Abstract
Abnormal protein homeostasis (proteostasis), dysfunctional mitochondria, and aberrant redox signalling are often associated in neurodegenerative disorders, such as Huntington's (HD), Alzheimer's and Parkinson's diseases. It remains incompletely understood, however, how changes in redox signalling affect proteostasis mechanisms, including protein degradation pathways and unfolded protein responses (UPR). Here we address this open question by investigating the interplay between redox signalling and proteostasis in a mouse model of HD, and by examining the in vivo effects of the mitochondria-targeted antioxidant MitoQ. We performed behavioural tests in wild-type and R6/2 HD mice, examined markers of oxidative stress, UPR activation, and the status of key protein degradation pathways in brain and peripheral tissues. We show that R6/2 mice present widespread markers of oxidative stress, with tissue-specific changes in proteostasis that were more pronounced in the brain and muscle than in the liver. R6/2 mice presented increased levels of cytosolic and mitochondrial chaperones, particularly in muscle, indicating UPR activation. Treatment with MitoQ significantly ameliorated fine motor control of R6/2 mice, and reduced markers of oxidative damage in muscle. Additionally, MitoQ attenuated overactive autophagy induction in the R6/2 muscle, which has been associated with muscle wasting. Treatment with MitoQ did not alter autophagy markers in the brain, in agreement with its low brain bioavailability, which limits the risk of impairing neuronal protein clearance mechanisms. This study supports the hypotheses that abnormal redox signalling in muscle contributes to altered proteostasis and motor impairment in HD, and that redox interventions can improve muscle performance, highlighting the importance of peripheral therapeutics in HD.
Keywords: Mitochondria, Huntington's disease, Oxidative stress, Antioxidant, Neurodegeneration, Unfolded protein response
Abbreviations: ALS, amyotrophic lateral sclerosis; BSA, bovine serum albumin; HD, Huntington's disease; Htt, huntingtin; mHtt, mutant huntingtin; MitoQ, mitoquinone or MitoQ10; mtDNA, mitochondrial DNA; UPRmt, mitochondrial unfolded protein response; ROS, reactive oxygen species; SDHA, succinate dehydrogenase complex, subunit A; SOD2, superoxide dismutase 2; TFAM, mitochondrial transcription factor A; UPS, ubiquitin-proteasome system; WT, wild-type
Graphical abstract
Highlights
-
•
R6/2 mice display behavioural impairments, oxidative stress and altered proteostasis.
-
•
R6/2 mice show tissue-specific cytosolic & mitochondrial unfolded protein responses.
-
•
R6/2 muscle has increased oxidative markers, ubiquitination, and autophagy activation.
-
•
MitoQ reduces autophagy in R6/2 muscle but not brain, where bioavailability is lower.
-
•
MitoQ ameliorates fine motor control through reduction of oxidative damage in muscle.
1. Introduction
Neurodegenerative disorders often present common pathological mechanisms, including abnormal protein homeostasis (proteostasis), mitochondrial dysfunction, and aberrant redox signalling [1,2]. Physiological redox signalling involves the reversible oxidation of specific protein residues by reactive oxygen species (ROS), such as the H2O2 generated from superoxide produced in mitochondria and by NADPH oxidases [3,4]. The interplay between redox signalling and proteostasis is a subject under active discussion. Disturbed proteostasis and the associated proteotoxic stress may trigger a loss of redox homeostasis [5]. Conversely, aberrant redox signalling may compromise the normal activity of key proteostasis effectors such as molecular chaperones [6,7] and protein degradation pathways, including the ubiquitin-proteasome system (UPS) [8] and autophagy [9]. In the context of neurodegeneration, it remains incompletely understood how changes in redox signalling modulate proteostasis, particularly cytosolic and mitochondrial unfolded protein responses (UPR), and also how targeted interventions that prevent localized ROS production in mitochondria affect proteostasis. A better understanding of this interplay should contribute to develop effective treatments.
Huntington's disease (HD) is a monogenic neurodegenerative disorder that has been increasingly recognized as a systemic disease, due to the presence of several peripheral pathologies in HD patients [10,11]. HD is caused by a CAG repeat expansion in the first exon of the huntingtin (Htt) gene, determining the formation of a mutant Htt protein (mHtt) with an elongated N-terminal polyglutamine stretch. mHtt is prone to proteolytic cleavage, misfolding and aggregation [11,12]. Key pathogenic mechanisms in HD present similarities with other neurodegenerative disorders. The accumulation and aggregation of mHtt disturbs the proteostasis network [13], mHtt induces abnormalities in mitochondrial dynamics and quality control [14], and oxidative biomarkers are increased in HD patients and animal models [[15], [16], [17]], suggesting aberrant redox signalling. The well-defined genetic cause and the pathological features similar to other neurodegenerative disorders support the selection of HD as a model-disease to study the interplay between redox signalling and proteostasis. Related to this interplay, we recently showed an increase of mHtt aggregation upon enhancement of mitochondrial superoxide generation in a HD cell model [18].
Mitochondrial superoxide is a major source of ROS, playing a key role in redox signalling [19,20]. MitoQ10 (MitoQ) or mitoquinone is a lipophilic, positively-charged chain-breaking antioxidant that accumulates in mitochondria [21], where it may contribute to attenuate maladaptive ROS-induced ROS release cycles [22]. MitoQ has been tested in models of neurodegenerative disorders, including spinocerebellar ataxia [23], amyotrophic lateral sclerosis (ALS) [24], Parkinson's [25] and Alzheimer's diseases [26], where it was found to reduce oxidative damage and ameliorate disease phenotypes. However, in these previous studies the effects of MitoQ on cellular proteostasis were not addressed. Moreover, while ROS are early inducers of autophagy [9] and non-targeted antioxidants, such as N-acetylcysteine and vitamin E, inhibited autophagy in cellular and small-organism HD models [27], it remains uncertain what is the impact of mitochondria-targeted antioxidants on proteostasis mechanisms, including autophagy. In this context, the mitochondria-targeted antioxidant MitoQ is an interesting model-drug to assess the effects of ROS modulation on proteostasis.
In the present study we investigate the interplay between redox signalling and proteostasis in the R6/2 HD mouse model and wild-type controls. We characterize the behavioural phenotype of the HD mice and examine the association with markers of oxidative stress, activation of unfolded protein responses, and the status of key protein degradation pathways in the brain and peripheral tissues. We find that while R6/2 mice present widespread markers of oxidative stress, changes in proteostasis are more pronounced in the brain and muscle than in the liver. We test the in vivo effects of MitoQ and find that it ameliorates fine motor control by reducing markers of oxidative damage, particularly in the muscle where it shows higher bioavailability. MitoQ also attenuates ROS-induced muscle autophagy, without changing autophagy markers in the brain, where MitoQ has lower bioavailability. These findings have important implications for understanding the molecular pathogenesis of neurodegenerative disorders and the therapeutic potential of mitochondria-targeted antioxidants.
2. Material and methods
2.1. Animals and treatment
Male wild-type (WT) B6CBAF1/J mice and male transgenic R6/2 mice (B6CBA-Tg(HDexon1)62Gpb/3J) expressing exon 1 of the human huntingtin gene with 120 ± 5 CAG were obtained from Charles River (Barcelona, Spain). R6/2 mice are an HD model with a rapid onset of symptoms, being the most frequently used in the pre-clinical settings [28,29]. Mice arrived at 4 weeks old and were housed in groups of 5 animals under controlled environment (12 light/dark cycle, 21±1 °C) with food and water ad libitum. Experiments were planned and performed according to the 3Rs principle (Replacement, Reduction and Refinement), which comprise the reduction of animal suffering and the number of mice used [30]. Sample size (n = 5, per group) was chosen according to calculation of the standardised effect size using rotarod data of WT and R6/2 mice described in the literature [31], for a significance level of 5% and a power of 90% (http://www.biomath.info/power/). All procedures were previously approved by the Institutional Animal Care and Use Committee from the Centre for Neuroscience and Cell Biology of the University of Coimbra and by the Portuguese Directorate-General for Food and Veterinary, and performed according to the European Union guidelines.
Treatment with 500 μM MitoQ complexed β-cyclodextrin (MitoQ Inc, Auckland) or vehicle (0.12% w/v β-cyclodextrin, Sigma-Aldrich) were administered ad libitum in drinking water, starting at 5 weeks of age. This treatment regime was previously shown safe and effective in mice [23,32]. Drug renewal, mouse weighing, and welfare monitoring were performed twice a week until the end of the experiments. Animals were euthanized by cervical dislocation at 11 weeks of age. Brain, liver and quadriceps muscle were immediately extracted, snap-frozen and stored at -80 °C.
2.2. Behavioural tests
Behavioural assays were performed between 5 and 11 weeks of age, in a sound-attenuated room under controlled temperature and low-intensity light. Mice were acclimated to the room in their home cages, for at least 2 h prior to testing, and were handled by the same person during the tests. The apparatuses were cleaned with 10% ethanol between animals.
2.2.1. Grasping strength
Grasping strength was performed as previously described with minor adaptations [33]. Mice were allowed to grasp with their forepaws a metal grid, fixed to a 300 g weight on top of an electronic scales, while being held by the tail with increasing firmness, until they loosened the grid. The weight (g) change recorded in the scales was divided by the animal weight (g) and expressed as the grasping strength. This assay was repeated 10 times for each animal and the computed result was the average of the 5 trials with the highest values.
2.2.2. Open field
Individual mice were gently positioned in the centre of an open field arena (38 × 38 cm) and allowed to move freely for 15 min, while being video-recorded. The ANY-MAZE software (Stoelting Co.) was used for mouse detection and tracking [34].
2.2.3. Paw clasping
Clasping was assessed in mice suspended by the tail for 30 s [31]. Mice were scored as positive when at least one event of fore and/or hindlimb clasping was observed in the 30 s period.
2.2.4. Pole test
The pole test was performed with a previously described modification for the R6/2 model [31]. Mice were placed head downward on the top of a vertical pole (50 cm in height and 1 cm in diameter) whose base was positioned in the home cage. The time required for mice to descend and reach the floor with all four paws was recorded. The task was repeated 3 times with 60 s rest in between. Each repetition had a maximum waiting period of 120 s. For each mouse, the lowest descending time out of the 3 repetitions was used for statistical analysis [33].
2.2.5. Rotarod
Mice were placed on the cylinder at a constant rotation (4 rpm) (rotarod LE8200, Panlab). After 2 s at 4 rpm, the cylinder was progressively accelerated from 4 to 40 rpm over 5 min. Mice were assayed individually: if they fell before starting the acceleration, the assay could be repeated until a maximum of 3 trials. The assay ended when the mouse fell off the rod and the latency to fall was recorded [33].
2.3. Quantification of tissue MitoQ content
MitoQ content was quantified by liquid chromatography-tandem mass spectrometry (LC-MS-MS) in brain, liver and skeletal muscle. Tissue samples of ~ 50 mg in 250 μL of 95% acetonitrile and 0.1% formic acid were spiked with 50 pmol of d15-MitoQ and processed with a bead-based homogenizer (Precellys Evolution, Bertin Instruments - 6800 rpm, 3 × 20 s and pauses of 30 s). The homogenate was centrifuged at 16,000×g for 10 min and the supernatant collected. The pellet was re-extracted (resuspended in 95% acetonitrile and 0.1% formic acid and homogenised) and re-centrifuged once. Pooled supernatants were filtered (0.22 μm) and evaporated under a stream of nitrogen. Dried material was then resuspended in 100 μL of 60% acetonitrile/0.1% formic acid with vortexing and sonication. MitoQ standards were prepared in liver samples from untreated WT mice, spiked with 0–500 pmol MitoQ and 50 pmol of d15-MitoQ. MitoQ quantification was performed in a LC/MS/MS system comprising a Waters Quattro Ultima triple-quadrupole mass spectrometer, a binary pump (Model 1585; Jasco), an HTC-PAL autosampler (CTC-Analytics) and a Luna 5μm Phenyl–Hexyl column (1 × 50 mm, 5 μm) with a Phenyl–Hexyl guard column (2 × 4 mm; both from Phenomenex) as previously described [32].
2.4. Protein extraction
Brain (cortex), liver and muscle samples were homogenised (Precellys Evolution - 6800 rpm, 3 × 20s and pauses of 30s) in RIPA buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 0.25% sodium deoxycholate; pH 7.4) containing protease inhibitors. Muscle samples required an additional 20 s of sonication to ensure homogenisation. Homogenates were incubated for 1 h in ice and centrifuged at 12,000×g for 10 min. When processing samples for Htt quantification, the centrifugation force was reduced to 300×g to avoid depositing Htt oligomers/aggregates in the pellet. Supernatants were collected and frozen. Protein was quantified by the Bradford protein assay (Bio-Rad).
2.5. Dot blot
For the Dot blot technique protein extracts were spotted in nitrocellulose membrane using a Bio-Dot Microfiltration Apparatus (Bio-Rad). To quantify carbonyl groups, sample derivatization was performed as we previously described [18]. Briefly, protein extracts containing 20 μg of protein were incubated with 200 μL of 12% sodium dodecyl sulfate (SDS) and 400 μL of 20 mM 2,4-dinitrophenylhydrazine hydrochloride (TCI Europe) for 30 min in the dark. After neutralization with 300 μL of 2 M Tris with 18% β-mercaptoethanol, samples containing 1 μg of protein were spotted on the membrane. To quantify ubiquitinated proteins, protein extracts containing 20 μg of protein were denatured at 95 °C for 5min in presence of 2% of SDS before spotting on the membrane. To quantify Htt, protein extracts containing 20 μg of protein were directly spotted on the membrane without derivatization or denaturation.
Spotted membranes were blocked with 5% bovine serum albumin (BSA) in PBST (phosphate buffer solution with 0.05% Tween 20) and incubated with primary antibodies (Htt: 1:1000 anti-N-terminal-Htt, CH00146, Coriell Institute; 1:1000 mEM48, MAB5374, Sigma-Aldrich; ubiquitin: 1:1000 anti-ubiquitin, sc-8017, Santa Cruz Biotechnology; carbonyl groups: 1:1000 anti-dinitrophenol, MAB2223, Sigma-Aldrich). After washing, membranes were incubated with suitable horseradish peroxidase-conjugated secondary antibodies (1:4000 anti-mouse, G-21040; or 1:4000 anti-rabbit, G-21234, Life Technologies) for 1 h, and imaged using a Chemiluminescent kit (Novex ECL, Life Technologies) and a ChemiDoc MP Imaging system (Bio-Rad). Membrane Coomassie staining was used for loading control. Densitometric analyses were performed with Image J (https://imagej.nih.gov/ij/).
2.6. Western blot
Western blot was performed as we previously described with some modifications [35]. Briefly, protein extracts were denatured at 95 °C for 5min in NuPAGE LDS sample buffer (Invitrogen) supplemented with NuPAGE sample reducing agent (Invitrogen). Protein extracts were loaded (25 μg) into 4–12% Bis-Tris gels (Invitrogen), electrophoresed at 200 V for 30 min and electroblotted to PVDF membranes using an iBlot-2 dry transfer device (Invitrogen). Membranes were blocked in PBST containing 5% of BSA and then incubated with primary and respective horseradish peroxidase-conjugated secondary antibodies. Detection and quantification were performed as described in section 2.5. Membrane Coomassie staining was used as loading control. Primary Antibodies: anti-Hsp60 (1:1000, sc-13115), anti-Hsp70 (1:1000, sc-66049), Hsp90 (1:1000, sc-13119), anti-SOD2 (1:500, sc-137254) and TFAM (1:750, sc-23588) from Santa Cruz Biotechnology; anti-SDHA (1:2000, ab14715) from Abcam; anti-SQSTM1/p62 (1:500, #5114) and anti-LC3A/B (1:1000, #4108) from Cell Signaling Technology; and anti-Hsp40 (1:1000, PA5-17382) from Invitrogen. Secondary Antibodies: horseradish peroxidase-conjugated anti-mouse (1:4000, G-21040) and anti-rabbit (1:4000, G-21234) from Invitrogen.
2.7. Immunoprecipitation and blue-native electrophoresis
Htt was detected in native conditions with the mEM48 antibody in brain and muscle extracts previously enriched by immunoprecipitation. Briefly, samples containing 200 μg of total protein were incubated (1 h) with SureBeads Protein G Magnetic beads (Bio-Rad) previously adsorbed (30 min) with anti-N-term-Htt (1:100, CH00146, Coriell Institute). Protein elution was performed in non-denaturing conditions using 20 mM glycine buffer (pH 2.0) [36]. After pH neutralization with 1 M phosphate buffer (pH 7.4), samples were supplemented with NativePAGE sample buffer (Invitrogen) and 10x Coomassie G-250 5% (w/v). Samples were loaded in a polyacrylamide gel (4% stacking, 4–10% resolving gradient) and run with the blue-native PAGE technique [37]: anode buffer (50 mM bis-tris, pH 7.0), cathode buffer (50 mM tricine, 15 mM bis-tris, pH 7.0). The run started with the cathode buffer containing 0.02% Coomassie blue G-250, which was replaced by a cathode buffer without Coomassie blue G-250 at the middle of the run. Proteins were transferred into PVDF membranes (overnight, 30 V, 4 °C; Mini Trans-blot Cell, Bio-Rad) in electroblotting buffer (50 mM tricine, 7.5 mM imidazole; pH 7.0). Membranes were then washed with methanol to reduce the Coomassie G-250 staining and blocked in PBST containing 5% of BSA. Htt was labelled using the mEM48 antibody (1:1000, MAB5374, Sigma-Aldrich). Detection and quantification were performed as described in section 2.5.
2.8. Statistical analysis
Data are from a minimum of 5 mice per condition and are plotted as mean ± standard error of the mean (SEM), unless otherwise stated in figure legends. The Shapiro-Wilk normality test was used to assess data distributions. Two-way ANOVA was used to test for interactions and main effects of the factors: genotype (WT, R6/2) vs. treatment (vehicle, MitoQ); and of the factors: tissue (brain, liver, muscle) vs. treatment. When two-factor interactions were significant, multiple comparisons among groups were performed with the Sidak's post-Hoc test. Correlations between normally distributed data were assessed with the Pearson correlation test. For all analyses, a P value under 0.05 was taken as statistically significant. Data analyses were performed with Prism 8.0 (GraphPad).
3. Results
3.1. R6/2 mice show impaired behaviour and MitoQ ameliorates loss of fine motor control
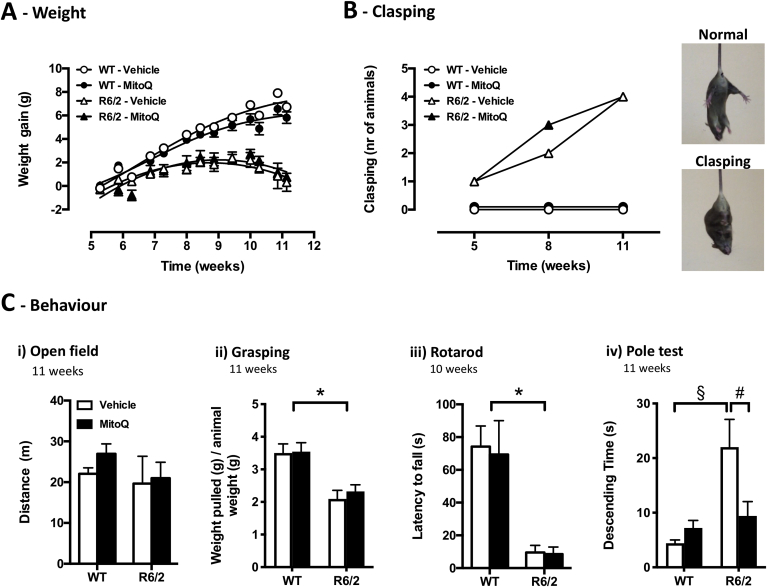
To investigate the role of aberrant ROS signalling in HD pathogenesis and, particularly, the therapeutic potential of treatment with a mitochondria-targeted antioxidant (MitoQ), we compared R6/2 HD mice with age-matched WT controls and tested for genotype × treatment interactions and main effects. The treatment regime with MitoQ (500 μM ad libitum) vs. vehicle control was selected based on previous in vivo data on MitoQ safety and efficacy [23,32]. We first characterized the HD phenotype of R6/2 mice by comparing their weight gain over time with that of age-matched WT controls, and by performing behavioural tests [31,38].
R6/2 mice exhibited a progressive HD-like phenotype, characterized by reduced weight gain and motor dysfunction, as previously described [39,40]. While WT mice gained weight steadily over time, R6/2 mice gained weight only until 8 weeks of age (Fig. 1A) and showed a progressive increase of paw clasping scores (Fig. 1B). This motor dysfunction of R6/2 mice was not so severe as to impair spontaneous locomotion in the open field test, where R6/2 mice travelled similar distances to WT mice even at 11 weeks (Fig. 1Ci). However, the R6/2 mice behavioural impairments manifested with lower grasping strength per animal weight, shorter latency to fall from a rotating rod, and increased descending time in a vertical pole (Fig. 1Cii-iv).
Fig. 1.
Weight and behavioural phenotypes of R6/2 mice and the effect of MitoQ. (A) Weight gain over time in wild-type (WT; circles) and R6/2 (triangles) mice under vehicle (white) or MitoQ (500 μM; black) treatment conditions. (B)Left, paw clasping scores at 5, 8 and 11 weeks; Right, representative images of normal paw positioning vs. paw clasping under tail-suspension. (C)i) Travelled distance in the open-field test; ii) weight pulled per mouse weight in the grasping strength test; iii) latency to fall from the accelerating rotarod; iv) descending time in the pole test. n = 5; *P < 0.05, genotype effect; §P < 0.05 vs. WT-vehicle; #P < 0.05 vs. R6/2-vehicle.
When compared with vehicle treatment (control), treatment with MitoQ evoked no changes in weight gain or clasping phenotypes (Fig. 1A,B). Also, MitoQ did not alter the grasping strength or the latency to fall (Fig. 1Ci-iii). However, MitoQ significantly reduced the time required by R6/2 mice to descend the vertical pole (Fig. 1Civ). These results indicate that treatment with MitoQ is only partially effective in rescuing HD phenotypes, but may hold potential to ameliorate loss of fine motor control in HD, as suggested by improved performance in the vertical pole test.
3.2. MitoQ reduces oxidative stress biomarkers, particularly in the R6/2 muscle
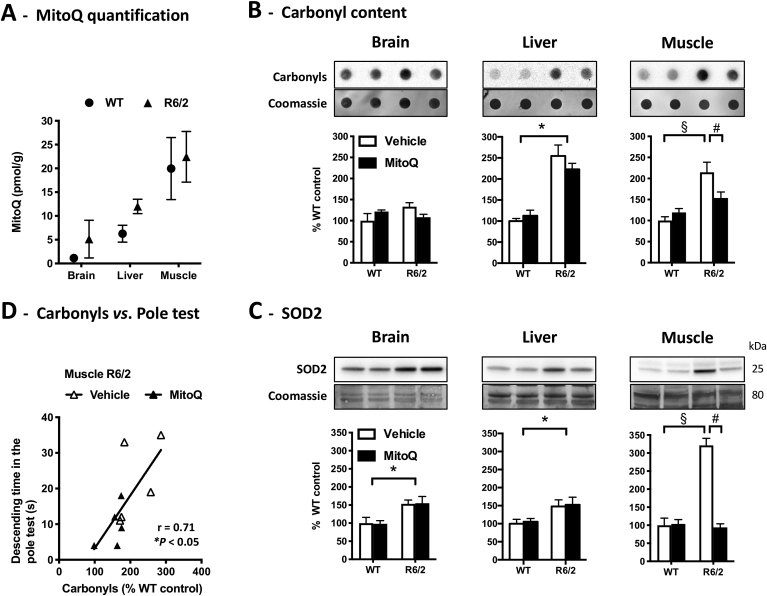
To characterize the molecular pathogenesis in the HD mice, to assess MitoQ distribution, and to test for MitoQ effects upon oxidative stress markers, we analysed samples from R6/2 and WT tissues. Brain, muscle, and liver were chosen given that HD is a neurodegenerative disorder with muscular atrophy and metabolic dysfunction [10,41]. The muscle of R6/2 and WT mice presented the highest levels of MitoQ, about twice the levels found in the liver, whereas the brain presented the lowest amounts, indicating a low brain bioavailability of MitoQ (Fig. 2A).
Fig. 2.
MitoQ tissue distribution and effects on oxidative stress biomarkers. (A) Quantification of MitoQ levels in brain, liver and muscle of MitoQ-treated WT and R6/2 mice by LC-MS-MS; n = 3. (B, C) Oxidative stress biomarkers in brain, liver and muscle of WT and R6/2 mice treated with vehicle or MitoQ: protein carbonyl groups measured by dot blot; SOD2 levels measured by western-blot. Data are in percentage of the mean of WT-vehicle (control); n = 5; *P < 0.05, genotype effect; §P < 0.05 vs. WT-vehicle; #P < 0.05 vs. R6/2-vehicle. (D) Correlation between the descending time in the pole test and muscle carbonyl content for R6/2 mice.
We measured protein carbonylation as a marker of oxidative damage [42] and levels of mitochondrial superoxide dismutase (SOD2), a marker of the antioxidant response to oxidative stress [43]. R6/2 mice presented higher levels of protein carbonyls and of SOD2 than WT mice: carbonyls were significantly elevated in liver and muscle (Fig. 2B), whereas SOD2 levels were significantly elevated in all tissues analysed (Fig. 2C). Focusing on R6/2 mice, treatment with MitoQ induced an overall decrease in protein carbonyls (tissue × treatment interaction: F(2,24) = 0.615, P = 0.549; treatment effect: F(1,24) = 7.74, P = 0.010). However, the effect of MitoQ was particularly evident in muscle, where it reduced not only protein carbonyls but also SOD2 levels (Fig. 2B,C). These results suggest that MitoQ, a mitochondria-targeted chain-breaking antioxidant [21], reduces oxidative stress in R6/2 mice, particularly in muscle where the compound has higher bioavailability. This raises the mechanistic hypothesis that MitoQ improves fine motor performance in the pole test by reducing oxidative damage in the R6/2 muscle. To test this hypothesis, we plotted levels of protein carbonyls in R6/2 mice against the respective scores in the pole test (Fig. 2D). The correlation between both values strongly suggests that the levels of carbonyls in muscle predict pole test performance. Moreover, these data support the mechanistic hypothesis that high muscle bioavailability of MitoQ reduces oxidative damage to muscle and thereby ameliorates fine motor control in R6/2 mice.
3.3. R6/2 mice have increased levels of mitochondrial and cytosolic chaperones
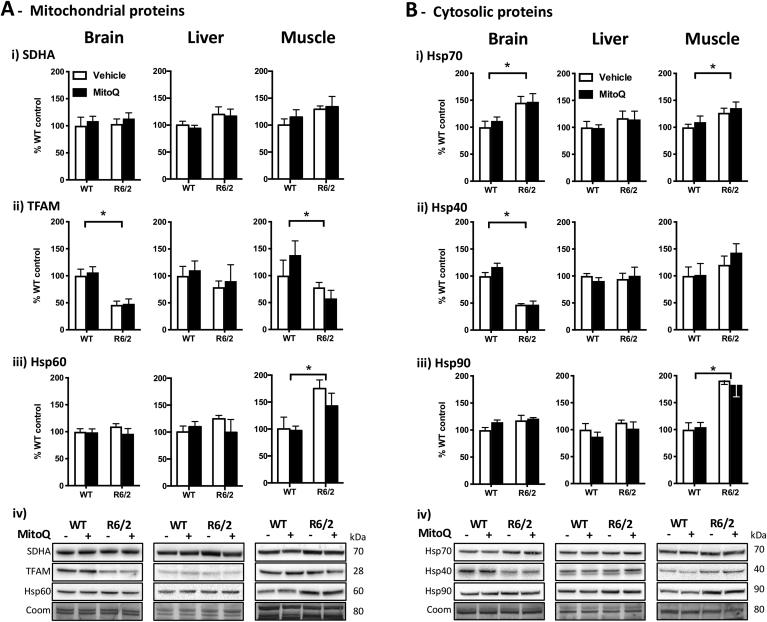
To examine the molecular changes in proteostasis effectors in the HD mice, and also attending to the mitochondrial targeting of MitoQ [21], we measured key mitochondrial biomarkers and molecular chaperones. Concerning mitochondria, we focused on SDHA (succinate dehydrogenase complex, subunit A), TFAM (mitochondrial transcription factor A), and Hsp60 (mitochondrial chaperone) as indicators of mitochondrial mass, biogenesis, and proteostasis, respectively [[44], [45], [46]]. Concerning cytosolic chaperones, we focused on Hsp90, Hsp70 and its co-chaperone Hsp40 as key effectors of the proteostasis network involved in protein stabilization, refolding, or targeting for degradation [47].
The levels of SDHA were similar in R6/2 and WT mice, independently of the tissue (Fig. 3Ai), which suggests no major differences in mitochondrial mass between genotypes. In contrast with similar SDHA levels, however, TFAM and Hsp60 levels were significantly altered (Fig. 3A). The brain and muscle of R6/2 mice displayed lower levels of TFAM (Fig. 3Aii), suggesting a reduction of the mitochondrial DNA transcription and, consequently, of the mitochondrial biogenesis [48]. Regarding mitochondrial proteostasis, the muscle of R6/2 mice presented higher levels of the Hsp60 chaperone (Fig. 3Aiii), indicating an activation of the mitochondrial unfolded protein response (UPRmt), which is a transcriptional response of the cell to a mitochondrial stress [49].
Fig. 3.
Levels of mitochondrial biomarkers and chaperones in WT and R6/2 mice. Western blot quantifications in brain, liver and muscle samples from WT and R6/2 mice treated with vehicle or MitoQ. (A) Mitochondrial biomarkers: i) SDHA, succinate dehydrogenase complex, subunit A; ii) TFAM, mitochondrial transcription factor A, iii) Hsp60; and iv) representative blots. (B) Cytosolic chaperones: i) Hsp70; ii) Hsp40; iii) Hsp90; and iv) representative blots. Coom – Coomassie staining loading control. Data are in percentage of the mean of WT-vehicle (control); n = 5; *P < 0.05, genotype effect.
Concerning the cytosolic proteostasis network, the brain and muscle from R6/2 mice presented significant differences from WT mice in the levels of molecular chaperones (Fig. 3B). This shows that the R6/2 brain and muscle are under proteotoxic stress, unlike the liver where the lack of differences in chaperone levels suggests no activation of an unfolded protein response. Specifically, the R6/2 brain and muscle presented higher levels of Hsp70 and, interestingly, the co-chaperone Hsp40 was significantly decreased in the R6/2 brain, whereas the Hsp90 chaperone was significantly elevated in the R6/2 muscle (Fig. 3B). These results suggest that there is altered mitochondrial and cytosolic proteostasis in R6/2 mice, with an activation of the unfolded protein responses. While treatment with MitoQ evoked no significant changes in the levels of chaperones, we cannot exclude the hypothesis that reducing mitochondrial oxidative damage with MitoQ may attenuate the proteotoxic stress in the mitochondria (MitoQ treatment effect upon Hsp60 levels across R6/2 tissues: F(1,24) = 3.22, P = 0.086).
3.4. R6/2 mice show increased protein ubiquitination and autophagy induction in muscle, which is decreased by MitoQ
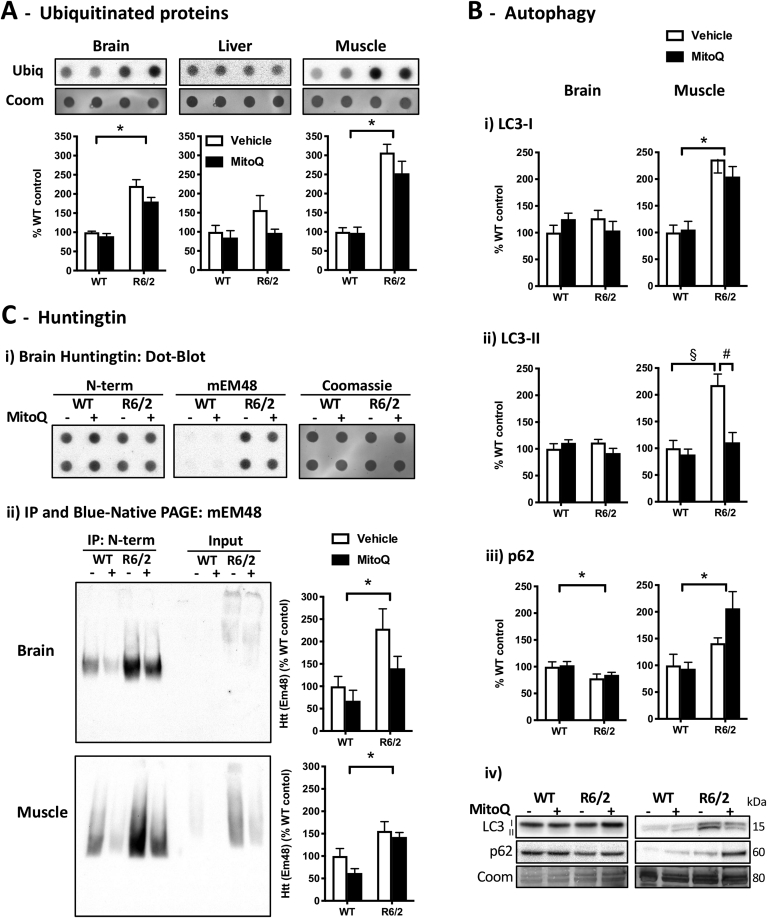
Following evidence for oxidative stress and altered chaperone levels in R6/2 mice, we further investigated how changes in redox signalling might influence the proteostasis network, now focusing on protein degradation pathways. Pursuing the hypothesis that the UPS and autophagy pathways are under redox regulation [8,50], we examined the levels of ubiquitinated proteins and autophagy markers in R6/2 and WT mice, and how these were affected by treatment with the mitochondria-targeted antioxidant MitoQ.
The levels of ubiquitinated proteins were significantly elevated in the brain and muscle, but not in the liver of R6/2 mice when compared with WT mice (Fig. 4A). This tissue-specific profile agrees with the observed changes in molecular chaperones (Fig. 3B) and indicates that the proteotoxic stress is higher in the R6/2 brain and muscle than in the liver. We next studied autophagy markers, particularly LC3-I, LC3-II and p62. In the process of autophagosome formation, LC3-I converts into the lipidated form LC3-II, which is a marker of completed autophagosomes [51]. The autophagic adaptor p62 functions as a link between LC3 and ubiquitinated substrates, and is itself a substrate for autophagic degradation [51,52].
Fig. 4.
Levels of ubiquitinated proteins, autophagy biomarkers and huntingtin. Proteostasis markers in tissue samples from WT and R6/2 mice treated with vehicle or MitoQ. (A) Dot blot quantification of ubiquitinated proteins (ubiq); Coom, Coomassie staining loading control. (B) Western blot quantification of autophagy biomarkers: i) LC3-I; ii) LC3-II; iii) p62; and iv) representative blots. (C) Quantification of huntingtin (Htt): i) dot blot of native (non-denatured) brain extracts using an N-terminal (N-term) Htt antibody or the anti-Htt mEM48 antibody; ii) immunoprecipitation (IP) with anti-N-terminal Htt in brain and muscle samples, followed by blue-native page using the mEM48 antibody. Data are in percentage of the mean of WT-vehicle (control). n = 5; *P < 0.05, genotype effect; &P < 0.05, treatment effect; §P < 0.05 vs. WT-vehicle, #P < 0.05 vs. R6/2-vehicle.
The levels of LC3-I and LC3-II in the R6/2 brain were not significantly different from WT, albeit the decrease in p62 levels in the R6/2 brain suggests there could be a modest increase in its autophagic degradation (Fig. 4B). In the brain, the levels of autophagic markers were unaffected by treatment with MitoQ, in agreement with its reduced brain bioavailability (Fig. 2A). In contrast, the levels of LC3-I and LC3-II were significantly elevated in the R6/2 muscle in comparison with WT (Fig. 4B), suggesting more autophagosomes. In the R6/2 muscle, treatment with MitoQ decreased LC3-II levels without decreasing LC3-I and p62 levels (Fig. 4B, R6/2 muscle: MitoQ vs. vehicle), whereas in WT muscle MitoQ did not change autophagy markers (Fig. 4B, WT muscle: MitoQ vs. vehicle). Taken together with increased carbonyl levels in the R6/2 muscle (Fig. 2B), these results suggest that there is a ROS-mediated autophagy induction in the R6/2 muscle, which is attenuated by the antioxidant MitoQ resulting in decreased LC3-I to LC3-II conversion.
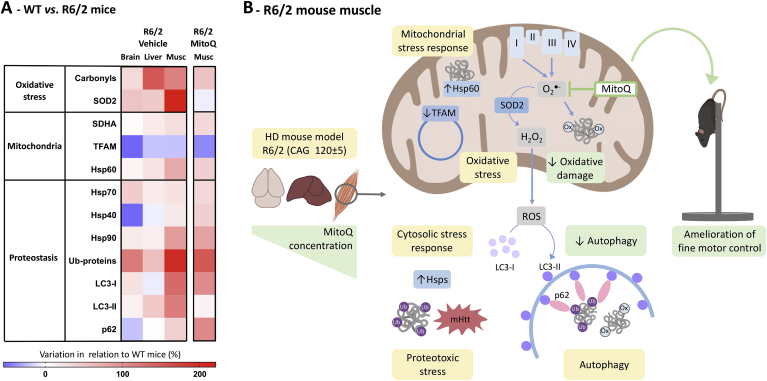
Given the observed changes in protein degradation pathways in the R6/2 brain and muscle, we next examined the levels of Htt in these tissues and whether these were modulated by MitoQ. To detect Htt, we first performed dot-blots using brain samples in native (non-denatured) conditions. In these native dot-blots the anti-N-terminal Htt antibody labelled both WT and R6/2 samples, whereas the mEM48 antibody preferentially labelled the R6/2 samples (Fig. 4Ci). Indeed, although mEM48 was generated by using the first 256 amino acids of Htt without the polyQ tract, mEM48 preferentially recognizes Htt aggregates [[53], [54], [55]]. To quantify Htt, we performed an immunoprecipitation with the anti-N-terminal antibody and ran the enriched samples in blue-native PAGE (with Coomassie blue G-250 to confer a net negative charge to proteins while preserving their native state [37]). In the blue-native PAGE, mEM48 yielded a significantly stronger labelling in the immunoprecipitated samples of R6/2 vs. WT mice (Fig. 4Cii). Treatment with MitoQ induced no significant changes in the overall labelling achieved with mEM48. These results suggest that treatment with MitoQ does not significantly alter the levels of Htt. As summarized in Fig. 5, the amelioration of fine motor performance in R6/2 mice treated with MitoQ is likely due to a combination of reduced oxidative damage and attenuation of ROS-induced muscle autophagy.
Fig. 5.
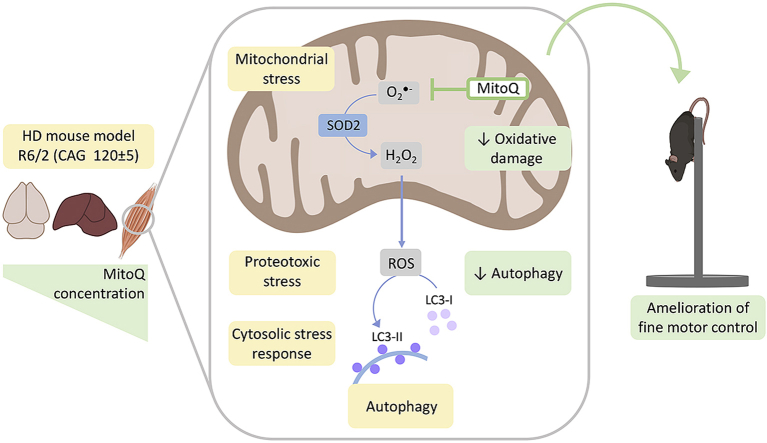
Interplay between ROS signalling and proteostasis: Effects of MitoQ. (A) Heat map summarising the biomarker profile in tissues from R6/2 HD mice in relation to WT under vehicle-treatment conditions, and the effects of MitoQ treatment in R6/2 muscle. (B) Mechanistic pathways for MitoQ effects in the R6/2 mouse muscle. The mitochondria-targeted antioxidant MitoQ accumulates in mitochondria where it interacts with ROS, such as superoxide [21], reducing overly active redox signalling pathways and thereby attenuating oxidative damage and the redox-regulated induction of autophagy [9].
4. Discussion
The bidirectional interplay between redox signalling and proteostasis is an active field of research. Disturbances in these processes are associated with neurodegenerative disorders and may hold potential for therapeutic targeting. Here we investigate this interplay and perform the first in vivo testing of the mitochondria-targeted antioxidant MitoQ in HD mice. We found widespread oxidative stress and alterations in the proteostasis network that differ across HD mice tissues. Data highlight the relevance of peripheral HD pathology, particularly in muscle where MitoQ may have beneficial effects.
4.1. R6/2 mice model systemic HD with widespread oxidative stress and tissue-specific alterations in the proteostasis network
HD is primarily known as a neurodegenerative disorder. However, patients often present weight loss, metabolic dysfunction, skeletal muscle atrophy, and osteoporosis [41,56], highlighting the systemic nature of HD [10,11]. Recent studies with neurons, muscle cells, and hepatocytes derived from HD stem cells support the tissue-specificity of molecular HD phenotypes [57]. Thus, to address HD as a systemic disease, we studied the brain, skeletal muscle, and liver from R6/2 HD mice.
Impaired redox signalling is a hallmark of HD, with patients and animal models exhibiting oxidative damage to proteins, lipids, and DNA [7,[15], [16], [17],58]. In the present study, R6/2 mice with 11 weeks presented increased levels of the mitochondrial antioxidant enzyme SOD2 in the brain, liver, and muscle, indicating a widespread response to oxidative stress. Protein carbonyls were also elevated in R6/2 liver and muscle, suggesting that muscle dysfunction contributes to the motor behaviour phenotypes. In the R6/2 brain, however, carbonyls were not yet significantly elevated, possibly because the brain susceptibility to oxidative damage [59] was still compensated by its effective antioxidant defences and proteostatic machinery that handle oxidized proteins [60].
The proteostasis network includes molecular chaperones and protein clearance pathways, such as the UPS and autophagy. Under physiological conditions, the proportion of central chaperones such as Hsp70, Hsp40 and Hsp90 is thought to remain relatively constant in all tissues. Recent data, however, show that the proteostasis network can be tailored to specific proteomes, being controlled by neuronal activity and inter-tissue communication [61]. Here, we measured protein levels of central chaperones and found them significantly altered in the brain and muscle of R6/2 mice, together with increased ubiquitinated proteins. These alterations in proteostasis were tissue-specific, being absent in the liver. Accordingly, elevations in chaperone mRNAs were previously found in skeletal muscle, but not in liver from R6/2 mice [62]. We also found pronounced changes in autophagy markers in the R6/2 muscle, which together with increased SOD2 and carbonyls support the hypothesis that an interplay between abnormal redox signalling and proteostasis in muscle contributes to motor impairment. These data support skeletal muscle pathology as a hallmark of HD [29] and a therapeutic target for redox and proteostasis modulators.
4.2. R6/2 mice display mitochondrial and cytosolic unfolded protein responses
Mitochondrial stress conditions, such as those triggered by excessive ROS or the accumulation of misfolded proteins, can elicit a transcriptional response called the UPRmt [63]. The UPRmt promotes mitochondrial recovery by decreasing general translation while inducing mitochondrial proteases and chaperones, such as Hsp60 [49,64]. It remains uncertain whether the UPRmt is activated in HD. Contrasting data from HD cell models either support UPRmt activation – by showing increases in mitochondrial chaperone levels [65]; or suggest that mHtt inhibits the UPRmt – by showing impaired mRNA stability of the ABCB10 transporter whose deletion decreases mitochondrial chaperone levels [66]. Given the tissue-specificity of proteostatic responses [61], studies in animal models may help clarify the UPRmt status in HD.
Our data in R6/2 mice suggest an activation of the UPRmt specifically in muscle, characterized by increased levels of Hsp60 and decreased levels of TFAM. Increased Hsp60 contributes to the UPRmt by promoting protein folding in mitochondria, whereas decreased TFAM may contribute to the UPRmt by limiting mitochondrial protein synthesis [45]. Two complementary mechanisms may explain decreased TFAM levels in the UPRmt: (1) increased TFAM degradation by the mitochondrial LON protease [67,68]; and (2) repression of NRF1-mediated TFAM transcription [69]. We also observed decreased TFAM in R6/2 brain samples, which is compatible with the transcriptional attenuation detected in brain samples of HD patients [70], and could be part of a pro-survival unfolded protein response. In fact, the experimental activation of the UPRmt was found protective in disease models [71,72], including HD cell models where UPRmt activation by GRP75 knockdown reduced mHtt aggregation [65].
Recent studies suggest that the UPRmt can coordinate with UPRs in other compartments, namely via changes in lipid biosynthesis that signal to the nucleus and induce the expression of cytosolic chaperones [64,65]. Here, we found increased levels of Hsp70 in the brain and muscle of R6/2 mice, together with increased ubiquitinated proteins, suggesting the activation of a cytosolic UPR in response to mHtt proteotoxicity. Despite the increase in Hsp70, its activity may be compromised in the R6/2 brain where we also found decreased levels of Hsp40. Indeed, Hsp40 is a limiting factor for a trimeric chaperone complex, including Hsp70 and Hsp110, which disaggregates mHtt fibrils [73]. Previous studies in R6/2 mice have found age-dependent decreases in the levels of brain chaperones [74,75], their sequestration into mHtt aggregates [76,77], and a phenotypic rescue with Hsp40 gene therapy [[78], [79], [80]].
In the R6/2 muscle, in addition to increased Hsp70 we found increased Hsp90, supporting the hypothesis that mHtt can induce the proteostasis network in peripheral tissues such as skeletal muscle [62]. Interestingly, it was recently proposed that Hsp90 and Hsp70 interact with the ATP-gated ion channel P2RX7 to mediate autophagy induction in myoblasts [81]. This raises the hypothesis that increased cytosolic chaperones in the R6/2 muscle might play a role in autophagy induction in this tissue, which has been associated with muscle wasting [82].
4.3. Treatment with MitoQ ameliorates fine motor control and attenuates ROS-induced muscle autophagy in R6/2 mice
Oxidative stress is associated with neurodegenerative diseases [2], including HD [15], however, classical antioxidants have failed to benefit HD patients [[83], [84], [85]]. This could be due to lack of selectivity or disruption of redox signalling [85], being possible that antioxidants that selectively target mitochondrial ROS might provide clinical benefit [3,4]. Accordingly, the mitochondria-targeted antioxidant MitoQ reduced oxidative damage and ameliorated disease phenotypes in animal models of Parkinson's [25], Alzheimer's [26,86], ALS [24], and spinocerebellar ataxia [23]. Concerning HD models, MitoQ was tested in vitro in STHdhQ111 cells where it reduced markers of oxidative damage and the mutant-huntingtin-induced mitochondrial toxicity and synaptic damage [87]. While these studies suggest protective effects for MitoQ, its impact on proteostasis was not assessed. Here, we investigated the interplay between redox signalling and proteostasis in R6/2 mice, using MitoQ as a redox-modulator and assessing its in vivo therapeutic potential in HD.
Treatment with MitoQ was only partially effective against the behavioural phenotypes of R6/2 mice, but significantly improved their fine motor control in the pole test. The pole test is a complex task that has been previously used to monitor motor and sensorimotor impairments in rodents [38]. Since pole test performance correlated with the levels of muscle carbonyls and these were significantly decreased by MitoQ, together with SOD2 levels, these data suggest that MitoQ ameliorates fine motor control by reducing oxidative damage in muscle. Indeed, the high bioavailability of MitoQ in muscle contrasts with its low brain bioavailability, suggesting peripheral rather than central nervous system effects. These findings support the hypothesis that redox interventions can improve muscle performance [88] and highlight the importance of peripheral therapeutics in HD.
The interplay between redox signalling and proteostasis is a topic under active discussion. ROS can modulate proteostasis effectors and, in turn, imbalances of proteostasis regulate redox homeostasis. Interestingly, this bidirectional regulation may occur across neighbouring cells and tissues, with proteostasis imbalances in neurons affecting the redox state of muscle cells and vice versa [5]. Although ROS can induce and regulate the activity of molecular chaperones [6,7] and the UPS [8,49], we found no significant treatment effect with MitoQ upon the levels of molecular chaperones, which remained altered in R6/2 tissues together with elevated levels of ubiquitinated proteins. However, the treatment with MitoQ significantly attenuated ROS-induced autophagy in the R6/2 muscle, where MitoQ has higher bioavailability.
ROS are early inducers of autophagy [9,50], and untargeted antioxidants are known to inhibit autophagy [27]. Recent in vitro studies in C2C12 myoblasts treated with the mitochondria-targeted MitoQ (≤0.5 μM) found that MitoQ attenuated autophagy by suppressing mitochondrial ROS production, which reduced mTOR and ULK1 phosphorylation, expression of Atg proteins, and the conversion of LC3-I to LC3-II [89]. In the present in vivo study, using a previously reported safe and effective dose [23,32], we found that MitoQ reduced the elevated LC3-II levels in the R6/2 muscle, without decreasing its p62 levels, and also without changing autophagy markers in WT muscle. This suggests that MitoQ reduces ROS-mediated autophagy induction in the R6/2 muscle. Importantly, this effect of MitoQ was selective for muscle, where MitoQ has higher bioavailability, and MitoQ did not change autophagic markers in the brain, where it has lower bioavailability. Thus, although autophagy is an important pathway to clear misfolded proteins, its excessive ROS-mediated induction in muscle appears maladaptive, so that selective autophagy attenuation with MitoQ might be beneficial to tackle peripheral HD symptoms.
5. Conclusion
Data from this study support the hypothesis that abnormal redox signalling in muscle contributes to altered proteostasis and motor impairment in HD. Moreover, this first in vivo study with the mitochondria-targeted redox modulator MitoQ in a HD mouse model shows that it can rescue fine motor control through its antioxidant effects at peripheral level. The high bioavailability of MitoQ in muscle and the attenuation of ROS-induced autophagy in this tissue could be advantageous to ameliorate muscle wasting, whereas low brain bioavailability of MitoQ limits the risk of impairing neuronal protein clearance mechanisms. This study also suggests that chaperone-mediated cytosolic and mitochondrial unfolded protein responses are active in the R6/2 HD mice and unimpaired by MitoQ treatment. MitoQ thus holds potential as an adjuvant treatment for peripheral HD symptoms. Future studies are now warranted with mitochondria-targeted antioxidants in slow progressive HD models to examine the potential long-term amelioration of disease phenotypes. Moreover, these data suggest that mitochondria-targeted antioxidants warrant further investigation in the context of neuromuscular disorders.
Author contributions
BRP performed the majority of experiments, data analysis, and literature search. Behavioural experiments: BRP, AID, PMC. Molecular Biology: BRP. Manuscript writing: BRP and JMAO. Experimental design, data analysis, and discussion: BRP, AID, PMC, PIM, MPM, JMAO.
Declaration of competing interest
MPM holds patents in the development of MitoQ as a therapy.
Acknowledgements
Work in JMAO's lab was supported by the FCT – Fundação para a Ciência e a Tecnologia – (P2020-PTDC/NEU-NMC/0412/2014; 3599-PPCDT; UID/QUI/50006/2019) and the European Union (POCI-01-0145-FEDER-016577). BRP and AID acknowledge FCT for PostDoc fellowships (SFRH/BPD/102259/2014 and SFRH/BPD/84473/2012). Work in MPM's lab was supported by the Medical Research Council UK (MC_U105663142) and by a Wellcome Trust Investigator award (110159/Z/15/Z).
References
- 1.Gan L., Cookson M.R., Petrucelli L., La Spada A.R. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 2018;21:1300–1309. doi: 10.1038/s41593-018-0237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niedzielska E., Smaga I., Gawlik M., Moniczewski A., Stankowicz P., Pera J., Filip M. Oxidative stress in neurodegenerative diseases. Mol. Neurobiol. 2016;53:4094–4125. doi: 10.1007/s12035-015-9337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 4.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirstein J., Morito D., Kakihana T., Sugihara M., Minnen A., Hipp M.S., Nussbaum-Krammer C., Kasturi P., Hartl F.U., Nagata K., Morimoto R.I. Proteotoxic stress and ageing triggers the loss of redox homeostasis across cellular compartments. EMBO J. 2015;34:2334–2349. doi: 10.15252/embj.201591711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McArdle A., Pollock N., Staunton C.A., Jackson M.J. Aberrant redox signalling and stress response in age-related muscle decline: role in inter- and intra-cellular signalling. Free Radic. Biol. Med. 2019;132:50–57. doi: 10.1016/j.freeradbiomed.2018.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorolla M.A., Rodriguez-Colman M.J., Vall-llaura N., Tamarit J., Ros J., Cabiscol E. Protein oxidation in Huntington disease. Biofactors. 2012;38:173–185. doi: 10.1002/biof.1013. [DOI] [PubMed] [Google Scholar]
- 8.Segref A., Kevei E., Pokrzywa W., Schmeisser K., Mansfeld J., Livnat-Levanon N., Ensenauer R., Glickman M.H., Ristow M., Hoppe T. Pathogenesis of human mitochondrial diseases is modulated by reduced activity of the ubiquitin/proteasome system. Cell Metabol. 2014;19:642–652. doi: 10.1016/j.cmet.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Filomeni G., De Zio D., Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mielcarek M. Huntington's disease is a multi-system disorder. Rare Dis. 2015;3 doi: 10.1080/21675511.2015.1058464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R., Wild E.J., Tabrizi S.J. Huntington disease. Nat. Rev. Dis. Primers. 2015;1 doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 12.Tabrizi S.J., Ghosh R., Leavitt B.R. Huntingtin lowering strategies for disease modification in Huntington's disease. Neuron. 2019;101:801–819. doi: 10.1016/j.neuron.2019.01.039. [DOI] [PubMed] [Google Scholar]
- 13.Soares T.R., Reis S.D., Pinho B.R., Duchen M.R., Oliveira J.M.A. Targeting the proteostasis network in Huntington's disease. Ageing Res. Rev. 2019;49:92–103. doi: 10.1016/j.arr.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guedes-Dias P., Pinho B.R., Soares T.R., de Proenca J., Duchen M.R., Oliveira J.M. Mitochondrial dynamics and quality control in Huntington's disease. Neurobiol. Dis. 2016;90:51–57. doi: 10.1016/j.nbd.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Paul B.D., Snyder S.H. Impaired redox signaling in Huntington's disease: therapeutic implications. Front. Mol. Neurosci. 2019;12:68. doi: 10.3389/fnmol.2019.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal S., Fox J., Thyagarajan B., Fox J.H. Brain mitochondrial iron accumulates in Huntington's disease, mediates mitochondrial dysfunction, and can be removed pharmacologically. Free Radic. Biol. Med. 2018;120:317–329. doi: 10.1016/j.freeradbiomed.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorolla M.A., Reverter-Branchat G., Tamarit J., Ferrer I., Ros J., Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic. Biol. Med. 2008;45:667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Pinho B.R., Reis S.D., Hartley R.C., Murphy M.P., Oliveira J.M.A. Mitochondrial superoxide generation induces a parkinsonian phenotype in zebrafish and huntingtin aggregation in human cells. Free Radic. Biol. Med. 2019;130:318–327. doi: 10.1016/j.freeradbiomed.2018.10.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angelova P.R., Abramov A.Y. Functional role of mitochondrial reactive oxygen species in physiology. Free Radic. Biol. Med. 2016;100:81–85. doi: 10.1016/j.freeradbiomed.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Smith R.A., Hartley R.C., Cocheme H.M., Murphy M.P. Mitochondrial pharmacology. Trends Pharmacol. Sci. 2012;33:341–352. doi: 10.1016/j.tips.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stucki D.M., Ruegsegger C., Steiner S., Radecke J., Murphy M.P., Zuber B., Saxena S. Mitochondrial impairments contribute to Spinocerebellar ataxia type 1 progression and can be ameliorated by the mitochondria-targeted antioxidant MitoQ. Free Radic. Biol. Med. 2016;97:427–440. doi: 10.1016/j.freeradbiomed.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Miquel E., Cassina A., Martinez-Palma L., Souza J.M., Bolatto C., Rodriguez-Bottero S., Logan A., Smith R.A., Murphy M.P., Barbeito L., Radi R., Cassina P. Neuroprotective effects of the mitochondria-targeted antioxidant MitoQ in a model of inherited amyotrophic lateral sclerosis. Free Radic. Biol. Med. 2014;70:204–213. doi: 10.1016/j.freeradbiomed.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh A., Chandran K., Kalivendi S.V., Joseph J., Antholine W.E., Hillard C.J., Kanthasamy A., Kanthasamy A., Kalyanaraman B. Neuroprotection by a mitochondria-targeted drug in a Parkinson's disease model. Free Radic. Biol. Med. 2010;49:1674–1684. doi: 10.1016/j.freeradbiomed.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McManus M.J., Murphy M.P., Franklin J.L. The mitochondria-targeted antioxidant MitoQ prevents loss of spatial memory retention and early neuropathology in a transgenic mouse model of Alzheimer's disease. J. Neurosci. 2011;31:15703–15715. doi: 10.1523/JNEUROSCI.0552-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Underwood B.R., Imarisio S., Fleming A., Rose C., Krishna G., Heard P., Quick M., Korolchuk V.I., Renna M., Sarkar S., Garcia-Arencibia M., O'Kane C.J., Murphy M.P., Rubinsztein D.C. Antioxidants can inhibit basal autophagy and enhance neurodegeneration in models of polyglutamine disease. Hum. Mol. Genet. 2010;19:3413–3429. doi: 10.1093/hmg/ddq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figiel M., Szlachcic W.J., Switonski P.M., Gabka A., Krzyzosiak W.J. Mouse models of polyglutamine diseases: review and data table. Part I. Mol. Neurobiol. 2012;46:393–429. doi: 10.1007/s12035-012-8315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zielonka D., Piotrowska I., Marcinkowski J.T., Mielcarek M. Skeletal muscle pathology in Huntington's disease. Front. Physiol. 2014;5:380. doi: 10.3389/fphys.2014.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Festing M.F., Altman D.G. Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J. 2002;43:244–258. doi: 10.1093/ilar.43.4.244. [DOI] [PubMed] [Google Scholar]
- 31.Ochaba J., Monteys A.M., O'Rourke J.G., Reidling J.C., Steffan J.S., Davidson B.L., Thompson L.M. PIAS1 regulates mutant huntingtin accumulation and Huntington's disease-associated phenotypes in vivo. Neuron. 2016;90:507–520. doi: 10.1016/j.neuron.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Cuenca S., Cocheme H.M., Logan A., Abakumova I., Prime T.A., Rose C., Vidal-Puig A., Smith A.C., Rubinsztein D.C., Fearnley I.M., Jones B.A., Pope S., Heales S.J., Lam B.Y., Neogi S.G., McFarlane I., James A.M., Smith R.A., Murphy M.P. Consequences of long-term oral administration of the mitochondria-targeted antioxidant MitoQ to wild-type mice. Free Radic. Biol. Med. 2010;48:161–172. doi: 10.1016/j.freeradbiomed.2009.10.039. [DOI] [PubMed] [Google Scholar]
- 33.Rial D., Castro A.A., Machado N., Garcao P., Goncalves F.Q., Silva H.B., Tome A.R., Kofalvi A., Corti O., Raisman-Vozari R., Cunha R.A., Prediger R.D. Behavioral phenotyping of Parkin-deficient mice: looking for early preclinical features of Parkinson's disease. PLoS One. 2014;9 doi: 10.1371/journal.pone.0114216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguiar A.S., Jr., Speck A.E., Amaral I.M., Canas P.M., Cunha R.A. The exercise sex gap and the impact of the estrous cycle on exercise performance in mice. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-29050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guedes-Dias P., de Proenca J., Soares T.R., Leitao-Rocha A., Pinho B.R., Duchen M.R., Oliveira J.M. HDAC6 inhibition induces mitochondrial fusion, autophagic flux and reduces diffuse mutant huntingtin in striatal neurons. Biochim. Biophys. Acta. 2015;1852:2484–2493. doi: 10.1016/j.bbadis.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Sousa M.M., Steen K.W., Hagen L., Slupphaug G. Antibody cross-linking and target elution protocols used for immunoprecipitation significantly modulate signal-to noise ratio in downstream 2D-PAGE analysis. Proteome Sci. 2011;9:45. doi: 10.1186/1477-5956-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittig I., Braun H.P., Schagger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 38.Brooks S.P., Dunnett S.B. Tests to assess motor phenotype in mice: a user's guide. Nat. Rev. Neurosci. 2009;10:519–529. doi: 10.1038/nrn2652. [DOI] [PubMed] [Google Scholar]
- 39.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 40.Ferrante R.J. Mouse models of Huntington's disease and methodological considerations for therapeutic trials. Biochim. Biophys. Acta. 2009;1792:506–520. doi: 10.1016/j.bbadis.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll J.B., Bates G.P., Steffan J., Saft C., Tabrizi S.J. Treating the whole body in Huntington's disease. Lancet Neurol. 2015;14:1135–1142. doi: 10.1016/S1474-4422(15)00177-5. [DOI] [PubMed] [Google Scholar]
- 42.Fedorova M., Bollineni R.C., Hoffmann R. Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom. Rev. 2014;33:79–97. doi: 10.1002/mas.21381. [DOI] [PubMed] [Google Scholar]
- 43.Miao L., St Clair D.K. Regulation of superoxide dismutase genes: implications in disease. Free Radic. Biol. Med. 2009;47:344–356. doi: 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincent A.E., Rosa H.S., Pabis K., Lawless C., Chen C., Grunewald A., Rygiel K.A., Rocha M.C., Reeve A.K., Falkous G., Perissi V., White K., Davey T., Petrof B.J., Sayer A.A., Cooper C., Deehan D., Taylor R.W., Turnbull D.M., Picard M. Subcellular origin of mitochondrial DNA deletions in human skeletal muscle. Ann. Neurol. 2018;84:289–301. doi: 10.1002/ana.25288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Picca A., Lezza A.M. Regulation of mitochondrial biogenesis through TFAM-mitochondrial DNA interactions: useful insights from aging and calorie restriction studies. Mitochondrion. 2015;25:67–75. doi: 10.1016/j.mito.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Venkatesh S., Suzuki C.K. HSP60 takes a hit: inhibition of mitochondrial protein folding. Cell. Chem. Biol. 2017;24:543–545. doi: 10.1016/j.chembiol.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 47.Reis S.D., Pinho B.R., Oliveira J.M.A. Modulation of molecular chaperones in Huntington's disease and other polyglutamine disorders. Mol. Neurobiol. 2017;54:5829–5854. doi: 10.1007/s12035-016-0120-z. [DOI] [PubMed] [Google Scholar]
- 48.Kang I., Chu C.T., Kaufman B.A. The mitochondrial transcription factor TFAM in neurodegeneration: emerging evidence and mechanisms. FEBS Lett. 2018;592:793–811. doi: 10.1002/1873-3468.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Amico D., Sorrentino V., Auwerx J. Cytosolic proteostasis networks of the mitochondrial stress response. Trends Biochem. Sci. 2017;42:712–725. doi: 10.1016/j.tibs.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Scherz-Shouval R., Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem. Sci. 2011;36:30–38. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. third ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marx V. Autophagy: eat thyself, sustain thyself. Nat. Methods. 2015;12:1121–1125. doi: 10.1038/nmeth.3661. [DOI] [PubMed] [Google Scholar]
- 53.Gutekunst C.A., Li S.H., Yi H., Mulroy J.S., Kuemmerle S., Jones R., Rye D., Ferrante R.J., Hersch S.M., Li X.J. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J. Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H., Li S.H., Cheng A.L., Mangiarini L., Bates G.P., Li X.J. Ultrastructural localization and progressive formation of neuropil aggregates in Huntington's disease transgenic mice. Hum. Mol. Genet. 1999;8:1227–1236. doi: 10.1093/hmg/8.7.1227. [DOI] [PubMed] [Google Scholar]
- 55.Wang C.E., Tydlacka S., Orr A.L., Yang S.H., Graham R.K., Hayden M.R., Li S., Chan A.W., Li X.J. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington's disease. Hum. Mol. Genet. 2008;17:2738–2751. doi: 10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Burg J.M., Bjorkqvist M., Brundin P. Beyond the brain: widespread pathology in Huntington's disease. Lancet Neurol. 2009;8:765–774. doi: 10.1016/S1474-4422(09)70178-4. [DOI] [PubMed] [Google Scholar]
- 57.Ooi J., Langley S.R., Xu X., Utami K.H., Sim B., Huang Y., Harmston N.P., Tay Y.L., Ziaei A., Zeng R., Low D., Aminkeng F., Sobota R.M., Ginhoux F., Petretto E., Pouladi M.A. Unbiased profiling of isogenic Huntington disease hPSC-derived CNS and peripheral cells reveals strong cell-type specificity of CAG length effects. Cell Rep. 2019;26:2494–2508. doi: 10.1016/j.celrep.2019.02.008. e2497. [DOI] [PubMed] [Google Scholar]
- 58.Sadagurski M., Cheng Z., Rozzo A., Palazzolo I., Kelley G.R., Dong X., Krainc D., White M.F. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. J. Clin. Investig. 2011;121:4070–4081. doi: 10.1172/JCI46305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cobley J.N., Fiorello M.L., Bailey D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018;15:490–503. doi: 10.1016/j.redox.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garbarino V.R., Orr M.E., Rodriguez K.A., Buffenstein R. Mechanisms of oxidative stress resistance in the brain: lessons learned from hypoxia tolerant extremophilic vertebrates. Arch. Biochem. Biophys. 2015;576:8–16. doi: 10.1016/j.abb.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sala A.J., Bott L.C., Morimoto R.I. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 2017;216:1231–1241. doi: 10.1083/jcb.201612111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neueder A., Gipson T.A., Batterton S., Lazell H.J., Farshim P.P., Paganetti P., Housman D.E., Bates G.P. HSF1-dependent and -independent regulation of the mammalian in vivo heat shock response and its impairment in Huntington's disease mouse models. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-12897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shpilka T., Haynes C.M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018;19:109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 64.Melber A., Haynes C.M. UPR(mt) regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;28:281–295. doi: 10.1038/cr.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim H.E., Grant A.R., Simic M.S., Kohnz R.A., Nomura D.K., Durieux J., Riera C.E., Sanchez M., Kapernick E., Wolff S., Dillin A. Lipid biosynthesis coordinates a mitochondrial-to-cytosolic stress response. Cell. 2016;166:1539–1552 e1516. doi: 10.1016/j.cell.2016.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu Z., Liu F., Liu C., Jin B., Jiang Y., Tang M., Qi X., Guo X. Mutant huntingtin inhibits the mitochondrial unfolded protein response by impairing ABCB10 mRNA stability. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2019;1865:1428–1435. doi: 10.1016/j.bbadis.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsushima Y., Goto Y., Kaguni L.S. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM) Proc. Natl. Acad. Sci. U. S. A. 2010;107:18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rainbolt T.K., Saunders J.M., Wiseman R.L. Stress-responsive regulation of mitochondria through the ER unfolded protein response. Trends Endocrinol. Metab. 2014;25:528–537. doi: 10.1016/j.tem.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 69.Mohrin M., Shin J., Liu Y., Brown K., Luo H., Xi Y., Haynes C.M., Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347:1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aprile-Garcia F., Tomar P., Hummel B., Khavaran A., Sawarkar R. Nascent-protein ubiquitination is required for heat shock-induced gene downregulation in human cells. Nat. Struct. Mol. Biol. 2019;26:137–146. doi: 10.1038/s41594-018-0182-x. [DOI] [PubMed] [Google Scholar]
- 71.Sorrentino V., Romani M., Mouchiroud L., Beck J.S., Zhang H., D'Amico D., Moullan N., Potenza F., Schmid A.W., Rietsch S., Counts S.E., Auwerx J. Enhancing mitochondrial proteostasis reduces amyloid-beta proteotoxicity. Nature. 2017;552:187–193. doi: 10.1038/nature25143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Labbadia J., Brielmann R.M., Neto M.F., Lin Y.F., Haynes C.M., Morimoto R.I. Mitochondrial stress restores the heat shock response and prevents proteostasis collapse during aging. Cell Rep. 2017;21:1481–1494. doi: 10.1016/j.celrep.2017.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scior A., Buntru A., Arnsburg K., Ast A., Iburg M., Juenemann K., Pigazzini M.L., Mlody B., Puchkov D., Priller J., Wanker E.E., Prigione A., Kirstein J. Complete suppression of Htt fibrillization and disaggregation of Htt fibrils by a trimeric chaperone complex. EMBO J. 2018;37:282–299. doi: 10.15252/embj.201797212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hay D.G., Sathasivam K., Tobaben S., Stahl B., Marber M., Mestril R., Mahal A., Smith D.L., Woodman B., Bates G.P. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 75.Yamanaka T., Miyazaki H., Oyama F., Kurosawa M., Washizu C., Doi H., Nukina N. Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J. 2008;27:827–839. doi: 10.1038/emboj.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vilchez D., Saez I., Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 2014;5:5659. doi: 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]
- 77.Hosp F., Gutierrez-Angel S., Schaefer M.H., Cox J., Meissner F., Hipp M.S., Hartl F.U., Klein R., Dudanova I., Mann M. Spatiotemporal proteomic profiling of Huntington's disease inclusions reveals widespread loss of protein function. Cell Rep. 2017;21:2291–2303. doi: 10.1016/j.celrep.2017.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Popiel H.A., Takeuchi T., Fujita H., Yamamoto K., Ito C., Yamane H., Muramatsu S., Toda T., Wada K., Nagai Y. Hsp40 gene therapy exerts therapeutic effects on polyglutamine disease mice via a non-cell autonomous mechanism. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bason M., Meister-Broekema M., Alberts N., Dijkers P., Bergink S., Sibon O.C.M., Kampinga H.H. Astrocytic expression of the chaperone DNAJB6 results in non-cell autonomous protection in Huntington's disease. Neurobiol. Dis. 2019;124:108–117. doi: 10.1016/j.nbd.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 80.Park S.K., Arslan F., Kanneganti V., Barmada S.J., Purushothaman P., Verma S.C., Liebman S.W. Overexpression of a conserved HSP40 chaperone reduces toxicity of several neurodegenerative disease proteins. Prion. 2018;12:16–22. doi: 10.1080/19336896.2017.1423185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Young C.N., Sinadinos A., Lefebvre A., Chan P., Arkle S., Vaudry D., Gorecki D.C. A novel mechanism of autophagic cell death in dystrophic muscle regulated by P2RX7 receptor large-pore formation and HSP90. Autophagy. 2015;11:113–130. doi: 10.4161/15548627.2014.994402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.She P., Zhang Z., Marchionini D., Diaz W.C., Jetton T.J., Kimball S.R., Vary T.C., Lang C.H., Lynch C.J. Molecular characterization of skeletal muscle atrophy in the R6/2 mouse model of Huntington's disease. Am. J. Physiol. Endocrinol. Metab. 2011;301:E49–E61. doi: 10.1152/ajpendo.00630.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caron N.S., Dorsey E.R., Hayden M.R. Therapeutic approaches to Huntington disease: from the bench to the clinic. Nat. Rev. Drug Discov. 2018;17:729–750. doi: 10.1038/nrd.2018.133. [DOI] [PubMed] [Google Scholar]
- 84.Hersch S.M., Schifitto G., Oakes D., Bredlau A.L., Meyers C.M., Nahin R., Rosas H.D., Huntington Study Group C.-E.I. Coordinators, the CREST-E study of creatine for Huntington disease: a randomized controlled trial. Neurology. 2017;89:594–601. doi: 10.1212/WNL.0000000000004209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar A., Ratan R.R. Oxidative stress and Huntington's disease: the good, the bad, and the ugly. J. Huntingtons Dis. 2016;5:217–237. doi: 10.3233/JHD-160205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ng L.F., Gruber J., Cheah I.K., Goo C.K., Cheong W.F., Shui G., Sit K.P., Wenk M.R., Halliwell B. The mitochondria-targeted antioxidant MitoQ extends lifespan and improves healthspan of a transgenic Caenorhabditis elegans model of Alzheimer disease. Free Radic. Biol. Med. 2014;71:390–401. doi: 10.1016/j.freeradbiomed.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 87.Yin X., Manczak M., Reddy P.H. Mitochondria-targeted molecules MitoQ and SS31 reduce mutant huntingtin-induced mitochondrial toxicity and synaptic damage in Huntington's disease. Hum. Mol. Genet. 2016;25:1739–1753. doi: 10.1093/hmg/ddw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reid M.B. Redox interventions to increase exercise performance. J. Physiol. 2016;594:5125–5133. doi: 10.1113/JP270653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim J.H., Choi T.G., Park S., Yun H.R., Nguyen N.N.Y., Jo Y.H., Jang M., Kim J., Kim J., Kang I., Ha J., Murphy M.P., Tang D.G., Kim S.S. Mitochondrial ROS-derived PTEN oxidation activates PI3K pathway for mTOR-induced myogenic autophagy. Cell Death Differ. 2018;25:1921–1937. doi: 10.1038/s41418-018-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]