Significance
LEC1 is a central, transcriptional regulator of seed development, because it regulates diverse developmental processes at different stages, including embryo morphogenesis, photosynthesis, hormone biosynthesis and signaling, and the massive accumulation of seed storage macromolecules. We show that LEC1 acts in combination with the seed transcription factors (TFs), AREB3, bZIP67, and ABI3, and that different TF combinations regulate distinct gene sets. We show further that TF binding sites are closely clustered in the genome and contain enriched DNA sequence motifs that are bound by TFs and that distinct DNA motif sets recruit different TF combinations to binding site clusters. Our findings provide insights into the gene regulatory networks that govern seed development.
Keywords: cis-regulatory module, maturation, photosynthesis
Abstract
The LEAFY COTYLEDON1 (LEC1) transcription factor is a central regulator of seed development, because it controls diverse biological programs during seed development, such as embryo morphogenesis, photosynthesis, and seed maturation. To understand how LEC1 regulates different gene sets during development, we explored the possibility that LEC1 acts in combination with other transcription factors. We identified and compared genes that are directly transcriptionally regulated by ABA-RESPONSIVE ELEMENT BINDING PROTEIN3 (AREB3), BASIC LEUCINE ZIPPER67 (bZIP67), and ABA INSENSITIVE3 (ABI3) with those regulated by LEC1. We showed that LEC1 operates with specific sets of transcription factors to regulate different gene sets and, therefore, distinct developmental processes. Thus, LEC1 controls diverse processes through its combinatorial interactions with other transcription factors. DNA binding sites for the transcription factors are closely clustered in genomic regions upstream of target genes, defining cis-regulatory modules that are enriched for DNA sequence motifs that resemble sequences known to be bound by these transcription factors. Moreover, cis-regulatory modules for genes regulated by distinct transcription factor combinations are enriched for different sets of DNA motifs. Expression assays with embryo cells indicate that the enriched DNA motifs are functional cis elements that regulate transcription. Together, the results suggest that combinatorial interactions between LEC1 and other transcription factors are mediated by cis-regulatory modules containing clustered cis elements and by physical interactions that are documented to occur between the transcription factors.
The ability of plants to make seeds has conferred strong selective advantages to the angiosperms that, in part, explain their dominance within the plant kingdom (1). The seed habit requires that a novel, biphasic mode of development occurs at the earliest stage of the sporophytic life cycle. During the early, morphogenesis phase, the embryo and endosperm initially undergo regional specification into functional domains. The embryo develops further with the establishment of the shoot–root axis and differentiation of embryonic tissue and organ systems (2). Photosynthesis is initiated later during the morphogenesis phase, often in both the embryo and endosperm (3). During the maturation phase which follows morphogenesis, morphogenetic processes in the embryo are arrested; storage macromolecules, particularly proteins and lipids, accumulate and are stored; the embryo becomes desiccation tolerant; and seed germination is actively inhibited. The maturation phase is unique to seed plants, suggesting that this phase has been inserted into a continuous period of embryonic followed by postembryonic morphogenesis, characteristic of nonseed plants (4, 5). Relatively little is known of the gene regulatory networks that have enabled the maturation phase to be integrated into the angiosperm life cycle.
LEC1 is a central regulator of seed development that controls distinct developmental processes at different stages of seed development (reviewed in ref. 6). Analyses of loss- and gain-of-function mutants showed that LEC1 is a major regulator of the maturation phase that is required for storage macromolecule accumulation, the acquisition of desiccation tolerance, and germination inhibition during seed development (7, 8). However, LEC1 also appears to function during the morphogenesis phase. LEC1 mRNA is detected in the zygote within 24 h after fertilization, loss-of-function mutations indicate that LEC1 is required to maintain embryonic suspensor and cotyledon identities, and LEC1 is also involved in regulating genes that underlie photosynthesis and chloroplast biogenesis (9, 10). It is not known how LEC1 is able to regulate the diverse developmental processes that occur during both the morphogenesis and maturation phases.
LEC1 is an atypical transcription factor (TF) subunit: a NF-YB subunit whose canonical role is to interact with NF-YC and NF-YA subunits to form a NF-Y TF that binds CCAAT DNA sequences (9, 11, 12). The LEC1-type NF-YB subunit is found only in plants, and it confers seed-specific functions (13). LEC1 also interacts physically with other TFs to regulate a variety of developmental processes (reviewed in ref. 6).
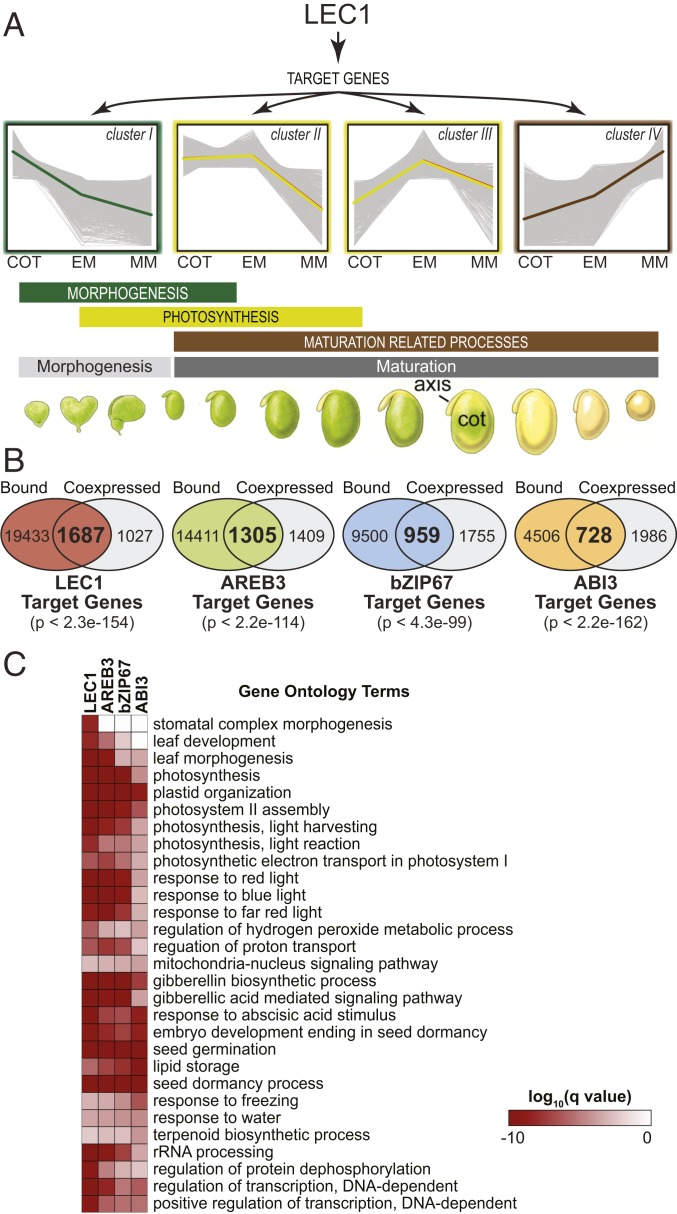
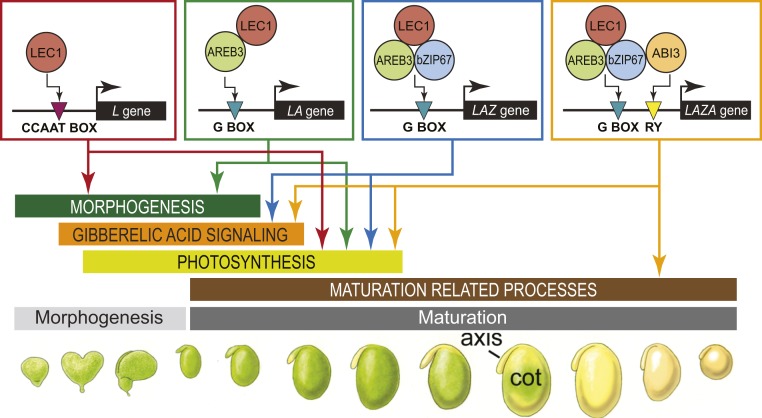
We showed previously that LEC1 sequentially transcriptionally regulates distinct gene sets at different stages of seed development in Arabidopsis and soybean (10). As summarized in Fig. 1A, LEC1 regulates genes involved in growth and morphogenesis, photosynthesis, and maturation during the morphogenesis, transition from morphogenesis to maturation, and maturation phases, respectively. We showed further that LEC1 genomic binding sites are enriched for different DNA sequence motifs, the CCAAT, G box, RY, and BPC1 motifs. Different LEC1 target gene sets were enriched for distinct combinations of these DNA motifs, opening the possibility that LEC1 interacts with other TFs to regulate different gene sets.
Fig. 1.
Identification of LEC1, AREB3, bZIP67, and ABI3 target genes in soybean early maturation embryos. (A) Overview of LEC1’s role in controlling distinct gene sets and developmental processes at different stages of seed development. (B) Target genes directly regulated by LEC1, AREB3, bZIP67, and ABI3 in soybean embryos at the EM stage. Venn diagrams show the overlap between bound genes (colored) and coexpressed genes (gray) for LEC1, AREB3, bZIP67, and ABI3. Statistical significance of the overlap between bound genes and coexpressed genes is indicated (hypergeometric distribution). (C) Heatmap showing the q value significance of GO terms for LEC1, AREB3, bZIP67, and ABI3 target genes. The GO terms listed are the top 5 enriched biological process GO terms for each TF. A comprehensive list of overrepresented GO terms is given in Dataset S1.
In Arabidopsis, substantial information is available about the involvement of LEC1 and other TFs, including LEC1-LIKE, LEC2, ABA INSENSITIVE3 (ABI3), FUSCA3 (FUS3), WRINKLED1, MYB115/118, ABI4, ABI5, AGAMOUS-LIKE15, and a number of BASIC LEUCINE ZIPPER (bZIP) TFs, in regulating the maturation phase, and genetic studies generally place LEC1 atop the regulatory hierarchy (reviewed in refs. 14–17). The LEC1-NF-YC dimer interacts physically with the bZIP67 TF and binds with a G box-like but not a CCAAT DNA motif to activate maturation genes, such as the CRUCIFERIN C, FATTY ACID DESATURASE3 (FAD3), and DELAY OF GERMINATION1 (DOG1) (18–20). LEC1 also operates synergistically with LEC2 and ABI3, 2 B3 domain TFs that bind RY-like motifs, to promote maturation gene expression (21–24). LEC2 interacts physically with LEC1 through its B2 domain, but no direct physical interactions between LEC1 and ABI3 have been reported (22).
Here, we show that LEC1 regulates distinct developmental processes at different stages by acting combinatorially with other TFs, specifically bZIP67, ABA-RESPONSIVE ELEMENT BINDING PROTEIN3 (AREB3), a TF closely related to bZIP67, and ABI3. We showed that 1) LEC1 alone and LEC1 in combination with AREB3 primarily regulate genes involved in morphogenesis; 2) LEC1 and AREB3, LEC1, AREB3, and bZIP67, and LEC1, AREB3, bZIP67, and ABI3 regulate genes involved in photosynthesis; and 3) all 4 TFs regulate maturation genes. We also show that the binding sites for these TFs are closely clustered in the genome, and they are enriched for DNA motifs that correspond to annotated cis elements known to be bound by the 4 TFs. These results suggest that LEC1 functions combinatorially with AREB3, bZIP67, and ABI3 to regulate distinct gene sets and diverse developmental processes.
Results
Identification of AREB3, bZIP67, and ABI3 Target Genes in Developing Soybean Embryos.
We hypothesized that LEC1 may act in combination with other TFs to regulate distinct gene sets at different stages of development, in part, because LEC1 has been shown to interact with a number of other TFs (reviewed in ref. 6). Based on their functions in Arabidopsis, we focused on 3 TFs: 1) bZIP67, a TF that interacts physically with the LEC1-NF-YC dimer to regulate maturation genes (19, 20); 2) AREB3, a TF closely related to and partially redundant functionally with bZIP67 that is expressed earlier in seed development than bZIP67 (25); and 3) the B3 domain TF, ABI3, a maturation regulator that interacts with bZIP TFs and, by extension, potentially with LEC1 (26–28).
We identified target genes directly regulated by AREB3, bZIP67, and ABI3 in soybean embryos at the early maturation (EM) stage (23 d after pollination) that corresponds to the transition from morphogenesis to maturation phases to compare the TFs’ functions with LEC1. We used the chromatin immunoprecipitation–DNA sequencing (ChIP-Seq) strategy described by Pelletier et al. (10) to identify genes bound by AREB3-1 (Glyma.04G124200), AREB3-2 (Glyma.06G314400), bZIP67 (Glyma.13G317000), ABI3-1 (Glyma.08G357600), and ABI3-2 (Glyma.18G176100) (29, 30). The AREB3-1 and AREB3-2 homeologs and ABI3-1 and ABI3-2 homeologs are recognized by the AREB3 and ABI3 antibodies, respectively. Binding sites for these TFs were located primarily at the transcription start site, as we found previously for LEC1, and each TF bound the 1-kb upstream region of between 21,120 and 5,234 genes (SI Appendix, Fig. S1 and Dataset S1). Experiments with antibodies against 2 different peptides each from AREB3, bZIP67, and ABI3 confirmed the specificity of the ChIP experiments, and data analysis followed ENCODE guidelines (Dataset S2) (31–33). Our data analysis methods differed slightly from that reported previously; therefore, we also present results for the LEC1-1 (Glyma.07G268100) and LEC1-2 (Glyma.17G005600) homeologs using previously reported primary data (10, 34).
Because only a fraction of the genes bound by a TF are transcriptionally regulated by that TF (35), we defined target genes regulated by these TFs as those that are both bound and coexpressed with the TF. All 4 of the TFs are expressed predominantly in embryos (SI Appendix, Fig. S2), and we used the Harada-Goldberg Soybean Seed Development Laser Capture Microdissection RNA-Seq Dataset (GEO accessions, GSE57606, GSE46096, and GSE99109) (36–38) to identify coexpressed genes as those whose mRNA levels accumulated at a 5-fold or higher level in embryo subregions compared with seed coat subregions (q < 0.01). As summarized in Fig. 1B, we identified 1,687, 1,305, 959, and 728 target genes, respectively, for LEC1, AREB3, bZIP67, and ABI3 and showed that the overrepresentation of bound and coexpressed genes was statistically significant (P < 2.3 × 10−154, P < 2.2 × 10−114, P < 4.3 × 10−99, and P < 2.2 × 10−162, respectively, Dataset S1). These TF target gene numbers are within the range reported for other plant TFs (39).
Gene Ontology (GO) representation analysis indicated that there was extensive overlap in the biological functions of the 4 TFs (Fig. 1C and Dataset S1), particularly processes related to morphogenesis, photosynthesis, GA biosynthesis and signaling, lipid storage, and seed dormancy. The results indicate that AREB3, bZIP67, and ABI3 TFs regulate developmental processes that are closely related to those controlled by LEC1.
LEC1 Operates in Combination with AREB3, bZIP67, and ABI3 to Regulate the Expression of Genes Involved in Distinct Developmental Processes in Soybean Embryos.
Deciphering combinatorial interactions among the 4 transcription factors.
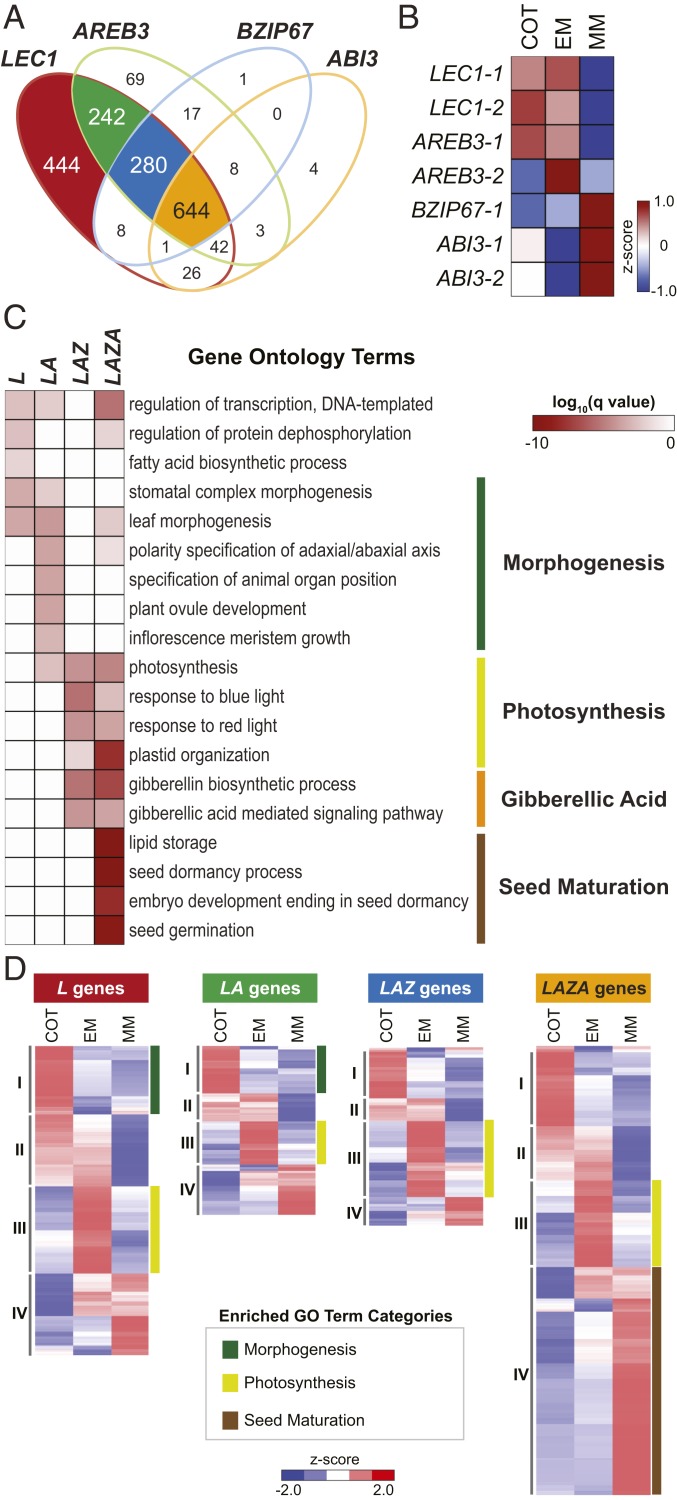
Because LEC1, AREB3, bZIP67, and ABI3 regulate genes involved in similar biological processes, we asked if they acted in coordination to regulate seed gene transcription by comparing their target genes. Fig. 2A shows that there was significant overlap in the target genes regulated by the 4 TFs. Of 1,687 LEC1 target genes, 1,243 (74%) were also targeted by at least 1 of the other TFs (Dataset S3). The vast majority of target genes were grouped into 4 categories: 1) those regulated by LEC1 alone (L genes); 2) LEC1 and AREB3 (LA genes); 3) LEC1, AREB3, and bZIP67 (LAZ genes); and 4) all 4 TFs (LAZA genes), with the largest number of target genes falling into the latter category. Thus, LEC1 appears to regulate gene transcription primarily in combination with AREB3, bZIP67, and ABI3.
Fig. 2.
Overlap between LEC1, AREB3, bZIP67, and ABI3 target genes indicate combinatorial interactions among transcription factors. (A) Venn diagram showing the overlap between LEC1, AREB3, bZIP67, and ABI3 target genes. Four major groups were identified: genes targeted by LEC1 (L genes, red); genes targeted by LEC1 and AREB3 (LA genes, green); genes targeted by LEC1, AREB3, and bZIP67 (LAZ genes, blue); and genes targeted by LEC1, AREB3, bZIP67, and ABI3 (LAZA genes, orange). Gene lists are given in Dataset S3. (B) LEC1, AREB3, bZIP67, and ABI3 mRNA accumulation in soybean embryos at the COT, EM, and MM stages. mRNA levels were obtained from the Harada Embryo mRNA-Seq Dataset (GEO accession no. GSE99571, ref. 79). (C) Heatmap shows the q value significance of GO terms for L, LA, LAZ, and LAZA gene sets. GO terms listed are the top 5 most enriched biological process GO terms for each gene set. A comprehensive list of overrepresented GO terms is given in Dataset S1. (D) Hierarchical clustering of L, LA, LAZ, and LAZA gene sets. Heatmaps show relative embryo mRNA levels at the COT, EM, and MM stages. The major enriched developmental processes associated with each cluster are indicated. Gene lists and GO term enrichments for each cluster are given in Dataset S3.
Combinatorial control of developmental processes.
We obtained insight into the biological processes regulated by different combinations of TFs by performing GO representation analysis on the different target gene sets. We were surprised to find that target genes regulated by different TF combinations were highly enriched for distinct GO term sets (Fig. 2C and Dataset S3). Specifically, 1) L and LA genes were most significantly overrepresented for GO terms related to morphogenesis, such as leaf morphogenesis, stomatal complex morphogenesis, polarity specification of adaxial/abaxial axis, and specification of organ position; 2) LA, LAZ, and LAZA genes were highly enriched for GO terms related to photosynthesis; 3) LAZ and LAZA genes were enriched for gibberellic acid (GA) biosynthesis and signaling; and 4) LAZA genes were overrepresented for GO terms related to maturation.
We asked if the accumulation of AREB3, bZIP67, and ABI3 mRNAs could explain LEC1’s ability to control the onset of the developmental processes temporally. Fig. 2B shows that LEC1, AREB3, and ABI3 mRNAs accumulate early in embryo development, whereas bZIP67 mRNA accumulates primarily at the midmaturation (MM, 40 to 45 d after pollination) stage. These results suggest that the TFs’ mRNA accumulation patterns alone do not explain the temporal regulation of biological processes.
During embryo development, morphogenetic events are largely initiated before the onset of photosynthesis which, in turn, is followed by the maturation phase. To determine if the different TF combinations underlie the temporal regulation of these biological processes, we used clustering analysis to identify L, LA, LAZ, and LAZA mRNAs that accumulate at different stages of seed development. As shown in Fig. 2D, each target gene set exhibited 4 different expression patterns, with clusters I, II, III, and IV containing mRNAs that accumulated primarily at the 1) cotyledon (COT, 15 d after pollination) stage; 2) COT and EM stages; 3) EM stage; and 4) MM stage, respectively. L and LA genes were fairly evenly distributed among the 4 different clusters, whereas LAZ and LAZA genes were enriched in cluster III and cluster IV, respectively. We found that genes involved in morphogenesis, photosynthesis, and maturation were enriched in particular clusters: 1) L and LA genes involved in morphogenesis; 2) L, LA, LAZ, and LAZA genes involved in photosynthesis; and 3) LAZA genes involved in maturation were enriched in cluster I, cluster III, and cluster IV, respectively. These results emphasize that the genes that underlie specific developmental processes during seed development are precisely regulated temporally, regardless of which TF sets are involved in their regulation.
We determined which TF combinations regulate gene sets previously defined to be involved in either maturation or photosynthesis to validate the GO term enrichment analysis (10). Virtually all of the maturation genes bound by 1 of the TFs were bound by all 4 TFs, and orthologs of most of these genes were down-regulated in Arabidopsis lec1 and/or abi3 mutants (SI Appendix, Fig. S3). Genes involved in the light reactions of photosynthesis were bound by between 1 and 4 of the TFs, and many were affected by the Arabidopsis lec1 mutation (SI Appendix, Fig. S4). These results emphasize the importance of LEC1 and/or ABI3 in controlling maturation and photosynthesis genes. Additionally, L and LA genes involved in morphogenesis included known regulators of morphogenetic processes, PHABULOSA, ASYMMETRIC LEAVES1, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN13, and TCP1. Among the genes related to GA biosynthesis and signaling, many of the LAZ genes encode proteins that promote GA synthesis, such as GA REQUIRING1 (GA1), GA3, GA4, and GA20 OXIDASE (GA20OX), and GA signaling, such as SLEEPY2 and GIBBERELLIN-INSENSITIVE DWARF1. By contrast, proteins encoded by the LAZA genes, REPRESSOR OF GA1-3-LIKE2, PHYTOCHROME INTERACTING FACTOR3 (PIF3), and PIF3-LIKE5, negatively affect GA signaling, although others promote GA synthesis, such as GA20OX. Thus, LEC1 may act in both positive and negative feedback loops to control GA responses during embryo development.
Physical interactions between the 4 transcription factors.
Combinatorial interactions among the TFs could indicate that they interact physically. In Arabidopsis, several of the 4 TFs have been shown to form complexes (18–20, 26–28, 40, 41). We obtained evidence indicating physical interactions between the soybean orthologs of LEC1 and bZIP67, LEC1 and AREB3, AREB3 and bZIP67, and bZIP67 and ABI3, as occurs in Arabidopsis (SI Appendix, Fig. S5). These results may indicate that the LA, LAZ, and LAZA genes are regulated by TF complexes.
LEC1, AREB3, bZIP67, and ABI3 Binding Sites Are Clustered and Contain Distinct Sets of DNA Sequence Motifs.
Identification of cis-regulatory module.
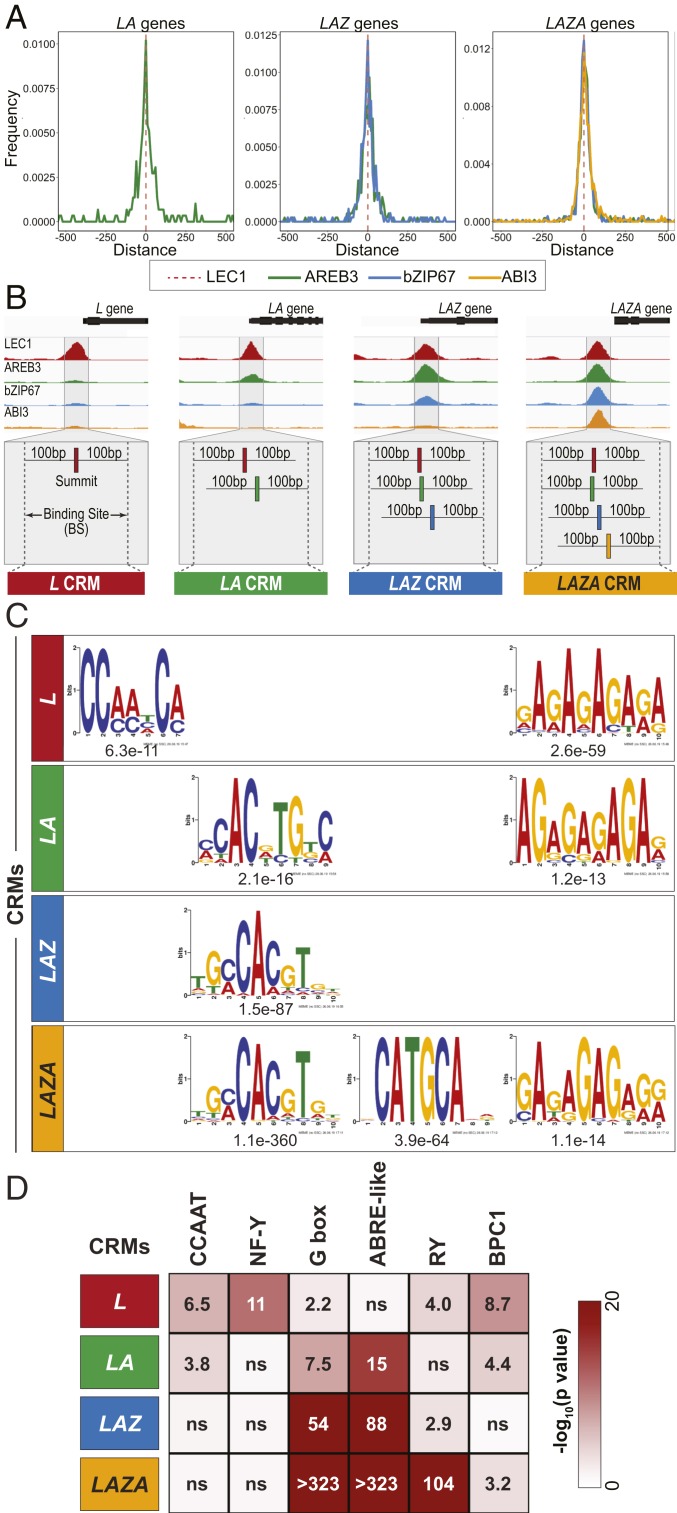
We determined the organization of LEC1, AREB3, bZIP67, and ABI3 binding sites in the upstream regions of target genes to obtain insight into the mechanisms by which LEC1 works in combination with the other TFs to regulate different gene sets. We plotted the distance between the summit of the LEC1 ChIP-Seq peak, which approximates the TF binding site, and the ChIP-Seq peak summits of AREB3, bZIP67, and ABI3 (42). As shown in Fig. 3A, the TF binding sites in LA, LAZ, and LAZA target genes were in very close proximity to each other. Measurements showed that the median distance between peak summits for the different TFs was between 25 and 53 bp, indicating that the binding sites are clustered. We hypothesized that the binding site clusters represent cis-regulatory modules (CRMs) or high occupancy target regions, genomic regions at which multiple, distinct TFs bind productively to regulate gene transcription (43–45). Therefore, we designated these binding site clusters as CRMs and used published criteria (46) to operationally define CRMs as genomic regions whose boundaries are extended by 100 bp on each side of the terminal ChIP peak summits within a cluster, although we also apply this term to L genes with single binding sites (Fig. 3B). Median CRM sizes for L, LA, LAZ, and LAZA genes were 201, 227, 235, and 240 bp, respectively.
Fig. 3.
Clustered binding sites for LEC1, AREB3, bZIP67, and ABI3 define cis-regulatory modules. (A) Distance between the positions of the ChIP peak summits of AREB3 (green), bZIP67 (blue), and ABI3 (yellow) and the LEC1 ChIP peak summit (dotted red line) for LA, LAZ, and LAZA genes. (B) Diagrammatic representation of the strategy used to define CRMs. (B, Upper) Genome browser representation of ChIP-Seq reads for LEC1 (red), AREB3 (green), bZIP67 (blue), and ABI3 (yellow) in the upstream genomic regions of L (Glyma.13G031500), LA (Glyma.08G106400), LAZ (Glyma.01G057700), and LAZA (Glyma.01G186200) genes. (B, Lower) CRMs are defined as the genomic region whose boundaries extend 100 bp beyond the terminal ChIP peak summits of a cluster. LA, LAZ, and LAZA CRMs were computed by merging the binding sites of 1) LEC1 and AREB3; 2) LEC1, AREB3, and bZIP67; and 3) LEC1, AREB3, bZIP67, and ABI3, respectively. L CRMs were defined as LEC1 binding sites of L genes. CRM genomic coordinates are listed in Dataset S4. (C) Position weight matrices of DNA sequence motifs discovered de novo in L, LA, LAZ, and LAZA CRMs and their relative enrichment as indicated by their associated E values. De novo discovered motifs and DNA motifs in the Arabidopsis DAP-Seq (67) or Human HOCOMOCOv11 (80) databases most closely related to the discovered motifs are listed in Dataset S4. (D) DNA motif enrichment in CRM regions. Heatmaps depict the statistical significance of the enrichment of annotated DNA motifs most closely related to de novo discovered motifs in L, LA, LAZ, and LAZA CRM regions, relative to the normal distribution of a population of randomly generated regions. Bonferroni-adjusted P values are listed, with a significance threshold of 0.01, with ns denoting no significant difference. Frequencies at which DNA motifs were identified in CRMs are shown in SI Appendix, Fig. S7.
Enriched DNA motifs in cis-regulatory modules.
To investigate how different TF combinations are recruited to target genes, we identified overrepresented DNA sequence motifs within the CRMs that may serve as TF binding sites. We used de novo DNA motif discovery algorithms to identify the enriched DNA sequence motifs in L, LA, LAZ, and LAZA CRMs that are shown as position weight matrices in Fig. 3C and Dataset S4. L CRMs contained overrepresented CCAAT-like motifs, consistent with the observation that LEC1 is an atypical subunit of the NF-Y complex that binds CCAAT DNA sequences (11, 12). LA and LAZ CRMs were enriched for G box-like motifs, such as G box- and ABRE-like elements, although the specific position weight matrices identified in LA and LAZ CRMs differed. bZIP TFs, such as bZIP67 and AREB3, bind G-box motifs (47, 48). LAZA CRMs contained overrepresented G box-like motifs that were similar to those found in LAZ CRMs, and RY-like motifs. RY motifs are bound by B3 domain transcription factors, such as ABI3 (49). CCAAT-like motifs were not enriched in LA, LAZ, and LAZA CRMs even though LEC1 was bound at these CRMs. CRMs for the L, LA, and LAZA target gene sets were also enriched for BPC1 motifs that are bound by BASIC PENTACYSTEIN TFs that act as transcriptional activators and repressors (50, 51).
To validate the DNA motif discovery analyses, we conducted find individual motif occurrences-receiver operating characteristics-area under the curve (FIMO ROC-AUC) and Homer hypergeometric analyses. The former analysis measures the extent to which DNA motifs in CRMs exhibit similarities to the de novo discovered position weight matrices, whereas the latter measures the percent of CRMs that contain DNA motifs that are exact matches with annotated cis elements most closely related to the discovered position weight matrices. Both analyses provided independent evidence in support of the DNA motif discovery results (Fig. 3D and SI Appendix, Figs. S6 and S7). Together, these findings indicate that the binding sites of different TFs are clustered in the upstream regions of target genes, and they suggest that DNA sequence motifs may represent functional cis elements that recruit TFs to CRMs. The number and distribution of DNA motifs in CRMs varied greatly, even among those bound by the same set of TFs, indicating that there is no easily discernible arrangement of DNA motifs for L, LA, LAZ, and LAZA genes (SI Appendix, Fig. S8).
Binding Site Clusters Are Functional cis-Regulatory Modules.
Analysis of cis-regulatory module function.
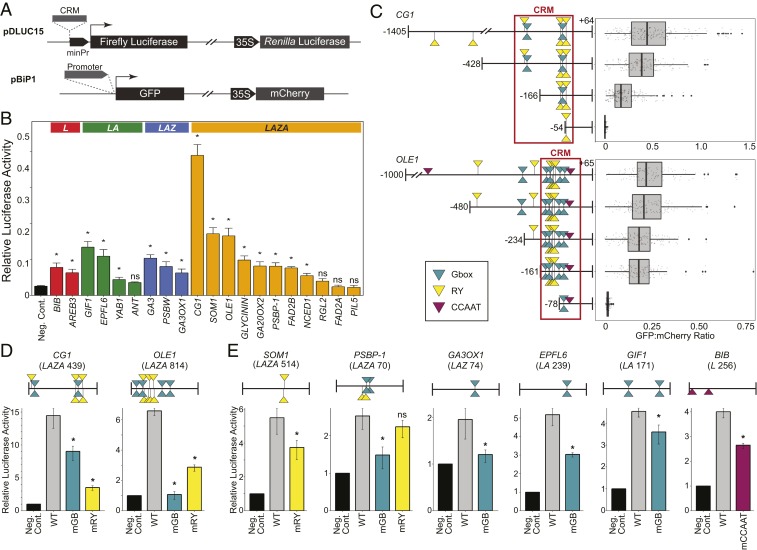
To test the hypothesis that the clustered TF binding sites represent functional CRMs, we determined whether 20 CRMs were sufficient to activate transcription. Functional cis elements are generally within 50 bp of ChIP peak summits and, therefore, they should be contained within the CRMs (46). Each CRM was inserted upstream of a 35S minimal promoter fused with the FIREFLY LUCIFERASE gene in a plasmid that also contained a 35S:RENILLA LUCIFERASE gene, as diagrammed in Fig. 4A. The CRM activity of the constructs was assessed using transient assays with cotyledon protoplasts from EM-stage embryos. Protoplasts have been used extensively to investigate developmental gene expression (reviewed in ref. 52), consistent with our control experiments showing that seed-specific promoters were active in embryo cotyledon but not in leaf protoplasts (SI Appendix, Fig. S9). Transfection of the CRM constructs into embryo cotyledon protoplasts demonstrated that 16 of 20 CRMs were sufficient to induce reporter gene activity at a significantly higher level than the negative control lacking a CRM insert (Fig. 4B). Results of these gain-of-function experiments suggest that L, LA, LAZ, and LAZA CRMs are functional CRMs containing cis elements that are sufficient to activate the transcription of LEC1 target genes during seed development.
Fig. 4.
Functional analysis of cis-regulatory modules and cis elements in soybean embryo cotyledon cells. (A, Top) Schematic diagram of the dual-luciferase vector used for the CRM assays which contains a 35S minimal promoter fused with a FIREFLY LUCIFERASE gene and a 35S:RENILLA LUCIFERASE gene. CRMs were inserted immediately upstream of the 35S minimal promoter. (A, Bottom) The vector containing promoterless GFP and 35S:mCHERRY genes were used for the 5′-deletion assays. The 5′-upstream regions were fused with the GFP gene. (B) CRMs effect on minimal promoter activity in soybean embryo cotyledon protoplasts at the EM stage as measured by the ratio of firefly to Renilla luciferase activities. Average value of 3 assays with SEs are plotted. Asterisks denote statistically significant differences relative to the negative control (Neg. Cont., no CRM insert), whereas ns indicates no significant difference (P < 0.05, paired, one-tailed t tests). (C) The 5′-deletion analyses of CG1 and OLE1 gene upstream regions. Diagrams of CG1 and OLE1 5′ upstream regions that were fused with the GFP gene and used for embryo cotyledon cell transient assays. Teal, yellow, and magenta symbols indicate the positions of G box-like, RY-like, and CCAAT-like DNA motifs, respectively, with a FIMO score greater than 2.4. Positions relative to the transcription start site are indicated. Box plots show the ratios of GFP to mCherry activities for at least 150 transfected protoplasts. (D) Regulatory activities of CG1 and OLE1 CRMs containing mutations in all detectable G box-like (mGB) or RY-like (mRY) DNA motifs in embryo cotyledon transient assays. (D, Upper) Diagrams of CG1 and OLE1 CRMs with motif positions indicated. (D, Lower) The ratios of firefly to Renilla luciferase activities with SEs are shown (n = 3). Asterisks indicate significant activity ratio differences for mutant relative to wild-type (WT) CRMs (P < 0.05, one-tailed t tests). (E) Regulatory activities of LAZA (SOM1 and PSBP-1), LAZ (GA3OX1), LA (EPFL6 and GIF1), and L (BIB) CRMs with mutations in G box-like (mGB), RY-like (mRY), or CCAAT-like (mCCAAT) DNA motifs in embryo cotyledon cell transient assays. Data are presented as in D.
Defining cis elements.
Because most of the tested CRMs activated transcription, we next asked if the overrepresented DNA sequence motifs are functional cis elements. We focused initially on 2 LAZA genes encoding the α′ subunit of the storage protein β-conglycinin (CG-1, Glyma.10G246300), and the lipid body protein oleosin1 (OLE1, Glyma.20G196600). A 5′-deletion series of the upstream region of each gene that was fused with the GREEN FLUORESCENCE PROTEIN (GFP) reporter gene in a plasmid that also contained 35S:mCHERRY was generated (Fig. 4A). The CG-1 CRM contained 4 G box-like and 5 RY-like motifs. As shown in Fig. 4C, deletion of the 2 5′-most G box-like and 1 RY-like motif caused a significant reduction in promoter activity relative to wild type, whereas deletion of all but 2 RY-like motifs eliminated detectable promoter activity. For the OLE1 CRM which contains 8 G box-like and 7 RY-like motifs, deletion of all G box-like and RY-like motifs upstream of the CRM caused only a modest reduction in promoter activity, but deletion of 6 G box-like and all 7 RY-like motifs within the CRM essentially eliminated detectable promoter activity. These results indicate that the enriched DNA motifs are required to activate transcription of the LAZA genes.
To test more stringently the hypothesis that the enriched DNA motifs are involved in controlling LAZA gene transcription, we specifically mutagenized the G box-like and RY-like motifs in the CG-1 and OLE1 CRMs and assessed their ability to activate transcription in embryo protoplasts using the dual luciferase assay. Both of these CRMs were sufficient to activate the minimal promoter in transient assays in embryo cotyledon protoplasts (Fig. 4A). Fig. 4D shows that mutating all of the G box-like or RY-like motifs in the CG1 CRM caused a reduction in promoter activity relative to wild type, with the RY-like motif mutations more severely compromising promoter activity. Mutating the OLE1 CRM motifs also caused a reduction in promoter activity, although mutations of the G box-like motifs more strongly diminished promoter activity than did mutations in RY-like motifs. The results indicate that G box-like and RY-like motifs within the CRMs play key roles in controlling LAZA gene transcription.
We also determined if overrepresented motifs in the CRMs of 2 additional LAZA genes, 1 LAZ gene, 2 LA genes, and 1 L gene were involved in controlling promoter activity. As shown in Fig. 4E, mutations in RY-like motifs caused a reduction in promoter activity relative to wild type of 1 LAZA gene, SOMNUS1 (SOM1, Glyma.12G205700), but RY-like motif mutations did not significantly alter the promoter activity mediated by the PHOTOSYSTEM II SUBUNIT P-1 (PSBP1, Glyma.02G282500) CRM. Rather, mutation of the G box-like motifs in the PSBP1 CRM caused promoter activity reduction. Promoter activity was reduced relative to wild type in constructs with mutations of G box-like motifs in the CRMs of the LAZ gene, GA3 OXIDASE 1 (GA3OX1, Glyma.04G071000) and the LA genes, EPIDERMAL PATTERNING FACTOR-LIKE 6 (EPFL6, Glyma.14G203100) and GRF1-INTERACTING FACTOR 1 (GIF1, Glyma.10G164100), and of the CCAAT motif in the L gene, BALDIBIS (BIB1, Glyma.12G070300). Although the motif mutations resulted in a significant decrease in promoter activity relative to wild type, in most cases they did not reduce activity to the level of constructs lacking a CRM, suggesting that other cis elements are present in the CRMs. Together, our results suggest that CRMs contain enriched DNA sequence motifs that act as functional cis elements that are bound by specific TF combinations.
Discussion
LEC1 Regulates Distinct Gene Sets at Different Developmental Stages by Interacting with Specific Combinations of Transcription Factors.
The rationale for this study is to determine how LEC1, a central regulator of seed development, is able to regulate diverse developmental processes at different stages of seed development. We have shown that LEC1 interacts with different combinations of the TFs AREB3, bZIP67, and ABI3 to regulate distinct gene sets, and it is likely that other TFs also interact with these TFs during seed development (21). Gene expression is often dictated by combinatorial interactions among functionally active and distinct TFs in plant and animal cells (reviewed in refs. 53–55). For example, DNA binding experiments with 27 different Arabidopsis TFs showed that 63% of the target genes are bound by more than 1 TF, with 8% being bound by 8 or more TFs (39). LEC1 is a NF-Y TF subunit, and a comprehensive study of 154 TFs in human cells showed that 48 operate combinatorially with NF-Y. Thus, NF-Y TFs and their subunits may be particularly likely to coordinate their activities with other TFs.
A key finding, summarized in Fig. 5, is that LEC1 interacts with AREB3, bZIP67, and ABI3 in different combinations to regulate distinct developmental processes: 1) L and LA genes; 2) LA, LAZ, and LAZA genes; 3) LAZ and LAZA genes; and 4) LAZA genes are primarily involved in morphogenesis, photosynthesis, GA synthesis and signaling, and maturation, respectively. Our finding that LEC1, bZIP67, and ABI3 are involved in regulating maturation genes is consistent with other reports showing that LEC1 acts synergistically with bZIP67 to promote the expression of CRUCIFERIN C, FAD3, and DOG1 and that LEC1 and ABI3 operate synergistically to regulate the OLE1 gene in Arabidopsis (18–21).
Fig. 5.
Model for LEC1 combinatorial interactions with AREB3, bZIP67, and ABI3 in the control of soybean seed development. Combinatorial interactions of LEC1 with AREB3, bZIP67, and ABI3 TFs account for LEC1’s ability to regulate different gene sets and diverse developmental processes during seed development. LEC1 interacts with NF-YC and NF-YA subunits to form a NF-Y TF that binds CCAAT motifs of genes involved in morphogenesis and photosynthesis. We propose that a LEC1-NF-YC dimer binds AREB3 or an AREB3-bZIP67 heterodimer to form complexes that bind G box-like motifs of genes involved in 1) morphogenesis and photosynthesis and 2) photosynthesis and GA signaling, respectively. We further propose that an AREB3-bZIP67 heterodimer binds both the LEC1-NF-YC dimer and ABI3 to form a complex that binds G box-like and RY-like motifs in genes involved with photosynthesis, GA signaling, and seed maturation.
Our studies establish that LEC1, AREB3, bZIP67, and ABI3 act combinatorially to globally regulate genes involved in maturation and other developmental processes (Fig. 2). Moreover, our studies also provide primary evidence that photosynthetic gene sets are regulated by LEC1 in combination with AREB3 and/or bZIP67 during seed development, as we suggested previously (10). These results indicate that transitions in biological processes during seed development are mediated by qualitative changes in the TF combinations in a cell, as shown for other developmental systems (reviewed in ref. 54).
Combinatorial interactions among LEC1, AREB3, bZIP67, and ABI3 are likely to result, in part, from their ability to interact physically and form a complex. In Arabidopsis, the LEC1-NF-YC2 dimer binds with bZIP67 (18–20). Because bZIP67 and AREB3 are closely related and they heterodimerize, the soybean homologs of both TFs are also likely to bind the LEC1-NF-YC dimer and form complexes (SI Appendix, Fig. S5; refs. 40 and 41). Although LEC1 and ABI3 operate synergistically to regulate maturation genes, no direct physical interaction between the TFs has been reported. Because Arabidopsis ABI3 binds bZIP TFs, AREB3 and/or bZIP67 are likely to serve as a bridge linking LEC1 and ABI3 in a complex (26–28). Given our findings indicating that soybean and Arabidopsis homologs of LEC1, AREB3, bZIP67, and ABI3 interact similarly (SI Appendix, Fig. S5), these results suggest that the TFs form complexes that regulate distinct gene sets during seed development.
Clustering of L, LA, LAZ, and LAZA mRNAs showed that genes involved in morphogenesis, photosynthesis, and maturation are expressed predominately at the COT, EM, and MM stages, respectively, regardless of which TF combination is involved in their regulation (Fig. 2D). This finding suggests strongly that combinatorial interactions among LEC1, AREB3, bZIP67, and ABI3 permit distinct developmental processes to be rigidly regulated temporally during seed development. Others have shown that a given combination of transcription factors can generate multiple and distinct spatial and temporal expression patterns (46, 56, 57).
The major temporal shift during seed development is the transition from the morphogenesis to maturation phase, and ABI3 and bZIP67 appears to be the key TFs associated with this transition. Among the 4 TFs, ABI3 is uniquely associated with the activation of maturation genes, and others have established the importance of ABI3 in controlling maturation in Arabidopsis (reviewed in refs. 14–17). Based on the bZIP67 mRNA accumulation pattern (Fig. 2B), bZIP67 may trigger the onset of the maturation phase although it is difficult to predict bZIP TF activity, because it is regulated posttranslationally (41). Thus, developmental function reflects qualitative changes in the combination of TFs that are present in a cell.
LEC1’s combinatorial interactions with AREB3, bZIP67, and ABI3 suggest that it may act as a pioneer TF. Pioneer TFs are able to bind DNA binding sequences associated with nucleosomes or compacted chromatin and increase chromatin accessibility, thereby promoting the recruitment of other TFs to the target sites (58, 59). LEC1 was recently characterized as a pioneer TF that reprograms the negative regulator of flowering, FLC, from a silenced to an active state during embryogenesis (60), and NF-Ys act as pioneer TFs in human cells (61, 62). Because LEC1 in combination with AREB3, bZIP67, and ABI3 does not appear to act as a NF-Y TF and bind CCAAT, it will be important to determine if LEC1 in association with other TFs has pioneer TF activity.
LEC1, AREB3, bZIP67, and ABI3 Bind Functional cis Elements to Control Specific Developmental Programs during Seed Development.
The striking arrangement of TF binding sites in the upstream regions of LEC1, AREB3, bZIP67, and ABI3 target genes explains, in part, the combinatorial interactions that occur between LEC1 and the other TFs (Fig. 3). We defined CRMs based on the close proximity of the binding sites of between 2 and 4 TFs, although we also used the term to describe genes bound only by LEC1.
A defining characteristic of CRMs is that they contain cis elements, short DNA sequences that, when recognized and bound by a TF, lead to transcriptional changes of the associated gene (43–45). Although not comprehensive, our results provide strong evidence that the CRMs contain functional cis elements. Gain-of-function experiments showed that 16 of 20 CRMs are sufficient to activate a minimal promoter, indicating that the CRMs contain functional cis elements (Fig. 4). Studies with human stem cells showed that between 9 and 25% of TF binding sites, defined as ChIP peaks, were sufficient to activate a minimal promoter (63). Consistent with these observations, others have shown that mutation of 4 soybean maturation genes, CG1, GLYCININ (Glyma.03G163500), KUNITZ TRYPSIN INHIBITOR1 (Glyma.01G095000), and LECTIN1 (Glyma.02G012600), in regions defined as CRMs in this study reduced transgene expression levels in developing seeds (64–66). These findings also demonstrate that results obtained with transgenic plants are reproduced in embryo protoplast transient assays.
The enriched DNA motifs in CRMs, CCAAT-like, G box-like, and RY-like motifs, correspond to annotated cis elements known to be bound by LEC1, AREB3, bZIP67, and ABI3 (Fig. 3 and SI Appendix, Figs. S6 and S7; refs. 47–49 and 67). Mutation of enriched motifs within CRMs compromised the CRM’s ability to enhance minimal promoter activity (Fig. 4), indicating that many of the enriched DNA motifs represent functional cis elements. Consistent with this conclusion, others have shown the importance of the G box-like and RY-like motifs in controlling the Arabidopsis OLE1 gene (21). Moreover, our results showing that L, LA, LAZ, and LAZA genes are overrepresented for 1) CCAAT-like, 2) G box-like, and 3) G box-like and RY-like DNA motifs, respectively, suggest that specific sets of DNA motifs are responsible for recruiting different TF combinations to the CRMs (Fig. 3). Together, these results provide insight into the basis of the combinatorial interactions between LEC1, AREB3, bZIP67, and ABI3 by showing that enriched DNA motifs in CRMs represent functional cis elements, and the recruitment of LEC1 along with AREB3, bZIP67, and/or ABI3 is determined, in part, by the specific set of cis elements present in the CRMs.
Although these results significantly advance our understanding of the TF networks that regulate seed development, several questions remain to be resolved. For example, although L, LA, LAZ, and LAZA gene sets are involved in diverse developmental processes, each contains genes that are expressed primarily at different seed development stages (Fig. 2). The temporal patterns are not explained by the DNA motifs in the CRMs. It is possible that other yet to be identified TFs bind the CRMs to dictate temporal expression patterns. Alternatively, “motif grammar,” the specific arrangement and/or spacing of cis elements in CRMs, may account for temporal expression differences. Others have shown that motif grammar explains temporal and spatial gene expression patterns (46, 56, 68). Another question stems from the observation that several CRMs are able to enhance transcription, even when all of the discernible DNA motifs are mutagenized, suggesting the presence of cryptic cis elements in the CRMs (Fig. 4). Latent specificity, the modification of DNA recognition specificity through combinatorial interactions between TFs, offers a potential explanation for this observation (39, 69). Understanding the regulatory logic that controls seed gene expression requires further studies to identify distinct TFs that act combinatorially with LEC1, AREB3, bZIP67, and ABI3 and to decipher the motif grammar governing CRM activity.
Materials and Methods
ChIP-Seq.
Soybean plants were grown and seeds were harvested for ChIP experiments as described by Pelletier et al. (10).
ChIP assays were performed, with modifications, as described previously (10) using peptide antibodies against AREB3, bZIP67, and ABI3 that were generated as described in SI Appendix, SI Materials and Methods. DNA sequencing libraries were prepared using the NuGEN Ovation Ultralow System V2, and DNA fragments were size selected by electrophoresis and sequenced at 50-bp single-end reads using an Illumina HiSEq. 4000 sequencing system.
ChIP-Seq data were analyzed as described previously (10), with the modifications described in SI Appendix, SI Materials and Methods. Briefly, sequencing reads were aligned using Bowtie v0.12.7 (70) and reproducible ChIP-Seq peaks were identified using MACS2 v2.1.0.20140616 (71) and the Irreproducible Discovery Rate pipeline (72) (https://github.com/nboley/idr). Antibody specificity was established by showing extensive overlap in genes bound by a given TF using antibodies generated against 2 separate peptides for each TF, and quality assessment of the ChIP-Seq data followed ENCODE guidelines (73) as summarized in Dataset S2. Because the data analysis pipeline was modified, we reanalyzed ChIP-Seq data for LEC1 ChIP-Seq experiments in EM-stage embryos (10). GO enrichment and hierarchical clustering analyses were performed as described (10). LEC1, AREB3, bZIP67, and ABI3 overlapping binding sites were used to define L, LA, LAZ, and LAZA CRMs using the bedtools merge function (74), as described in Fig. 3B and SI Appendix, SI Materials and Methods. De novo DNA motif discovery analysis of CRMs was performed using the MEME-ChIP tool from the MEME suite v5.0.5 (75), and DNA motif enrichment analysis was performed using the motifEnrich tool from HOMER (76) (homer.ucsd.edu/homer/motif/index.html) and the ROCR R package v1.0.2 (77), as detailed in SI Appendix, SI Materials and Methods.
Transient Assays in Embryo Cotyledon Protoplasts.
Transient assays in protoplasts isolated from soybean embryo cotyledons and Arabidopsis leaves were performed as described by Yoo et al. (78) with the modifications described in SI Appendix, SI Materials and Methods. Plasmids used in the transient assays were constructed as detailed in SI Appendix, SI Material and Methods, and primers used for DNA manipulations are listed in Dataset S5. Activities of 5′-deletion constructs were evaluated by measuring GFP and mCherry activity in transfected soybean embryo protoplasts using fluorescence filters GFP-3035B and TXrRED-4040B (Semrock) with an Eclipse E600 microscope (Nikon) and calculating GFP to mCherry ratios as described in SI Appendix, SI Material and Methods.
Measurements of firefly and Renilla luciferase activities in CRM gain-of-function experiments with soybean embryo cotyledon protoplasts were made using the Dual-Luciferase Reporter Assay System (Promega) with a TriStar2 LB942 multiplate reader (Berthold) as described in SI Appendix, SI Material and Methods.
Constructs for bimolecular fluorescence complementation assays were created by fusing open reading frames for the TFs with the amino or carboxyl terminus of the citrine fluorescence protein. Constructs were transfected into Arabidopsis leaf protoplasts as described in SI Appendix, SI Material and Methods. Citrine fluorescence signal was detected using the GFP-3035B filter of an Eclipse E600 microscope (Nikon).
Data Availability.
Data are available at Gene Expression Omnibus under the following accessions: AREB3-EM (GSE101672), bZIP67-EM (GSE101672), ABI3-EM (GSE101649), AREB3B-EM (GSE140699), bZIP67B-EM (GSE140701), and ABI3B-EM (GSE140700).
Supplementary Material
Acknowledgments
We thank Bo Liu for advice about antibody production and testing, Dan Runcie for advice about statistical analyses, Savithramma Dinesh-Kumar and Samuel Hazen for providing plasmids containing the citrine and dual-luciferase constructs, respectively, and Sharon Belkin for the soybean seed diagrams. This work was supported by a grant from the National Science Foundation Plant Genome Research Program (to R.B.G. and J.J.H.). L.J. was supported by a Coordination for the Improvement of Higher Level Personnel grant (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Brazil, No. 99999.013505/2013-00).
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE101672, GSE101649, GSE140699, GSE140701, and GSE140700).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918441117/-/DCSupplemental.
References
- 1.Steeves T. A., The evolution and biological significance of seeds. Can. J. Bot. 61, 3550–3560 (1983). [Google Scholar]
- 2.Palovaara J., de Zeeuw T., Weijers D., Tissue and organ initiation in the plant embryo: A first time for everything. Annu. Rev. Cell Dev. Biol. 32, 47–75 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Puthur J. T., Shackira A. M., Saradhi P. P., Bartels D., Chloroembryos: A unique photosynthesis system. J. Plant Physiol. 170, 1131–1138 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Harada J. J., “Seed maturation and control of germination” Advances in Cellular and Molecular Biology of Plants, Larkins B. A., Vasi I. K., Eds. (Cellular and Molecular Biology of Seed Development, Kluwer Academic Publishers, Dordrecht, 1997), vol. 4, pp. 545–592. [Google Scholar]
- 5.Vicente-Carbajosa J., Carbonero P., Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49, 645–651 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Jo L., Pelletier J. M., Harada J. J., Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant Biol. 61, 564–580 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Meinke D. W., A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258, 1647–1650 (1992). [DOI] [PubMed] [Google Scholar]
- 8.West M., et al. , LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6, 1731–1745 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lotan T., et al. , Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Pelletier J. M., et al. , LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. U.S.A. 114, E6710–E6719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvenzani V., et al. , Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One 7, e42902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnesutta N., Saad D., Chaves-Sanjuan A., Mantovani R., Nardini M., Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the LEC1 family of NF-Y subunits in plants. Mol. Plant 10, 645–648 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Kwong R. W., et al. , LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15, 5–18 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braybrook S. A., Harada J. J., LECs go crazy in embryo development. Trends Plant Sci. 13, 624–630 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Fatihi A., et al. , Deciphering and modifying LAFL transcriptional regulatory network in seed for improving yield and quality of storage compounds. Plant Sci. 250, 198–204 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Lepiniec L., et al. , Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 31, 291–307 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Santos-Mendoza M., et al. , Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54, 608–620 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Bryant F. M., Hughes D., Hassani-Pak K., Eastmond P. J., Basic LEUCINE ZIPPER TRANSCRIPTION FACTOR67 transactivates DELAY OF GERMINATION1 to establish primary seed dormancy in Arabidopsis. Plant Cell 31, 1276–1288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendes A., et al. , bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25, 3104–3116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto A., et al. , Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58, 843–856 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Baud S., et al. , Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed. Plant Physiol. 171, 1099–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulard C., et al. , LEC1 (NF-YB9) directly interacts with LEC2 to control gene expression in seed. Biochim. Biophys. Acta. Gene Regul. Mech. 1861, 443–450 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Braybrook S. A., et al. , Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 3468–3473 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reidt W., et al. , Gene regulation during late embryogenesis: The RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 21, 401–408 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Bensmihen S., Giraudat J., Parcy F., Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J. Exp. Bot. 56, 597–603 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Alonso R., et al. , A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21, 1747–1761 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura S., Lynch T. J., Finkelstein R. R., Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Lara P., et al. , Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 278, 21003–21011 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Pelletier J., et al. , Identification of GLYMA.06G314400 and GLYMA.13G317000 binding sites in soybean early maturation embryos. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101672. Deposited 20 July 2017.
- 30.Pelletier J., et al. , Identification of GLYMA.08G357600 binding sites in soybean early maturation embryos. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101649. Deposited 19 July 2017.
- 31.Pelletier J., et al. , Identification of GLYMA.06G314400 binding sites in soybean early maturation embryos. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140699. Deposited 19 November 2019.
- 32.Pelletier J., et al. , Identification of GLYMA.13G317000 binding sites in soybean early maturation embryos. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140701. Deposited 19 November 2019.
- 33.Pelletier J., et al. , Identification of GLYMA.08G357600 binding sites in soybean mid-maturation embryos II. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140700. Deposited 19 November 2019.
- 34.Pelletier J., et al. , Identification of LEC1 binding sites in soybean embryos at 3 developmental stages. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99882. Accessed 9 June 2017.
- 35.Farnham P. J., Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 10, 605–616 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada J. J., et al. , Gene expression changes in the development of the soybean seed-cotyledon stage. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57606. Accessed 13 May 2014.
- 37.Harada J. J., et al. , Gene expression changes in the development of the soybean seed-early maturation stage. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46096. Accessed 16 April 2013.
- 38.Harada J. J., et al. , Gene expression changes in the development of the soybean seed mid-maturation (B1) stage. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99109. Accessed 19 May 2017.
- 39.Heyndrickx K. S., Van de Velde J., Wang C., Weigel D., Vandepoele K., A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. Plant Cell 26, 3894–3910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arabidopsis Interactome Mapping Consortium , Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakoby M., et al. ; bZIP Research Group , bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Johnson D. S., Mortazavi A., Myers R. M., Wold B., Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Davidson E. H., et al. , A genomic regulatory network for development. Science 295, 1669–1678 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Gerstein M. B., et al. ; modENCODE Consortium , Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The modEncode Consortium et al. , Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zinzen R. P., Girardot C., Gagneur J., Braun M., Furlong E. E., Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Izawa T., Foster R., Chua N. H., Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230, 1131–1144 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Kim S. Y., Chung H.-J., Thomas T. L., Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 11, 1237–1251 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Mönke G., et al. , Seed-specific transcription factors ABI3 and FUS3: Molecular interaction with DNA. Planta 219, 158–166 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Meister R. J., et al. , Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 37, 426–438 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Xiao J., et al. , Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49, 1546–1552 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Sheen J., Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475 (2001). [PMC free article] [PubMed] [Google Scholar]
- 53.Bemer M., van Dijk A. D. J., Immink R. G. H., Angenent G. C., Cross-Family transcription factor interactions: An additional layer of gene regulation. Trends Plant Sci. 22, 66–80 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Peter I. S., Regulatory states in the developmental control of gene expression. Brief. Funct. Genomics 16, 281–287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reményi A., Schöler H. R., Wilmanns M., Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 11, 812–815 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Ouyang Z., Zhou Q., Wong W. H., ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 106, 21521–21526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swanson C. I., Evans N. C., Barolo S., Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell 18, 359–370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mayran A., Drouin J., Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 293, 13795–13804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaret K. S., Carroll J. S., Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao Z., et al. , Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551, 124–128 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Oldfield A. J., et al. , Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 55, 708–722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherwood R. I., et al. , Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 32, 171–178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barakat T. S., et al. , Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell 23, 276–288.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lessard P. A., Allen R. D., Fujiwara T., Beachy R. N., Upstream regulatory sequences from two beta-conglycinin genes. Plant Mol. Biol. 22, 873–885 (1993). [DOI] [PubMed] [Google Scholar]
- 65.De Paiva G. R., “Transcriptional regulation of seed protein genes,” PhD Dissertation, University of California, Los Angeles, CA (1994).
- 66.Yadegari R., “Regional specification and cellular differentiation during early plant embryogenesis,” PhD Dissertation, University of California, Los Angeles, CA (1996).
- 67.O’Malley R. C., et al. , Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnone M. I., Davidson E. H., The hardwiring of development: Organization and function of genomic regulatory systems. Development 124, 1851–1864 (1997). [DOI] [PubMed] [Google Scholar]
- 69.Slattery M., et al. , Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270–1282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y., et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Q., Brown J. B., Huang H., Bickel P. J., Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011). [Google Scholar]
- 73.Landt S. G., et al. , ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Machanick P., Bailey T. L., MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sing T., Sander O., Beerenwinkel N., Lengauer T., ROCR: Visualizing classifier performance in R. Bioinformatics 21, 3940–3941 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Yoo S. D., Cho Y. H., Sheen J., Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Harada J. J., et al. , Gene expression changes during embryo and seed maturation, quiescence and germination in soybean. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99571. Accessed 1 June 2017.
- 80.Kulakovskiy I. V., et al. , HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-seq analysis. Nucleic Acids Res. 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available at Gene Expression Omnibus under the following accessions: AREB3-EM (GSE101672), bZIP67-EM (GSE101672), ABI3-EM (GSE101649), AREB3B-EM (GSE140699), bZIP67B-EM (GSE140701), and ABI3B-EM (GSE140700).