Significance
Circadian rhythms are evolutionarily conserved biological features of most organisms that receive temporal cues from the sun. These rhythms orchestrate important biological processes, such as sleep/wake cycles and metabolism. NOC is a rhythmically expressed protein that, when genetically deleted, confers mice resistance to high-fat diet-induced obesity and hepatic steatosis. Here, we show that NOC is a regulator of intracellular NADP(H) levels and the oxidative stress response. We also show that NOC localizes to the mitochondria and the cytoplasm (including both cytosolic and endoplasmic reticulum-bound pools) and that this is under circadian control. Due to the existence of hundreds of NAD(H)- and NADP(H)-dependent enzymes, understanding the spatiotemporal regulation of these metabolites is critical to human health and disease.
Keywords: circadian, oxidative stress, Nocturnin, mitochondria, NADPH
Abstract
An intimate link exists between circadian clocks and metabolism with nearly every metabolic pathway in the mammalian liver under circadian control. Circadian regulation of metabolism is largely driven by rhythmic transcriptional activation of clock-controlled genes. Among these output genes, Nocturnin (Noct) has one of the highest amplitude rhythms at the mRNA level. The Noct gene encodes a protein (NOC) that is highly conserved with the endonuclease/exonuclease/phosphatase (EEP) domain-containing CCR4 family of deadenylases, but highly purified NOC possesses little or no ribonuclease activity. Here, we show that NOC utilizes the dinucleotide NADP(H) as a substrate, removing the 2′ phosphate to generate NAD(H), and is a direct regulator of oxidative stress response through its NADPH 2′ phosphatase activity. Furthermore, we describe two isoforms of NOC in the mouse liver. The cytoplasmic form of NOC is constitutively expressed and associates externally with membranes of other organelles, including the endoplasmic reticulum, via N-terminal glycine myristoylation. In contrast, the mitochondrial form of NOC possesses high-amplitude circadian rhythmicity with peak expression level during the early dark phase. These findings suggest that NOC regulates local intracellular concentrations of NADP(H) in a manner that changes over the course of the day.
Circadian clocks and metabolism are closely intertwined with extensive circadian control of most metabolic pathways in the mammalian liver. These daily rhythms coordinate and optimize nutrient uptake, utilization, and storage with daily rhythms in food intake and energy expenditure (1–3). One of the metabolically relevant outputs of the circadian clock is NOC, gene name Noct, formerly Ccrn4l. In mice, Noct mRNA is broadly expressed in cells throughout the body but with a particularly high-amplitude rhythm in the liver (4, 5) with peak expression in the early night. Mice lacking NOC (Noct−/−) are protected from high-fat diet-induced obesity and hepatic steatosis but are not more active and do not eat less (4, 6). The mechanism for this protection is not fully understood, but the Noct−/− mice have deficits in the transit of lipids from intestinal enterocytes into the circulation (7) and have increased reliance on lipids for fuel when fed a high-fat diet (4). Somewhat paradoxically, the Noct−/− mice exhibit increased mRNA expression encoding rate-limiting enzymes in cholesterol and triglyceride synthesis pathways in the liver during the night with greater bile acid production and transiently higher levels of plasma triglycerides and lipoproteins in the night (4).
NOC belongs to the large EEP family of proteins with closest similarity to the CCR4 family of deadenylases, which are exonucleases that remove poly(A) tails from mRNAs (8, 9). However, recombinant NOC exhibits weak or no deadenylase activity in vitro (10–12), and analysis of wild-type (WT) and Noct−/− mouse livers found only minor differences in the lengths of poly(A) tails (4, 13). Moreover, recent crystal structures of the catalytic domain of NOC (11, 12) revealed that while the catalytic residues are completely conserved, the substrate-binding residues are divergent, suggesting that poly(A) might not be the biologically relevant substrate.
Here, we use in vitro and in vivo approaches to demonstrate that NOC can remove the 2′-phosphate group from nicotinamide adenine dinucleotide phosphate NADP(H) to generate NAD(H), confirming the recent discovery by Estrella et al. (14). Furthermore, we have found that NOC’s intracellular localization is rhythmic, suggesting that the circadian clock exerts control over local NADP(H) levels within specific regions of the cell at different times of day.
Results
NOC Is a NADP(H) Phosphatase and Regulates Cellular Oxidative Stress Response.
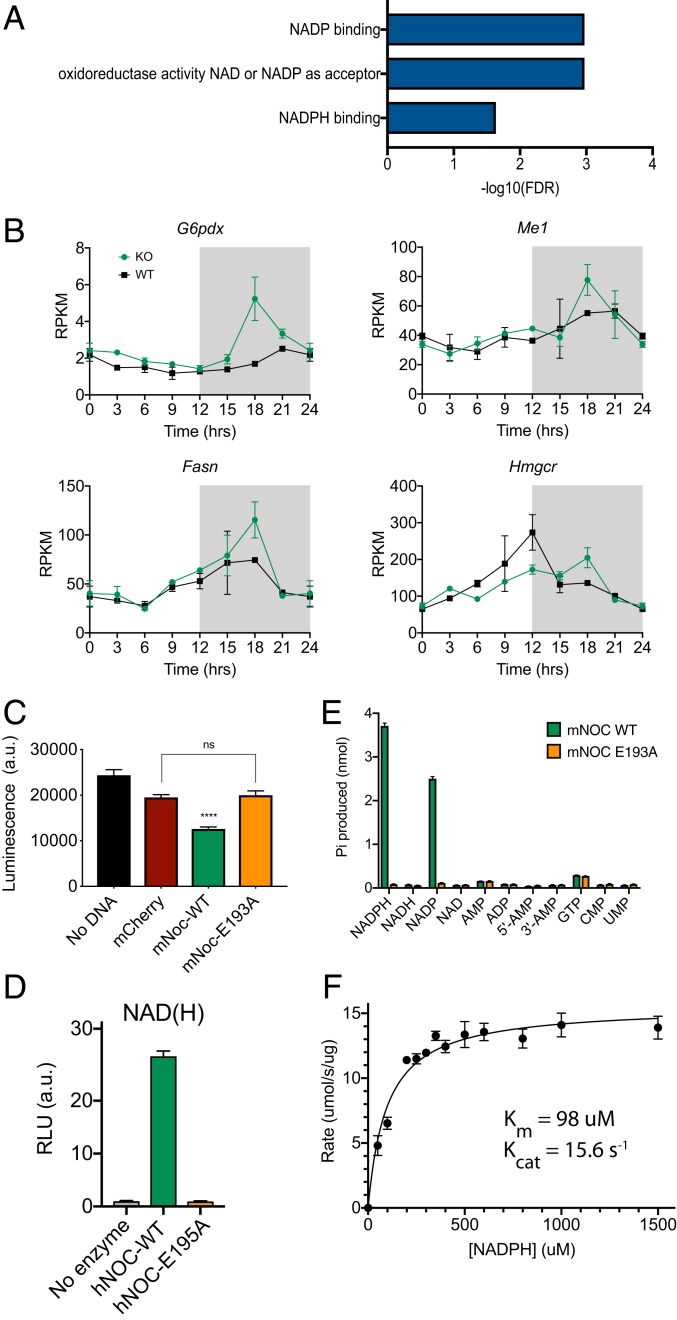
In a previous examination of mRNA expression from WT (Noct+/+) and KO (Noct−/−) mouse livers collected every 3 h over a 24-h day/night cycle, we observed rhythmic mRNAs that had altered rhythmic profiles between the two genotypes. These experiments showed that the KO mice had increased amplitudes of a group of rhythmic mRNAs that peak in the middle of the dark phase, a time when NOC protein levels are high (4). Among these changes were significantly increased amplitudes in rhythms of mRNAs encoding enzymes involved in cholesterol and triglyceride syntheses, resulting in higher nighttime peaks. Although these mRNAs seemed like good candidates for mRNA targets of deadenylase activity, we found that the poly(A) tail lengths of these mRNAs did not show dramatic increases (11), suggesting that these mRNA expression changes were not due to stabilization via poly(A) tail regulation. In order to gain insight into the mechanism of these changes, we reexamined the mRNA-sequencing (mRNA-seq) data by comparing expression differences between the WT and the KO livers in the middle of the night without regard to rhythmicity. Because the peak of many of these rhythms is at Zeitgeber 18 ([ZT18] where ZT is the time of the light:dark cycle in hours where ZT0 is lights on and ZT12 is lights off), we focused on mRNAs that were significantly increased at ZT18 in the KO mice. Among this group of 54 mRNAs, gene ontology analysis (15) revealed significant enrichment in terms NADP, NADPH, and oxidoreductase activity, NAD, or NADP as the acceptor (Fig. 1A). These genes included those involved in NADPH generation from NADP+ (for example, Gpdx1 and Me1) and enzymes that require NADPH as a cofactor for activity (for example, Fasn and Hmgcr) (Fig. 1B).
Fig. 1.
NOC dephosphorylates NADP(H) to produce NAD(H) and orthophosphate. (A) Gene ontology analysis of mRNAs up-regulated in the Noct−/− mouse livers at ZT18 reveals significant enrichment (false discovery rate [FDR] > −log10 = 1) in functional categories related to NADP(H) and NAD(H). (B) Examples of mRNA expression profiles from these enriched categories include mRNAs that encode proteins that produce NADPH from NADP+ (Top row: G6pdx and Me1) and those that use NADPH as a cofactor (Bottom row: Fasn and Hmgcr). Gray shading indicates the dark phase. mRNA expression data are from a previous mRNA-seq experiment (4), and values for ZT0 are replotted as ZT24 to aid visualization. (C) Cellular NADP+ and NADPH levels were measured from HEK 293A cells overexpressing mCherry, mNOC-WT, or mNOC-E193A (catalytically dead mutant) using the enzymatic-cycling NADP(H) Glo Assay (bioluminescent measurements are in arbitrary units [a.u.]). mNOC-WT and mNOC-E193A groups were individually compared to the mCherry negative control using Student’s t test, n = 3, and ****P < 0.0001. (D) NADH production was measured from 0.3 μM of recombinant human NOC or the E195A mutant (catalytically dead) incubated with 50 μM of NADPH for 60 min at 22 °C. The products were diluted 1/200, and NADH production was measured using the NAD(H) Glo Assay, n = 6. (E) Production of orthophosphate from NADPH by 0.35 μM of recombinant mouse NOC or the E193A mutant, incubated with 50 μM of the listed substrates for 60 min at 22 °C was measured using the malachite green assay, n = 6. (F) Initial rates of orthophosphate production were plotted against different concentrations of NADPH and fitted by the Michaelis–Menten equation, n = 6. Recombinant human NOC (0.1 μM) was incubated with NADPH for 5 min at 22 °C.
These gene expression changes suggested that the KO mice have increases in key NADP(H)-requiring enzymes, and therefore, we wondered whether NADP(H) levels might be affected by NOC. To test this directly, we expressed either WT NOC or NOC-E193A [a mutant that is predicted to be catalytically dead based on the crystal structure (11, 12, 14)] in HEK 293A cells and measured total NADP(H) levels using the bioluminescent, enzymatic-cycling NADP(H)-Glo Assay (this measurement does not distinguish between NADP+ and NADPH). Overexpression of the WT enzyme caused a significant reduction in total NADP(H) levels compared to the mCherry and E193A negative controls (Fig. 1C), demonstrating that NOC can regulate NADP(H) levels in a cell-autonomous manner. These findings suggested that NOC may play a direct role in metabolism by regulating the levels of nicotinamide dinucleotides.
Because NADPH is a dinucleotide containing adenine in a configuration similar to poly(A), we wondered whether NOC could use NADPH as a substrate thereby altering NADPH levels directly. This idea was confirmed by a recent publication where the crystal structure of the catalytic domain of NOC bound to NADPH was also reported (14). Similarly, we found that incubation of recombinant human NOC with NADPH resulted in significant production of NADH. This effect was dependent on the conserved metal-binding glutamic acid residue (Fig. 1D; hNOC-E195 is analogous to mNOC-E193), indicating that NOC must remove the 2′ phosphate from NADPH. This was confirmed by measurements of orthophosphate release from NADPH in vitro by the recombinant NOC using the malachite green assay (Fig. 1E). We further showed that recombinant mouse NOC catalyzed the release of orthophosphate specifically from NADPH and to a lesser extent NADP+ with no activity on other nucleotides tested (Fig. 1E). This activity is conserved in human NOC, which has an estimated Km of 98 μM and an estimated Kcat of 15.6 s−1 (Fig. 1F). Therefore, NOC is a phosphatase that dephosphorylates NADPH and NADP+ at the 2′-carbon position of the ribose ring with a preference for NADPH.
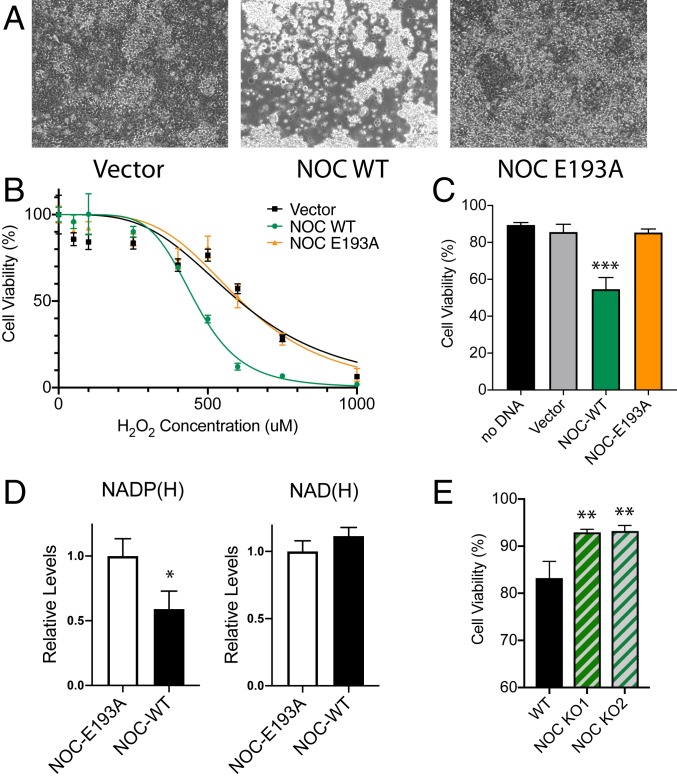
As a molecule with one of the highest reductive potentials in the cell, NADPH is used as the reducing equivalent by thioredoxin and glutathione reductases to regenerate reduced thioredoxin and glutathione in order to regulate the levels of intracellular reactive-oxygen species (ROS). To test whether NOC’s NADP(H) phosphatase activity plays a role in the regulation of cellular response to oxidative stress, HEK 293 cells were transfected with plasmids expressing either WT NOC or the E193A catalytically dead mutant and exposed to 500 μM H2O2-induced oxidative stress. At 500 μM of H2O2, NOC-overexpressing cells were qualitatively more susceptible to cytotoxicity and cell death as captured by phase-contrast microscopy (Fig. 2A and SI Appendix, Fig. S1A). Furthermore, using a quantitative orthogonal redox indicator of cell viability and cytotoxicity, the AquaBluer assay NOC-overexpressing cells were shown to be significantly more susceptible to H2O2 treatment with an EC50 of 482 μM compared to EC50 values of 693 and 680 μM for the vector control and NOC-E193A, respectively (Fig. 2B). Sensitivity to H2O2 at ∼500 μM was additionally confirmed by the increased percentage of trypan blue staining of cells that are overexpressing NOC (Fig. 2C). As the levels of NADP(H) are greatly reduced with minimal to no change in NAD(H), the sensitivity to oxidative stress in NOC-overexpressing cells is likely due to a reduction in NADP(H) (Fig. 2D). To test whether loss of NOC would confer a protective effect, we generated two independent clonal CRISPR-mediated Noct−/− HEK cell lines (SI Appendix, Fig. S1 B and C). Both KO cell lines are more protected against H2O2-mediated oxidative stress (Fig. 2E), suggesting that NOC does play a physiological role in regulating oxidative stress response.
Fig. 2.
NOC regulates H2O2-induced oxidative stress response. (A) Phase contrast-microscopy images (10× objective) of HEK 293 cells transfected with a vector, mNoc-WT, or mNoc-E193A that have been treated with 500 μM of H2O2 for 24 h at 37 °C. (B) Quantification of the AquaBluer cytotoxicity assay for H2O2-treated HEK 293 cells overexpressing mNOC-WT or mNOC-E193A, plotted as a function of H2O2 concentration, n = 4. (C) Quantification of cell viability by trypan blue staining at 500-μM H2O2, n = 4 and ***P < 0.001. (D) Quantification of the levels of total NADP(H) (Left) and NAD(H) (Right) in HEK cells overexpressing mNOC-WT or mNOC-E193A, n = 3 and *P < 0.05. (E) Quantification of the cell viability by trypan blue staining for WT and two independent CRISPR-mediated Noct−/− HEK 293 cell lines that have been treated with 500 μM of H2O2 and incubated for 24 h at 37 °C, n = 3 and **P < 0.01.
Mitochondrial and Cytoplasmic Isoforms of NOC Are Generated by Alternative Translation Initiation Sites.
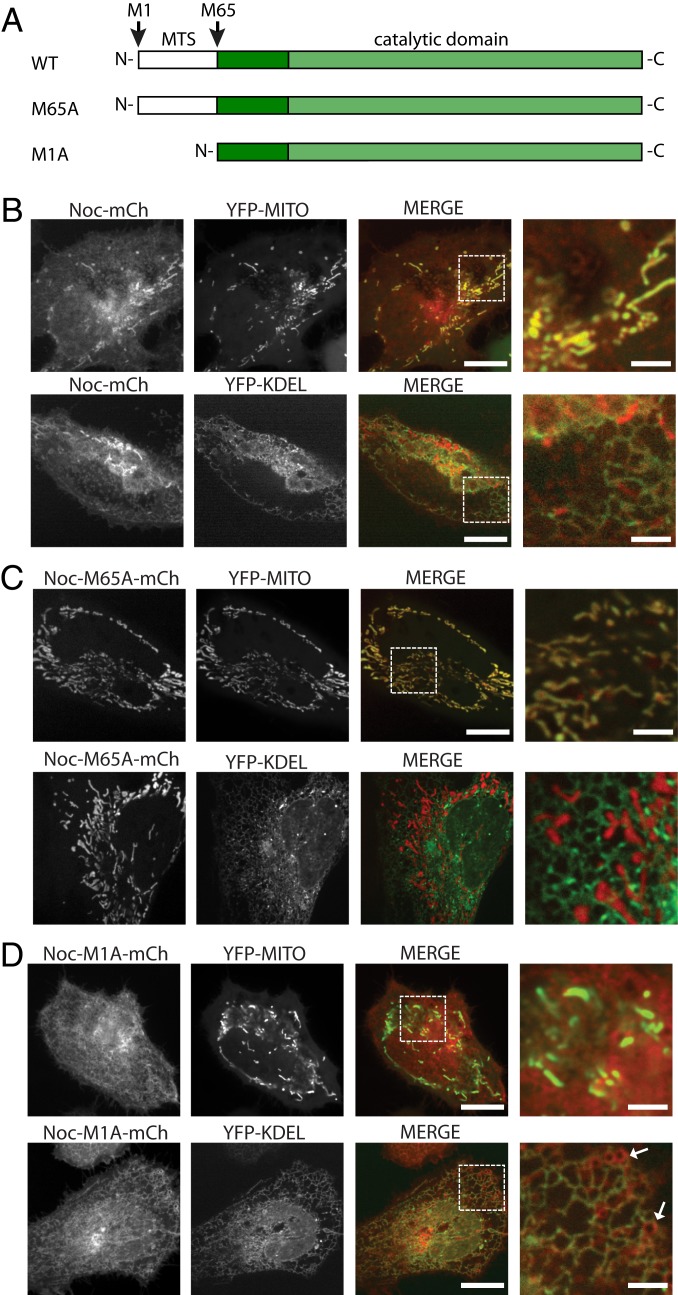
One of the interesting but understudied aspects of nicotinamide dinucleotides is the variation in local subcellular concentrations and their specific contributions to physiology (16). We have previously reported the existence of two isoforms of NOC that are generated by two in-frame translation initiation sites (17, 18). Initiation at Met1 produces a long isoform that encodes a mitochondrial-targeting signal (MTS) resulting in mitochondrial localization (followed by cleavage of the MTS), whereas initiation at Met65 produces a cytoplasmic short isoform (Fig. 3A). Because both translation initiation sites have a nonoptimal Kozak consensus sequence, we hypothesized that the two isoforms may be generated by leaky translation initiation. To test this, the Noct coding sequence, including −10 bp of the upstream endogenous Kozak consensus sequence, was cloned from mouse liver cDNA into a C-terminal mCherry-tagged expression vector. Live-cell imaging in HeLa cells expressing WT mNOC-mCherry reveals both cytoplasmic NOC as well as the previously reported mitochondria-localized isoform of NOC (Fig. 3B). By forcing translation initiation to Met1 in the mNoc-M65A-mCherry mutant, the long isoform was confirmed as the mitochondria-localized NOC (Fig. 3C), supporting our recently published work (17, 18). In contrast, the short isoform (M1A) was present in the cytoplasm in a highly reticular pattern resembling membranes with much of it colocalized with the YFP-KDEL endoplasmic reticulum (ER) marker (Fig. 3D). In addition, NOC-M1A could also be observed on additional membrane-like structures that were not labeled with YFP-KDEL, including small vesicular structures close to the ER (Fig. 3D, white arrows).
Fig. 3.
Cytoplasmic and mitochondrial isoforms of NOC are regulated by alternative translation initiation sites. (A) Cartoon representation of the coding sequences of WT and mutant NOC (mouse numbering). (B) Live-cell imaging of HeLa cells overexpressing mNOC-mCherry (shown in red) with either the YFP-MITO or the YFP-KDEL marker (shown in green). Colocalization can be seen with both markers. The rightmost column is the zoomed-in image of the area within the dotted white square in the merged column. The scale bars are 20 and 5 μM for the nonzoomed and the zoomed-in images, respectively. (C) Live-cell imaging of HeLa cells overexpressing mNOC-M1A-mCherry with either the YFP-MITO or the YFP-KDEL marker. The short isoform of NOC colocalizes with the YFP-KDEL but not the YFP-MITO marker. (D) Live-cell imaging of HeLa cells overexpressing mNOC-M65A-mCherry with either the YFP-MITO or the YFP-KDEL marker. The long isoform of NOC colocalizes with the YFP-MITO but not the YFP-KDEL marker. White arrows point to YFP-KDEL-negative membranous structures.
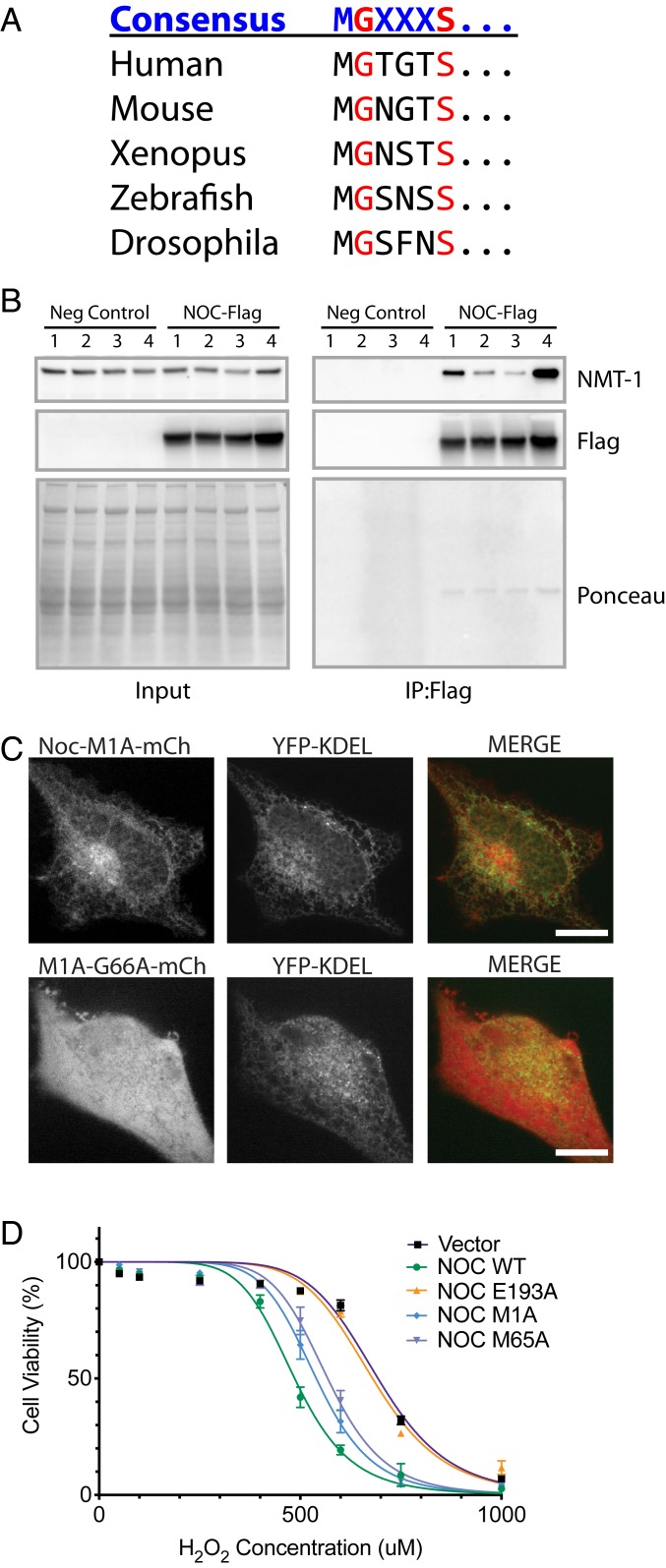
Because the short isoform of NOC does not contain an ER-targeting signal peptide, we reasoned that it does not enter the ER lumen but, instead, may associate with the ER membrane from the outside. Upon closer inspection of the N terminus of the short isoform, we noted that this region contains the MGXXXS consensus motif for the posttranslational modification called N-terminal glycine myristoylation, and this motif is conserved from human to Drosophila (Fig. 4A). Furthermore, in a previous coimmunoprecipitated mass spectroscopy experiment, we identified one of the enzymes that adds the myristoyl modification (glycylpeptide N-tetradecanoyltransferase 1 or [NMT1]) as a top NOC-interacting protein (58% coverage, 150 peptides, and >3,000-fold enriched vs. control). Thus, we hypothesized that the short isoform of NOC associates with the ER and other membranes via the hydrophobic myristoylated Gly66 residue. We validated the NMT1 association by mNOC-Flag immunoprecipitation from liver lysates of mice overexpressing mNOC-Flag specifically in the liver and found that NMT1 consistently coimmunoprecipitated with mNOC-Flag (Fig. 4B). Furthermore, mutation of the conserved glycine (NOC-M1A/G66A) prevented ER localization of the short isoform of NOC resulting in diffuse cytosolic localization (Fig. 4C). Therefore, we conclude that N-terminal glycine myristoylation is required for NOC to interact with the ER and other membranes in the cytoplasm.
Fig. 4.
Cytoplasmic NOC interacts with ER and other membranes via N-terminal glycine myristoylation. (A) Conservation of the N-terminal glycine myristoylation consensus motif of the short isoform of NOC. (B) Immunoblots of NMT1 (Top), Flag (Middle), and Ponceau S staining (Bottom) of the input and immunoprecipitated (IP) fractions from liver lysates of mice overexpressing mNOC-3xFlag specifically in the liver compared to no-transgene-negative (Neg) controls (WT mice without the mNOC-3xFlag transgene). Liver samples from four different mice are shown. (C) Live-cell imaging of HeLa cells overexpressing mNOC-M1A-mCherry or mNOC-M1A/G66A-mCherry (shown in red) and the YFP-KDEL marker (shown in green). (D) Quantification of the AquaBluer cytotoxicity assay for HEK cells overexpressing mNoc or the listed mutants that have been treated with 500 μM of H2O2 for 24 h at 37 °C, n = 4.
We tested whether the ER localization of NOC might play a role in the unfolded protein response due to changes in the cellular reductive environment by measuring XBP1 mRNA splicing (19) after treatment of the cells with DTT, but WT and NOC KO HEK cell lines had indistinguishable responses (SI Appendix, Fig. S2 A and B). Even though there are no observable differences in XBP1 mRNA splicing, H2O2 treatment of HEK cells overexpressing either the cytoplasmic or the mitochondrial isoform of NOC gives an intermediate phenotype compared to the mNOC-WT in the AquaBluer cytotoxicity assay, indicating that they both contribute to the oxidative stress response (Fig. 4D).
Subcellular Localization of NOC Is Regulated by the Circadian Clock.
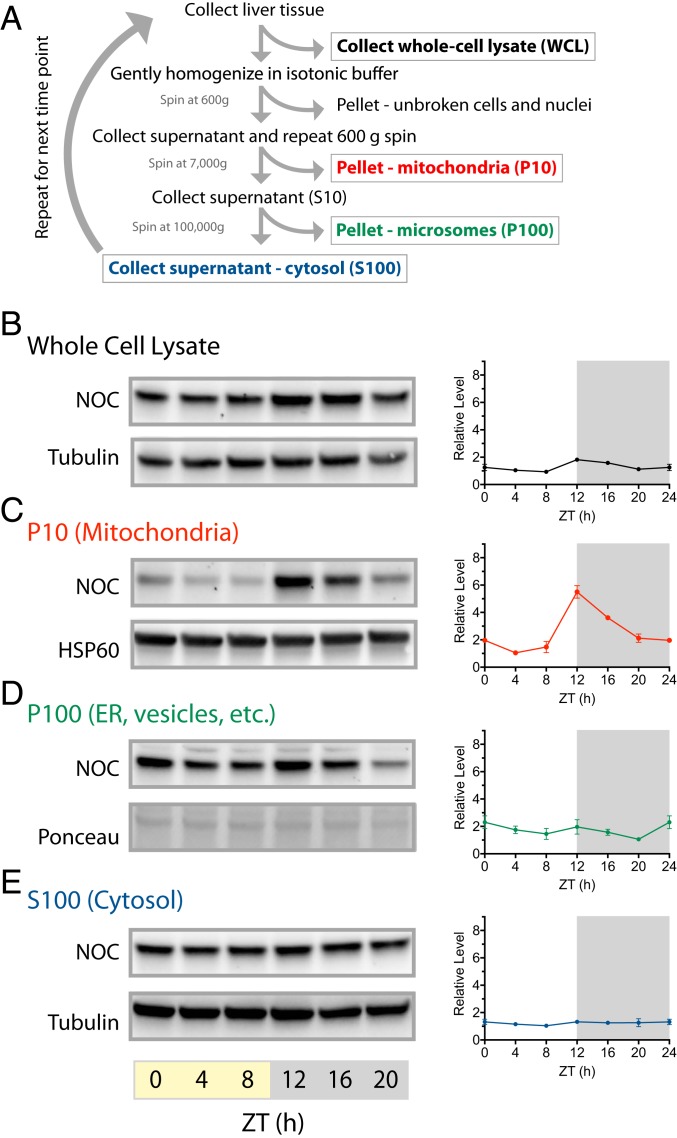
Noct mRNA expression is robustly circadian with high levels during the night and low levels during the day. Given the existence of two different isoforms of NOC, we wondered whether subcellular localization of NOC is temporally regulated by the circadian clock. Because the MTS is cleaved near the second methionine upon import into the mitochondria, the two isoforms of NOC cannot be resolved by size on SDS-PAGE (SI Appendix, Fig. S3A). Therefore, to address the spatiotemporal steady-state levels of NOC, we collected mouse livers every 4 h throughout the 12:12 LD cycle and performed subcellular fractionation (Fig. 5A and SI Appendix, Fig. S4 A and B). In agreement with previously published reports (20, 21), Western blot of the whole-cell lysate showed NOC’s circadian profile with an ∼twofold amplitude of oscillation, peaking at ZT12 (Fig. 5B and SI Appendix, Fig. S4B). Intriguingly, the levels of NOC in the mitochondria-associated P10 fraction are robustly rhythmic with ∼sixfold amplitude of oscillation that peaks at ZT12 (Fig. 5C), closely resembling the high-amplitude oscillation of Noct mRNA. In contrast, NOC levels in the microsome-associated P100 fraction exhibits only low-amplitude changes over the course of the day (Fig. 5D), and the levels of NOC in the cytosolic S100 fraction are constant throughout the 24-h cycle (Fig. 5E).
Fig. 5.
Mitochondrial NOC is specifically regulated by the circadian clock. (A) Schematic of the subcellular fractionation of mouse liver tissue. (B) Representative immunoblot of NOC (Top) and α-tubulin (Bottom) in the whole-cell lysate from ZT0-20. Quantification was performed using three replicates per timepoint (SI Appendix, Fig. S4B). (C) Representative immunoblot of NOC (Top) and HSP60 (Bottom) in the mitochondria-associated P10 fraction from ZT0-20. Quantification was performed using three replicates per timepoint (SI Appendix, Fig. S4B). (D) Representative immunoblot of NOC (Top) and Ponceau S staining (Bottom) in the P100 fraction from ZT0-20. Quantification was performed using three replicates per timepoint (SI Appendix, Fig. S4B). (E) Representative immunoblot of NOC (Top) and α-tubulin (Bottom) in the soluble S100 fraction from ZT0-20. Quantification was performed using three replicates per timepoint (SI Appendix, Fig. S4B).
In order to test whether changes in NADP(H) levels in the NOC KO mice resulted in observable changes in the redox status in vivo, we measured global protein glutathionylation in cytosolic and mitochondrial fractions at two times of day. Indeed, we observed an increase in the levels of protein glutathionylation in the NOC KO animals that is dependent on both the time and the subcellular compartment. In the mitochondria-enriched P10 fraction, there is a greater increase in glutathionylation at ZT12 when NOC levels are normally high (SI Appendix, Fig. S4 C, Top). However, in the liver S100 cytosolic fraction where NOC levels are normally constitutive, there is a general increase in glutathionylation in the KO at both ZT4 and ZT12 (SI Appendix, Fig. S4 C, Bottom).
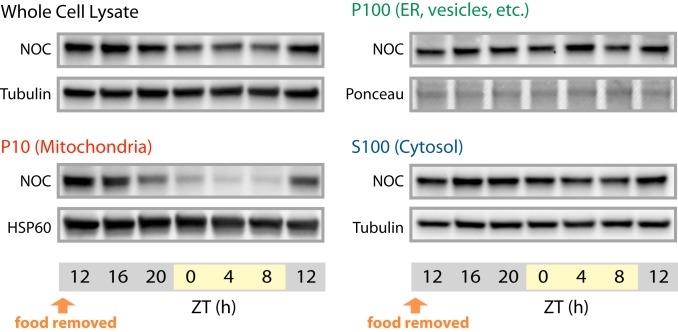
We wondered whether the NOC localization is being controlled by food intake or by redox stress. To test the latter, we fractionated HEK293 cells after 2-h treatment with either H2O2 or DTT. However, no significant change in localization was observed compared to PBS-treated cells (SI Appendix, Fig. S3B). Because NOC’s mitochondrial localization coincides with the onset of food intake and because the phase of NOC rhythmic expression can be shifted by feeding (20), we tested whether the rhythmic localization would be lost in liver tissue harvested from food-deprived mice. Food was removed from WT mice at ZT12, and livers were collected and fractionated at 4-h intervals. Western blot analysis of NOC in the different subcellular fractions revealed the same temporal pattern of mitochondrial localization as previously shown in ad libitum-fed mice (Fig. 6). Therefore, the rhythmic control of NOC expression and localization is not being driven by food intake. Overall, these results suggest that the circadian rhythmicity of NOC is being driven either by the hepatic circadian clock or by an unknown systemic signal and that the absence of NOC leads to a time-dependent change in the redox status in the mitochondria but a constitutive change in the cytosol.
Fig. 6.
Rhythmicity of NOC is not driven by food. Representative immunoblot of NOC from the WCL, P10, P100, and S100 subcellular fractions at the indicated times. α-Tubulin was used as the loading control for WCL and S100 fractions. HSP60 and Ponceau S stain were used as loading for P10 and P100 fractions, respectively.
Discussion
Most organisms that are under the influence of the geological day/night cycles have evolved endogenous circadian timekeeping mechanisms to anticipate daily changes in the environment. As the environmental conditions undergo regular changes in a 24-h cycle, so do the metabolic requirements of these organisms. It is well known that, in mammals, metabolism is linked to the core circadian clock through rhythmic transcriptional activation of clock-controlled genes (1–3). However, an additional layer of regulation can be achieved through circadian changes in metabolites, such as NAD+ levels, that confer rhythmicity to the enzymatic activity of many metabolic enzymes (22).
Here, we show that the clock-controlled protein NOC is a NADP(H) phosphatase with an estimated Km of 98 μM and Kcat of 15.6 s−1. To date, the only other mammalian NADP(H) phosphatase that has been identified is MESH1 [although this paper is not yet published but is archived in bioRxiv (23)], which has a Km in the same range (110 μM) as NOC. Our reported Km is substantially lower than the 331 μM recently reported by Estrella et al. (14), possibly due to different reaction buffer conditions. The intracellular concentration of NADP(H) is difficult to measure precisely, but it has recently become clear that subcellular compartments may have distinct local concentrations (24). Furthermore, most attention on NADP(H) and NAD(H) has been focused on the relative oxidation/reduction status rather than total levels. Changes in total NADP(H) may have profoundly different consequences than changes in the NADP+/NADPH ratio (16).
We confirmed the existence of two isoforms of NOC (mitochondrial and cytoplasmic) that are generated by alternative translation initiation sites and showed that both isoforms are produced when the endogenous Kozak sequence 10 bp upstream of the first methionine was cloned into an expression plasmid, suggesting that leaky translation initiation could potentially contribute to the generation of the two isoforms. We favor translation initiation regulation as the mechanism that regulates these two forms because previous studies have shown only one Noct mRNA in mice in Northern blot experiments (5). However, we cannot rule out the possibility that these two forms could arise from alternate transcription start sites since there could be other mRNAs that may have escaped detection; and in Drosophila, multiple isoforms of Noct mRNA can be found (25). Therefore, it is possible that alternative transcription initiation sites or posttranscriptional regulation may play a role in vivo.
Furthermore, we have shown that the short isoform of NOC localizes to the cytosol, ER, and other membranes and plays a role in oxidative stress response. Membrane localization is likely mediated by N-terminal glycine myristoylation. The purpose of this lipidation event is unclear, but myristoylation has been shown in other cases to be important for both localization and regulation of protein activity or function (26). Also, many NADPH-requiring cytochrome P450 enzymes are anchored to the ER membrane with the catalytic domain exposed to the cytosol (27), suggesting that the regulation of NADPH concentration near the ER membrane may be important.
More strikingly, we showed that the expression of the mitochondrial isoform undergoes large amplitude diurnal regulation, whereas the cytosolic isoform is constitutively expressed. Based on the 4-h interval collection under LD 12:12 and ad libitum-fed conditions, NOC protein levels peak at ZT12. Under the assumption that only the mitochondrial isoform of NOC is circadian and the rest are being constitutively expressed, we estimated the relative amount of each isoform by comparing the ratio of NOC at ZT12 to ZT4 in the whole-cell lysate to the ratio of NOC at ZT12 to ZT4 in the P10 fraction. Based on this, we estimated that the relative amount of the cytoplasmic isoform (including both membrane bound and cytosol) is approximately the same as that of the mitochondrial isoform at ZT12 but is constitutively there throughout the circadian cycle (SI Appendix, Fig. S5). Moreover, the daily rhythm in NOC mitochondrial localization is independent of rhythmic food signals (Fig. 6). The fact that NOC levels are differentially regulated in separate organellar compartments raises an interesting question. Namely, do NADP(H) levels undergo circadian regulation specifically in the mitochondria as a result of NOC’s catalytic activity, and if so, for what purpose?
In the current study, we found that knocking out NOC in mice leads to a constitutive increase in protein glutathionylation in the cytosol and a time-dependent increase in the mitochondria, which correlates with the subcellular compartment-specific localization of NOC. Even though the function of the posttranslational modification of glutathionylation is not fully understood, there is a body of literature suggesting that glutathionylation protects proteins from oxidative damage and is indirectly regulated by NADP(H) levels (28). Because glutathionylation is not rhythmic in the WT background, it is possible that NOC’s role is to help maintain homeostatic levels of protein glutathionylation. More importantly, it suggests that the spatiotemporal regulation of NOC plays a role in cellular compartment-specific redox status.
It is well known that NADPH is required by thioredoxin and glutathione reductases to regenerate reduced thioredoxin and glutathione in order to regulate the levels of ROS (29). The thioredoxin reductase variant that is encoded by the TrxR2 gene is localized to the mitochondria (30). It is possible that circadian redox cycles of thioredoxin due to rhythmic NADPH levels could explain the observation of the circadian rhythms of mitochondrial PrxIII-SO2 levels that have been reported (31). Alternatively, as opposed to reducing ROS levels, NADPH can also be used by NADPH oxidases to generate ROS as signaling molecules (32). Circadian regulation of ROS production could be a mechanism to balance the signaling function with the destructive effects of ROS. Future direct measurement of the levels of ROS in animals lacking NOC will be of interest in this regard.
A change in NADP(H) levels in the mitochondria is likely to have broad metabolic consequences. Mice with decreased levels of mitochondrial NADP(H) caused by specific deletion of the mitochondrial form of NAD kinase (NADK2; the enzyme catalyzing the opposite reaction of NOC) have many negative metabolic features on high-fat diets, including severe hepatic steatosis as compared to WT mice (33). The phenotypes of these mice are, in many respects, opposite of those found in the mice lacking NOC and, therefore, increases in mitochondrial NADP(H) may contribute to the mechanism by which Noct KO mice on high-fat diets are protected from hepatic steatosis and other metabolic problems (6).
Even though the total NAD(H) pool in the mitochondria is much larger than NADP(H) due to the huge abundance of NAD+, NADPH and NADH levels in the mitochondria have been shown to be approximately equimolar (34). It is possible that NADPH de-phosphorylation is a mechanism for generating antiphasic rhythms in mitochondrial NADPH and NADH. Because NADH is the substrate of the electron transport chain complex I, higher levels of mitochondrial NADH generated from NADPH de-phosphorylation could potentially explain our previous report (18) that WT MEFs have higher spare respiratory capacity compared to NOC KO MEFs.
Lastly, it is well known that measurements of local concentrations of NAD(H) and NADP(H) in subcellular compartments are extremely difficult to make, largely due to the difficulty in maintaining their native redox potential and total levels during subcellular fractionation. Bioinformatic analysis has predicted the existence of more than 300 NAD(H)- or NADP(H)-requiring enzymes, some of which are extremely abundant, and any of which can influence the resulting concentration measurements (16). Therefore, future studies that can employ methods of rapid mitochondrial isolation throughout the circadian cycle, such as the MitoTag system (34, 35), will be of great importance in understanding the communication between circadian rhythms and mitochondrial metabolism.
To summarize, NOC is an evolutionarily conserved NADPH phosphatase that links circadian rhythms to metabolism, likely through both the compartmentalization and the temporal regulation of NADP(H). Importantly, this may be one of the mechanisms by which cells temporally organize biochemical reactions where NAD(H)- and NADP(H)-dependent reactions are regulated by having a circadian-controlled enzyme that directly controls the ratio of NADP(H)/NAD(H).
Materials and Methods
Detailed methods, including protein purification, circadian tissue collection, metabolite measurement, cytotoxicity assay, live-cell imaging, and subcellular fractionation, are available in the SI Appendix. All animal work was approved by the UT Southwestern Medical Center institutional animal care and use committee.
Data Availability.
The mRNA-seq data reported in this paper have been deposited the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE105413). All other data and methods are presented within this paper or in the accompanying SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the C.B.G. Laboratory for discussion and critical reading of the paper. mCherry2-N1 was a gift from Michael Davidson (Addgene plasmid # 54517; http://www.addgene.org/54517/; RRID:Addgene_54517). This work was supported by NIH Grants R01 GM112991 and R35 GM127122 (to C.B.G.); NIH Grant R01 GM113079; and NIH Cell and Molecular Biology Training Program Grant T32 GM008203. J.L. is a Sowell Family Scholar in Medical Research, and C.B.G. is a Distinguished Scholar in Neuroscience.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The mRNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE105413). All other data and methods are presented within this paper or in the accompanying SI Appendix.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913712117/-/DCSupplemental.
References
- 1.Green C. B., Takahashi J. S., Bass J., The meter of metabolism. Cell 134, 728–742 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass J., Takahashi J. S., Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panda S., Circadian physiology of metabolism. Science 354, 1008–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stubblefield J. J., et al. , Temporal control of metabolic amplitude by nocturnin. Cell Rep. 22, 1225–1235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., et al. , Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev. Biol. 1, 9 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green C. B., et al. , Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 104, 9888–9893 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douris N., et al. , Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr. Biol. 21, 1347–1355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godwin A. R., Kojima S., Green C. B., Wilusz J., Kiss your tail goodbye: The role of PARN, Nocturnin, and angel deadenylases in mRNA biology. Biochim. Biophys. Acta 1829, 571–579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baggs J. E., Green C. B., Nocturnin, a deadenylase in Xenopus laevis retina: A mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13, 189–198 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Garbarino-Pico E., et al. , Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA 13, 745–755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abshire E. T., et al. , The structure of human Nocturnin reveals a conserved ribonuclease domain that represses target transcript translation and abundance in cells. Nucleic Acids Res. 46, 6257–6270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estrella M. A., Du J., Korennykh A., Crystal structure of human Nocturnin catalytic domain. Sci. Rep. 8, 16294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima S., Gendreau K. L., Sher-Chen E. L., Gao P., Green C. B., Changes in poly(A) tail length dynamics from the loss of the circadian deadenylase Nocturnin. Sci. Rep. 5, 17059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estrella M. A., et al. , The metabolites NADP+ and NADPH are the targets of the circadian protein Nocturnin (Curled). Nat. Commun. 10, 2367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J., Bardes E. E., Aronow B. J., Jegga A. G., ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman R. P., Calvo S. E., Mootha V. K., Spatiotemporal compartmentalization of hepatic NADH and NADPH metabolism. J. Biol. Chem. 293, 7508–7516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le P. T., et al. , A novel mouse model overexpressing Nocturnin results in decreased fat mass in male mice. J. Cell. Physiol. 234, 20228–20239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onder Y., et al. , The circadian protein nocturnin regulates metabolic adaptation in Brown adipose tissue. iScience 19, 83–92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ron D., Walter P., Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Sinturel F., et al. , Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169, 651–663.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu S., et al. , The circadian deadenylase Nocturnin is necessary for stabilization of the iNOS mRNA in mice. PLoS One 6, e26954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peek C. B., et al. , Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342, 1243417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding C.-K. C., et al. , Mammalian stringent-like response mediated by the cytosolic NADPH phosphatase MESH1. bioRxiv: 10.1101/325266 (17 May 2018). [DOI] [Google Scholar]
- 24.Tao R., et al. , Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods 14, 720–728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grönke S., Bickmeyer I., Wunderlich R., Jäckle H., Kühnlein R. P., Curled encodes the Drosophila homolog of the vertebrate circadian deadenylase Nocturnin. Genetics 183, 219–232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udenwobele D. I., et al. , Myristoylation: An important protein modification in the immune response. Front. Immunol. 8, 751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neve E. P., Ingelman-Sundberg M., Intracellular transport and localization of microsomal cytochrome P450. Anal. Bioanal. Chem. 392, 1075–1084 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Ghezzi P., Protein glutathionylation in health and disease. Biochim. Biophys. Acta 1830, 3165–3172 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Ren X., et al. , Redox signaling mediated by thioredoxin and glutathione systems in the central nervous system. Antioxid. Redox Signal. 27, 989–1010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmgren A., Lu J., Thioredoxin and thioredoxin reductase: Current research with special reference to human disease. Biochem. Biophys. Res. Commun. 396, 120–124 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Kil I. S., et al. , Circadian oscillation of sulfiredoxin in the mitochondria. Mol. Cell 59, 651–663 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Ray P. D., Huang B. W., Tsuji Y., Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 24, 981–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K., et al. , Deficiency of the mitochondrial NAD kinase causes stress-induced hepatic steatosis in mice. Gastroenterology 154, 224–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayraktar E. C., et al. , MITO-Tag Mice enable rapid isolation and multimodal profiling of mitochondria from specific cell types in vivo. Proc. Natl. Acad. Sci. U.S.A. 116, 303–312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W. W., Freinkman E., Wang T., Birsoy K., Sabatini D. M., Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166, 1324–1337.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mRNA-seq data reported in this paper have been deposited the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE105413). All other data and methods are presented within this paper or in the accompanying SI Appendix.