Abstract
Exosomes mediate cell-cell crosstalk in cancer progression by transferring a variety of biomolecules, including long noncoding RNAs (lncRNAs). Long non-coding RNA urothelial carcinoma-associated (UCA1) is a well-known lncRNA associated with the development and progression of various cancers, including colorectal cancer (CRC). However, the presence of UCA1 in exosomes and the roles and clinical values of exosomal UCA1 in CRC remain unknown. In this study, we systematically analyzed the expression profiles of exosomal lncRNAs in CRC patients using a high-throughput microarray assay. Then, we evaluated the UCA1 expression levels in a series of CRC tissues and the serum exosomes of CRC patients using quantitative real-time PCR. The roles of UCA1 on CRC in vitro and in vivo were investigated by MTT, colony formation, Transwell, quantitative real-time PCR, flow cytometry, and western blotting. The miRNA binding sites of UCA1 were predicted using the miRcode online database, and miR-143 was validated to target UCA1 by dual-luciferase activity assay and AGO2 RNA immunoprecipitation. Finally, the role of exosome-mediated UCA1 was further investigated by co-culturing with CRC cells. This study showed that UCA1 was upregulated in CRC tissues and functioned as an oncogene in CRC. Loss-of-function investigations showed that inhibition of UCA1 suppressed CRC cell proliferation and metastasis in vivo and in vitro. Mechanistically, UCA1 was identified as a miR-143 sponge. We also found that MYO6 was a direct target of miR-1205, which functioned as an oncogene in CRC. Moreover, UCA was also upregulated in the serum exosomes of CRC patients and could transfer UCA1 to CRC cells to increase their abilities of cell proliferation and migration. In conclusion, these data suggest that UCA1 could be an oncogene for CRC and may serve as a candidate target for new therapies in human CRC.
Keywords: exosome, UCA1, long noncoding RNA, CRC, ceRNA, metastasis
Introduction
Colorectal carcinoma (CRC) ranks third in terms of incidence and remains the second leading cause of cancer-related mortality worldwide.1 Early-stage CRC patients could be cured by surgery.2 Unfortunately, chemotherapy and surgical resection are not effective for patients with advanced CRC.3 Despite recent progresses in the therapeutic strategies for CRC, including targeted therapies and immunotherapy, the prognosis for advanced-stage patients is still far from satisfactory.4 Thus, further understanding of the molecular mechanisms of CRC may help identify potential tumor biomarkers and develop novel therapeutic targets.
Long non-coding RNAs (lncRNAs), with a length >200 nt, were once considered as junk DNA and transcriptional noise.5 Emerging evidence demonstrates that lncRNAs are evolutionarily conserved and that their expression is strictly regulated, playing critical roles in regulating gene expression.6 Notably, a large body of recent evidence indicates that lncRNAs are involved in the initiation and progression of CRC.7,8 Yu et al.9 found a novel lncRNA, u50535, which was greatly overexpressed in CRC tissues and was associated with poor prognosis in CRC patients. Wu et al.10 established and validated NEAT1 as a new diagnostic biomarker in CRC, and NEAT1 expression was associated with different prognostic outcomes. Recently, some studies showed that UCA1, initially discovered in bladder cancer and located at chromosome 19p13.12, is aberrantly expressed in various cancers, including CRC.11, 12, 13 Although many previous reports have documented that dysregulated expressions of UCA1 have been identified in CRC and play important roles in the pathogenic process of CRC; the precise molecular mechanisms—especially how cancer cells transfer UCA1 to surrounding cells, thereby promoting cancer progression—remain largely unknown.
Exosomes are small membrane-derived vesicles with a diameter of approximately 30–150 nm.14 It has been well established that a number of molecules regulate exosome secretion.15,16 Exosomes can regulate the physiological and pathological functions of various cancers as mediators of cell-to-cell communication by transferring nucleic acids, including mRNAs, microRNA (miRNAs), and lncRNAs;17,18 however, there is little evidence for the expression profile and potential function of secreted lncRNAs in CRC. In this study, we systematically analyzed the expression profiles of exosomal lncRNAs in CRC patients using a high-throughput microarray assay. We found that UCA1 was overexpressed in CRC tissues and highly enriched in plasma exosomes from patients with CRC. Moreover, UCA1 could function as a competing endogenous RNA (ceRNA) to regulate the expression of MYO6 by competing for miR-143 binding. We further showed that upregulation of circulating exosomes of HCC patients could transfer UCA1 to CRC cells to promote cell proliferation and migration.
Results
Expression Profiles of Peripheral Plasma Exosomal lncRNA in Human CRCs
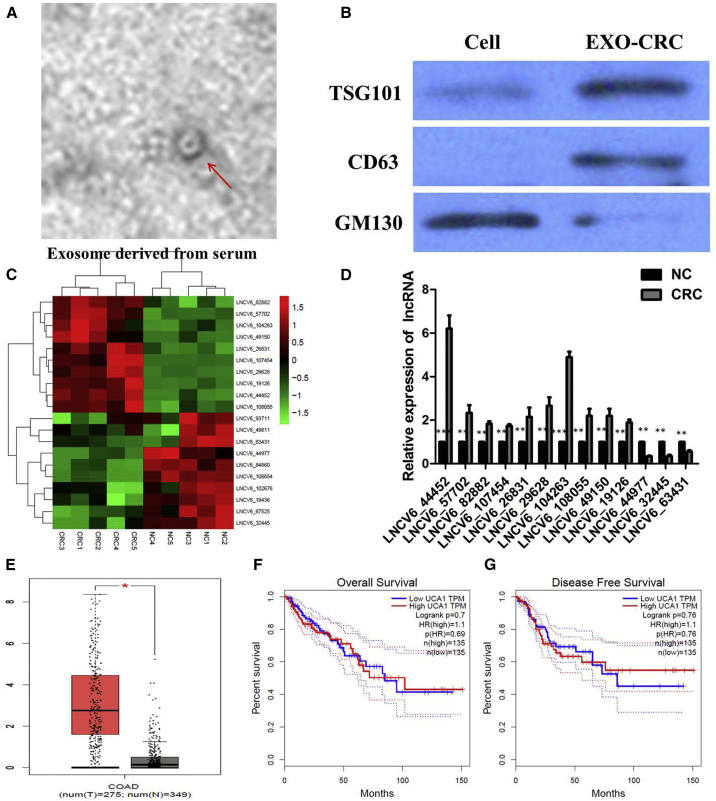
Transmission electron microscopy (TEM) showed that plasma exosomes have a diameter of 100–150 nm with a spherical-shaped morphology (Figure 1A). The nanoparticle tracking analysis (NTA) results showed a similar size distribution, and the peak size range was 80–135 nm. A western blot analysis showed that the proteins CD63 and Tsg101 were positively expressed in exosomes and that GM130 was mainly present in the cell lysate (Figure 1B). To identify exosomal lncRNAs, we extracted plasma exosomal RNA from 5 CRC patients and 5 healthy individuals and analyzed the expression profile using the ceRNA array mentioned earlier. As a result, 569 lncRNAs were upregulated, whereas 475 lncRNAs were downregulated based on the log2 (fold changes) ≥ 2, p < 0.01, and false discovery rate (FDR) < 0.05. Among these differently expressed genes, we identified the most top 10 upregulated and downregulated lncRNAs between CRC patients and negative control (NC) individuals (Figure 1C), which were then subjected to validation by quantitative real-time PCR. Their levels were consistently upregulated and downregulated in the plasma of CRC patients compared with the plasma of NC individuals (p < 0.01; Figure 1D).
Figure 1.
Expression Profiles of Peripheral Plasma Exosomal lncRNA in Human CRCs
(A) Transmission electron microscopy images of plasma exosomes of CRC patients. (B) Western blotting analysis of the exosomal markers Tsg101, CD63, and Alix in plasma exosomes of CRC patients and healthy control. (C) Microarray analysis of lncRNA profile in the NC group (n = 5) and CRC group (n = 5). (D) Expression levels of the most top 10 upregulated lncRNAs in CRC patients and NC individuals were determined by quantitative real-time PCR assay. (E) UCA1 expression in CRC and normal samples from the TCGA colon adenocarcinoma (COAD) dataset. (F) Kaplan-Meier analyses of the correlations between UCA1 expression and overall survival (OS) of COAD patients from the TCGA COAD dataset. (G) Kaplan-Meier analyses of the correlations between UCA1 expression and disease-free survival (DFS) of COAD patients from the TCGA COAD dataset. Log-rank test was used to calculate p values. All tests were performed at least three times. Data were expressed as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
UCA1 Was Upregulated in CRC Tissues and Cell Lines
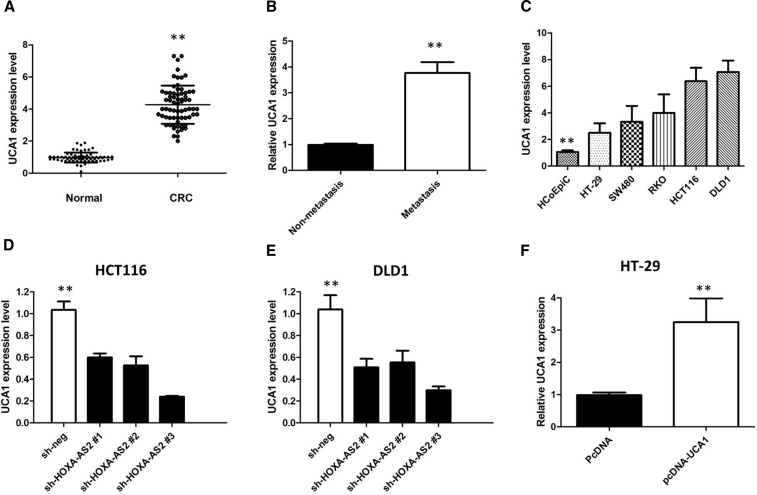
Among the upregulated exosomal lncRNAs in CRC patients, we analyzed the colon adenocarcinoma (COAD) dataset from The Cancer Genome Atlas (TCGA) database and found that the level of UCA1 was significantly higher in 275 COAD tissues than 349 normal tissues (p < 0.05; Figure 1E). However, Kaplan-Meier survival analysis from TCGA COAD datasets suggested that UCA1 expression is not significantly associated with worse overall survival (OS) (log-rank test, p = 0.69, Figure 1F) and disease-free survival (DFS) of COAD patients (log-rank test, p = 0.76; Figure 1G). Moreover, we used a qPCR assay to analyze the expression of UCA1 in our cohort of 68 pairs of CRC tumor tissues. We first found that UCA1 was upregulated in CRC tissues when compared with normal tissues (p < 0.001; Figure 2A). Analyses of the correlation between UCA1 expression and clinicopathological traits showed that UCA1 expression is much higher in advanced-stage tumors (p = 0.007), in tumors with distant metastasis (p = 0.003; Figure 2B), and in tumors >5 cm compared with tumors ≤5 cm (p = 0.005). There is no significant correlation between UCA1 expression and age (p = 0.807) or gender (p = 0.808) (Table 1). These data suggest that high UCA1 expression may have an important role in CRC progression and metastasis.
Figure 2.
UCA1 Was Up-regulated in CRC Tissues and Cell Lines
(A) Quantitative real-time PCR showed the expression levels of UCA1 in CRC tissues and adjacent noncancerous tissues. (B) Quantitative real-time PCR showed the expression level of UCA1 in metastasis CRC tissues. (C) Quantitative real-time PCR showed the expression level of UCA1 in CRC cell lines. (D) UCA1 siRNAs were used to enhance the efficiency of UCA1 knockdown in HCT116 cells. (E) UCA1 siRNAs were used to enhance efficiency of DLD1 knockdown in HCT116 cells. (F) Ectopic overexpression of UCA1 by transfecting HT-29 cells with pcDNA-UCA1 expression vector. All tests were performed at least three times. Data were expressed as mean ± SD. **p < 0.01.
Table 1.
Correlation between UCA1 Expression and Clinicopathologic Characteristics of CRC Patients
| Clinicopathological Features | Overall (n = 68) | UCA1 |
p Value | |
|---|---|---|---|---|
| High (n = 35) | Low (n = 33) | |||
| Age, in years: more than median | 31 | 15 | 16 | 0.807 |
| Gender: male | 33 | 16 | 17 | 0.808 |
| Tumor size: >5 | 24 | 18 | 6 | 0.007 |
| Tumor stages: T3 and T4 | 28 | 20 | 8 | 0.005 |
| Metastatic status: present | 12 | 11 | 1 | 0.003 |
| Neoadjuvant therapy: yes | 12 | 9 | 3 | 0.111 |
To explore the expression of UCA1 in CRC cells, quantitative real-time PCR was performed in five human CRC cell lines (HT-29, SW480, RKO, HCT116, and DLD1) and a normal colonic epithelial cell line (HCoEpiC). As shown in Figure 2C, UCA1 expression was higher in all CRC cells than that in HCoEpiC cells (p < 0.01). Among these CRC cells, HCT116 and DLD1 cells showed the highest UCA1 level, and the lowest expression of UCA1 was observed in HT-29 cells.
To investigate the functional role of UCA1 in CRC cells, we performed loss- or gain-of-function experiments. First, three independent UCA1-specific siRNAs (respectively si-UCA1#1, si-UCA1#2, and si-UCA1#3) and empty vectors (si-NC) were designed and transfected into HCT116 and DLD1 cells. We detected UCA1 expression at 48 h post-transfection by quantitative real-time PCR analysis to analyze knockdown efficiency and revealed that si-UCA1#3 had a higher efficiency of interference than si-UCA1#1 and #2 (p < 0.01; Figures 2D and 2E), so we chose si-UCA1#3 subsequently for the following experiments. Meanwhile, we induced ectopic overexpression of UCA1 by transfecting HT-29 cell lines with pcDNA-UCA1 expression vector (p < 0.01; Figure 2F).
UCA1 Promotes CRC Cell Proliferation In Vitro
MTT assays showed that the silencing of UCA1 expression by si-UCA1#3 significantly inhibited the cell proliferation rates of HCT116 and DLD1 cells compared with controls (p < 0.01; Figures 3A and 3B), whereas the results of MTT assays showed that the growth of HT-29 cells transfected with pCDNA-UCA1 was significantly increased compared with that of control cells (p < 0.01; Figure 3C). Furthermore, colony formation assays also indicated that clonogenic survival was significantly decreased following silencing of UCA1 in HCT116 and DLD1 cells, and clonogenic survival was significantly increased in HT-29 cells transfected with pCDNA-UCA1 (p < 0.01; Figures 3E and 3F).
Figure 3.
UCA1 Promotes CRC Cells Proliferation In Vitro
(A) MTT assay showed that knockdown of UCA1 inhibited cell proliferation of HCT116 cells. (B) MTT assay showed that knockdown of UCA1 inhibited cell proliferation of DLD1 cells. (C) MTT assay showed that overexpression of UCA1 promoted cell proliferation of HT-29 cells. (D) Colony formation assay showed that knockdown of UCA1 inhibited cell proliferation of HCT116 cells. (E) Colony formation assay showed that knockdown of UCA1 inhibited cell proliferation of DLD1 cells. (F) MTT assay showed that overexpression of UCA1 promoted cell proliferation of HT-29 cells. All tests were performed at least three times. Data were expressed as mean ± SD. **p < 0.01.
Downregulation of UCA1 Promotes G1 Arrest and Causes Apoptosis in CRC Cells In Vitro
To assess whether the pro-proliferative effects of UCA1 on the CRC cells are mediated by promoting cell-cycle progression, we examined cell cycling in CRC cells by flow cytometry. We proved that knockdown of UCA1 caused significant G1-phase cell-cycle arrest of HCT116 and DLD1, and cell-cycle checkpoint proteins were greatly inhibited (p < 0.01; Figures 4A, 4B, 4D, and 4E). Enhanced UCA1 expression increased the S-phase percentage and decreased the G0/G1 phase percentage of HT-29 cells (p < 0.01; Figures 4C and 4F). Moreover, flow-cytometric analysis was utilized to investigate whether apoptosis regulation is a potential contributing factor to cell-growth progress induced by UCA1. The results demonstrated that the apoptotic percentage of UCA1-silenced HCT116 and DLD1 cells was obviously increased (p < 0.01; Figures 5A and 5B). As expected, the cell apoptosis was markedly decreased in HT-29 cells by pcDNA-UCA1 (p < 0.01; Figure 5C).
Figure 4.
Downregulation of UCA1 Promotes G1 Arrest in CRC Cells In Vitro
(A) The flow cytometry assay showed that HCT116 cells transfected with si-UCA1#3 had cell-cycle arrest at the G1-G0 phase in comparison with control cells. (B) The flow cytometry assay showed that DLD1 cells transfected with si-UCA1#3 had cell-cycle arrest at the G1-G0 phase in comparison with control cells. (C) The flow cytometry assay showed that HT-29 cells transfected with pcDNA 3.1-UCA1 had cell-cycle arrest at the S phase in comparison with control cells; (D) UCA1 knockdown altered cell cycle proteins of HCT116 cells; (E) UCA1 knockdown altered cell-cycle proteins of DLD1 cells. (F) UCA1 overexpression altered cell-cycle proteins of HT-29 cells. All tests were performed at least three times. Data were expressed as mean ± SD. **p < 0.01.
Figure 5.
Downregulation of UCA1 Causes Apoptosis in CRC Cells In Vitro
(A) The flow cytometry assay showed that HCT116 cells transfected with si-UCA1#3 had a higher apoptotic rate in comparison with control cells. (B) The flow cytometry assay showed that DLD1 cells transfected with si-UCA1#3 had a higher apoptotic rate in comparison with control cells. (C) The flow cytometry assay showed that HT-29 cells transfected with pcDNA 3.1-UCA1 had a lower apoptotic rate in comparison with control cells. (D) The Transwell assay showed that knockdown of UCA1 inhibited the migratory and invasive abilities of HCT116 cells. (E) The Transwell assay showed that knockdown of UCA1 inhibited the migratory and invasive abilities of DLD1 cells. (F) The Transwell assay showed that ectopic expression of UCA1 promoted cell invasion and migration of HT-29 cells. All tests were performed at least three times. Data were expressed as mean ± SD. **p < 0.01.
UCA1 Promotes CRC Cell Invasion and Metastasis In Vitro and In Vivo
We further explored the physiological relevance of UCA1 to the CRC cell motility. The Transwell assay demonstrated that UCA1 knockdown significantly impeded the migratory and invasive abilities of HCT116 and DLD1 cells compared with s-NC control (Figures 5D and 5E). In addition, ectopic expression of UCA1 dramatically promoted cell invasion and migration in HT-29 cells (Figure 5F). Thus, UCA1 significantly increased the migration and invasion potential of CRC cells in vitro.
To further confirm the aforementioned findings in vivo, a lung metastasis model was introduced by inoculating HCT116 cells with modified UCA1 expression directly into the tail veins of nude mice. Luciferase signals were monitored to observe the location and growth of tumor xenografts in the lung. As a result, at 35 days, luciferase signals were lower in the UCA1-knockdown group compared with those in the control group, and UCA1 depletion significantly reduced the lung metastasis burden of HCT116 cells (Figures 6A and 6B). All of these findings suggest a crucial role of UCA1 in promoting CRC cell invasion and metastasis.
Figure 6.
Knockdown of UCA1 Suppressed the Tumor Growth of CRC In Vivo
(A) Luciferase signals are lower in the UCA1 knockdown group compared with those in control group. (B) UCA1 depletion significantly reduced the lung metastasis burden of HCT116 cells. (C) UCA1 knockdown inhibits HCT116 tumor growth in vivo. The tumor volume curve of nude mice was analyzed. (D) The tumor weights of nude mice were measured. (E) The expression level of UCA1 in tumors of nude mice was detected by quantitative real-time PCR. (F) IHC revealed that xenograft tumors derived from HCT116 cells with UCA1 knockdown had lower expression of Ki67. Data were expressed as mean ± SD. **p < 0.01.
Knockdown of UCA1 Suppressed the Tumor Growth of CRC In Vivo
To determine whether UCA1 could affect tumorigenesis, HCT116 cells with stable UCA1 (sh-UCA1) and empty-vector-transfected HCT116 cells were inoculated into nude mice. Compared with those in the vector control, the tumors formed in the sh-UCA1 group were dramatically smaller. Consistently, the tumor growth in the sh-UCA1 group was significantly slower than that in the control group (p < 0.01; Figure 6C). Remarkably, the average tumor weight was obviously lower in the sh-UCA1 group compared with that in the empty-vector group (p < 0.01; Figure 6D). Quantitative real-time PCR analysis of the UCA1 expression was then performed using the xenograft tumor tissues. The results showed that the levels of UCA1 expression in tumor tissues formed from sh-UCA1 cells were lower than those of the tumors formed in the control group (p < 0.01; Figure 6E). Results of immunohistochemistry (IHC) revealed that xenograft tumors derived from HCT116 cells with UCA1 knockdown had lower expression of Ki67 than the sh-NC group (Figure 6F). These results indicate that UCA1 is significantly associated with the tumor growth of CRC in vivo.
UCA1 Functions as a ceRNA via Direct Sponging of miR-143
Up to now, accumulating evidence indicated that lncRNAs exerted the function by interacting with miRNAs. Therefore, to investigate the effect of UCA1 on the expression of miRNAs, the bioinformatics predicton analysis was performed by using the miRcode online database. As shown in Figure 7A, miR-143 harbors the complementary binding sequence of UCA1. In order to further validate the interaction, the UCA1 sequence containing the putative or mutated miR-143 binding site was cloned into the downstream of the luciferase reporter gene, generating wild-type (WT)-UCA1 or mutant (MUT)-UCA1 luciferase reporter plasmids. Then, the effect of miR-143 on WT-UCA1 or MUT-UCA1 luciferase reporter systems was determined. With transfected miR-143 mimics, the luciferase activities of WT-UCA1 luciferase reporter vector were significantly decreased, while the luciferase activities in the MUT group showed no observable change (p < 0.01; Figure 7B). In a further RNA immunoprecipitation (RIP) experiment, UCA1 and miR-143 simultaneously existed in the production precipitated by anti-AGO2 (Figure 7C), suggesting that miR-143 is a UCA1-targeting miRNA. These outcomes indicated that the interaction of UCA1 and miR-143 was realized by the putative binding site.
Figure 7.
UCA1 Functions as a ceRNA via Directly Sponging of miR-143
(A) miRcode online database results showed the sequence of UCA1 with highly conserved putative miR-143 binding sites. (B) miR-143 mimic considerably reduced the luciferase activity of the WT-UCA1 luciferase reporter vector compared with negative control, while miR-143 mimic did not pose any impact on the luciferase activity of MUT-UCA1-transfected cells. (C) UCA1 and miR-143 simultaneously existed in the production precipitated by anti-AGO2. (D) Expression levels of miR-143 in different CRC cell lines were determined by quantitative real-time PCR. (E) Silencing of UCA1 increased the expression level of miR-143 in HCT116 and DLD1 cells. All tests were performed at least three times. Data were expressed as mean ± SD. **p < 0.01.
Next, we measured the levels of miR-143 expression in various CRC cell lines. As shown in Figure 7D, the expression of miR-143 was obviously decreased in HCT116 and DLD1 cells, indicating the opposite result to UCA1 expression. Subsequently, the effect of UCA1 on miR-143 expression was also observed in HCT116 and DLD1 cells. The results manifested that miR-143 expression was elevated in HCT116 and DLD1 cells after the silencing of UCA1 (Figure 7E). All these results suggested that UCA1 could sponge miR-143 to suppress its expression.
MYO6 Was a Direct Target of miR-143
TargetScan online software demonstrated that MYO6 was a candidate target of miR-143 (Figure 8A). To further validate the inference, MYO6-WT or MYO6-MUT 3′ UTR luciferase reporter vector was conducted. MYO6-WT or MYO6-MUT was co-transfected with miR-143 mimics or NC into HEK293T cells. The relative luciferase activity was remarkably reduced in cells co-transfected with the MYO6-WT luciferase reporter and miR-143 mimic than in the NC cells. However, inhibitory effects were abolished when 3′ UTRs that contained both MUT-binding sites were co-transfected with miR-143, confirming that MYO6 is a target of miR-143 (p < 0.01; Figure 8B).
Figure 8.
MYO6 Was a Direct Target of miR-143
(A) Bioinformatics analysis revealed the predicted binding sites between MYO6 and miR-143. (B) Luciferase reporter assay demonstrated that miR-143 mimics significantly decreased the luciferase activity of MYO6-WT in HEK293T cells. (C) miR-143 expression was significantly downregulated in CRC tissues compared with adjacent normal tissues by quantitative real-time PCR analysis. (D) IHC analysis was performed to examine the expression levels of MYO6 in CRC tissues and adjacent normal tissues. (E) Enforced expression of miR-143 significantly diminished protein expression of MYO6 in HCT116 cells. (F) MYO6 expression in CRC and normal samples from the TCGA COAD dataset. (G) Kaplan-Meier analyses of the correlations between UCA1 expression and OS of COAD patients from the TCGA COAD dataset. All tests were performed at least three times. Data were expressed as mean ± SD. **p < 0.01.
Furthermore, quantitative real-time PCR was performed to detect the expression of miR-143 in tumor tissues and adjacent normal tissues from 68 patients suffering from CRCs. The miR-143 was significantly lower in CRC tissues compared with adjacent normal tissues (p < 0.01; Figure 8C). The IHC results showed that MYO6 expression in CRC specimens was significantly upregulated compared with that in the adjacent normal tissues (53/68 vs. 12/68; p < 0.001; Figure 8D). Subsequently, the actual impacts of miR-143 on MYO6 expression were detected in HCT116 cells by immunoblotting assays. Our results showed that enforced expression of miR-143 significantly diminished protein expression of MYO6 in HCT116 cells (Figure 8E). Taken together, miR-143 downregulated the expression of MYO6 by directly targeting the 3′ UTR region of MYO6 in CRC cells. Furthermore, we analyzed the COAD dataset from the TCGA database and found that the level of MYO6 was significantly higher in 275 COAD tissues than in 349 normal tissues (p < 0.05; Figure 8F). Kaplan-Meier survival analysis from TCGA COAD datasets suggested that MYO6 expression is significantly associated with worse OS (log-rank test, p = 0.018; Figure 8G).
Circulating Exosomes Promoted CRC Cell Proliferation and Migration through the Transfer of UCA1
Increasing evidence has shown that exosomes are an important mediator of information delivery between donor cells and recipient cells, including lncRNAs. In this study, we determined whether circulating exosomes could mediate the transfer of UCA1 to the target cells (HCT116 and DLD1) and affect their behaviors. Therefore, we isolated exosomes from the serum of CRC patients and healthy people. Quantitative real-time PCR results showed that UCA1 was highly expressed in the exosomes of CRC patients compared with that in healthy people (p < 0.01; Figure 9A). TEM was performed to observe the morphology of exosomes, the particles that had round-shaped vesicles with a double-layer membrane structure and diameters of about 100 nm that could transfer UCA1 to HCT116 and DLD1 cells by coculture (p < 0.01; Figure 9B). To confirm that UCA1-associated exosomes were taken up by CRC cells, exosomes prepared from the serum of CRC patients were labeled with PKH67. Exosome uptake by Huh7 cells was observed in a time-dependent manner at 37°C (p < 0.01; Figure 9C). Moreover, cell viability and the abilities of colony formation and migration were significantly enhanced by treatment with exosomes from CRC patients (p < 0.01; Figures 9D–9G). CRC cell treatment with exosomes from CRC patients downregulated the expression of miR-143 but increased MYO6 expression (Figures 9H and 9I). Taken together, these results suggest that exosomes from the serum of CRC patients could promote cell proliferation and migration by transmitting UCA1 to HCT116 and DLD1 cells.
Figure 9.
Circulating Exosomes Promoted CRC Cell Proliferation and Migration through the Transfer of UCA1
(A) Exosomal UCA1 expression levels in CRC patients and healthy people. (B) The expression levels of UCA1 after treated with PBS (Control), exosomes of healthy control (Normal-Exo), and exosomes of CRC patients (CRC-Exo). (C) HCT116 cells were incubated with PKH67-labeled CRC exosomes for the indicated times at 4°C or 37°C. Quantification of exosome uptake is shown. (D) HCT116 cells were treated with PBS, Normal-Exo, and HCC-Exo, and then cell viability was assessed by MTT at 24 h, 48 h, and 72 h. (E) DLD1 cells were treated with PBS, Normal-Exo, and HCC-Exo, and then cell viability was assessed by MTT at 24 h, 48 h, and 72 h. (F) Colony formation abilities were measured by colony formation assay. (G) Transwell migration and invasion assays were used to detect the migration and invasion abilities. (H) The expression of miR-143 was detected by quantitative real-time PCR. (I) The protein levels of MYO6 were measured by western blot. All tests were performed at least three times. Data were expressed as mean ± SD. **p < 0.01.
Discussion
In this study, the peripheral plasma exosomal lncRNA UCA1 was found to be aberrantly upregulated in CRC by high-throughput microarray assay. We first found that UCA1 was significantly upregulated in the CRC tissues and CRC cell lines. In addition, increased expression of UCA1 in CRC patients is associated with increased tumor size and advanced stage. Our subsequent studies demonstrate that UCA1 knockdown decreased cell proliferation and caused a dramatic decrease in CRC cell-colony formation, whereas UCA1 overexpression has the opposite results. In addition, UCA1 knockdown promoted significant arrest in the G0/G1-phase and an obvious increase in CRC cell apoptosis. These observations of tumor growth were verified in a mouse xenograft model. Based on bioinformatics analysis, it was assumed that the UCA1/miR-143/MYO6 axis plays a pivotal role in CRC progression. Specifically, we also showed mechanistically that UCA1 promotes the progression of CRC by functions as a miR-143 sponge to modulate MYO6 expression. Moreover, UCA1 was also upregulated in serum exosomes of CRC patients and could transfer UCA1 to CRC cells to increase their abilities of cell proliferation and migration. These findings suggest that UCA1 might be a novel clinical molecular marker for the treatment of CRC patients.
A number of recent papers have revealed that lncRNAs play an important role in the pathogenesis of cancer. Cancer-associated lncRNAs and investigation of their molecular and biological functions are important in providing new insights into the diagnosis and treatment of cancer, including CRC. The lncRNA UCA1 is generally regarded as an oncogene, and it is usually highly expressed in a variety of cancers, including CRC.19, 20, 21, 22 Our functional experiments demonstrated that UCA1 had an important role in cell proliferation and the cell cycle of CRC cells. UCA1 knockdown could suppress CRC cell proliferation in vitro and growth in vivo. As one of the most important malignant tumor hallmarks, tumor metastasis is a complex and multistep process influenced by genetic and epigenetic changes. We revealed that UCA1 contributed to the metastasis of CRC both in vitro and in vivo.
In recent years, emerging evidence proposed that lncRNAs mainly act as a miRNA sponge to exert their post-transcriptional functions as ceRNAs, which is more effective than the traditional anti-miRNA approach.23 Accordingly, we performed RIP and luciferase assays and found that the mechanism by which UCA1 promotes tumor progression of CRC in vitro is mediated by inhibiting miR-143 expression, thus, influencing downstream gene MYO6 expression. Moreover, miRNAs are the most widely studied non-coding RNAs and also can act as oncogenes or tumor suppressor genes.24 In this study, bioinformatics analysis showed that miR-143 interacted with the 3′ UTR of MYO6 and suppressed MYO6 expression at the post-transcriptional level, which was confirmed by the results of the luciferase reporter assay. We found that the miR-143 was significantly lower in CRC tissues compared with adjacent normal tissues and that the MYO6 expression was significantly higher in CRC tissues.
Mounting evidence indicates that exosomes are critical mediators of communication and information transfer between tumor cells and surrounding cells and that cancer-derived exosomes can enrich proteins, mRNAs, miRNAs, and lncRNAs, which may horizontally transfer to recipient cells and result in a phenotypic effect. Inspired by these studies, we hypothesized that extracellular UCA1 promoted CRC progression through incorporation into exosomes. To validate this hypothesis, we isolated exosomes from the serum of CRC patients and found that UCA1 was highly expressed in the exosomes of CRC patients and that the exosomes could transfer UCA1 to CRC cells to affect the cell viability, the ability of colony formation, and the ability of migration of CRC cells by downregulating miR-143. These results suggest that circulating exosomes could promote tumor growth and metastasis by transmitting UCA1 to CRC cells.
Taken together, the evidence indicates that UCA1 performed a pivotal function in the tumor progression of CRC by packaging into exosomes. We found that UCA1 affects the proliferation and apoptosis of CRC cells by functioning as a ceRNA to regulate MYO6 expression by sponging miR-143.
Materials and Methods
Patients and Sample Collection
Pairs of fresh CRC tissues and adjacent normal tissues were collected from 68 CRC patients at Sixth People’s Hospital of Dalian City, Dalian, China, between January 2010 and January 2018. Tissues were immediately snap-frozen in liquid nitrogen and stored at −80°C until total RNA was extracted. For exosome purification, whole blood samples were collected from these 68 CRC patients and healthy control. Fresh plasma samples (3 mL) were collected in ethylenediamine tetra-acetic acid tubes from each of the subjects. These samples were centrifuged at 3,000 × g for 10 min at 4°C and then stored at −80°C. The specimens were evaluated according to the World Health Organization’s classification criteria. Disease progression was classified using the CRC guidelines outlined in the seventh edition of the American Joint Committee on Cancer’s staging manual. Patients who underwent chemotherapy, radiotherapy, or any other adjuvant treatment before surgery were excluded from the study. The study was approved by the research ethics committees of Sixth People’s Hospital of Dalian City and Southwest Forestry University, and written informed consent was obtained from all patients.
Plasma Exosome Isolation
Exosome extraction was performed essentially as described before.25 First, the samples were centrifuged twice at 3,000 × g and 10,000 × g for 20 min at room temperature to remove cells and other debris in the plasma. The supernatants were then centrifuged at 100,000 × g for 30 min at 4°C to remove microvesicles that were larger than exosomes, harvested, and again centrifuged at 10,000 × g for 70 min at 4°C. Subsequently, the supernatants were gently decanted, and the exosome sediments were re-suspended in phosphate-buffered saline (PBS). Concentration of exosomes was determined using the bicinchoninic acid (BCA) method as recommended by the manufacturer (Thermo Scientific, Waltham, MA, USA).
TEM
The exosome suspension was diluted to 0.5 mg/mL with PBS and then spotted onto a glow-discharged copper grid placed on a filter paper and dried for 10 min by exposure to infrared light. Next, the exosome samples were stained with one drop of phosphotungstic acid (1% aqueous solution) for 5 min and dried for 20 min by exposure to infrared light. Finally, the exosomes were visualized under a transmission electron microscope (HT7700, Hitachi, Tokyo, Japan) at 100 keV.
NTA
Briefly, the exosomes were re-suspended in PBS and filtered with a syringe filter (Millipore). Then, the samples were diluted until individual nanoparticles could be tracked. The size distribution of the exosomes was evaluated using a NanoSight NS300 instrument (Malvern Instruments, Worcestershire, UK).
Western Blotting
Western blotting was performed to assess the levels of these markers. Total proteins were extracted from exosome samples using lysis buffer. Each sample (40 μg) was loaded onto a 12% sodium dodecyl sulfate-polyacrylamide gel and then transferred to a polyvinylidene fluoride membrane (Roche, Mannheim, Germany). These membranes were immersed in 2% bovine serum albumin at room temperature for 1 h and incubated with the following primary antibodies: anti-Tsg101 (1:1,000, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-CD63 (1:2,000, Abcam, Cambridge, UK), and anti-GM130 (1:1,000, Abcam, Cambridge, UK) followed by subsequent incubation with appropriate secondary antibodies after washing with PBS. Lastly, protein bands were visualized by incubating with an electro-chemiluminescence reagent.
Exosome Labeling
The PKH67 Green Fluorescent Cell Linker Kit (Sigma) was used to label exosomes according to the manufacturer’s protocol. Briefly, exosome pellets were re-suspended in 500 μL Diluent C. In addition, 2 μL PKH67 was mixed with 1 mL Diluent C, followed by mixing with the exosome suspension and incubation for 4 min. To stop the labeling reaction, an equal volume of 1% BSA was added into the mixed liquor. Then, the labeled exosomes were ultracentrifuged at 100,000 × g for 1 h, washed with 1× PBS, and ultracentrifuged again.
Exosomal Uptake into Recipient Cells
To detect exosomal uptake into recipient cells, Huh7 cells were grown on sterile glass coverslips in 24-well plates at a density of 1 × 104 cells per well for cultivation for 24 h. Then, the slides were washed three times using 1× PBS, and PKH67-labeled exosomes were diluted in DMEM solution and added into each well. Cells were cultured for 1 h at 4°C or for 1 h, 2 h, and 4 h at 37°C. The slides were washed three times using 1× PBS and fixed with 3.7% formaldehyde solution at room temperature for 15 min followed by washing three times using 1× PBS. After that, DAPI was used to stain the nuclei, and the slides were covered with coverslips and observed using a confocal laser scanning microscope (LSM710; Carl Zeiss, Oberkochen, Germany).
Agilent Methods for ceRNA Array
Total RNA of exosomes was extracted from plasma using TRIzol LS Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Total RNA from exosomes was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and purified using a RNeasy Mini Kit (QIAGEN, Hilden, Germany). RNA samples of each group were then used to generate fluorescently labeled cRNA targets for the human ceRNA array v.1.0 (4 × 180 K; Shanghai Biotechnology, Shanghai, China). The labeled cRNA targets were then hybridized with the slides. After hybridization, slides were scanned on the Agilent Microarray Scanner (Agilent Technologies, Santa Clara, CA, USA). Data were extracted using Feature Extraction software v.10.7 (Agilent Technologies). Raw data were normalized by the quantile algorithm of the limma package in the R program. Microarray experiments were performed at Shanghai Biotechnology following the protocol of Agilent Technologies. Ratios between CRC patients and normal (NC) subjects were calculated. Genes with a fold change of at least 2 were selected for further analysis. The selected parent genes of the lncRNA were grouped into functional categories based on the Gene Ontology database (http://geneontology.org/), and functional pathways (Kyoto Encyclopedia of Genes and Genomes) were also analyzed using the online enrichment analysis tool of Shanghai Biotechnology.
TCGA Dataset Analysis
The data and the corresponding clinical information of patients were collected from the TCGA database (http://cancergenome.nih.gov/). We used the edgeR package of R packages to perform the difference analysis (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) and used the pheatmap package of R packages to perform the cluster analysis (https://cran.r-project.org/web/packages/pheatmap/index.html). The sva R package was used to remove the batch effect. Genes with adjusted p values <0.05 and absolute fold changes (FCs) >1.5 were considered differentially expressed genes. Kaplan-Meier survival curves were drawn to analyze the relationships between genes and OS in the survival package. The corresponding statistical analysis was performed and graphics were created in R software (R v.3.3.2).
Cell Lines and Culture Conditions
The human CRC cell lines (HCT116, DLD1, SW480, RKO, and HT-29) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The human colonic epithelial cell line HCoEpiC and the 293T cell line were obtained from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI 1640 or DMEM (GIBCO, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (10% FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (GIBCO) in humidified air at 37°C with 5% CO2.
RNA Extraction and Quantitative Real-Time PCR Analyses
Total RNA was extracted from tissues or cultured cells using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). For quantitative real-time PCR, RNA was reverse transcribed to complementary DNA (cDNA) by using a PrimeScript RT Reagent Kit (Takara, Dalian, China). Real-time PCR analyses were performed with SYBR Green (Takara, Dalian, China). Results were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The relative expression of UCA1 was calculated and normalized using the 2−ΔΔCt method relative to GAPDH.
Cell Transfection
To construct UCA1 overexpression plasmid, the full length of the UCA1 cDNA sequence was amplified, cloned into pcDNA3.1 vector (Invitrogen), and sequenced and named as pcDNA3.1-UCA1 (UCA1). Three specific small interference RNAs (siRNAs) targeting UCA1 (si-UCA1#1, si-UCA1#2, and si-UCA1#3) and scrambled siRNA control (si-NC) were obtained from GenePharma (Shanghai, China). miR-143 mimic (miR-143) and scrambled mimic control (miR-NC) were purchased from RiboBio (Guangzhou, China). All these plasmids and oligonucleotides were transfected into cells by Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s instructions.
MTT Assay
The Cell Proliferation Reagent Kit I (MTT) (Roche, Basel, Switzerland) was used to assess cell proliferation. Transfected cells were plated in each well of a 96-well plate and assessed every 24 h according to the manufacturer’s instructions. The plate was then read at 490 nm using a plate reader.
Colony Formation Assay
Cells were seeded into 6-well plates at 200 cells per well. Approximately 14 days later, the number of macroscopic colonies was counted, and images of the representative colonies were obtained.
Flow-Cytometric Analysis
Cells for cell-cycle analysis were stained with propidium oxide using the CycleTEST PLUS DNA Reagent Kit (BD Biosciences) following the manufacturer’s protocol and analyzed by FACScan. The percentages of the cells in G0-G1, S, and G2-M phases were counted and compared.
Transfected cells were harvested after transfection by trypsinization. After the double staining with fluorescein isothiocyanate (FITC)-annexin V and propidium iodide was performed with the FITC Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s recommendations, the cells were analyzed by flow cytometry (FACScan; BD Biosciences) equipped with Cell Quest software (BD Biosciences). Cells were discriminated into viable cells, dead cells, early apoptotic cells, and apoptotic cells, and then the relative ratio of early apoptotic cells was compared with control transfection from each experiment.
Transwell Migration Assays
Tumor cell in vitro migration assays were carried out using the Transwell system (24-well inserts, 8.0 μm; Corning, Corning, NY, USA). After specific treatment, tumor cells were detached and re-suspended in serum-free medium. Cells were cultured on the top chambers for 18 h, and growth medium was added to the bottom wells. In some cases, specific antibodies or inhibitors were added into both chambers. Quantification was performed by counting crystal-violet-stained migrated cells in five 200× high-power fields (HPFs) of every top chamber.
Tumor Xenograft Experiments
For the in vivo metastasis model, representative mice were injected with modified UCA1 expressing HCT116 cells (n = 5 per group). The luciferase signal intensity from days 7 to 35 is on equivalent scales in the lung metastasis model. All the mice were killed after 6 weeks, and the lungs were subjected to immunohistochemical analysis and H&E staining. Metastatic progression was monitored and quantified using an IVIS-100 system (Caliper Life Sciences, Boston, MA, USA).
For the xenograft model of HCT116 cells, 4-week-old male BALB/c nude mice were housed under standard conditions and cared for according to protocols. Cells were harvested and re-suspended in serum-free medium at a concentration of 1 × 107 cells per 0.2 mL. Each mouse (n = 4 per group) was inoculated subcutaneously in the right flank with cells stably transduced with shUCA1 or shControl. Tumor size was monitored every 3 days, and mice were euthanized after 4 weeks. All animal studies were approved by the Animal Ethics Committee of Southwest Forestry University, and animal experiments were performed at Southwest Forestry University.
Luciferase Reporter Assay
HEK293T cells (5 × 103) were seeded into 96-well plates and co-transfected with corresponding plasmids and miRNA mimics or inhibitors using the Lipofectamine 2000 transfection reagent. Luciferase activity was measured using the dual-luciferase reporter assay system (Promega, Madison, WI, USA) after 48 h of incubation according to the manufacturer’s instructions. Independent experiments were performed in triplicate. Relative luciferase activity was normalized to the Renilla luciferase internal control.
RIP Assay
The RIP assay was performed using an EZ-Magna RiP Kit (Millipore, Billerica, MA, USA) in accordance with the manufacturer’s instructions. Cells were lysed at 70%–80% confluence in RIP lysis buffer and then incubated with magnetic beads conjugated with human anti-Ago2 antibody (Millipore) and normal mouse immunoglobulin G (IgG) control (Millipore) in RIP buffer. The RNAs in the immunoprecipitates were isolated with TRIzol Reagent and analyzed by quantitative real-time PCR.
IHC
For each patient sample, three paraffin sections of 5 μm were prepared, one for hematoxylin and eosin (H&E) staining and the other two for immunohistochemical staining. PBS instead of primary antibodies was used for negative control, and the breast cancer tissue was used for positive control. Sections were dewaxed using xylene, followed by hydration with ethanol solutions and the addition of EDTA for antigen retrieval. Later, sections were blocked with normal goat serum for 30 min to eliminate non-specific binding. Sections were incubated with anti-human MYO6 polyclonal antibody (1:1,000; ab124805; Abcam, Cambridge, MA, USA). Sections were then incubated with biotin-labeled secondary antibodies for 30 min at room temperature, followed by staining with diaminobenzidine (DAB). Finally, the sections were counterstained with hematoxylin. The result of staining was determined by two doctors who did not know the clinical condition of the patients. The proportions of positive cells of 0%, 1%–5%, 6%–25%, 26%–75%, and 76%–100% were assigned with scores of 0, 1, 2, 3, and 4, respectively. Scores of 0–2 were considered as negative expression, and scores of 3–4 were considered as positive expression.
Statistical Analysis
Results are presented expressed as mean ± SD (standard deviation). Student’s t test was performed to measure the difference between two groups, and differences between more than two groups were assessed using one-way ANOVA. p < 0.05 was considered significant.
Availability of Data and Materials
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request, but no information infringing on the privacy of the participants will be given.
Author Contributions
Yunpeng Luan and Xiang Li conceived and designed the project and wrote the manuscript. Yunpeng Luan, Yunqi Luan, Rong Zhao, Yizhuo Hao, Burakovaov .Oleg.Vladimir, Lu Jia and Yanmei Li performed most of the experiments. Lili Liu collected and analyzed the clinical and pathological data. All of the authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Key Laboratory of Forest Biotechnology in Yunnan, Southwest Forestry University; the National Natural Science Foundation of China (NSFC) (31860254, 81560799, 8176150582) and Advanced and Characteristic Key Biological Disciplines of Yunnan Province (project serial code: 50097505). The thesis of “Scientific and Technological Innovation Team Construction Project for Protection and Utilization of Under-forest Biological Resources (project serial code: 51400605)” was completed through joint support.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Graham L.D., Pedersen S.K., Brown G.S., Ho T., Kassir Z., Moynihan A.T., Vizgoft E.K., Dunne R., Pimlott L., Young G.P. Colorectal Neoplasia Differentially Expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2:829–840. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L., Jiang C., Xu C., Xue H., Zhou H., Gu L., Liu Y., Xu Q. Down-regulation of long non-coding RNA RP11-708H21.4 is associated with poor prognosis for colorectal cancer and promotes tumorigenesis through regulating AKT/mTOR pathway. Oncotarget. 2017;8:27929–27942. doi: 10.18632/oncotarget.15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y., Yang Y., Wang F., Moyer M.P., Wei Q., Zhang P., Yang Z., Liu W., Zhang H., Chen N. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 8.Weakley S.M., Wang H., Yao Q., Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J. Surg. 2011;35:1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X., Yuan Z., Yang Z., Chen D., Kim T., Cui Y., Luo Q., Liu Z., Yang Z., Fan X. The novel long noncoding RNA u50535 promotes colorectal cancer growth and metastasis by regulating CCL20. Cell Death Dis. 2018;9:751. doi: 10.1038/s41419-018-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y., Yang L., Zhao J., Li C., Nie J., Liu F., Zhuo C., Zheng Y., Li B., Wang Z., Xu Y. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol. Cancer. 2015;14:191. doi: 10.1186/s12943-015-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue M., Chen W., Li X. Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer. J. Cancer Res. Clin. Oncol. 2016;142:1407–1419. doi: 10.1007/s00432-015-2042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y.T., Wang Y.F., Lai J.Y., Shen S.Y., Wang F., Kong J., Zhang W., Yang H.Y. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/β-catenin signaling pathway. Cancer Sci. 2016;107:1581–1589. doi: 10.1111/cas.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bian Z., Jin L., Zhang J., Yin Y., Quan C., Hu Y., Feng Y., Liu H., Fei B., Mao Y. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci. Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Yuan X., Shi H., Wu L., Qian H., Xu W. Exosomes in cancer: small particle, big player. J. Hematol. Oncol. 2015;8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., Battistelli C., Alonzi T., Weisz A., Tripodi M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Beckham C.J., Olsen J., Yin P.N., Wu C.H., Ting H.J., Hagen F.K., Scosyrev E., Messing E.M., Lee Y.F. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J. Urol. 2014;192:583–592. doi: 10.1016/j.juro.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 17.Bae S., Brumbaugh J., Bonavida B. Exosomes derived from cancerous and non-cancerous cells regulate the anti-tumor response in the tumor microenvironment. Genes Cancer. 2018;9:87–100. doi: 10.18632/genesandcancer.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang Z., Zhao J., Xie W., Sun Q., Wang H., Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR-184 expression. Cancer Med. 2017;6:2897–2908. doi: 10.1002/cam4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y., Sun Q., Zhao L., Wu J., Chen X., Wang Y., Zang W., Zhao G. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med. Oncol. 2016;33:88. doi: 10.1007/s12032-016-0804-2. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z.Q., He C.Y., Hu L., Shi H.P., Li J.F., Gu Q.L., Su L.P., Liu B.Y., Li C., Zhu Z. Long noncoding RNA UCA1 promotes tumour metastasis by inducing GRK2 degradation in gastric cancer. Cancer Lett. 2017;408:10–21. doi: 10.1016/j.canlet.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Tuo Y.L., Li X.M., Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur. Rev. Med. Pharmacol. Sci. 2015;19:3403–3411. [PubMed] [Google Scholar]
- 23.Lin P., Wen D.Y., Li Q., He Y., Yang H., Chen G. Genome-wide analysis of prognostic lncRNAs, miRNAs, and mRNAs forming a competing endogenous RNA network in hepatocellular carcinoma. Cell. Physiol. Biochem. 2018;48:1953–1967. doi: 10.1159/000492519. [DOI] [PubMed] [Google Scholar]
- 24.Christensen L.L., Holm A., Rantala J., Kallioniemi O., Rasmussen M.H., Ostenfeld M.S., Dagnaes-Hansen F., Øster B., Schepeler T., Tobiasen H. Functional screening identifies miRNAs influencing apoptosis and proliferation in colorectal cancer. PLoS ONE. 2014;9:e96767. doi: 10.1371/journal.pone.0096767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller L., Mitsuhashi M., Simms P., Gooding W.E., Whiteside T.L. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci. Rep. 2016;6:20254. doi: 10.1038/srep20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request, but no information infringing on the privacy of the participants will be given.