffracting crystals were obtained only for the isolated Rec1 domain, but not for full-length ShkA/c-di-GMP, and the structure was determined to 1.8-Å resolution (Fig. 3 A and B and SI Appendix, Table S1). The structure of the Rec1 domain and of the ligand are very well resolved (SI Appendix, Fig. S4A). The asymmetric unit contains 2 monomers, each with a bound monomeric c-di-GMP ligand, which are virtually identical and superimpose closely with the corresponding domain in full-length ShkA (SI Appendix, Fig. S4B). In addition, an intercalated c-di-GMP dimer is observed that mediates a 2-fold symmetry contact between the 2 Rec1 monomers (SI Appendix, Fig. S4C) that is most likely a crystal artifact. Indeed, the isothermal titration calorimetry (ITC) data of full-length ShkA are consistent with a 1:1 stoichiometry (SI Appendix, Fig. S5 A–E).
Monomeric c-di-GMP is bound to the α4-β5-α5 face of Rec1 (Fig. 3 A and B), that is, to the surface that is involved in the dimerization of canonical Rec domains. The Watson–Crick edges of both guanine bases form H-bonds with the backbone of the exposed edge of the β5 edge and the β5-α5 loop. In addition, Y338 and R344 are forming stacking and cation–π interactions with the guanyl bases, respectively. S347 is H-bonded to a guanyl base, and R324 forms an ionic interaction with a c-di-GMP phosphate. All interacting ShkA residues are highly conserved in related sequences (Fig. 3C).
In full-length ShkA, the c-di-GMP binding site is occupied by the Rec1-Rec2 linker (Figs. 1C and 3D). Specifically, residues shown to be engaged in c-di-GMP binding (Y338, R324, and R344) are interacting with a conserved DDR motif of the linker region. Thus, we reasoned that if the DDR segment were to compete with c-di-GMP binding, its deletion should increase c-di-GMP affinity. Indeed, affinity measurements by ITC and by a fluorescence competition assay (FCA) showed that the DHp-CA-Rec1 construct (residues 1 to 366) had a lower dissociation constant compared to the wild-type or to the DHp-CA-Rec1-DDR truncation construct (residues 1 to 379) still containing the DDR segment (Fig. 3E). Why this difference is considerably more pronounced in the fluorescence data is not clear.
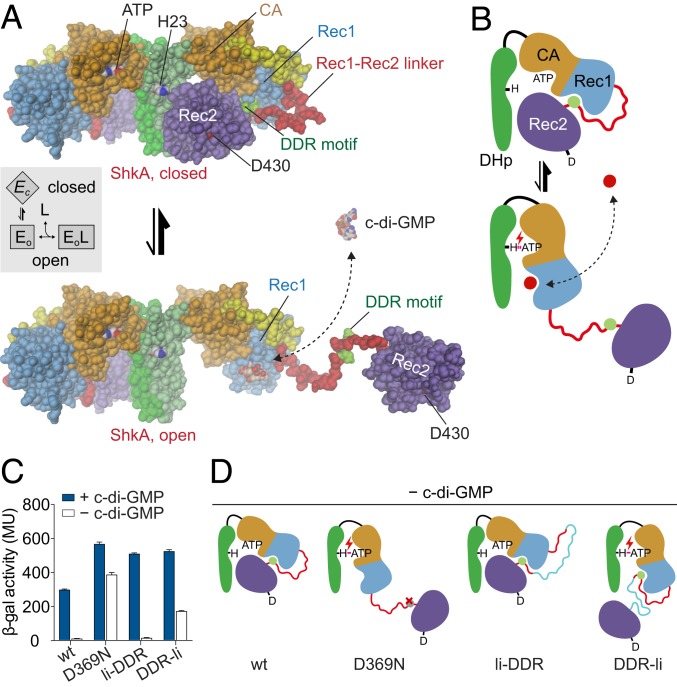
Taking together all data on structure and dynamics, we propose a “ligand-coupled conformational equilibrium” (LCCE) model as the mechanism underlying c-di-GMP–mediated ShkA activation (Fig. 4 A and B). In the absence of c-di-GMP, the enzyme is in a dynamic equilibrium between a closed, autoinhibited state (Ec) and an ensemble of open, inhibition-relieved states (Eo) that are characterized by a liberated Rec2 domain. In the absence of the ligand, this equilibrium would be populated largely on the Ec side, in line with the NMR results and the constitutive inactivity of the enzyme (12). C-di-GMP–mediated ShkA activation would then proceed by conformational selection (20) of the Eo states with their unobstructed Rec1 binding site, shifting the equilibrium in a dose-dependent fashion to these active states.
Fig. 4.
Mechanistic model of ShkA activation by c-di-GMP induced change of conformational equilibrium. (A) ShkA is in a dynamic equilibrium between a closed, autoinhibited conformation as seen in the full-length crystal structure (Upper) and an ensemble of open, catalytically active conformations with liberated Rec2 domains. An arbitrarily modeled open conformation is shown at the bottom, full activity probably requires both subunits to be open. C-di-GMP can bind only to the open conformation, because, in the closed conformation, the binding site on Rec1 is obstructed by the DDR motif (light-green). (Inset) Corresponding thermodynamic LCCE model. (B) Schematic representation of the mechanism shown in A. C-di-GMP (red circle) competes with the DDR motif (light-green circle) for Rec1 binding resulting in Rec2 liberation allowing catalysis (e.g., autophosphorylation as indicated). (C) In vivo transcription activity of ShkA variants assayed in a strain harboring a spmX′-′lacZ reporter. Mutation D369N affects a crucial Rec1-Rec2 linker tethering interaction, mutants li-DDR and DDR-li have an elongated Rec1-Rec2 linker with the insertion placed N or C terminal to the DDR motif as indicated in the scheme (D). Strains rcdG0 ΔshkA (−c-di-GMP) and ΔshkA (+c-di-GMP) expressed the indicated shkA alleles from plasmid pQF. Note that no inducer (cumate) was present since leaky expression from the PQ5 promoter was sufficient to complement the ΔshkA phenotype. Mean values and SDs are shown (n = 3). (D) Schematic representation of wild-type and mutant ShkA to demonstrate their phenotypes. In absence of c-di-GMP, mutations D369N and DDR-li both are predicted to liberate the Rec2 domain, which is in line with the data in C.
The regulatory model was tested in vivo by analyzing the transcriptional activity of the ShkA-ShpA-TacA pathway with a β-Gal assay (9). In this assay, plasmid-borne variants of shkA were expressed in a C. crescentus shkA deletion background that harbors a plasmid-borne translational fusion between the TacA target gene spmX and lacZ (spmX′-′lacZ). This allowed following the activity of the ShkA-ShpA-TacA pathway directly by reading out the activity of β-Gal, encoded by lacZ. The assay was performed in the wild-type and in a c-di-GMP minus strain [rcdG0 strain (21)]. While in absence of c-di-GMP (rcdG0 strain) wild-type ShkA showed no activity, introducing D369N or placing an insertion between the DDR motif and Rec2 (but not between Rec1 and DDR) rendered ShkA active (Fig. 4C). Thus, the results fully corroborate the regulatory model, as illustrated in the scheme (Fig. 4D). Additional support for the model is provided by analytical ultracentrifugation sedimentation velocity (SV-AUC) experiments (SI Appendix, Fig. S6) that show a decrease in the sedimentation coefficient upon ligand addition, indicative of a more open structure. In the following, the LCCE model (Fig. 4 A, Inset) is scrutinized and its thermodynamic parameters are determined by quantitative functional investigations.
ShkA Autophosphorylation Is c-Di-GMP–Dependent, Reversible, and Noncooperative.
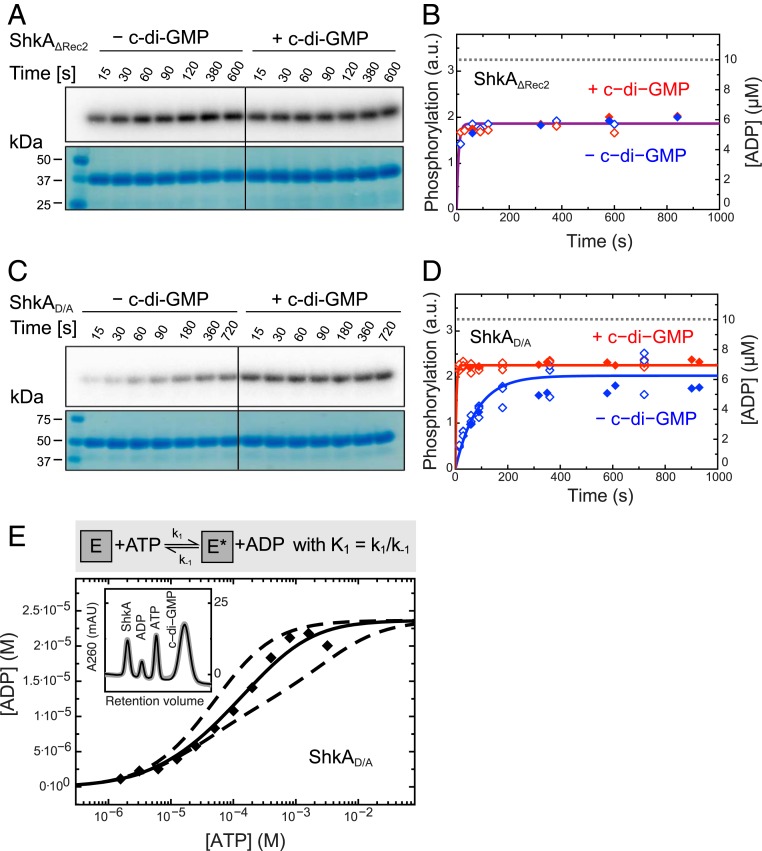
To determine the reaction kinetics of the full-length ShkA and mutants thereof, we acquired progress curves of enzyme net phosphorylation (E*) by autoradiography and of ATP to ADP turnover by on-line ion exchange chromatography (oIEC) (SI Appendix, Methods) quantification. First, we determined the kinetics of autophosphorylation separately using phosphotransfer-deficient variants that had the phospho-acceptor D430 mutated (ShkAD/A) or the Rec2 domain deleted (construct DHp-CA-Rec1 or, in short, ShkAΔRec2). As found previously (12) and consistent with the proposed regulatory model, ShkAΔRec2 is constitutively active (Fig. 5 A and B), since it lacks the obstructing Rec2 domain. ShkAΔRec2 attains very quickly (in less than 15 s) a stable phosphorylation state with or without the ligand; that is, it proceeds with an autophosphorylation rate constant k1 > 1/(15 s) = 0.07 s−1. For ShkAD/A, the same fast kinetics is observed, but only in presence of c-di-GMP (Fig. 5 C and D and SI Appendix, Fig. S7A), which demonstrates that under this condition the mutant is fully active. In the absence of the ligand, the turnover number is strongly reduced (k1,-c-di-GMP = 0.012 s−1) but, within error limits, phosphorylation is obtained to almost the same degree as for the activated enzyme. The observations are consistent with the regulatory LCCE model (Fig. 4) under the assumption that the samples had reached equilibrium after the about 15 min before start of the enzymatic reaction. No attempts were made to determine the kinetics of the conformational equilibrium or of c-di-GMP binding, but the ITC spikes induced upon ligand binding didn’t exhibit unusual broadening (width < 300 s) (SI Appendix, Fig. S10 A–D). Since the initial reaction velocity is at least 20 times larger in the presence of c-di-GMP (Fig. 5D), a lower boundary of 20 can be estimated for the conformational equilibrium constant K = [Ec]/[Eo].
Fig. 5.
ShkA autophosphorylation is reversible and, in the presence of Rec2, controlled by c-di-GMP. (A and C) ShkAΔRec2 and ShkAD/A phosphorylation after indicated times of incubation as measured by autoradiography using [γ-32P]-ATP, in the absence and presence of c-di-GMP as indicated. SDS/PAGE gels are shown below and indicate equal loading. For additional data, see SI Appendix, Fig. S7A. (B and D) Time course of ShkAΔRec2 and ShkAD/A phosphorylation (hollow symbols) and ADP production (filled symbols), in the absence (blue) and presence (red) of c-di-GMP. Phosphorylation values were calculated from the band intensities in A and C; ADP concentrations were determined by oIEC. Solid lines represent exponential fittings of the autoradiograph data [for ShkAD/A: k1,-c-di-GMP = (0.012 ± 0.002)s−1]. The dashed line indicates the employed enzyme concentration. (A–D) [ShkA variant]: 10 μM, [c-di-GMP]: 25 µM, [ATP]: 200 µM. (E) Equilibrium concentration of ADP as a function of initial ATP concentration as determined by oIEC. [ShkAD/A]: 20 μM, [c-di-GMP]: 50 μM. The continuous line is the fit of the simple reversible, noncooperative bi-bi reaction shown on the top to the data with K1 = k1/k−1 = 0.13 and a refined [ShkAD/A] of 23 μM, (see also SI Appendix, Methods). Reactions with positive or negative cooperativity would yield the dashed curves (calculated with equilibrium constants for second phosphorylation event of 1.3 and 0.013, respectively) that do not fit the data. (Inset) Representative oIEC chromatogram.
To obtain more quantitative information about the autophosphorylation reaction, we measured ATP to ADP turnover by oIEC. Fig. 5 B and D show the ADP progress curves (Fig. 5 B and D, filled symbols) of the phosphotransfer-deficient ShkA mutants that turn out to be congruent with the appropriately scaled phosphorylation curves (Fig. 5 B and D, open symbols). This indicates stable histidine phosphorylation at the employed condition as verified by long-term measurements (SI Appendix, Fig. S8A). Intriguingly, for both mutants, the progress curves (Fig. 5 B and D) indicate that the phosphotransfer-deficient mutants do not proceed to full quantitative modification (i.e., did not reach the level corresponding to the enzyme concentration, 10 μM). We reasoned that this may be due to reversibility of the autophosphorylation reaction, and that only a high ATP substrate concentration would shift the equilibrium completely to the product state. Indeed, upon ATP titration, the equilibrium ADP concentration was increased and reached the expected value close to the enzyme concentration indicating complete phosphorylation (Fig. 5E). A simple reversible bi-bi reaction model (SI Appendix, Methods) reproduced the data well and yielded an equilibrium constant K1 = k1/k−1 of 0.13 for activated ShkAD/A. Enzyme titration (SI Appendix, Fig. S8B) confirmed the result.
C-di-GMP Shifts Conformational ShkA Equilibrium to the Active State.
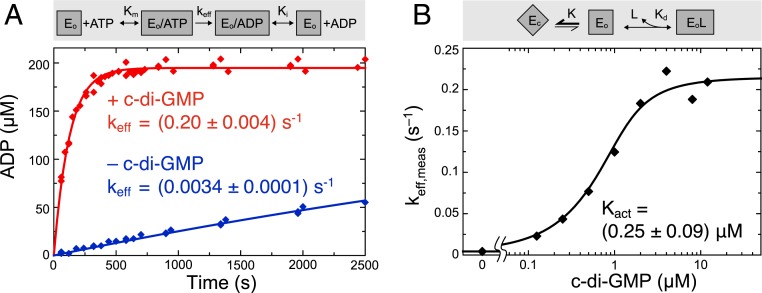
Full-length ShkA is effectively an ATPase, converting ATP to ADP via phospho-enzyme intermediates. Progress curves of ATP turnover acquired under various conditions by oIEC (Fig. 6A and SI Appendix, Fig. S9C) were fitted with a competitive product inhibition Michaelis–Menten model (SI Appendix, Fig. S9A) to derive the enzymatic parameters. The Km and Ki values are both about 60 μM, a value that agrees well with dissociation constants of AMPPNP and ADP obtained by FCA, while the ITC measurements agreed less, probably due to the intrinsic limitations of this method for low-affinity binders (SI Appendix, Fig. S10 A–D). Interestingly, the FCA measurements clearly showed that c-di-GMP has no significant effect on mononucleotide affinity (SI Appendix, Fig. S10 E–H). In the presence of c-di-GMP, ShkA is rather active (keff = 0.20 s−1) (Fig. 6A), considering that the reaction involves 3 individual steps (autophosphorylation, phosphotransfer, dephosphorylation) and requires several domain rearrangements (Discussion). Importantly, low but significant activity (keff = 0.0034 s−1) (Fig. 6A) is observed also in absence of the activator, which corroborates the LCCE model and allows an estimation of the conformational equilibrium constant to K = 58. Assuming the same K, the microscopic autophosphorylation rate of activated ShkAD/A can be calculated to k1 = k1,nonact × K = 0.7 s−1. Regarding wild-type ShkA phosphorylation, we confirmed that it gets boosted by c-di-GMP, but the phosphorylation level is clearly lower than that of the phosphotransfer-deficient ShkAD/A mutant (SI Appendix, Fig. S7B), which can be attributed to the dephosphorylation activity of the former.
Fig. 6.
C-di-GMP activation profile of wild-type ShkA is consistent with an LCCE model. (A) ADP production in the presence (red symbols) and absence of 25 μM c-di-GMP (blue) as measured by oIEC at room temperature. The reaction was started by addition of 200 μM ATP to 10 μM ShkA in reaction buffer. The data were fitted (solid line) with a competitive product inhibition Michaelis–Menten model (gray box; shown fully in SI Appendix, Fig. S9A) with Km and Ki determined by ATP and ADP titration (SI Appendix, Fig. S9C). (B) C-di-GMP activation profile obtained by acquisition of ADP progress curves at various c-di-GMP concentrations (SI Appendix, Fig. S9B) to yield keff,meas. The data were fitted to the LCCE model shown in the gray box (SI Appendix, Methods).
To quantitatively describe the activating effect of c-di-GMP, progress curves were acquired at various c-di-GMP concentrations (SI Appendix, Fig. S9B) and fitted with the Michaelis–Menten model. The resulting keff,meas values yielded a sigmoidal activation profile with Kact = 0.25 μM, as shown in Fig. 6B. This agrees reasonably well with the (apparent) dissociation constant of c-di-GMP as determined by ITC (1.5 μM in the presence of AMPPNP) (SI Appendix, Fig. S5B). The microscopic dissociation constant in the absence of the competing DDR motif would then be considerably smaller, namely Kd = Kact/(1 + K) = 4 nM (SI Appendix, Methods). This value is smaller than that observed for the DHp-CA-Rec1 construct by FCA and ITC (80 nM and 300 nM, respectively) (Fig. 3E and SI Appendix, Fig. S5 F–H), which may be due to residual binding competition by the truncated Rec1-Rec2 linker.
In summary, the enzymatic characterization shows that, in the presence of c-di-GMP, ShkA efficiently catalyses ATP turnover by enabling autophosphorylation and subsequent phosphotransfer and dephosphorylation. The data fully support the regulatory LCCE model and thus confirm that the c-di-GMP activation profile is governed not only by the affinity of the ligand to its binding site, but also by the free-energy difference between the conformational states.
Discussion
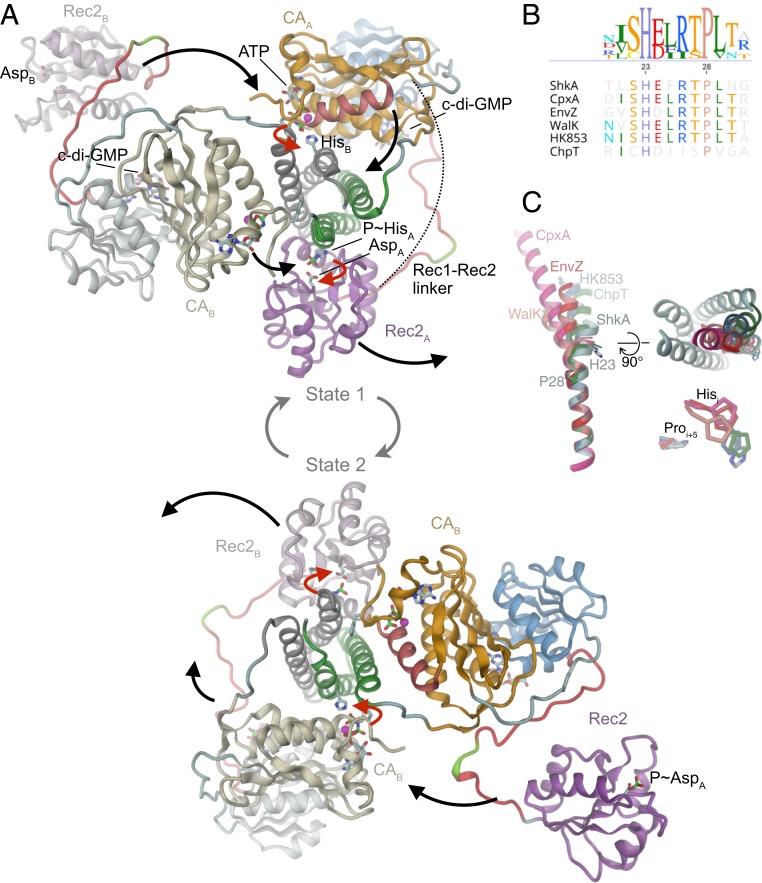
The structural, dynamic, and functional results presented in this study consistently show that activation of ShkA proceeds via the c-di-GMP–mediated liberation of a locked, autoinhibited state (Fig. 4). Thereby, c-di-GMP competes with the tethering of a domain linker (Fig. 3 A and D) to unleash the C-terminal domain allowing the enzyme to step through the catalytic cycle that involves large domain motions (Fig. 7). C-di-GMP interference with protein–protein interactions as a regulatory mechanism was also proposed for a YajQ-like transcription factor/coactivator complex (22), but structural information is missing.
Fig. 7.
Proposed domain arrangements during the catalytic cycle of ShkA. (A) During the catalytic cycle, the enzyme adopts 2 asymmetric dimer states that are structurally identical, but with domain roles interchanged. Subunits are distinguished by color hue (“A”: strong, “B”: weak), the active His and Asp and ligands are shown in full. State 1 (Upper): CA/ATP and Rec2 of chain A are poised for trans-autophosphorylation and cis-phosphotransfer, respectively. After concerted execution of the reactions (red arrows), CA/ADP and P∼Rec2 dislodge and CA/ATP, Rec2 of chain B engage with the DHp histidines (black arrows) to yield state 2 (Lower). The competent DHp/CA and DHp/Rec2 domain arrangements have been modeled based on the structures of CpxA [4BIW (18)] and ChpT/CtrA(Rec) [4QPJ (37)], respectively. In canonical HHK, CA would be linked directly to Rec2 (dashed line). (B) Part of DHp α1 sequence alignment of selected pfam HisKA family members. (C) Overlay of DHp α1 helix structures of the HKs of B after superposition of C-terminal parts. The structures with reported autophosphorylation competent domain arrangement (CpxA, 4BIW; WalK, 5C93; EnvZ, 4KP4) and those in phosphotransfer or dephosphorylation/arrangement (HK853, 3DGE; ChpT, 4QPJ) and ShkA form structural clusters distinguished by the degree of helix kinking. (Right) The kinase competent structures have the N-terminal part of α1 bent toward the center of the DHp bundle (ShkA in gray).
In ShkA, c-di-GMP binds to the edge strand (β5) of the Rec1 β-sheet and the following β5-α5 loop. The Watson–Crick edges of both guanines form H-bonds with the main chain (Fig. 3B), reminiscent of the backbone interactions in a β-sheet. The same kind of interaction occurs in the CckA/c-di-GMP complex, where 1 of the guanines binds to the β-sheet edge of the CA domain (16). In the accompanying study (12), Y338 was identified as a c-di-GMP binding residue in a targeted alanine scan. In addition to Y338A, several other mutants (R324A, I340A, S347A, and Q351A) showed severely impaired activity in vivo (12). Since all these residues contribute to c-di-GMP binding (Fig. 3B), this can be attributed to an impaired activation by the cellular c-di-GMP pool and suggests that the affinity of wild-type ShkA to c-di-GMP is finely tuned to its raise in concentration during cell cycle progression (21).
A mutation of R344, the remaining c-di-GMP binding residue, showed a wild-type–like phenotype. This may be explained by the dual role of R344 being involved in c-di-GMP binding as well as Rec1-Rec2 linker tethering (Fig. 3 A and D), resulting in opposing effects on activation upon mutation. Although R324 also has such a dual role, its strong phenotype suggests that this residue contributes more to c-di-GMP binding than to linker tethering. These observations demonstrate that identification of ligand binding residues is not straightforward if they are also involved in opposing activities like protein–protein interactions. However, mutation of the DDR motif or inserting a loop between the motif and the Rec2 domain clearly resulted in constitutively active enzyme and, thus, confirmed the importance of Rec2 tethering for autoinhibition (Fig. 4C).
The NMR studies on full-length ShkA using specific isotope labeling of the isoleucine methyl groups provided atomic scale information of structural and dynamic changes upon ligand binding (Fig. 2). Binding of c-di-GMP, but not AMPPNP, changed the dynamics of Rec2 substantially and exclusively, leading to the conclusion that the mobility of the Rec2 domain is restricted in the absence of c-di-GMP but is drastically enhanced upon addition of the ligand such that its relaxation times become comparable to those of an individual protein of this size. The binding of c-di-GMP to Rec1 therefore releases Rec2 allosterically.
In order to relate the mechanistic “activation by domain liberation” model of ShkA with the function of this hybrid kinase, comprehensive enzymatic data were acquired. The specially developed oIEC method proved highly valuable as it enabled the efficient recording of quantitative enzyme progress curves. We found, using phosphotransfer-deficient mutants, that ShkA autophosphorylation is clearly reversible. Under various titration regimes, the equilibrium concentrations were consistent with a simple reversible bi-bi reaction with the equilibrium largely on the side of the reactants (Fig. 5E and SI Appendix, Fig. S8B). Thus, the degree of phosphorylation depends on the mononucleotide concentrations, and a high ADP concentration promotes enzyme dephosphorylation by back transfer of the phosphoryl group onto ADP. Reversibility of the HK autophosphorylation reaction was originally demonstrated for CheA (23) (see also ref. 24 and references therein) and KinA (25), and later for NRII (26). Although largely neglected in the literature, this is probably a general feature of ATP-driven histidine phosphorylation. It should be tested whether it can explain the ADP dependence of the kinase reaction as observed in vitro for HK853 (27) and other HKs (28). The rate constant of autophosphorylation is with k1 = 0.7 s−1 at least 30 times faster than measured for other HKs (16, 29). Possibly, some of these enzymes were partly impaired by truncation or not fully activated. ATP turnover of full-length ShkA proceeds somewhat slower, which demonstrates that phosphotransfer and P∼Asp hydrolysis are not much slower. To answer whether the latter reaction is entirely due to the intrinsic lability of the phospho-aspartate or catalyzed by docking of the Rec2 domain to the DHp bundle (4, 19) requires further investigations. Although dephosphorylation of ShkA has not been investigated in vivo and is not part of the current model of cell cycle control (30), it may be controlled by yet unknown factors that, for example, affect the DHp bundle geometry as in DesK (31). Additionally, the counteracting dephosphorylation activity may be relevant for concentration robust signaling (32).
Evidence has accumulated that HKs form asymmetric dimers (9) that can catalyze in concert both autophosphorylation and phosphotransfer and a structural model has been put forward for CpxA (33). The corresponding model for activated (i.e., c-di-GMP–complexed) ShkA is shown in Fig. 7 A, Upper, where domains have been placed on the basis of homologous structures with competent autophosphorylation or phosphotransfer domain arrangement. Canonical autophosphorylation and phosphotransfer mechanisms can be anticipated, since the functional interfaces show no severe clashes or polarity mismatch. Both CA and Rec2 of subunit A (CAA, Rec2A) (Fig. 7A, bright colors) would be engaged in concerted catalysis resulting in a net transfer of a phosphoryl group from CAA bound ATP to the active aspartate of Rec2A (Fig. 7A, red arrows), while the corresponding domain of subunit B (Fig. 7A, tinted colors) would be in stand-by mode. Subsequently, both domains would dislodge (Fig. 7A, black arrows), with CAA moving to a near-by parking position and Rec2A becoming available for down-stream phosphoryl transfer (Fig. 7 A, Lower). Concurrently, the catalytic domains of subunit B would move to catalytically competent positions, resulting in the same structure as at the beginning, although 2-fold related and with the roles of the subunits interchanged (state 2). After concerted catalysis, equivalent domain rearrangements would bring back the enzyme to state 1.
In line with the left-handed connectivity of the 2 DHp helices, the model predicts that autophosphorylation occurs in trans (34). Due to the almost equal distance between either of the C-termini of the Rec1 domains and the N terminus of the competently docked Rec2 domain (about 60 Å), phosphotransfer may occur in trans or in cis as modeled. In any case, the Rec1-Rec2 linker of ShkA is long enough (27 residues) to allow simultaneous execution of autophosphorylation and phosphotransfer. Notably, also for canonical HHKs (without an intervening domain like Rec1), this appears possible, since their CA–Rec linkers appear long enough (23 ± 3 residues for the 23 HHKs in C. crescentus) to bridge the distance (again 60 Å) (dashed line in Fig. 7A).
Most likely the asymmetry is rooted in the inward kinking of the N-terminal part of DHp α1 helix upon interaction with the ATP complexed CA domain (Fig. 7C), which would be sterically incompatible with an analogous motion of α1 from the other subunit. This has been discussed and analyzed thoroughly by Bhate et al. (9) and corroborated by the recent structure and modeled dynamics of WalK (35). The DHp domain of ShkA belongs to the major HisKA pfam family (36) that exhibits a HxxxxP motif (Fig. 7B). Superposition of the C-terminal part of DHp α1 structures of this family (Fig. 7C) demonstrates kinking of α1 with a pivot close to the proline and shows that the N-terminal parts of α1 helices of autophosphorylation competent structures (CpxA, EnvZ, WalK) are tilted toward the helix bundle axis. A symmetric structure with both α1 helices tilted inwards is sterically probably not feasible, which would thus be the basis for the asymmetry of the functional dimer.
The proposed asymmetric catalysis is not equivalent with half-of-sites modification and is, therefore, not at odds with our finding of complete modification at high ATP concentration (Fig. 5E). Disengagement of the CA domain after the first phosphorylation event could easily allow the other CA domain to approach the second histidine for modification (compare states 1 and 2 in Fig. 7A). Intriguingly, however, CpxA shows a hemiphosphorylated crystal structure in the presence of ATP (33), although the nonphosphorylated histidine seems perfectly poised to react with CA bound ATP in the observed canonical kinase competent arrangement. Based on our thermodynamic results on ShkA, we predict that CpxA behaves similarly, with the equilibrium of the reversible autophosphorylation reaction being on the side of the reactants (ATP, His). This would explain why the substrate and not the product complex was observed, since the products (ADP, P∼His) would react back and ADP would stay in the active site assuming strong crystal lattice constraints.
Our analyses provide mechanistic and kinetic insight into how c-di-GMP competes with a protein–protein interaction to activate an HHK, and how ADP affects the autophosphorylation reaction. For a full mechanistic understanding of HHKs, more investigations are required to decipher the kinetics of the phosphotransfer and autodephosphorylation steps and to unravel the physiological advantage of combining HK core and cognate receiver domain on 1 polypeptide.
Materials and Methods
A detailed description of the materials and methods used in this study is provided in SI Appendix, including the following: Cloning, expression and protein purification, crystallization, data collection and X-ray structure determination, NMR experiments, ITC and FBC assay, SV-AUC, β-Gal transcription assay, radiometric phosphorylation assay, and nucleotide quantification by oIEC. Also given are the thermodynamic equations for reversible autophosphorylation and the LCCE model used for data fitting.
Supplementary Material
Acknowledgments
We thank the beamline staff at the Swiss Light Source in Villigen for expert help in data acquisition; T. Sharpe from the Biophysics facility at the Biozentrum Basel for expert biophysical support; A. Eberhardt and A. Roulier for technical assistance; and H. Pickersgill (Life Science Editors) for editorial assistance. This work was supported by Grant 31003A-166652 of the Swiss National Science Foundation (to T.S).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. E.A.G. is a guest editor invited by the Editorial Board.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6QRJ and 6QRL). Sequence-specific resonance assignments have been submitted to the Biological Magnetic Resonance Data Bank (accession no. 27882).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911427117/-/DCSupplemental.
References
- 1.Gao R., Stock A. M., Catalytically incompetent by design. Structure 17, 1038–1040 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez A. F., Barba-Ostria C., Silva-Jiménez H., Georgellis D., Organization and mode of action of two component system signaling circuits from the various kingdoms of life. Environ. Microbiol. 18, 3210–3226 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Buschiazzo A., Trajtenberg F., Two-component sensing and regulation: How do histidine kinases talk with response regulators at the molecular level? Annu. Rev. Microbiol. 73, 507–528 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Jacob-Dubuisson F., Mechaly A., Betton J.-M., Antoine R., Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 16, 585–593 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Wuichet K., Cantwell B. J., Zhulin I. B., Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13, 219–225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capra E. J., Laub M. T., Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Townsend G. E. 2nd, Raghavan V., Zwir I., Groisman E. A., Intramolecular arrangement of sensor and regulator overcomes relaxed specificity in hybrid two-component systems. Proc. Natl. Acad. Sci. U.S.A. 110, E161–E169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zschiedrich C. P., Keidel V., Szurmant H., Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428, 3752–3775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhate M. P., Molnar K. S., Goulian M., DeGrado W. F., Signal transduction in histidine kinases: Insights from new structures. Structure 23, 981–994 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diensthuber R. P., Bommer M., Gleichmann T., Möglich A., Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21, 1127–1136 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Moore J. O., Hendrickson W. A., An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure 20, 729–741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaczmarczyk A., et al. , Precise transcription timing by a second-messenger drives a bacterial G1/S cell cycle transition. bioRxiv:10.1101/675330 (20 June 2019).
- 13.Jenal U., Reinders A., Lori C., Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Galperin M. Y., Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lori C., et al. , Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523, 236–239 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Dubey B. N., et al. , Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking. Sci. Adv. 2, e1600823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biondi E. G., et al. , Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444, 899–904 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Mechaly A. E., Sassoon N., Betton J.-M., Alzari P. M., Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol. 12, e1001776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., et al. , A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases. Nat. Commun. 8, 2104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weikl T. R., Paul F., Conformational selection in protein binding and function. Protein Sci. 23, 1508–1518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abel S., et al. , Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the caulobacter cell cycle. PLoS Genet. 9, e1003744 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An S.-Q., et al. , Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS Pathog. 10, e1004429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borkovich K. A., Simon M. I., The dynamics of protein phosphorylation in bacterial chemotaxis. Cell 63, 1339–1348 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Greenswag A. R., Muok A., Li X., Crane B. R., Conformational transitions that enable histidine kinase autophosphorylation and receptor array integration. J. Mol. Biol. 427, 3890–3907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimshaw C. E., et al. , Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 37, 1365–1375 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Jiang P., Peliska J. A., Ninfa A. J., Asymmetry in the autophosphorylation of the two-component regulatory system transmitter protein nitrogen regulator II of Escherichia coli. Biochemistry 39, 5057–5065 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Casino P., Miguel-Romero L., Marina A., Visualizing autophosphorylation in histidine kinases. Nat. Commun. 5, 3258 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Noriega C. E., Schmidt R., Gray M. J., Chen L.-L., Stewart V., Autophosphorylation and dephosphorylation by soluble forms of the nitrate-responsive sensors NarX and NarQ from Escherichia coli K-12. J. Bacteriol. 190, 3869–3876 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueno T. B., Johnson R. A., Boon E. M., Optimized assay for the quantification of histidine kinase autophosphorylation. Biochem. Biophys. Res. Commun. 465, 331–337 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Kaczmarczyk A., et al. , Precise transcription timing by a second-messenger drives a bacterial G1/S cell cycle transition. bioRxiv:10.1101/675330 (20 June 2019).
- 31.Trajtenberg F., et al. , Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action. eLife 5, e1001479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Straube R., Analysis of network motifs in cellular regulation: Structural similarities, input-output relations and signal integration. Biosystems 162, 215–232 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Mechaly A. E., et al. , Structural coupling between autokinase and phosphotransferase reactions in a bacterial histidine kinase. Structure 25, 939–944.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Ashenberg O., Keating A. E., Laub M. T., Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. J. Mol. Biol. 425, 1198–1209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Y., et al. , Conformational dynamics of the essential sensor histidine kinase WalK. Acta Crystallogr. D Struct. Biol. 73, 793–803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn R. D., et al. , The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willett J. W., Herrou J., Briegel A., Rotskoff G., Crosson S., Structural asymmetry in a conserved signaling system that regulates division, replication, and virulence of an intracellular pathogen. Proc. Natl. Acad. Sci. U.S.A. 112, E3709–E3718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.