Significance
The cohesin complex both tethers together sister chromatids following DNA replication and ensures chromosome compaction and proper gene expression throughout interphase by folding chromosomes into loops and domains. How these very different activities of cohesin are controlled is largely unknown. The ESCO2 acetyltransferase modifies the SMC3 subunit of cohesin and is uniquely able to promote cohesion between sister chromatids, while the related ESCO1 acetyltransferase, which has the same catalytic activity, does not. Here we show that ESCO2 interacts with the DNA replication machinery through several highly conserved sequences in its unstructured tail. These interactions ensure that cohesin is modified precisely when sister chromatids are formed. This work provides a model to understand how cohesin activity is entrained to specific contexts.
Keywords: chromosome biology, DNA replication, cohesin, sister chromatid cohesion
Abstract
The tethering together of sister chromatids by the cohesin complex ensures their accurate alignment and segregation during cell division. In vertebrates, sister chromatid cohesion requires the activity of the ESCO2 acetyltransferase, which modifies the Smc3 subunit of cohesin. It was shown recently that ESCO2 promotes cohesion through interaction with the MCM replicative helicase. However, ESCO2 does not significantly colocalize with the MCM complex, suggesting there are additional interactions important for ESCO2 function. Here we show that ESCO2 is recruited to replication factories, sites of DNA replication, through interaction with PCNA. We show that ESCO2 contains multiple PCNA-interaction motifs in its N terminus, each of which is essential to its ability to establish cohesion. We propose that multiple PCNA-interaction motifs embedded in a largely flexible and disordered region of the protein underlie the unique ability of ESCO2 to establish cohesion between sister chromatids precisely as they are born during DNA replication.
Sister chromatids, the identical products of chromosome replication, are tethered together from the time they are made until cell division when they are segregated into daughter cells. Sister chromatid cohesion is mediated by cohesin, a protein complex that has the capacity to topologically entrap DNA. In addition to its role in tethering sister chromatids together, cohesin also plays critical roles in folding chromosomes into loops and domains throughout interphase, which in turn ensures normal transcription and thus proper development. How cohesin is regulated to result in these very distinct outcomes is not well understood. The activity of the cohesin complex is controlled in part by modulation of the stability of its interaction with chromatin. In higher eukaryotes, cohesin is associated with chromatin throughout interphase, but a small pool becomes more stably bound in a DNA replication-dependent manner, and persists into G2 (1). This observation and a wealth of genetic and biochemical data suggest that the association of cohesin with chromatin is stabilized by factors or activities associated with DNA replication (2, 3).
Acetylation of the SMC3 subunit of cohesin by members of the Eco1 family of acetyltransferases stabilizes cohesin binding (4–8). The founding member of the cohesin acetyltransferase family, the Eco1/Ctf7 protein of Saccharomyces cerevisiae, is required for sister chromatid cohesion (9, 10). Vertebrates express 2 homologs of Eco1, called ESCO1 and ESCO2 (11, 12). In addition to sequences highly conserved with yeast Eco1, both ESCO1 and ESCO2 have N-terminal extensions not found in the yeast protein (Fig. 1A). Although ESCO1 and ESCO2 have the same catalytic activity, they make distinct contributions to cohesin regulation. ESCO2 is uniquely able to promote cohesion between sister chromatids, though the majority of SMC3 acetylation is ESCO1 dependent (13, 14).
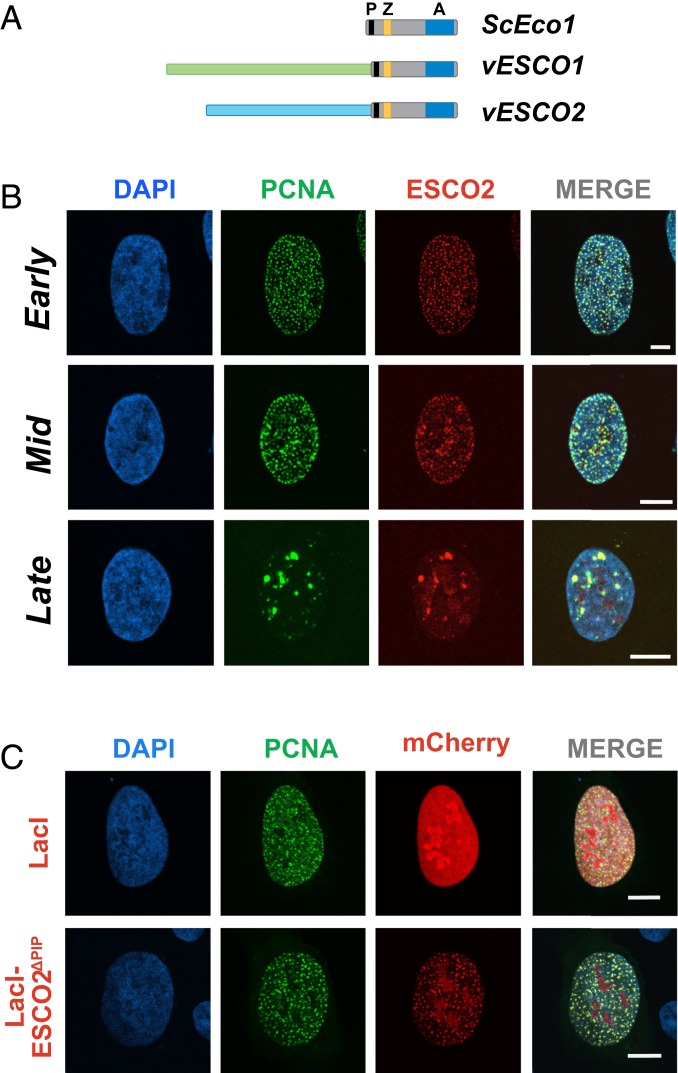
Fig. 1.
ESCO2 associates with sites of active DNA replication. (A) The Eco acetyltransferases. Shown are the S. cerevisiae Eco1 protein and the vertebrate homologs ESCO1 and ESCO2. All 3 proteins contain a highly conserved domain containing a PIP box (P; black), a zinc finger motif (Z; yellow), and the catalytic acetyltransferase domain (A; blue). The vertebrate proteins ESCO1 and ESCO2 have unique N-terminal extensions, with no apparent sequence homology to each other (light green and light blue). (B) ESCO2 colocalizes with PCNA in replication foci. Confocal micrographs of nuclei of U2OS cells cotransfected with mCherry-ESCO2 and GFP-PCNA. ESCO2 colocalized with PCNA in replication foci in patterns of early, mid, and late DNA replication. Sites of colocalization appear yellow in the merged images. (C) The PIP box in ESCO2 is dispensable for localization to replication foci. Confocal micrographs of U2OS cells cotransfected with GFP-PCNA and mCherry-ESCO2 in which the PIP box is deleted. Pearson’s correlation coefficient for red and green signal intensities R = 0.837, 0.754, and 0.591 for early, mid, and late images, respectively (B). R = 0.189 for lacI, and 0.837 for ΔPIP (C). Shown is a representative example of 3 independent experiments. (Scale bars, 10 μm.)
What confers to ESCO2 the unique ability to promote cohesion between sister chromatids? All Eco1 family members contain a conserved PCNA-interacting protein (PIP) box, which in budding yeast has been shown to promote association of Eco1 with the replication factor PCNA, ensuring its association with chromatin during DNA replication (15). Here we set out to define the elements in ESCO2 that underlie its ability to ensure sister chromatid cohesion. We find that ESCO2 colocalizes with PCNA at sites of active DNA replication, and that it does this independently of the conserved PIP box. We show that ESCO2 interacts with the replication machinery through 2 noncanonical PCNA-interaction motifs in its N-terminal tail. These motifs are embedded in a flexible region of the protein, and the spacing between these motifs varies significantly among ESCO2 proteins from different species. We conclude from these observations that ESCO2 activity is entrained to sites of DNA replication by multiple interactions with the replisome. We propose that the nature of these interactions ensures that ESCO2 retains interaction with this dynamic cellular machine.
Results
ESCO2 Localizes to Sites of DNA Replication.
We showed previously that ESCO2 function is critically dependent on a conserved PIP box sequence motif near the C terminus of the protein (Fig. 1A), suggesting that the ability of ESCO2 to promote cohesion may require direct interaction with PCNA, a critical replication processivity factor (13). To further explore the potential direct interaction between ESCO2 and PCNA, we coexpressed fluorescent derivatives of both proteins (GFP-PCNA, mCherry-ESCO2) in U2OS cells. Consistent with their direct interaction, the proteins were found colocalized in the nucleus (Fig. 1B). The proteins were found together in “replication foci,” well-documented sites of active DNA replication (16, 17). During S phase the pattern of replication foci progresses through a stereotypical spatiotemporal program: the pattern begins first with numerous small foci in euchromatic regions in early S phase, followed by accumulation of foci at heterochromatic regions around the nuclear rim in mid S, and finishing with localization to larger patches adjacent to the nucleoli. ESCO2 and PCNA were found colocalized in all of these patterns in fixed cells (Fig. 1B). These data are consistent with interaction of ESCO2 with PCNA at sites of active DNA replication.
The presence of ESCO2 at sites of DNA replication is consistent with the presence of the PIP box, which in fungal models is important in cohesion establishment (15). To test whether ESCO2 colocalizes with the replication machinery through the PIP box motif, we generated a derivative of ESCO2 in which the PIP box was deleted. Unexpectedly, we found that colocalization of PCNA and ESCO2 was unaffected by this mutation: both proteins were found in replication foci, in a manner indistinguishable from the wild-type controls (Fig. 1C). We conclude that ESCO2 interacts with the replication machinery independently of the PIP box.
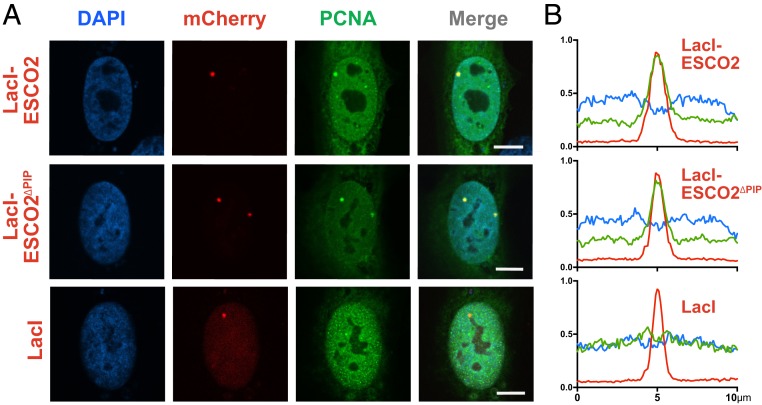
Our data suggested either that sequences other than the PIP box promote ESCO2 interaction with PCNA, or that ESCO2 is recruited to replication foci through proteins other than PCNA. Because coimmunoprecipitation experiments proved inconclusive, we used an alternative approach to test interaction of ESCO2 with PCNA in the nuclear context. To do this, we expressed ESCO2 fused to a fluorescent protein (mCherry) and the lac repressor DNA-binding domain in U2OS cells containing an array of lac operon operator repeats integrated in the genome. The lac repressor DNA-binding domain was recruited to the lac operator array, resulting in 1 or 2 bright mCherry nuclear foci in confocal images, as seen previously depending on the confocal plane (18, 19). GFP-labeled PCNA, when coexpressed with the ESCO2 fusion, was recruited to the same foci (Fig. 2 A and B). Control foci containing mCherry-lacI showed no enrichment for PCNA. A derivative of ESCO2 without the PIP box also recruited PCNA (Fig. 2 A and B). We conclude that ESCO2 interacts with PCNA through motifs other than the canonical PIP box.
Fig. 2.
ESCO2 interacts with PCNA outside of replication foci. (A) Tethered ESCO2 recruits PCNA. An mCherry-lacI-ESCO2 fusion protein, expressed in U2OS cells containing a stably integrated tandem array of lac operator sequences, is recruited to nuclear loci where it can be seen as 1 or 2 red foci in confocal micrographs. Cotransfected GFP-PCNA was recruited to the ESCO2 focus (Top), and this was unaffected by deletion of the PIP box in ESCO2 (Middle). PCNA was not enriched in the foci in cells containing mCherry-lacI (without ESCO2) (Bottom). (Scale bars, 10 μm.) (B) Normalized fluorescence intensity. The fluorescence intensity of lines drawn across the lac operator arrays from a number of cells treated as in A was averaged. Fluorescence of mCherry-lacI fusion is indicated by a red line, DAPI in blue, and GFP in green. n > 25 cells in each sample. Shown is a representative example of 3 independent experiments.
The ESCO2 N Terminus Contains Multiple Essential Motifs.
Reasoning that sequences important for interaction with the replication machinery are likely to be relatively well conserved, we compared the amino acid sequences of ESCO2 from a number of organisms (Fig. 3A). In general, the N termini are very poorly conserved, with an overall pairwise identity of 26.4%, and only 2.4% identity with the consensus. In contrast, the C termini, starting just after the PIP box, show 72.5% pairwise identity, and 36.1% identity to the consensus. We identified several short sequence elements that were nearly invariant among all of the species analyzed. These included motifs previously shown to be important for chromatin binding (boxes A and B) (13, 20), as well as a motif near the PIP box, which we call box C (Fig. 3B).
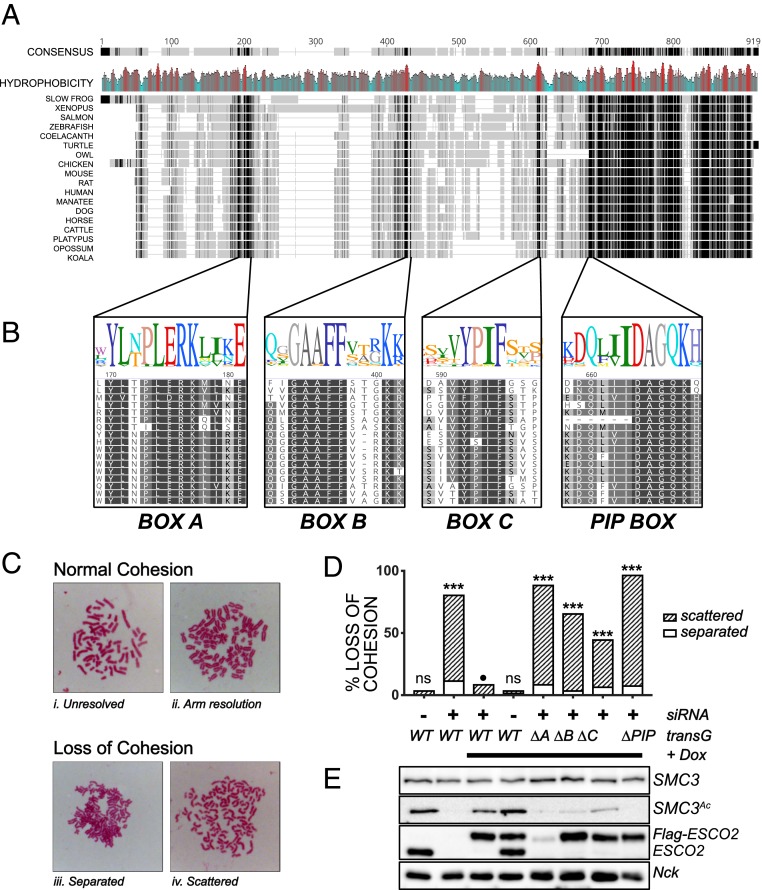
Fig. 3.
Sequences in the N terminus of ESCO2 are required for cohesion establishment. (A) The N terminus of ESCO2 contains several short, conserved motifs. Clustal Omega alignment of ESCO2 proteins from the indicated species is shown. The overall consensus is shown at Top, in which the grayscale indicates the degree of conservation (black is 100% and the lightest gray is <60%), and gaps are shown with a gray line. Mean hydrophobicity is also shown, with red indicating hydrophobic patches. Accession nos. are in SI Appendix, Table S1. (B) Enlargement of conserved motifs in the ESCO2 N terminus. The alignment of the 3 motifs, box A, box B, and box C, as well as the conserved PIP box, are shown. Sequence logos are colored according to the RasMol scheme (52). (C) Representative chromosome spreads as analyzed to score sister chromatid cohesion. Categories i and ii, in which sister chromatids were clearly tethered together, were considered normal cohesion, while chromatids that were well separated, as in categories iii (separated) and iv (scattered), were scored as loss of cohesion. (D) Cohesion assay. HeLa cells expressing siRNA-resistant FLAG-tagged derivatives of ESCO2 with the indicated mutations were treated with siRNA against ESCO2 to deplete endogenous transcripts and were scored for cohesion as shown in C (n ≥ 100/sample). ***P < 0.005 compared to wild-type (WT) control indicated by the black circle (Fisher’s exact test with Bonferonni’s correction for multiple comparisons; ns, not significant). Shown is a representative experiment; each mutant was tested at least 4 times independently (data from additional experiments are summarized in SI Appendix, Table S2). (E) Immunoblot showing expression of ESCO2 transgenes and SMC3 acetylation. Cell lysates from samples in D were probed with antibodies for the indicated proteins. SMC3Ac, NCK, and ESCO2 came from the same gel. NCK was used as a loading control. SMC3 and ESCO2 were analyzed separately. Tg, transgene; Dox, doxycyline (used to activate expression of transgenes).
The high level of conservation of boxes A, B, and C suggested that they might be critical for ESCO2 function. To test this, we performed cohesion assays, using a gene knockdown and rescue strategy. HeLa cells lacking a functional ESCO1 gene were engineered to express ESCO2 derivatives as inducible siRNA-resistant cDNA transgenes. Expression of endogenous ESCO2 was reduced by siRNA transfection, expression of flag-tagged ESCO2 derivatives was induced by addition of doxycycline, and mitotic spreads were analyzed for cohesion phenotypes (Fig. 3C). We tested each of the conserved motifs, boxes A, B, and C, as well as the conserved PIP box for their impact on cohesion. We found that deletion of any of these conserved motifs resulted in significant loss of cohesion (Fig. 3D and SI Appendix, Table S2). Therefore, each of the conserved motifs is required for full function of ESCO2.
We analyzed the level of SMC3 acetylation in cells expressing the mutant ESCO2 derivatives with deletion of the conserved motifs (Fig. 3E). Because the cells lack functional ESCO1, all acetylation of SMC3 in these samples is ESCO2 dependent (14). We found that in general the relative amount of SMC3 acetylation correlated with the amount of cohesion. For example, cells expressing the PIP box mutant of ESCO2 were strongly compromised in cohesion (>90% loss of cohesion), and had essentially undetectable levels of SMC3 acetylation. In contrast, the box C mutant was less defective in cohesion establishment, and SMC3 acetylation was reduced but detectable. All of the derivatives of ESCO2 were expressed at similar levels, with the exception of the box A mutant. The low levels of expression of this mutant may be due to its inability to bind MCM2-7 helicase, as interaction with the MCM helicase has recently been shown to protect ESCO2 from CUL4-DDB1–mediated degradation (21). We conclude from this experiment that all conserved motifs in the otherwise poorly conserved N terminus, boxes A, B, C, and the PIP box, are required for efficient cohesion establishment or maintenance.
Box C Ensures Association with Replication Foci.
The functional difference between ESCO2 and the related ESCO1 maps to their distinct N termini, as opposed to their conserved C-terminal acetyltransferase domains (14). This suggests that the N terminus of ESCO2 might be sufficient to promote localization to replication foci. To test this, we fused the ESCO2 N terminus directly to GFP (ESCO2N-GFP), eliminating the catalytic C terminus entirely (Fig. 4A), and coexpressed this fusion together with mRuby-PCNA. We found that ESCO2N-GFP, like the full-length protein, colocalized with PCNA at replication foci (Fig. 4B). We then tested derivatives of ESCO2N-GFP in which each of the conserved motifs was deleted. We found that ESCO2N-GFP with deletion of box A or box B still colocalized with PCNA at replication foci, but deletion of box C abrogated this localization (Fig. 4B). We also noted that localization of ESCO2N fusions to the nucleoli, weakly apparent in the full-length ESCO2 (Fig. 1B), was frequently more pronounced in the ESCO2N fusions. Partitioning of ESCO2 to nucleoli, while not well understood, has been seen previously, and is consistent with models suggesting that nucleolar function is disrupted in patients with Roberts syndrome, a developmental disorder that results from faulty ESCO2 function (22–24). We confirmed that the large nuclear patches represent nucleolar localization of ESCO2 by coexpression with a nucleolar resident protein (SI Appendix, Fig. S1).
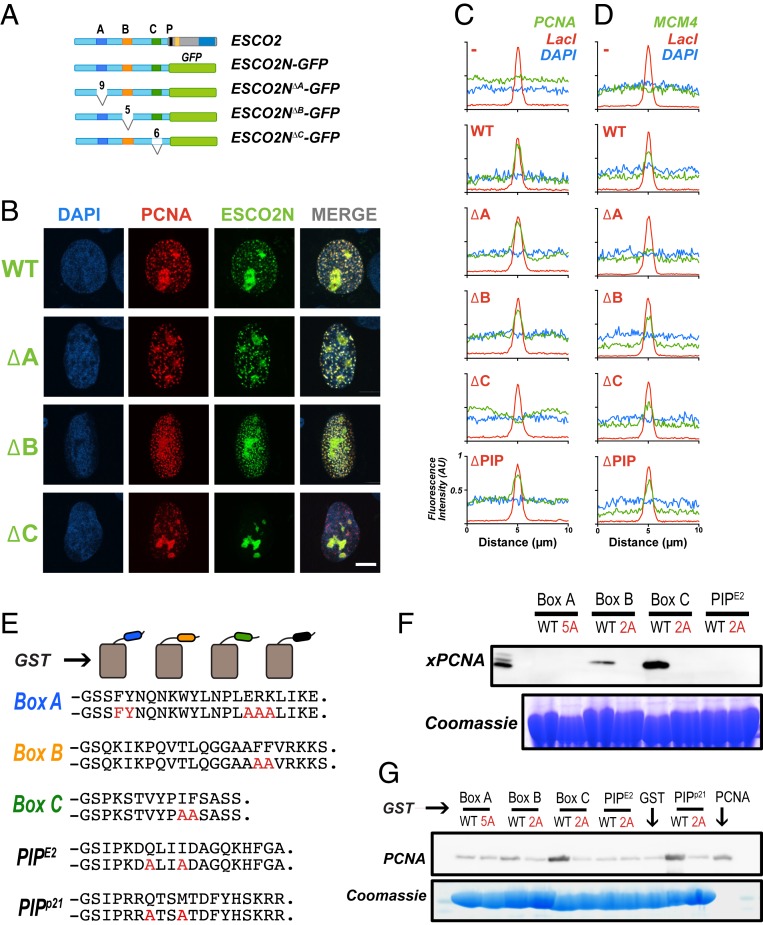
Fig. 4.
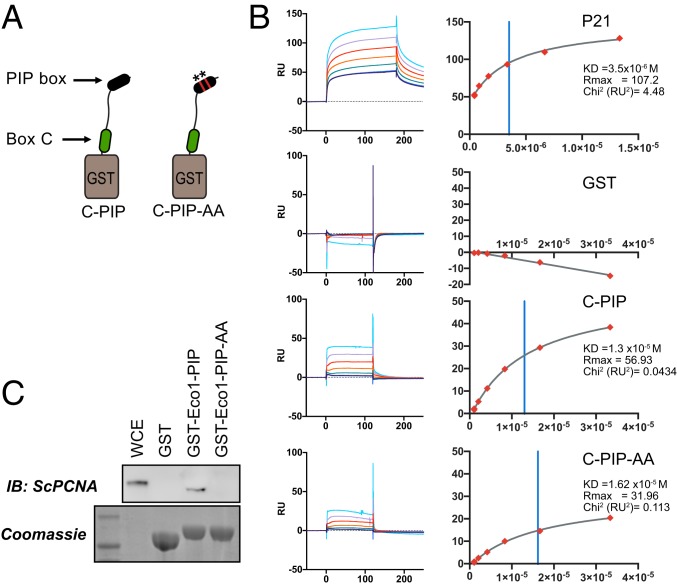
PCNA interacts with ESCO2 motifs in vivo and in vitro. (A) ESCO2N-GFP fusions. A cartoon depicting constructs in which the N-terminal 375 amino acids (a.a.) of ESCO2 are fused directly to eGFP. Motifs A, B, and C were deleted independently, as shown; numbers indicate the number of amino acids deleted (not drawn to scale). (B) Localization to replication foci. Confocal images of U2OS cells cotransfected with Ruby-PCNA and the GFP-fusion constructs. Colocalization is indicated in yellow in the merge image, as in Fig. 2. (C) Box C is critical for PCNA recruitment by tethered ESCO2. mCherry-lacI-ESCO2 (full-length) fusions with the deletions indicated in A were coexpressed with GFP-tagged PCNA as in Fig. 2, and the colocalization at nuclear foci was scored by fluorescence intensity profile analysis as in Fig. 2. (D) MCM4 recruitment to tethered ESCO2 is dependent upon box A. The experiment in C was repeated, only in this case the ESCO2 constructs were coexpressed with mEmerald-MCM4. Recruitment to tethered ESCO2 was scored as in Fig. 2. (E) Pull-down assay using GST-fusion proteins. Short peptide sequences including box A, box B, box C, or the ESCO2 PIP box motifs were expressed as GST-fusion proteins. A parallel set was made in which alanine substitutions were made at the invariant amino acids (shown in red). The PIP box from p21 was used as a positive control. (F) Coprecipitation from cell-free extracts. GST-fusion proteins shown in A were mixed with Xenopus egg extract and incubated with glutathione sepharose beads. The beads were washed and bound proteins were eluted and probed for PCNA by immunoblot. A duplicate gel was stained with Coomassie dye to detect the GST-fusion proteins. (G) Coprecipitation of purified proteins. The indicated GST-fusion proteins (E) were mixed with purified recombinant PCNA, pulled down with glutathione agarose beads, and analyzed as in F for PCNA.
The experiment in Fig. 4B suggested that box C is essential for interaction with the replication machinery and is consistent with a model in which ESCO2 interacts, perhaps directly, with PCNA through the box C motif. To confirm, we again used the assay in which the lacI-ESCO2 is tethered to an integrated array of lac operator repeats. Strikingly, the ability of immobilized ESCO2 to recruit PCNA was lost upon deletion of the C box, and unaffected by the absence of boxes A or B, or the PIP box (Fig. 4C).
As we were doing this work, 2 reports were published indicating that ESCO2 interacts directly with the MCM2-7 helicase through the box A motif, and suggesting that this interaction is essential for cohesion establishment (21, 25). We therefore tested our same panel of mutants to determine whether the conserved motifs in ESCO2 are important for interaction with MCM proteins in the tethering assay. Cells with integrated lac operator repeats were cotransfected with constructs encoding lacI-ESCO2 and mEmerald-MCM4, and assayed for their colocalization in nuclear foci. While wild-type ESCO2 was able to recruit MCM4, deletion of box A caused significant loss of interaction in this assay. The other mutants, including deletions of box B, box C, or the PIP box were all able to recruit MCM4 (Fig. 4D). We conclude that ESCO2 interacts with both the MCM helicase and with PCNA and that these interactions map to distinct motifs in the ESCO2 N terminus.
ESCO2 Contains Multiple PCNA-Interaction Motifs.
Although box A has been shown to promote interaction of ESCO2 with the MCM2-7 helicase, it was still not clear what functional role boxes B and C play in association with the replication machinery. We noted that the conserved sequences in both box B and box C contain invariant pairs of hydrophobic amino acids, FF (Phe-Phe) and IF (Ilu-Phe), respectively. As similar peptides are present in a number of noncanonical PCNA-interaction motifs, we tested the possibility that ESCO2 interacts with PCNA through 3 distinct motifs: box B, box C, and the PIP box. We expressed short peptides containing the conserved functional motifs as fusions with the GST protein (Fig. 4E). The fusion proteins were immobilized on beads, which were then incubated in cell extracts. PCNA bound specifically to the beads carrying GST fusions to both box B and box C (Fig. 4B). To our surprise, the previously characterized, functionally critical PIP box at residues 374 to 381 (QLIIDAGK, in which the conserved residues are underlined) did not bind to PCNA in this assay. PCNA bound most efficiently to the fusion with the box C peptide, and this binding was abolished when the conserved hydrophobic residues (IF) were changed to alanine (Fig. 4F). Similarly, PCNA bound to the box B peptide, and this interaction was disrupted by mutation of the FF motif to AA. These data suggest that box B and box C are noncanonical PIP boxes. We saw no strong evidence for interacting proteins other than PCNA in this assay (SI Appendix, Fig. S2).
To understand whether interaction of PCNA with GST fusions was likely to be direct, we purified PCNA and performed coprecipitation experiments with the GST-peptide fusions, including the well-characterized PIP motif from p21 as a positive control (26). PCNA readily coprecipitated with the box C peptide, and this interaction was disrupted by mutation of the conserved hydrophobic residues (Fig. 4G). We repeatedly saw weak interaction with the box B peptide, though this interaction was apparently less robust than that with box C. The interaction with the box C peptide was comparable to that of the p21 PIP box in the pull-down assay. Taken together the experiments in Fig. 4 F and G indicate that ESCO2 contains multiple PCNA-interaction motifs in its N terminus. Given that the canonical PIP box is required for full ESCO2 function, we suspect that it may also promote interaction with PCNA, though not in the assays shown in Fig. 4.
To test the possibility that the PIP box might promote interaction with PCNA in the context of the intact ESCO2 N-terminal region, we performed binding assays using surface plasmon resonance (SPR) in which the GST-peptide fusions were immobilized and probed with recombinant PCNA trimers (Fig. 5). Due to solubility problems in expressing the full ESCO2 N terminus, we limited our analysis to a fragment of ESCO2 including both box C and the PIP box (GST-C-PIP; amino acids 320 to 388 of human ESCO2). We tested whether 2 amino acid substitutions to disrupt conserved PIP box residues (Q374A, I377A) affected the ability to bind PCNA. Using this approach, we found that the PIP box made a modest contribution to the interaction in this assay (KD = 13 μM for GST-C-PIP and 16 μM for GST-C-PIP-AA). In both cases the interactions were weaker than that detected for p21 (3.5 μM). The data suggest that the PIP box in ESCO2 contributes slightly to PCNA binding, at least in the context of purified proteins.
Fig. 5.
Contributions of the canonical PIP motif to PCNA binding. (A) Generation of combined fusions. GST was fused to a peptide spanning amino acids 320 to 388 of human ESCO2 containing both box C and the downstream canonical PIP-like motif at amino acids 374 to 377. A derivative in which the PIP-like motif was mutated (ESCO2 Q374A, I377A) was also purified. (B) Surface plasmon resonance analysis. The GST-fusion peptides in A were used to test PCNA binding using SPR, in which GST fusions were bound by anti-GST antibody and recombinant trimeric PCNA was used as an analyte. Shown are the background-subtracted sensorgrams (Left) and binding curves (Right) for p21 (positive control), GST (negative control), and the indicated fusions. Binding constants are shown at Right. The χ2 residuals are represented as RU2, an indication of curve fitting. Vertical blue lines indicate KD. (C) Contribution of the Eco1 PIP box to PCNA interaction in S. cerevisiae. GST was fused to the first 33 amino acids of S. cerevisiae Eco1p (GST-Eco1-PIP) for use in a pull-down assay (as in Fig. 4). Disruption of the PIP motif by 2 amino acid substitutions (Q18A, L21A = GST-Eco1-PIP-AA) disrupted the ability to pull down PCNA from whole cell yeast extract, as indicated by immunoblot analysis. A parallel gel was Coomassie stained, showing the bead-associated GST fusions. WCE, whole cell yeast extract.
The identification of the PIP box in ESCO2 was originally based on its conservation with the PIP motif in Eco1 the budding yeast ortholog, which is critical for cohesion establishment (15). We were therefore somewhat surprised by the weakness of the interaction of the ESCO2 PIP box with PCNA. To clarify this discrepancy, we tested whether the conserved PIP residues in Eco1p of budding yeast indeed promote PCNA interaction (Fig. 5C). Using the same GST-fusion approach we found that indeed the conserved Q18 and I21 of Eco1 make critical contributions to PCNA binding in a pull-down assay. We do not currently know why the interaction between the vertebrate ESCO2 PIP box and PCNA is not as readily detected. Nonetheless, we have shown that ESCO2 contains multiple, separable, PCNA-interaction motifs. For simplicity, we will refer to box B, box C, and the PIP box as PIP1, PIP2, and PIP3, respectively, from here forward.
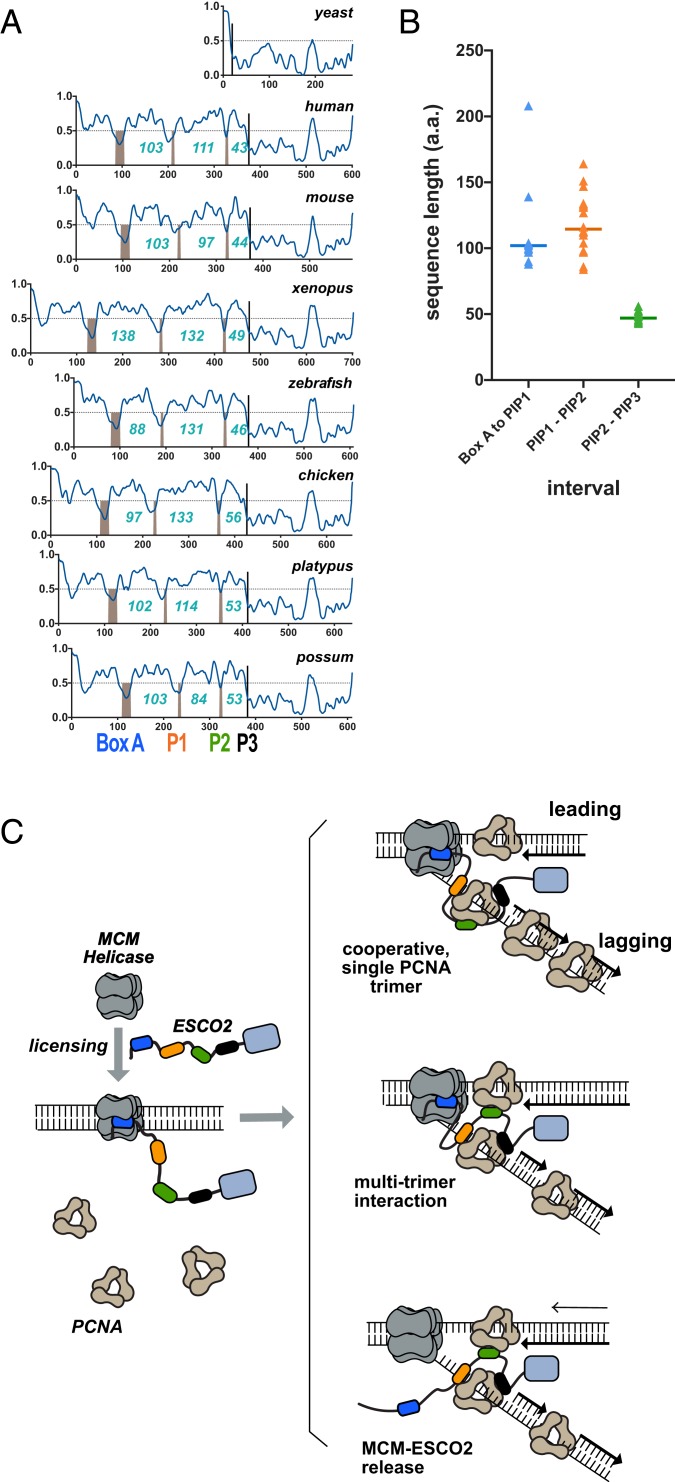
We further analyzed the ESCO2 proteins to better understand how the multiple PIP boxes in ESCO2 might function in context of the full-length protein. We analyzed ESCO2 protein sequences for propensity to adopt specific structures by using the Protein Disorder Prediction System (PrDOS) algorithm (27). This algorithm combines comparison of homologous proteins with amino acid content analysis to predict whether particular sequences are likely to fold into specific structures. Strikingly, all ESCO2 proteins are predicted to be largely disordered throughout their N termini, with clear exceptions at the conserved motifs: box A, PIP1, PIP2, and PIP3 (Fig. 5A). We found that the spacing between PIP1 and PIP2 varied significantly among species, with a median of 114 amino acids, and a range from 84 to 164 amino acids, while the spacing between PIP2 and PIP3 was relatively constant (median distance: 47 amino acids, range: 44 to 56) (Fig. 5B). The distance between box A and PIP1 was ∼100 amino acids for most species, with a few significant outliers among the species analyzed. We conclude that the essential functional motifs in the ESCO2 N terminus, box A, PIP1, PIP2, and PIP3, are in a floppy or disordered region of the protein, and that the spacing between these motifs, particularly between PIP1 and PIP2, is not critical to ESCO2 function. This may indicate that the disordered regions simply serve as flexible linkers, consistent with their poor sequence conservation (Fig. 3).
Discussion
In budding yeast, interaction between PCNA and Eco1 occurs through Eco1’s PIP motif, and this interaction is required for cohesion establishment (15). Here we show that vertebrate ESCO2, which ensures sister chromatid cohesion, contains 2 motifs in its N terminus that promote interaction with PCNA. This conclusion is based on colocalization of ESCO2 with PCNA at “replication factories,” the dependence of this interaction on specific sequences in ESCO2, and the ability of these same motifs to promote interaction with PCNA in cell-free extracts and with purified proteins. We find that PCNA interacts with each of these peptides in a sequence-specific manner. We have also shown, using a depletion and rescue approach, that each of these motifs is critical to the ability of ESCO2 to promote sister chromatid cohesion. Perhaps surprisingly, although it is essential for cohesion establishment, we were unable to detect a direct interaction between the conserved PIP box in vertebrate ESCO2 (PIP3) and PCNA.
Recent reports show convincingly that ESCO2 is recruited to chromatin by the MCM2-7 helicase and that this interaction is important for cohesin acetylation (21, 25). How then is MCM-dependent recruitment of ESCO2 compatible with the PCNA interaction(s) we report here? The MCM2-7 complex is loaded onto DNA in excess in G1, and only a small fraction of these loaded MCMs are activated to initiate DNA replication (28, 29). Indeed, although its activity is required for replication, the MCM helicase is not clearly enriched at sites of active replication nor is it found in replication foci (30–32). It is possible that ESCO2 is initially recruited to chromatin through interaction with the MCM helicase, and subsequently interacts with PCNA during active DNA replication. We do not know currently whether ESCO2 can interact simultaneously with both PCNA and the MCM helicase, or whether these interactions are mutually exclusive. Because PCNA is enriched on chromatin at sites of active DNA replication, we favor a model in which polyvalent interaction of ESCO2 with PCNA ensures SMC3 acetylation specifically at active replication forks, promoting cohesin stabilization precisely when and where sister chromatids are formed.
There is significant degeneracy among functional PIP boxes, and alternative PCNA-interaction motifs have also been identified (33–35). Our data demonstrate that 2 PCNA-interaction motifs, PIP1 and PIP2, mediate interaction between ESCO2 and PCNA. An initial report of the ESCO2-PCNA interaction implicated the PIP3 motif (15). We tested for, and were unable to detect, this interaction. The previously reported interaction may have been due to the presence of PIP1 and PIP2 in the region analyzed; a specific requirement for PIP3 was not reported (15). This is not to say that PIP3 is unimportant; ESCO2 deleted for the PIP3 box localizes normally to replication foci, but is profoundly compromised in cohesion establishment. It is possible that ESCO2 PIP3, which is conserved with the PIP box in S. cerevisiae Eco1, in fact promotes interaction with a replication protein other than PCNA. Indeed, a number of PIP-like motifs have been shown to promote such interactions (36), and some PIP-like motifs even have dual-binding specificity (37). Alternatively, interaction of PIP3 with PCNA may be enhanced in the context of the full-length protein, or seeded by prior PIP2 engagement. PIP1 and PIP2 can be categorized as noncanonical PIP boxes: they contain pairs of hydrophobic amino acids that mediate their interaction with PCNA (Fig. 3).
The interactions of ESCO2 with PCNA may be cooperative, ensuring robust binding through multiple low-affinity interactions. PCNA in its functional state is a ring-shaped homotrimer, in which the flexible interdomain connector loop, where PIP motifs interact, is exposed on the ring surface, and DNA is topologically entrapped within the ring (38). It is theoretically possible that a single ESCO2 molecule could interact simultaneously with each subunit of a single PCNA trimer, or it may interact with protomers from separate trimers, spanning 2 or 3 complexes simultaneously (Fig. 5C). In each of these models, we imagine that interaction with PCNA ensures that ESCO2 is associated with the lagging strand, which is enriched for PCNA, and strongly implicated in cohesion establishment (3, 39). The presence of multiple interaction motifs is a feature shared with other proteins that associate with PCNA, including Y family DNA polymerases, which participate in translesion synthesis, as well as poly ADP ribose glycohydrolase (PARG), which is essential for regulation of ADP ribose levels in response to DNA damage (40–42). ESCO2-PCNA interactions may ensure cohesin acetylation near the single-stranded DNA associated with DNA replication; single-stranded DNA has recently been proposed as an intermediate in cohesion establishment (43).
As in other proteins, the PCNA-interaction motifs in ESCO2 are embedded in an intrinsically disordered region (42, 44) (Fig. 6). The disordered nature and variable length of the spacers between the motifs may make an important contribution to how ESCO2 interacts with the replication machinery; the flexibility may reduce the entropic penalty of binding and enhance searching for multiple binding partners. Alternatively, the flexibility may allow ESCO2 to retain association with PCNA as it spins along the DNA helix, through multiple weak and dynamic interactions. Previous attempts to identify ESCO2-interacting proteins using affinity-based methods did not report identification of PCNA, suggesting that the interaction may indeed be weak once removed from chromatin (21, 25). ESCO2 may interact more strongly with PCNA once PCNA is DNA bound, as reported for other proteins (45, 46).
Fig. 6.
Structural disorder is an intrinsic property of the ESCO2 N terminus. (A) Functional motifs are embedded in the disordered region. The protein sequences of ESCO2 from multiple species were analyzed to detect regions of predicted disorder using the PrDOS algorithm. Sequences above the dotted horizontal line are predicted to be disordered. Conserved motifs, including box A, PIP1, PIP2, and PIP3, are predicted to be structured and are indicated by the gray bars. Numbers indicate the number of amino acids between each conserved motif. Accession nos. are shown in SI Appendix, Table S3. (B) Spacing between motifs. The number of amino acids between each of the conserved motifs in the ESCO2 N terminus are indicated in blue. Species included are the same as in A, except that Xenopus tropicalis ESCO2 was also included. (C) Models. ESCO2 is initially recruited to chromatin through interaction with the loaded MCM helicase (gray) through box A (blue). Subsequently, ESCO2 associates with sites of active DNA replication through multiple PCNA-interaction motifs (orange, green, and black lozenges). ESCO2 may interact with a single PCNA trimer, as shown at Top, or may interact simultaneously with more than one PCNA trimer (Middle). The interaction of ESCO2 with MCMs and PCNA may occur simultaneously, or ESCO2 may be released from the MCM complex to associate with PCNA. Multiple, flexible low-affinity interactions may ensure association of ESCO2 with the dynamic replisome.
The presence of multiple binding motifs embedded in a disordered region is consistent with allovalency, in which a single binding site on a receptor (PCNA in this case) can bind several motifs in the ligand, increasing affinity by increasing the effective local concentration of interaction motifs (47). In the context of the replication fork in which there are multiple PCNA complexes, this kind of interaction may achieve “fuzziness,” in which both partners have multiple interaction sites, no one particular interaction is favored, and intrinsically disordered regions retain conformational freedom (48, 49). Further experiments will be required to elucidate in detail the nature of the interaction between ESCO2 and the replication machinery.
Methods
Cell Culture.
HeLa (CCL-2) and U2OS (HTB-96) cells were obtained from the American Type Culture Collection (ATCC), and U2OS-LacO-I-SceI-TetO cells were obtained from Kerafast (ENH105-FP). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Corning) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals). Cells were maintained at 37 °C in a 5% CO2 atmosphere. Cells were transfected according to manufacturer’s instructions using Lipofectamine 2000 (Invitrogen) for plasmid DNA or Lipofectamine RNAiMAX (Invitrogen) for siRNA. Stable cell lines with doxycycline-inducible siRNA-resistant Flag-tagged ESCO2 cDNAs were generated using HeLa Flp-In T-Rex cells as previously described (14). The ESCO2 cDNA was cloned into a pcDNA5/FRT-based Flag-tag vector and cotransfected along with a plasmid expressing the FLP recombinase (pOG44, Invitrogen) using Lipofectamine 2000 (Invitrogen). Cells were selected in 200 μg mL−1 hygromycin B (Gold Biotechnology), colonies were isolated and expanded, and transgene induction was confirmed by immunoblot.
Fluorescence Analysis.
U2OS cells plated on coverslips were transfected with the indicated plasmids, incubated for 24 h, permeabilized for 5 min on ice in prepermeabilization buffer (20 mM Hepes 0.5% Triton X-100, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose), then fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde (PFA) and 0.1% Triton X-100 for 15 min at room temperature (RT). Cells were washed with antibody dilution buffer (AbDil; 20 mM Tris, 150 mM NaCl, 2% bovine serum albumin, 0.1% sodium azide, 0.1% Triton X-100 in PBS) and stained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei for 1 min at RT and washed again with PBS containing 0.1% Triton X-100. Coverslips were mounted on glass slides using Fluoromount-G (Electron Microscopy Sciences) mounting reagent and sealed with nail polish. Images were collected with a Nikon C2 confocal on a Ti-E motorized inverted microscope using a 60× 1.4 numerical aperture (n.a.) oil immersion objective lens. Colocalization was analyzed using the NIS Elements software to measure Pearson’s correlation coefficient of relevant fluorescence signals.
For the tethering assay, U2OS-LacO-I-SceI-TetO cells were plated on coverslips, transfected with indicated plasmids, and incubated for 24 h. Cells on coverslips were fixed (PBS containing 4% PFA and 0.1% Triton X-100) for 15 min at RT and washed with AbDil. Cells were stained with DAPI, mounted, and imaged as above. NIS Elements software was used to analyze fluorescent intensity along 10-μm lines centered on foci identified in the mCherry channel, and then scored for PCNA in cotransfected cells. Using Prism, the signal for each line scan was normalized from 0 to 1, and aggregate plots representing at least 25 mCherry foci were prepared. All experiments were repeated at least 3 times.
Immunofluorescence.
HeLa cells grown on coverslips were fixed with 4% paraformaldehyde in PBS containing 0.1% Triton X-100 at RT for 15 min. After fixation, cells were washed and incubated for 30 min in AbDil then incubated with appropriate primary antibody diluted in AbDil for 1 h at RT, washed, and probed with secondary antibody for 1 h at RT. Cells were washed and stained with DAPI to label nucleus for 1 min at RT and washed again with Tris-buffered saline (TBS) with 0.1% Triton X-100. Coverslips were mounted on glass slides and imaged as above.
Cohesion Assays.
For cohesion assays, cells with integrated siRNA resistant transgenes were transfected with siRNA against ESCO2 (Dharmacon J-025788-09, target: CGAGUGAUCUAUAAGCCAA) for 4 h, and cells were left to incubate for 48 h in doxycycline (2 μg/mL). The media were then supplemented with 0.2 μg/mL colcemid (AdipoGen) for 15 min, and cells were collected with trypsin ethylenediaminetetraacidic acid (EDTA), washed with PBS, and treated with hypotonic buffer (0.075 M KCl) at 37 °C for 15 min. Cells were collected at 2, 000 × g for 5 min, resuspended in ∼200 μL of hypotonic buffer, and 5 mL of ice-cold fix (3:1 methanol: acetic acid) was added, with gentle mixing. Fixed cells were stored in −20 °C. For slide preparation the samples were pelleted and resuspended in freshly prepared fix, then dropped onto slides, steamed over a hot water bath for 15 s, and dried on a slide warmer at 60 °C. The slides were stained with Giemsa (VWR) and coverslips were mounted with Permount (Fisher). Images were collected using a Zeiss Axio Imager Z.1, using 63× 1.4 n.a. oil immersion lens. Phenotypes were assigned to mitotic chromosome spreads as previously described (14). Samples were scored blind and each mutant was analyzed in at least 4 independent experiments. At least 100 cells were scored for each sample. Statistical analysis was performed using Fisher’s exact test, 2-tailed, with Bonferroni’s correction for multiple comparisons. All mutants were tested at least 4 times independently (SI Appendix, Table S2).
Coprecipitation Assays.
GST-peptide fusions were purified in lysis buffer (50 mM Tris 8.2, 500 mM KCl, 1 mM dithiothreitol (DTT), 1% Triton X-100) and left on glutathione sepharose 4B (GE Healthcare) beads following purification. The beads were washed extensively in nuclear extract buffer (NEB, 20 mM Hepes 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 2 mM DTT). The beads were then incubated with Xenopus egg extract (50), and mixed on a twirler at 4 °C for 1 to 2 h. The beads were washed 3 times in NEB, and bead-associated proteins were eluted by boiling the beads in sample buffer. To analyze the interactions between purified proteins, purified GST fusions were incubated with beads for 1 h at 4 °C. Bacterially expressed 6His-GFP-PCNA (>95% purity) was added and allowed to incubate for 2 h on a twirler at 4 °C. Beads were washed 3 times with low-salt buffer (50 mM Tris, 100 mM NaCl, 10 mM MgCl2, 0.05% Tween-20) prior to elution by boiling in SDS/PAGE sample buffer. Proteins eluted from the beads were analyzed by immunoblot and Coomassie stain. To prepare yeast whole cell extract, 100 mL of haploid culture (strain Y169; ref. 51) at 5 × 107 cells/mL was collected and cells were washed with water containing 0.2 mM phenylmethylsulfonyl fluoride (PMSF). Cells were pelleted and mixed with equal volume of beads (zirconium oxide, 0.5-mm diameter; Next Advance, Inc.) and 1 mL lysis buffer (25 mM Hepes pH 8.0, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 150 MM KCl, 15% glycerol containing 10 μg/mL each leupeptin, pepstatin, and chymostatin, and 0.2 mM PMSF) and disrupted in a 50-mL conical tube using a Bullet blender (Next Advance) at speed 10 for 15 min at 4 °C. The supernatant was collected and combined with a second 2-mL wash of the same beads in lysis buffer and spun at 24,000 rpm in a SW55 rotor for 60 min. The supernatant was collected, snap frozen in liquid nitrogen in 200 μL aliquots, and stored at −80 °C until use.
Surface Plasmon Resonance.
SPR was performed using a Biacore T200 instrument (GE Healthcare). Anti-GST antibody (laboratory made) was captured by covalent coupling to a Series S CM5 sensor chip (GE Healthcare, Cat. No. BR-1005-30, lot 10272742). The carboxymethylated dextran surface was activated by injection of 1-ethyl-3-(3-dimethylaminopropyl) carbomide hydrochloride (EDC) and ethanoloamide hydrochloride-NaOH pH 8.5 (NHS) from an Amine Coupling Kit (GE Healthcare Cat. No. BR-1000-50). The antibody was immobilized to 2 flow cells using 10 mM Na-acetate pH 5.0 yielding coupling levels of 11,900 to 15,500 response units (RU). Remaining NHS esters were blocked by injection of 1 M ethanolamine, pH 8.5. The binding surface was prepared by capturing GST fusions at a flow rate of 10 μL/min in NEB buffer (20 mM Hepes pH 8.0, 100 mM KCl, 0.2 mM EDTA, 20% glycerol) which led to capture levels between 300 and 800 RU; the control surface was not incubated with GST fusions. The method included 180 s of contact time to capture GST-fusion proteins followed by 180 s of dissociation time, then 180 s of analyte (PCNA) exposure, and 180 s dissociation. The GST fusions were stripped using regeneration with 10 mM glycine pH 2.2 between different analyte concentrations. Serial 2-fold dilutions of His-PCNA starting at 33 μM (trimer concentration) were used for all fusions except p21, which started at 13.3 μM; the lowest concentration was repeated in each run to confirm reproducibility. SPR results were analyzed using Biacore T200 Evaluation software. Sensorgrams were created using the surface bound fraction followed by normalization to baseline after GST-fusion capture, and both the reference well (without GST-fusion protein) and a blank analyte condition (running buffer) were subtracted. Affinity curves and KD were determined using a steady-state affinity model and graphs were prepared in Prism.
Gels and Immunoblots.
For immunoblots, protein samples were resolved on 7 to 15% gradient SDS/PAGE gels, transferred to nitrocellulose membranes, incubated with 5% milk in TBS, and probed with empirically determined concentrations of primary antibodies overnight at 4 °C. Horseradish peroxidase-labeled secondary antibodies were detected with chemiluminescent substrate (Licor Biosciences) and signals were collected using an Azure C600 CCD imager (Azure Biosystems).
Protein Sequence Analysis.
Protein alignments (Fig. 3) were done using Geneious 2019.0 (Biomatters) using the built-in Clustal Omega algorithm and default parameters. The sequence logos were generated in Geneious using RasMol colors. The hydrophobicity prediction graph was generated using a sliding 5-amino acid window.
Plasmids and Antibodies.
See SI Appendix, Tables S4 and S5, respectively, for plasmids and antibodies.
Data Availability.
All data associated with this paper are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01GM101250 and the Oklahoma Center for Adult Stem Cell Research, both to S.R.; a Pat and Don Capra Predoctoral Fellowship to D.B.; and a fellowship from the National Council for Scientific and Technological Development of Brazil (to E.M.L.D.S.). SPR analysis was performed at the Oklahoma Medical Research Foundation Biacore Facility, made possible by grant S10 OD025014-01. We are grateful to Padmaja Mehta-D’souza for assistance with SPR analysis, Paul Kaufman for providing the anti-PCNA antibody, Dean Dawson for careful review of the manuscript, and to all members of the Program in Cell Cycle and Cancer Biology for helpful discussions during the course of this work.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911936117/-/DCSupplemental.
References
- 1.Gerlich D., Koch B., Dupeux F., Peters J.-M., Ellenberg J., Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr. Biol. 16, 1571–1578 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Sherwood R., Takahashi T. S., Jallepalli P. V., Sister acts: Coordinating DNA replication and cohesion establishment. Genes Dev. 24, 2723–2731 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudra S., Skibbens R. V., Sister chromatid cohesion establishment occurs in concert with lagging strand synthesis. Cell Cycle 11, 2114–2121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., et al. , Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31, 143–151 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Sutani T., Kawaguchi T., Kanno R., Itoh T., Shirahige K., Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr. Biol. 19, 492–497 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Rolef Ben-Shahar T., et al. , Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321, 563–566 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Unal E., et al. , A molecular determinant for the establishment of sister chromatid cohesion. Science 321, 566–569 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Rowland B. D., et al. , Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol. Cell 33, 763–774 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Tóth A., et al. , Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320–333 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skibbens R. V., Corson L. B., Koshland D., Hieter P., Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13, 307–319 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou F., Zou H., Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol. Biol. Cell 16, 3908–3918 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellows A. M., Kenna M. A., Cassimeris L., Skibbens R. V., Human EFO1p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7p/Eco1p chromatid cohesion establishment domains. Nucleic Acids Res. 31, 6334–6343 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song J., et al. , Cohesin acetylation promotes sister chromatid cohesion only in association with the replication machinery. J. Biol. Chem. 287, 34325–34336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alomer R. M., et al. , Esco1 and Esco2 regulate distinct cohesin functions during cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 114, 9906–9911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moldovan G. L., Pfander B., Jentsch S., PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 23, 723–732 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Hozák P., Jackson D. A., Cook P. R., Replication factories and nuclear bodies: The ultrastructural characterization of replication sites during the cell cycle. J. Cell Sci. 107, 2191–2202 (1994). [DOI] [PubMed] [Google Scholar]
- 17.O’Keefe R. T., Henderson S. C., Spector D. L., Dynamic organization of DNA replication in mammalian cell nuclei: Spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 116, 1095–1110 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soutoglou E., Misteli T., Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320, 1507–1510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess R. C., Burman B., Kruhlak M. J., Misteli T., Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 9, 1703–1717 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higashi T. L., et al. , The prereplication complex recruits XEco2 to chromatin to promote cohesin acetylation in Xenopus egg extracts. Curr. Biol. 22, 977–988 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Minamino M., et al. , Temporal regulation of ESCO2 degradation by the MCM complex, the CUL4-DDB1-VPRBP complex, and the anaphase-promoting complex. Curr. Biol. 28, 2665–2672.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Vega H., et al. , Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 37, 468–470 (2005). [DOI] [PubMed] [Google Scholar]
- 23.van der Lelij P., et al. , The cellular phenotype of Roberts syndrome fibroblasts as revealed by ectopic expression of ESCO2. PLoS One 4, e6936 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu B., Lu S., Gerton J. L., Roberts syndrome: A deficit in acetylated cohesin leads to nucleolar dysfunction. Rare Dis. 2, e27743 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov M. P., et al. , The replicative helicase MCM recruits cohesin acetyltransferase ESCO2 to mediate centromeric sister chromatid cohesion. EMBO J. 37, e97150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruning J. B., Shamoo Y., Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-δ p66 subunit and flap endonuclease-1. Structure 12, 2209–2219 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Ishida T., Kinoshita K., PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460–W464 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahbubani H. M., Chong J. P., Chevalier S., Thömmes P., Blow J. J., Cell cycle regulation of the replication licensing system: Involvement of a Cdk-dependent inhibitor. J. Cell Biol. 136, 125–135 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter J., Newport J. W., Regulation of replicon size in Xenopus egg extracts. Science 275, 993–995 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Aparicio T., Megías D., Méndez J., Visualization of the MCM DNA helicase at replication factories before the onset of DNA synthesis. Chromosoma 121, 499–507 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Krude T., Musahl C., Laskey R. A., Knippers R., Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J. Cell Sci. 109, 309–318 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Kuipers M. A., et al. , Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J. Cell Biol. 192, 29–41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Biasio A., Blanco F. J., Proliferating cell nuclear antigen structure and interactions: Too many partners for one dancer? Adv. Protein Chem. Struct. Biol. 91, 1–36 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Xu H., Zhang P., Liu L., Lee M. Y., A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry 40, 4512–4520 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Gilljam K. M., et al. , Identification of a novel, widespread, and functionally important PCNA-binding motif. J. Cell Biol. 186, 645–654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boehm E. M., Washington M. T., R.I.P. To the PIP: PCNA-binding motif no longer considered specific: PIP motifs and other related sequences are not distinct entities and can bind multiple proteins involved in genome maintenance. BioEssays 38, 1117–1122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer R. R., et al. , MutLalpha and proliferating cell nuclear antigen share binding sites on MutSbeta. J. Biol. Chem. 285, 11730–11739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dieckman L. M., Freudenthal B. D., Washington M. T., “PCNA structure and function: Insights from structures of PCNA complexes and post-translationally modified PCNA” The Eukaryotic Replisome: A Guide to Protein Structure and Function, Subcellular Biochemistry, MacNeill S., Ed. (Springer, Dordrecht, The Netherlands, 2012), pp. 281–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibahara K., Stillman B., Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Masuda Y., et al. , Different types of interaction between PCNA and PIP boxes contribute to distinct cellular functions of Y-family DNA polymerases. Nucleic Acids Res. 43, 7898–7910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W., An overview of Y-family DNA polymerases and a case study of human DNA polymerase η. Biochemistry 53, 2793–2803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaufmann T., et al. , A novel non-canonical PIP-box mediates PARG interaction with PCNA. Nucleic Acids Res. 45, 9741–9759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murayama Y., Samora C. P., Kurokawa Y., Iwasaki H., Uhlmann F., Establishment of DNA-DNA interactions by the cohesin ring. Cell 172, 465–477.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Biasio A., et al. , p15PAF is an intrinsically disordered protein with nonrandom structural preferences at sites of interaction with other proteins. Biophys. J. 106, 865–874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Havens C. G., Walter J. C., Docking of a specialized PIP box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 35, 93–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomes X. V., Burgers P. M., Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19, 3811–3821 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein P., Pawson T., Tyers M., Mathematical modeling suggests cooperative interactions between a disordered polyvalent ligand and a single receptor site. Curr. Biol. 13, 1669–1678 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Olsen J. G., Teilum K., Kragelund B. B., Behaviour of intrinsically disordered proteins in protein-protein complexes with an emphasis on fuzziness. Cell. Mol. Life Sci. 74, 3175–3183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tompa P., Fuxreiter M., Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 33, 2–8 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Murray A. W., Cell cycle extracts. Methods Cell Biol. 36, 581–605 (1991). [PubMed] [Google Scholar]
- 51.Meyer R. E., et al. , Ipl1/Aurora-B is necessary for kinetochore restructuring in meiosis I in Saccharomyces cerevisiae. Mol. Biol. Cell 26, 2986–3000 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sayle R. A., Milner-White E. J., RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 20, 374 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this paper are included in the manuscript and SI Appendix.