Abstract
The dataset showed in this manuscript belongs to the investigation of the Southern-Brazilian geopropolis of stingless bees. Stingless bees are native species of insects from tropical areas; they produce honey, pollen and geopropolis that is composed of a mix of vegetal extracts, digestive enzymes, and mostly by soil. Used in folk medicine as antiseptic, antioxidant and antimicrobial agent, the composition is due to bee species, climate changes, local flora, and soil type. Moreover, the complex chemical content gives to the geopropolis a bioactive potential, with scavenging characteristics that is important to avoid free radical damages in the human health.
Regarding the importance of exploring new natural matrices sources with bioactive potential, the first approach of chemical characterization of geopropolis is indispensable. Thus, ten samples of Southern-Brazilian geopropolis were analyzed and the bioactive responses obtained were discussed in the accompanying article titled “Southern-Brazilian geopropolis: A potential source of polyphenolic compounds and assessment of mineral composition”. Furthermore, the physicochemical analysis of moisture and ash content, the yield of extraction, the reducing activity and free radical scavenging potential of ethanolic extracts, the antimicrobial activity, and the analysis of HPLC-ESI-MS/MS chromatograms are the main data presented in brief. The data can guide scientists in order to know methods and data for these samples.
Keywords: Stingless bess propolis, Melipona mondury, Melipona quadrifasciata, Melipona scutellaris, Melipona seminigra, Tetragonisca angustula, Antimicrobial potential, Chemical characterization
Specifications Table
| Subject | Agricultural and Biological Sciences |
| Specific subject area | Agricultural and Biological Sciences and Food Science |
| Type of data | Tables of sampling, moisture and ash content, extract yield, and ethanolic extracts characteristics Figures of antimicrobial activity and HPLC-ESI-MS/MS Chromatograms Tables and figures |
| How data were acquired | Moisture and ash content: oven SP-400 (SPlabor) and QUIMIS; Extracts yield: analytical scale AB204-S (Mettler Toledo); Reducing activity and free radical scavenging potential: Molecular absorption spectrophotometer UV/Vis (Spectro Vision); HPLC-ESI-MS/MS chromatograms: HPLC model 1200 Series (Agilent Technologies, Alemanha) coupled with mass spectrometer Q Trap 3200 (Applied Biosystems/MDS Sciex, Canada), and Analyst 1.6.2 software; Antimicrobial activity: ultrasonic bath (Unique 1400A); |
| Data format | Jpg images Analyzed Raw (supplementary material) Raw chromatograms |
| Parameters for data collection | All geopropolis samples were previously dried in oven 30 °C, 12 h For moisture and ash content were used raw geopropolis samples For reducing activity and free radical scavenging potential, extract yield, HPLC-ESI-MS/MS chromatograms and antimicrobial activity a solid-liquid extraction procedure was used |
| Description of data collection | The data was collected by measuring the absorbance (UV/Vis) The chromatograms were collected by using Analyst software in the HPLC-ESI-MS/MS system |
| Data source location | Federal University of Santa Catarina Laboratory of Food Chemistry, Florianópolis, Santa Catarina, Brazil The samples were collected during 2016–2017 at Santa Catarina state (further details below) |
| Data accessibility | Within the article and in supplementary material |
| Related research article | Ferreira B. L., Gonzaga L. V., Vitali L., Micke G. A., Maltez H. F., Ressureição C., Costa A. C. O., Fett R. Southern-Brazilian geopropolis: A potential source of polyphenolic compounds and assessment of mineral composition Food Research International 10.1016/j.foodres.2019.108683 |
Value of the Data
|
1. Data
The dataset in this article describes some physical and chemical characteristics of ten samples of geopropolis. Table 1 describes the sample collection with general information about bee species, the geographical location where the geopropolis were collected, also the code used to refer to each sample. The percentage of moisture and ash content are showed in Table 2. In Table 3, it is describing the yield of extraction regarding two different solvents and three different periods for each geopropolis sample, elsewhere the statistical standard deviation and analysis of means by Tukey's test (95%). Regarding the ethanol as solvent, Table 4 brings the reducing activity and the free radical scavenging potential of geopropolis samples in three different periods of extraction. The mass/charge relation of each polyphenolic compound indicating the parent íon and the quantification íon, in addition to the retention time is in the Table 5.
Table 1.
Samples of geopropolis, location, and reference codes.
| Stingless Bee Species | Code | Location | Latitude | Longitude | Altitude |
|---|---|---|---|---|---|
| Melipona mondury | MMS | Santa Rosa de Lima (C) | 28°02′21″ south | 49°7′40″ west | 240 m |
| MMI | Iporã do Oeste (A) | 26°8′8″ south | 53°53′5″ west | 557 m | |
| Melipona quadrifasciata | MQS | Santa Rosa de Lima (C) | 28°02′21″ south | 49°7′40″ west | 240 m |
| MQR | Rio do Sul (B) | 27°12′51″ south | 49°38′35″ west | 340 m | |
| MQF | Florianópolis (D) | 27°35′48″ south | 48°32′57″ west | 0 m | |
| MQI | Iporã do Oeste (A) | 26°8′8″ south | 53°53′5″ west | 557 m | |
| Melipona scutellaris | MSS | Santa Rosa de Lima (C) | 28°02′21″ south | 49°7′40″ west | 240 m |
| MSI | Iporã do Oeste (A) | 26°8′8″ south | 53°53′5″ west | 557 m | |
| Melipona seminigra | MSeS | Santa Rosa de Lima (C) | 28°02′21″ south | 49°7′40″ west | 240 m |
| Tetragonisca angustula | TAI | Iporã do Oeste (A) | 26°8′8″ south | 53°53′5″ west | 557 m |
 | |||||
Table 2.
Moisture and ash content of crude geopropolis samples.
| Samples | Moisture (%) | Ash content (%) |
|---|---|---|
| MMS | 3.51 ± 0.10 | 80.18 ± 2.50 |
| MMI | 2.60 ± 0.02 | 77.51 ± 0.73 |
| MQS | 3.79 ± 0.28 | 66.01 ± 0.58 |
| MQR | 4.77 ± 0.06 | 51.97 ± 0.76 |
| MQF | 3.65 ± 0.17 | 65.96 ± 0.68 |
| MQI | 3.32 ± 0.10 | 58.32 ± 0.38 |
| MSS | 3.23 ± 0.26 | 78.29 ± 0.30 |
| MSI | 3.21 ± 0.02 | 70.17 ± 1.23 |
| MSeS | 8.80 ± 0.19 | 71.48 ± 0.84 |
| TAI | 4.12 ± 0.11 | 2.23 ± 0.01 |
Data showed by percentage (mean ± standard deviation, n = 3). Raw data available in supplementary material 1.
Table 3.
Yield of extraction of geopropolis samples regarding pure ethanol and pure methanol as solvents over storage time.
| Samples | Ethanolic extraction |
Methanolic extraction |
||||
|---|---|---|---|---|---|---|
| 10 days | 20 days | 30 days | 10 days | 20 days | 30 days | |
| MMS | 10.91 ± 1.71a | 11.31 ± 1.14a | 9.70 ± 0.01a | 12.47 ± 0.01a | 14.96 ± 0.78b | 12.05 ± 0.59a |
| MMI | 64.77 ± 6.33a | 42.68 ± 3.48a | 29.96 ± 0.58a | 40.84 ± 2.31a | 46.56 ± 5.78a | 47.38 ± 4.62a |
| MQS | 179.52 ± 1.15a | 174.65 ± 5.64a | 173.02 ± 1.15a | 157.96 ± 0.01a | 158.79 ± 3.53a | 165.44 ± 8.23a |
| MQR | 338.83 ± 1.76a | 336.75 ± 7.06a | 350.05 ± 14.11a | 331.17 ± 0.58a | 329.54 ± 5.20a | 355.67 ± 13.28a |
| MQF | 229.38 ± 0.58a | 231.85 ± 0.58b | 233.91 ± 0.01c | 237.92 ± 4.10a | 239.57 ± 1.76a | 261.50 ± 2.34b |
| MQI | 185.80 ± 3.49a | 190.73 ± 11.63a | 203.48 ± 12.21a | 192.74 ± 2.32a | 212.01 ± 0.58b | 220.22 ± 1.74c |
| MSS | 29.73 ± 1.14a | 26.11 ± 0.57a | 25.31 ± 0.57a | 25.59 ± 1.17a | 36.32 ± 0.01b | 23.11 ± 0.01a |
| MSI | 59.48 ± 1.15a | 67.22 ± 7.49a | 64.77 ± 6.34a | 114.97 ± 1.76a | 136.97 ± 9.39a | 117.46 ± 0.59a |
| MSeS | 23.17 ± 0.01a | 21.32 ± 1.12a | 22.90 ± 1.12a | 15.78 ± 1.17a | 25.09 ± 0.23b | 10.80 ± 1.17c |
| TAI | 395.95 ± 8.78a | 398.43 ± 1.76a | 403.40 ± 1.76a | 196.37 ± 2.92a | 213.32 ± 0.01a | 202.16 ± 6.43a |
Data showed as mg g-1 89 (mean±standard deviation, n=3). Different letters in the same line regarding the same solvent indicate statistical difference according to Tukey's test (95%). Raw data available in supplementary material 2.
Table 4.
Reducing activity and the free radical scavenging potential of crude geopropolis samples in three different periods of extraction using ethanol as extractor agent.
| Samples | Days of extraction | Ethanolic extraction |
||
|---|---|---|---|---|
| Reducing activity (GAE mg 100g−1) | Free radical scavenging potential |
|||
| AAE mg 100g−1 | TE mg 100g−1 | |||
| MMS | 10 | 62.68 ± 0.77 | 75.83 ± 1.15 | 111.32 ± 1.68 |
| 20 | 65.40 ± 1.71 | 74.26 ± 2.83 | 109.01 ± 4.16 | |
| 30 | 65.79 ± 2.97 | 77.96 ± 0.44 | 114.44 ± 0.64 | |
| MMI | 10 | 479.15 ± 31.05 | 549.86 ± 10.41 | 806.95 ± 15.29 |
| 20 | 491.73 ± 2.90 | 528.79 ± 7.89 | 775.99 ± 11.59 | |
| 30 | 549.81 ± 13.31 | 602.88 ± 7.37 | 884.84 ± 10.83 | |
| MQS | 10 | 1023.37 ± 15.37 | 1164.56 ± 3.84 | 1709.16 ± 5.64 |
| 20 | 1021.44 ± 39.22 | 1149.70 ± 9.03 | 1687.31 ± 13.26 | |
| 30 | 1046.61 ± 8.71 | 1218.26 ± 13.30 | 1788.04 ± 19.53 | |
| MQR | 10 | 1067.96 ± 134.74 | 254.11 ± 3.42 | 372.88 ± 5.03 |
| 20 | 1258.21 ± 26.53 | 286.24 ± 1.32 | 420.08 ± 1.94 | |
| 30 | 1319.99 ± 4.49 | 325.15 ± 1.00 | 477.24 ± 1.46 | |
| MQF | 10 | 1651.45 ± 44.52 | 3057.62 ± 32.88 | 4487.67 ± 48.31 |
| 20 | 1748.59 ± 33.65 | 3390.98 ± 26.96 | 4977.41 ± 39.61 | |
| 30 | 2069.16 ± 261.20 | 3613.46 ± 22.16 | 5304.25 ± 32.56 | |
| MQI | 10 | 1555.32 ± 47.62 | 1624.30 ± 24.74 | 2382.00 ± 36.34 |
| 20 | 1508.78 ± 26.66 | 1665.46 ± 17.76 | 2442.45 ± 26.09 | |
| 30 | 1557.26 ± 42.88 | 1954.49 ± 6.93 | 2867.08 ± 10.18 | |
| MSS | 10 | 439.05 ± 10.60 | 977.36 ± 6.50 | 1434.12 ± 9.56 |
| 20 | 456.69 ± 23.34 | 1018.49 ± 19.83 | 1494.53 ± 29.13 | |
| 30 | 460.61 ± 30.17 | 1111.43 ± 4.11 | 1631.07 ± 6.04 | |
| MSI | 10 | 1652.80 ± 24.00 | 1927.42 ± 38.60 | 2827.34 ± 56.70 |
| 20 | 1395.28 ± 17.61 | 1780.25 ± 8.49 | 2611.13 ± 12.47 | |
| 30 | 1326.09 ± 53.26 | 2065.76 ± 11.47 | 3030.58 ± 16.86 | |
| MSeS | 10 | 67.83 ± 1.60 | 63.05 ± 1.58 | 92.54 ± 2.32 |
| 20 | 67.73 ± 0.60 | 64.96 ± 0.95 | 95.34 ± 1.40 | |
| 30 | 71.09 ± 4.58 | 70.56 ± 1.70 | 103.58 ± 2.50 | |
| TAI | 10 | 1301.95 ± 109.71 | 347.71 ± 9.84 | 510.39 ± 14.46 |
| 20 | 1231.68 ± 106.86 | 315.44 ± 12.76 | 462.99 ± 18.74 | |
| 30 | 1370.27 ± 129.01 | 349.03 ± 20.63 | 512.34 ± 30.31 | |
Data showed as mg 100 g−1 (mean ± standard deviation, n = 3). Raw data available in supplementary material 3.
Table 5.
Mass/charge relation of each polyphenolic compound analized in the geopropolis samples.
| Polyphenolic compound | Parent íon (m/z) - Q1 | Quantitative íon (m/z) - Q3 | Retention time (min) |
|---|---|---|---|
| Gallic ac | 168.908 | 125 | 3.98 |
| Protocatechuic ac | 152.921 | 109 | 6.95 |
| Mandelic ac | 150.996 | 107 | 7.86 |
| Catechin | 289.045 | 109 | 8.82 |
| 4-(Hydroxymethyl)benzoic ac | 150.967 | 107 | 8.84 |
| Chlorogenic ac | 353.155 | 191 | 9.19 |
| Epicatechin | 288.954 | 109 | 9.41 |
| Caffeic ac | 178.927 | 135 | 9.45 |
| Vanillic ac | 166.923 | 108 | 9.65 |
| Syringic ac | 196.939 | 121.1 | 10.01 |
| Epicatechin gallate | 441.6 | 168.9 | 10.15 |
| Fustin | 286.969 | 109 | 10.32 |
| Vanilin | 150.958 | 136 | 10.42 |
| p-Coumaric ac | 162.926 | 119.1 | 10.46 |
| 4-aminobenzoic ac | 135.995 | 91.9 | 10.47 |
| α-Methoxyphenylacetic ac | 164.976 | 121.1 | 10.51 |
| Taxifolin | 303.019 | 125.1 | 10.7 |
| Rutin | 609.242 | 300.1 | 10.72 |
| Ferulic ac | 192.957 | 134 | 10.73 |
| Syringaldehyde | 180.94 | 151 | 10.76 |
| Umbelliferone | 160.941 | 133.1 | 10.78 |
| Rosmarinic ac | 359.082 | 161 | 10.83 |
| Isoquercitrin | 463.155 | 300 | 10.83 |
| Quercetin | 300.968 | 151 | 10.84 |
| Sinapic ac | 223.011 | 148.8 | 10.87 |
| Salicylic ac | 136.942 | 93 | 10.99 |
| Escopoletin | 190.972 | 176 | 10.99 |
| Resveratrol | 226.999 | 142.9 | 11.14 |
| Naringin | 580.276 | 151 | 11.18 |
| Miricetrin | 316.995 | 151 | 11.24 |
| Aromadendrin | 287.004 | 125 | 11.29 |
| Coniferaldehyde | 177.015 | 162 | 11.31 |
| p-Anisic ac | 150.947 | 136 | 11.34 |
| Sinapaldehyde | 207.04 | 177 | 11.39 |
| Ellagic ac | 300.959 | 145 | 11.71 |
| Cinnamic ac | 146.952 | 102.9 | 11.8 |
| Eriodictyol | 186.97 | 151 | 11.85 |
| Kaempferol | 284.995 | 93 | 12.34 |
| Naringenin | 270.985 | 151.1 | 12.37 |
| Apigenin | 268.992 | 117.1 | 12.62 |
| Hispidulin | 298.957 | 284 | 12.72 |
| Galangin | 268.981 | 117 | 13.44 |
| Pinocembrin | 255.051 | 65 | 13.59 |
| Chrysin | 252.988 | 62.9 | 13.88 |
| Carnosol | 329.167 | 285.2 | 14.32 |
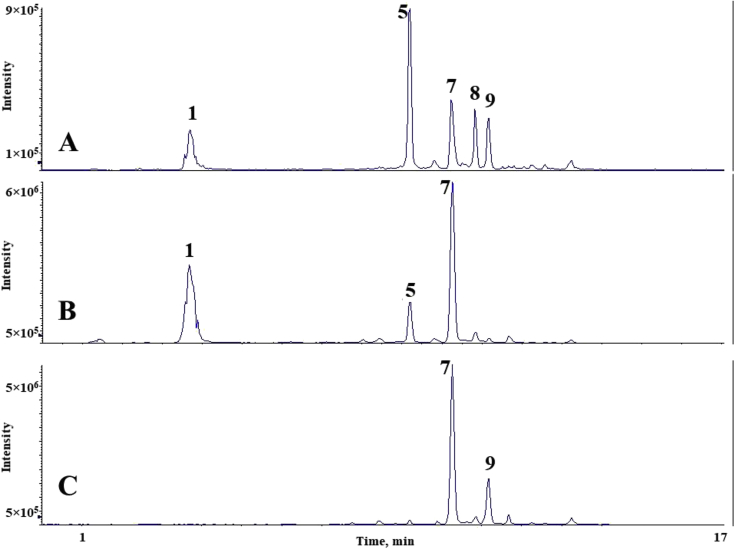
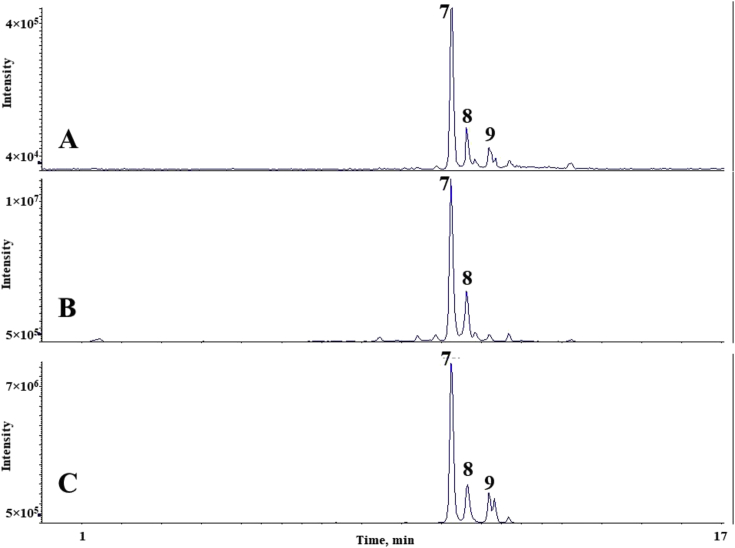
The chromatograms of polyphenolic analysis are in Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11. The analytical standards separation is represented in Fig. 1. Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11 are the geopropolis samples, regarding the use of three different strategies to access the polyphenolic composition of each: free polyphenolics, and bonded polyphenolics by using acid and alkaline hydrolysis. Finally, Fig. 12 showed the antimicrobial potential of geopropolis samples.
Fig. 1.
HPLC-ESI-MS/MS chromatograms of polyphenolic standards. *Polyphenolic compounds on a mix solution of standards: (1) Gallic ac. (2) Protocatechuic ac. (3) Mandelic ac. (4) Catechin, 4-(Hydroxymethyl)benzoic ac. (5) Vanillic ac, Caffeic ac, Chlorogenic ac, Epicatechin. (6) Syringic ac. (7) Vanilin, 4-aminobenzoic ac, p-Coumaric ac, α-Methoxyphenylacetic ac, Syringaldehyde, Taxifolin, Epicatechin gallate, Rutin. (8) Salicylic ac, Umbelliferone, Escopoletin, Ferulic ac, Sinapic ac, Rosmarinic ac, Isoquercitrin, Naringin, Fustin. (9) Aromadendrin, p-Anisic ac, Coniferaldehyde, Sinapaldehyde, Resveratrol, Miricetrin. (10) Cinnamic ac, Eriodictyol, Ellagic ac, Quercetin. (11) Galangin, Naringenin, Kaempferol. (12) Apigenin, Hispidulin. (13) Pinocembrin. (14) Chrysin. (15) Carnosol. The chemical structure of each polyphenolic compound are in the supplementary material of [4].
Fig. 2.
Polyphenolic profile of MMS sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 3.
Polyphenolic profile of MMI sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 4.
Polyphenolic profile of MQS sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 5.
Polyphenolic profile of MQR sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 6.
Polyphenolic profile of MQF sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 7.
Polyphenolic profile of MQI sample (HPLC-ESI-MS/MS chromatogram) (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 8.
Polyphenolic profile of MSS sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 9.
Polyphenolic profile of MSI sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 10.
Polyphenolic profile of MSeS sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 11.
Polyphenolic profile of TAI sample (HPLC-ESI-MS/MS chromatogram). (A) Free polyphenolic profile. (B) Polyphenolic profile after acid hydrolysis. (C) Polyphenolic profile after alkaline hydrolysis. Each peak can possess more than one polyphenolic compound, according to Fig. 1. The results regarding the polyphenolic quantification are in the original paper [4].
Fig. 12.
Antimicrobial potential of MQF sample. Sample MQF (100 (A) and 200 mg mL-1 (B)) showed inhibition halo formation surrounding the well containing S. aureus.
2. Experimental design, materials, and methods
Initially 10 geopropolis samples of Melipona mondury (n = 2), Melipona quadrifasciata (n = 4), Melipona scutellaris (n = 2), Melipona seminigra (n = 1) and Tetragonisca Angustula (n = 1) were collected in three cities of Santa Catarina State, Brazil: Santa Rosa de Lima, Rio do Sul, Iporã do Oeste and Florianópolis region characterized by the tropical climate.
Samples were dried in an oven at 30 °C for 12 h to avoid biological damages, subsequently grinded to standard the particle size and storage at −18 °C in the dark until the analysis moment.
2.1. Determination of moisture and ash content
The moisture (925.09) content was determined using 3 g of each geopropolis sample in porcelain caps previously dried, and then samples were placed in oven at 105 °C until constant weight [6]. Subsequently the residue of moisture content was reused to ash content (923.03) determination. The caps were heated in oven at 550 °C until constant weight [6]. Both datas were expressed in % (m/m) of moisture and % (m/m) of ash content for each geopropolis sample.
2.2. Extraction procedure, the yield of extraction and determination of reducing activity and the free radical scavenging potential
The details about the extraction procedure and the yield determination are available at [4]; topic 2.2.1. Briefly the extraction in two different solvents (pure methanol and pure ethanol) in a solid-liquid ratio of 3 g/10 mL were used for the determination of yield of extraction and the determination of reducing activity and the free radical scavenging potential of geopropolis samples in three different periods of extraction.
The determination of reducing activity was evaluated according to the capacity of extract to reduce the Folin-Ciocalteau reagent [8]. A hundred microliters of each geopropolis extract were added in a 10 mL glass tube with 2 mL of ultra-pure water, then 500 μL of Folin-Ciocalteau was added, and the reaction occurred after the addition of 1.5 mL of sodium carbonate (20% m/m). After 2 h, the absorbance was read in 765 nm, and the results evaluated in gallic acid equivalents (mg GAE 100−1g of sample) [8].
The free radical scavenging potential was determined according to the DPPH method. A methanolic DPPH solution (Abs 515nm 0.800) was added in cuvettes (2.9 mL) with 100 μL of each geopropolis sample. The absorbance was read after 30 min in the absence of light in 515 nm, and the results evaluated in ascorbic acid equivalents (mg AAE 100−1g of sample) and Trolox equivalent (mg TE 100−1g of sample) [2].
2.3. Polyphenolic composition by HPLC–ESI-MS/MS
For the polyphenolic determination showed in Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11, three extraction strategies were used to investigate the free and bonded phenolic compounds. First the free phenolic compounds were analyzed using a solid-liquid extraction regarding the methodology needs [1,7].
Second, to investigate the bonded polyphenolic compounds, an acid [7] and alkaline [5] hydrolysis were used in order to release this compounds to the solution.
The chromatographic separation occurred in an HPLC-ESI-MS/MS system, coupled with mass spectrometer. The details about the extraction method and the separation conditions are available in Ref. [4].
2.4. Antimicrobial potential
One gram of each geopropolis sample was extracted with 5 mL of methanol. Samples were extracted in the ultrasonic bath for 30 minutes (room temperature); after that, were kept under low temperature (5 ± 2 °C) for 24 h, after that were again sonicated for more 30 minutes. The supernatant was separated in a centrifuge and reduced under low pressure until complete solvent evaporation. Subsequently, 5 mL of DMSO have used to recovery the geopropolis samples, filtered in 0.45 μm polytetrafluoroethylene syringe filter and analyzed.
Mueller Hinton agar plates with available cells of Escherichia coli (ATCC: 25922), Staphylococcus aureus (ATCC: 25923) and Salmonella typhimurium (ATCC: 14028) in 105 CFU/mL cultivated in BHI broth were used to determinate the antimicrobial potential, according to agar diffusion method with wells technique [3].
The agar plates were perforated and 6–8 mm wells were performed. 30 μL of each geopropolis extracts (200 and 150 mg mL-1) were added in the wells followed by negative control (pure DMSO) and positive control (ciprofloxacin 0.05 mg mL−1). Petri plates were incubated at 37 °C for 24 h. The potential antimicrobial effect was attributed when observed halo formation surrounding geopropolis samples wells. Assays were performed in duplicate.
Acknowledgments
The authors are grateful to Universidade Federal de Santa Catarina, CAPES, and CPNq for financial support [132384/2016-7]. Furthermore, we are thankful for all geopropolis samples provided by beekeepers from FASC (Beekeepers Federated Association of Santa Catarina State).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2020.105109.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Biluca F.C. Phenolic compounds, antioxidant capacity and bioaccessibility of minerals of stingless bee honey (Meliponinae) J. Food Compos. Anal. 2017;63:89–97. out. [Google Scholar]
- 2.Brand-Williams W., Cuvelier M.E., Berset C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. jan. 1995;28(1):25–30. [Google Scholar]
- 3.Dimkić I. Phenolic profiles and antimicrobial activity of various plant resins as potential botanical sources of Serbian propolis. Ind. Crops Prod. 2016;94:856–871. 30 dez. [Google Scholar]
- 4.Ferreira B.L. Southern-Brazilian geopropolis: a potential source of polyphenolic compounds and assessment of mineral composition. Food Res. Int. 2019;126:108683. doi: 10.1016/j.foodres.2019.108683. dez. [DOI] [PubMed] [Google Scholar]
- 5.Nardini M. Detection of bound phenolic acids: prevention by ascorbic acid and ethylenediaminetetraacetic acid of degradation of phenolic acids during alkaline hydrolysis. Food Chem. 2002;79(1):119–124. out. [Google Scholar]
- 6.Official Methods of Analysis of AOAC INTERNATIONAL. vol. 18. AOAC INTERNATIONAL; Gaithersburg: 2005. [Google Scholar]
- 7.Schulz M. Chemical composition, bioactive compounds and antioxidant capacity of juçara fruit (Euterpe edulis Martius) during ripening. Food Res. Int. 2015;77:125–131. [Google Scholar]
- 8.Singleton V.L., Rossi J.A.J. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965;16:144–158. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.