Abstract
Purpose
Fanconi anemia complementation group I (FANCI) is a key protein in ribosome biogenesis and DNA repair. Here, we aimed to determine the clinical significance, prognostic value and biology functions of FANCI in lung adenocarcinoma (LUAD).
Methods
The expression of FANCI in LUAD tissue and its relationship with patient outcomes were assessed using bioinformatics analysis, as well as quantitative reverse-transcription PCR (qRT-PCR) and Western blot analysis of LUAD tissue and adjacent normal lung tissue. The chi-squared test and Cox regression analysis were used to analyze the clinical significance of FANCI expression. The biological effects of FANCI knockdown in human LUAD cell lines were investigated by analysis of proliferation, colony formation, cell cycle distribution, migration, and invasion in vitro, and monitoring of tumor xenograft growth in vivo. FANCI interactions with IMPDH2 and involvement in MEK/ERK/MMPs signaling were analyzed using co-immunoprecipitation assays, immunofluorescence microscopy, and Western blotting.
Results
FANCI was identified as a hub gene for LUAD. FANCI expression was upregulated in LUAD tissues compared with normal lung tissues and was positively associated with lymphatic metastasis, distant metastasis, and poor outcome. FANCI was also an independent prognostic factor in LUAD patients. Knockdown of FANCI in LUAD cell lines decreased their proliferation, migration, invasion, and cell cycle progression in vitro, and decreased the growth of xenografts in mice. Direct binding of FANCI to IMPDH2 decreased IMPDH2 degradation, regulated activation of MEK/ERK/MMPs signaling. Overexpression of IMPDH2 reversed the inhibitory effects of FANCI knockdown.
Conclusion
FANCI may act as an oncogene in LUAD by cooperating with IMPDH2 to promote cell proliferation via the MEK/ERK/MMPs pathway. These results identify FANCI as a potential prognostic biomarker and therapeutic target for LUAD.
Keywords: lung adenocarcinoma, FANCI, proliferation, metastasis, IMPDH2
Introduction
Lung adenocarcinoma (LUAD) is one of the most common and lethal malignant cancers worldwide.1 Despite advances in the diagnosis and treatment of LUAD, its prognosis remains poor.2 Therapies targeted at suppressing driver genes, including epidermal growth factor receptor (EGFR, HGNC ID: 8236) and anaplastic lymphoma kinase (ALK, HGNC ID: 427), have been shown to extend the survival time and improve the quality of life of LUAD patients. However, the emergence of resistance limits the clinical utility of such drugs. Hence, there is an urgent need to identify the molecular mechanisms associated with the progression of LUAD, thereby advancing the discovery of novel diagnostic/prognostic biomarkers and potential therapeutic targets.
Fanconi anemia (FA) is a rare syndrome caused by mutations in the FA family of proteins, leading to chromosome instability, bone marrow failure, and other pathologies, including cancer.3,4 The FA protein family consists of 15 members that regulate cell cycle progression and DNA damage repair.5 Several studies have investigated the role of FA proteins in the occurrence and development of cancer. For example, Yao et al showed that FANCF (HGNC ID: 3587; UniProt ID: Q9NPI8) plays a key role in the drug resistance of leukemia cells via regulation of DNA repair,6 while Park et al showed that deficiency of FANCD2 (HGNC ID: 3585, UniProt ID: Q9BXW9) leads to skin cancer.7 He et al found that inhibition of FANCF (HGNC ID: 3587, UniProt ID: Q9NPI8) reversed the resistance of ovarian cancer cells to adriamycin,8 and Dai et al suggested that knockdown of FANCL (HGNC ID: 20748, UniProt ID: Q9NW38) promotes the chemosensitivity of cisplatin-resistant lung cancer cells to cisplatin.9
Inosine monophosphate dehydrogenase type II (IMPDH2; HGNC ID: 6053, UniProt ID: P12268) catalyzes the conversion of inosine monophosphate (IMP) to xanthosine monophosphate and is a rate-limiting enzyme in the de novo biosynthesis of guanine nucleotides.10 Previous studies showed that IMPDH2 is highly expressed in several cancers, such as kidney and bladder cancer,11 and IMPDH2 has been shown to promote colorectal cancer cell proliferation by activating the AKT–mTOR (UniProt ID: Q96B36 and P42345, respectively) signaling pathway.12 High expression of IMPDH2 is associated with poor outcome in breast cancer13 and predicts poor prognosis in human hepatocellular carcinoma.14 IMPDH2 expression is regulated by proteins such as ANKRD9 (UniProt ID: Q96BM1)15 that directly bind to and promote the degradation of IMPDH2.
In the present study, we aimed to investigate the expression, biological functions, and mechanism of action of FANCI (HGNC ID: 25568; Uniport ID: Q9NVI1) in LUAD, as well its clinical significance. We show that FANCI promotes the development of LUAD and may be an independent prognostic factor and potential therapeutic target for this disease.
Materials and Methods
Bioinformatics Analysis of Hub Genes in LUAD
Two gene expression datasets (GSE2514 and GSE62113) of LUAD tissues and adjacent normal lung tissues were downloaded from the Gene Expression Omnibus database. After normalization of expression data, differentially expressed genes (DEGs) between tumor and normal tissue were identified using cutoff values of log fold-change (FC) >1 and P<0.05. DEGs common to both GSE2514 and GSE62113 were uploaded to the online website STRING, and the relationship between DEGs was visualized using Cytoscape. The top 25 genes based on degrees were used to construct a protein–protein interaction subnetwork.
Bioinformatics Analysis of FANCI Expression and Diagnostic Value
The expression of FANCI was analyzed using the Oncomine database with the GEPIA online tool. DEGs were selected using a threshold of log FC>1 and P<0.01. The relationship between FANCI expression and prognosis was analyzed using OncoLnc, with P<0.05 set as the cutoff for significance.
Patients and Tissue Collection
We enrolled a total of 80 patients with LUAD between January 2013 and January 2019 at Jingchu Center Hospital Affiliated to The Institute of Technology, Jingmen (Hubei, China). The Medical Ethics Committee of the hospital approved the study. All patients provided written informed consent, and that this was conducted in accordance with the Declaration of Helsinki. The inclusion criteria were: (i) a diagnosis of LUAD with complete pathological reports, (ii) both tumor and adjacent non-tumor tissues were available, (iii) staging information was complete, (iv) age ≤80 years, (v) home address and telephone number were available, and (vi) patients received only standard chemotherapy after collection of tissue samples. The exclusion criteria were: (i) a history of malignancies other than LUAD, (ii) antitumor therapy before collection of tissue samples, (iii) received targeted therapy or immunotherapy-related drugs during the follow-up period, (iv) withdrew from treatment, and (v) did not approve use of their tissues for this study. All samples were stored in liquid nitrogen before analysis.
Cell Culture and Knockdown/Overexpression Vectors
The human LUAD cell lines A549, H1299, PC9, H1975, and SPCA1 and the lung epithelial cell line BEAS-2B were purchased from American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Gibco) at 37°C in a 5% CO2 atmosphere. Three distinct short hairpin RNA (shRNA) sequences targeting human FANCI were synthesized (RIBO) and cloned into pSuper-retro-puro to generate pSuper-retro-FANCI-RNAi plasmids. The sequences were: RNAi#1, ATGTAAGCTCGGAGCTAATAT; RNAi#2, CATGTGGAAGGCACCATTATT; and RNAi#3, CAATTGCCACGAACGGTAATG. An IMPDH2 overexpression lentivirus was constructed by subcloning PCR-amplified full-length human IMPDH2 cDNA into the pMSCV retrovirus plasmid. Cells with stable IMPDH2 overexpression or FANCI knockdown were selected for 2 weeks with 0.5 μg/mL puromycin after transfection or infection.
Quantitative Reverse-Transcription PCR (qRT-PCR)
Total RNA was extracted from cell lines and frozen tissues using TRIzol reagent (Yeasen Shanghai, China) and cDNA was synthesized using a qPCR RT Master Mix Kit (Yeasen) according to the manufacturers’ instructions. qPCR was performed using a SYBR Green Assay Mix (Takara, Japan) and a CFX96 real-time PCR detection system (Bio-Rad). The 2−ΔΔT method was used to measure the relative expression of target genes. Glyceraldehyde 3-phosphate dehydrogenase GAPDH was amplified as an internal control. The primer sequences (Sangon Biotech, Shanghai, China) were:
FANCI forward: CCACCTTTGGTCTATCAGCTTC,
FANCI reverse: CAACATCCAATAGCTCGTCACC,
GAPDH forward: GGAGCGAGATCCCTCCAAAAT, and
GAPDH reverse: GGCTGTTGTCATACTTCTCATGG.
Western Blot Analysis
Total protein was extracted from cells using RIPA buffer (Boster, Wuhan, China) containing the protease inhibitor PMSF (Boster). Proteins were resolved by SDS-PAGE and transferred to PVDF membranes. The blots were blocked by incubation with 5% fat-free milk at room temperature for 2 h and then incubated overnight at 4°C with a 1:500 dilution of antibodies to the following proteins: FANCI (Santa Cruz Biotechnology, Dallas, TX, USA), IMPDH2, MEK1/2, ERK1/2, MMP2, MMP9, GAPDH (all Proteintech, Wuhan, China), phospho (p)-MEK1/2, and p-ERK1/2 (both Cell Signaling Technology, Danvers, MA, USA). The membranes were washed three times with TBST and then incubated for 2 h with horseradish peroxidase-conjugated rabbit or mouse secondary antibodies. After signal development, expression of proteins was analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Proliferation Assay
Aliquots of 5×103 cells/well were seeded into 96-well plates and incubated at 37°C for the indicated times. Cell Counting Kit-8 (CCK-8, Boster) solution (10 μL) was then added to each well, the plates were incubated for an additional 2 h, and absorbance at 450 nm was measured. All experiments were performed in three times.
Colony Formation Assay
Aliquots of 5×102 cells/well were seeded into 6-well plates and cultured for 2 weeks, with the medium replaced every 4 days. At the end of the incubation period, the cells were fixed in 4% paraformaldehyde for 15 min and then incubated in 1% crystal violet stain. Colonies were enumerated and photographed.
Cell Cycle Distribution Assay
Cells were incubated in DMEM medium without FBS for 24 h to synchronize cell growth, and the medium was then exchanged for DMEM with 10% FBS. After 48 h culture, the cells were fixed in 75% ethanol at −20°C for 24 h, washed with PBS three times, resuspended in propidium iodide (PI)-RNase A solution (Invitrogen, USA), and incubated at 37°C for 30 min. Cell cycle distribution was analyzed using a FACScan flow cytometer (BD Biosciences, San Jose, CA, USA).
Wound Healing Assay
Aliquots of 1×106 cells/well in DMEM medium without FBS were seeded into 6-well plates and grown to confluence. A 100 μL pipette tip was then used to scratch a wound in the cell monolayer, and floating cells were removed. The medium was exchanged to DMEM without FBS and the plates were incubated at 37°C. At the indicated times, the cells were observed using an inverted microscope, and the change in wound area was measured using ImageJ software.
Invasion Assay
Aliquots of 4×105 cells in 200 μL DMEM without FBS were seeded into the upper wells of Transwell chambers (Invitrogen, USA) coated with Matrigel (Invitrogen, USA). DMEM with 10% FBS (600 μL) was added to the lower chambers and the cells were incubated for 28 h. Invaded cells on the lower sides of the membrane were then fixed with paraformaldehyde and stained with 0.5% crystal violet. A total of five fields of view were visualized using an inverted microscope and photographed, and the number of invasive cells per field was counted.
Mouse Tumor Xenografts
Ten 6-week-old female BALB/c nude mice were purchased from Beijing Huafukang (Beijing, China). Aliquots of 1×107 A549 cells expressing negative control shRNA (NC) or FANCI-targeting shRNA (sh-FANCI) were suspended in 100 μL medium and injected subcutaneously into the right flanks of mice (n=5 per group). Tumor size and mouse weight were recorded weekly for 5 weeks. The mice were then sacrificed and the tumors were excised and weighed. The animal experiments were approved by the Animal Care and Use Committee of Jingchu Center Hospital Affiliated to The Institute of Technology, and was carried out following the Animal Protection Law of the People’s Republic of China-2009.
Immunoprecipitation Analysis
Cell lysates were incubated with anti-FANCI or anti-IMPDH2 primary antibody overnight at 4°C, and protein A/G-Sepharose beads (MCE, Wuhan, China) were added for an additional 4 h. The beads were collected and washed, and co-precipitated proteins were eluted, separated by SDS-PAGE, and detected by immunoblotting with anti-FANCI and anti-IMPDH2 primary antibodies as described above.
Immunofluorescence Microscopy
Cells were fixed with paraformaldehyde, blocked with 5% BSA (Boster), and incubated with mouse anti-FANCI and rabbit anti-IMPDH2 primary antibodies. The cells were washed with PBS and then incubated with FITC-conjugated anti-mouse (Boster) and Cy3-conjugated anti-rabbit (Boster) secondary antibodies. Nuclei were stained with 4′,6-diamidino-2-phenylindole (Boster). Finally, the cells were visualized by fluorescence microscopy and photographed.
Gene Set Enrichment Analysis (GSEA)
LUAD samples in GSE62113 were assigned to two groups based on high and low FANCI or IMPDH2 expression. GSEA software was employed to explore pathways regulated by FANCI and IMPDH2 by detecting enrichment of DEGs in a priori defined pathways between the high and low expression groups. P <0.05 was set as the threshold of significance.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 6.01 software. One-way ANOVA and Student’s t-test were used to evaluate differences between groups. The Kaplan–Meier method was used to estimate the overall survival of patients, and the log rank test was used to determine differences between groups. The significance of clinicopathological traits were analyzed using the chi-squared test and univariate and multivariate Cox regression analyses. P<0.05 was considered statistically significant.
Results
FANCI Is a Predicted Hub Gene for LUAD
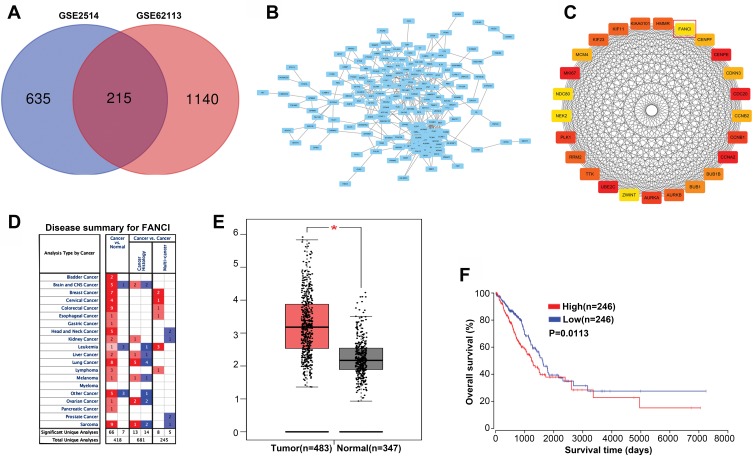
To identify genes that play a key role in the progression of LUAD, we analyzed two gene expression datasets, GSE2514 and GSE62113, from the Gene Expression Omnibus. We identified 215 genes that were expressed in LUAD tissues but not adjacent tissues in both datasets (Figure 1A). To understand the function of the DEGs, we constructed a protein–protein interaction network from the genes with the top 25 degrees. This analysis identified FANCI as a hub gene for LUAD, although its pathological role was unknown (Figure 1B and C). Analysis of Oncomine and GEPIA datasets verified that FANCI is highly expressed in LUAD tissues compared with adjacent tissues (Figure 1D and E, P<0.05), and analysis of TCGA datasets revealed that high expression of FANCI was associated with poor outcome in LUAD patients (Figure 1F, P=0.0113).
Figure 1.
FANCI is a predicted hub gene in LUAD. (A) Venn diagram showing individual and shared differentially expressed genes (DEGs) in the GSE2514 and GSE62113 genesets. (B) Protein–protein interaction network of shared DEGs visualized in Cytoscape. (C) Construction of a protein–protein interaction subnetwork from DEGs with the top 25 degree scores, with FANCI highlighted. (D) Expression of FANCI in various cancers identified using the Oncomine database. Red and blue indicate upregulated and downregulated genes, respectively. (E) Expression of FANCI in LUAD tissues and adjacent normal tissues identified using the GEPIA database. *P<0.05. (F) Relationship between prognosis and FANCI expression analyzed using the OncoLnc database.
FANCI Is Upregulated in LUAD Tissues and Its Expression Level Predicts Poor Prognosis
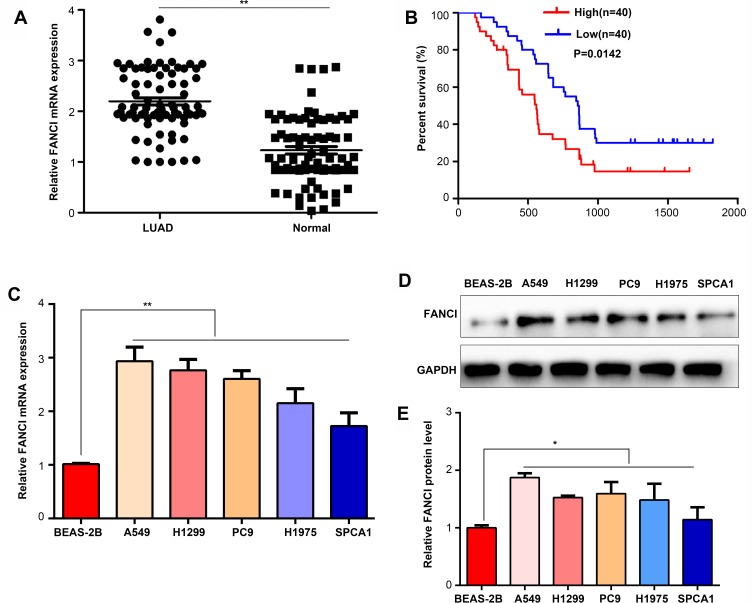
To verify the results obtained by bioinformatics, we performed qRT-PCR analysis of 80 pairs of LUAD tissues and adjacent tissues obtained from patients at our hospital. The results not only confirmed that FANCI expression was higher in LUAD tissues than in adjacent tissues (Figure 2A, P<0.05) but also showed that high FANCI expression was associated with a low overall survival rate (Figure 2B, P=0.0142). Similarly, FANCI mRNA and protein were both highly expressed in human LUAD cell lines (Figure 2C and D, P<0.05). We next assessed the clinicopathological significance of FANCI expression and discovered that it was positively associated with lymphatic metastasis (χ2=8.584, P=0.003) and distant metastasis (χ2=8.455, P=0.004; Table 1). Furthermore, Cox regression analysis demonstrated that FANCI expression was an independent factor for predicting patient outcome (Table 2, P<0.05), suggesting that FANCI may have utility as a prognostic biomarker in LUAD.
Figure 2.
FANCI is upregulated in LUAD tissues and cell lines. (A) qRT-PCR analysis of FANCI mRNA levels in 80 pairs of LUAD tissues and adjacent tissues. (B) Kaplan–Meier overall survival analysis of the 80 LUAD patients assigned to FANCI low and high expression groups. (C) qRT-PCR analysis of FANCI mRNA levels in human LUAD cell lines A549, H1299, PC9, H1975, SPCA1, and the lung epithelial cell line BEAS-2B. (D) Western blot analysis of FANCI protein levels in the indicated cell lines. (E) Relative statistics for the western blot result in (D). *P<0.05, **P<0.01.
Table 1.
Relationship Between FANCI Expression and Clinicopathological Traits in Patients with LUAD
| Clinicopathological Feature | n | FANCI Expression | X2 | P | |
|---|---|---|---|---|---|
| High | Low | ||||
| Gender | 0.228 | 0.633 | |||
| Male | 54 | 26 | 28 | ||
| Female | 26 | 14 | 12 | ||
| Age(years) | 0.464 | 0.496 | |||
| <60 | 47 | 22 | 25 | ||
| ≥60 | 33 | 18 | 15 | ||
| Turmor size (cm) | 0.802 | 0.37 | |||
| <3 | 42 | 19 | 23 | ||
| ≥3 | 38 | 21 | 17 | ||
| Lymphatic metastasis | 8.584 | 0.003** | |||
| N0 | 45 | 16 | 29 | ||
| N1–N3 | 35 | 24 | 11 | ||
| Disant metastasis | 8.455 | 0.004** | |||
| M0 | 41 | 14 | 27 | ||
| M1 | 39 | 26 | 13 | ||
Note: **P<0.01.
Table 2.
Prognostic Value of FANCI Expression in Patients with LUAD
| Clinicopathological Features | Univariate Analyses | Multivariate Analyses | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% (CI) | P value | HR | 95% (CI) | P value | ||
| Age (years) | ≤40 | 1.00 | 0.91–1.43 | 0.63 | |||
| >40 | 1.17 | ||||||
| Gender | Female | 1.00 | 0.85–1.50 | 0.42 | |||
| Male | 1.13 | ||||||
| Tumour size | ≤3 cm | 1.00 | 1.43–2.68 | 0.02* | 1.00 | 1.16–1.59 | 0.21 |
| >3 cm | 2.11 | 1.52 | |||||
| Lymphatic metastasis | N0 | 1.00 | 1.65–2.94 | <0.01** | 1.00 | 1.46–2.55 | 0.02* |
| N1–N3 | 2.37 | 2.11 | |||||
| Disant metastasis | M0 | 1.00 | 2.67–3.42 | <0.01** | 1.00 | 1.77–3.11 | <0.01** |
| M1 | 3.14 | 2.67 | |||||
| FANCI expression | Low | 1.00 | 1.8–2.89 | 0.01* | 1.00 | 1.41–2.37 | 0.03* |
| High | 2.31 | 2.07 | |||||
Note: *P<0.05, **P<0.01.
Knockdown of FANCI Inhibits LUAD Cell Proliferation in vitro
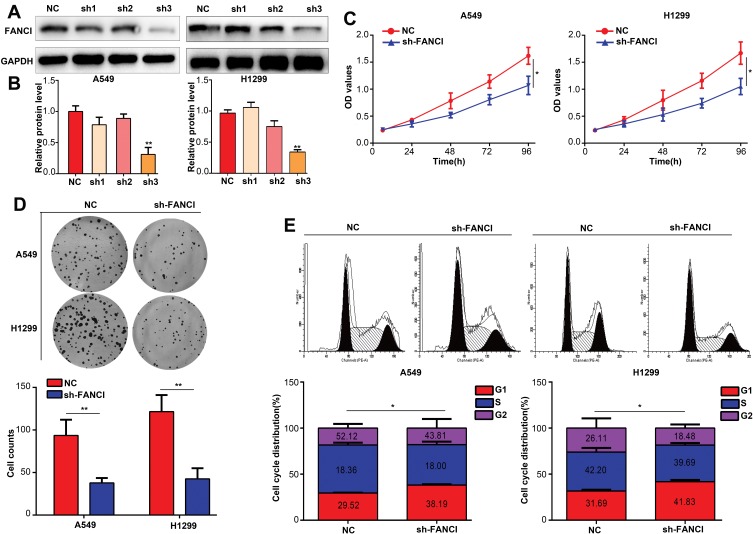
To assess the biological functions of FANCI in LUAD, we knocked down FANCI in the human LUAD cell lines A549 and H1299 cell using three FANCI-targeted shRNAs. Western blot analysis showed that sh3-FANCI was the most effective silencer of expression (Figure 3A and B), and this shRNA was selected for further experiments. Functional assays of A549 and H1299 cells demonstrated that FANCI knockdown decreased cell proliferation, as measured using the CCK-8 assay (Figure 3C, P<0.05), and suppressed colony formation (Figure 3D, P<0.05) compared with control cells expressing NC shRNA. In addition, cell cycle distribution analysis showed that the proportion of sh3-FANCI-expressing cells in G1 phase was significantly higher, while the proportion of cells in G2 phase was significantly lower, compared with control cells (Figure 3E, P<0.05). Taken together, these data showed that knockdown of FANCI in LUAD cells inhibited their proliferation and promoted cell cycle arrest in vitro.
Figure 3.
Knockdown of FANCI inhibits the proliferation of LUAD cells in vitro. (A, B) Western blot analysis of the knockdown efficiency of sh1-FANCI, sh2-FANCI, and sh3-FANCI shRNAs. (C–E) The effects of FANCI knockdown on LUAD cell proliferation (C) colony formation (D), and cell cycle distribution (E). *P<0.05, **P<0.01.
Knockdown of FANCI Decreases LUAD Cell Migration and Invasion in vitro
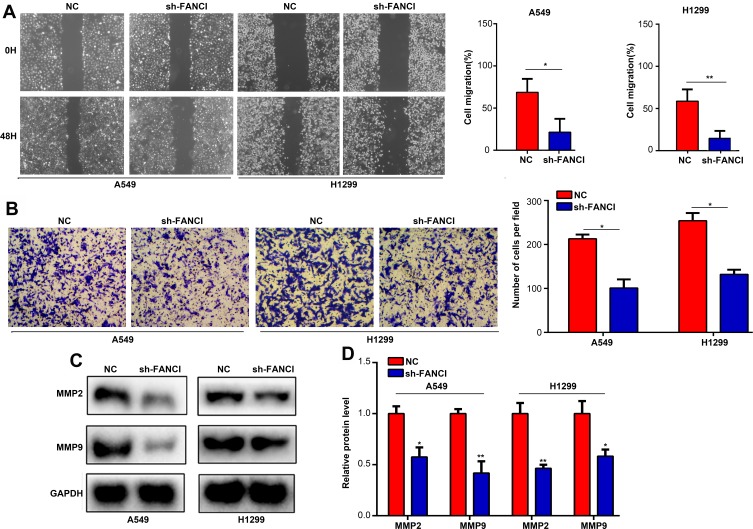
Next, we analyzed the effects of FANCI on LUAD cell migration and invasion using wound healing and Transwell invasion assays, respectively. The results showed that knockdown of FANCI significantly decreased the ability of A549 and H1299 cells to migrate (Figure 4A, P<0.05) and undergo invasion (Figure 4B, P<0.05), suggesting that FANCI may be required for the metastatic behavior of LUAD cells in vivo. Because MMPs are key proteins for tumor metastasis, we examined the expression of MMP2 and MMP9 in LUAD cells by Western blotting. These experiments showed that expression of both enzymes was significantly decreased in sh3-FANCI-expressing cells compared with control cells (Figure 4C and D, P<0.05). These data demonstrate a role for FANCI in promoting the migration and invasion of LUAD cells in vitro.
Figure 4.
Knockdown of FANCI reduces the migration and invasion of LUAD cells in vitro. (A–D) A549 cells and H199 cells expressing sh-NC or sh-FANCI were analyzed in wound healing migration assays (A), Transwell invasion assays (B), and by Western blot analysis of MMP2 (C) or MMP9 (D) protein levels. *P<0.05, **P<0.01.
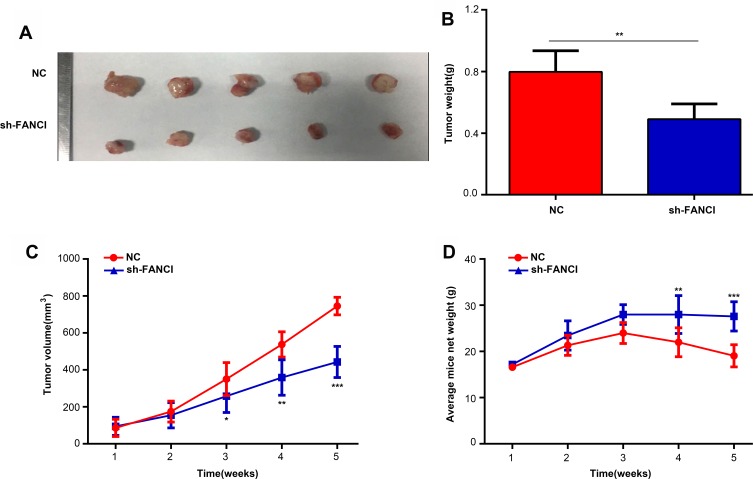
Knockdown of FANCI Decreases the Growth of A549 Tumor Xenografts in vivo
To determine whether FANCI affected the proliferation of LUAD cells in vivo, we injected control (NC) or sh3-FANCI-expressing A549 cells subcutaneously into groups of nude mice and monitored tumor growth and body weights for 5 weeks. As shown in Figure 5, tumors formed by FANCI knockdown cells were smaller than those formed by the control cells (Figure 5A and B, P<0.05), and the proliferation rate of cells in sh3-FANCI tumors was also slower than that of the control NC tumors (Figure 5C, P<0.05). The mice bearing sh3-FANCI tumors also lost weight at a slower rate than did the mice bearing control tumors (Figure 5D, P<0.05).
Figure 5.
Inhibition of FANCI suppresses the proliferation of A549 cells in vivo. (A, B) Appearance (A) and mean weights (B) of tumors formed by sh-NC- and sh-FANCI-expressing A549 cells excised at 5 weeks after injection. (C) The proliferation rate of sh-NC- and sh-FANCI-expressing tumors. (D) Weight change of mice harboring sh-NC- and sh-FANCI-expressing tumors. *P<0.05, **P<0.01, ***P<0.001.
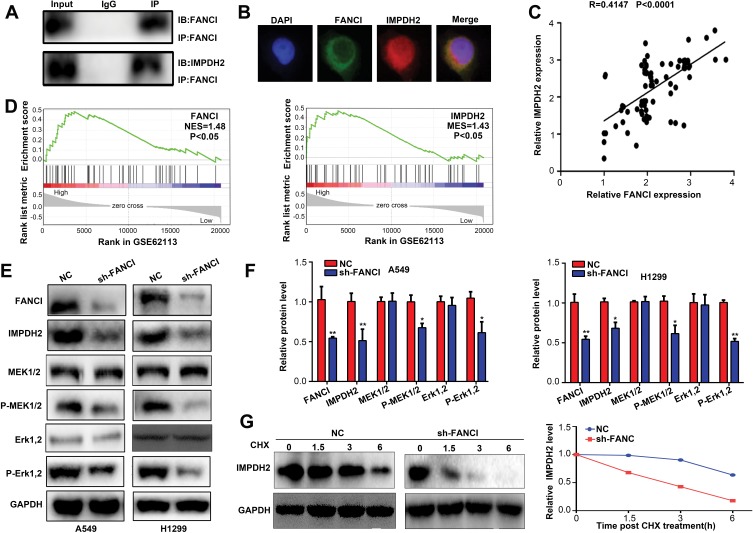
FANCI Cooperates with IMPDH2 in Regulating the MEK/ERK/MMPs Pathway
To explore the molecular mechanisms underlying FANCI function in LUAD, we employed the Genemania tool (https://genemania.org/) to identify proteins predicted to bind directly to FANCI (data not shown). Among the proteins identified, IMPDH2 has been demonstrated to act as an oncogene in various cancers, including lung cancer. Indeed, coimmunoprecipitation experiments and immunofluorescence microscopy demonstrated that FANCI and IMPDH2 directly interacted and were colocalized in LUAD cells (Figure 6A and B), suggesting that IMPDH2 may mediate the function of FANCI in these cells, at least in part. In support of this, FANCI knockdown in LUAD cells significantly decreased the expression of IMPDH2, and FANCI and IMPDH2 expression was significantly correlated in LUAD tissues (Figure 6C, R=0.4147, P<0.0001).
Figure 6.
FANCI cooperates with IMPDH2 in regulating MEK/ERK signaling. (A) Western blot analysis of FANCI and IMPDH2 immunoprecipitates probed with antibodies against FANCI. (B) Immunofluorescence microscopy of A549 cells co-stained with antibodies against FANCI and IMPDH2. (C) Co-expression analysis of FANCI and IMPDH2 in LUAD tissues at our samples. (D) GSEA of GSE62113 datasets showing significant enrichment of MEK/ERK pathway genes in LUAD tissues expressing high levels of FANCI and IMPDH2. (E, F) Western blot analysis of FANCI, IMPDH2, MEK1/2, p-MEK1/2, ERK1/2 and p-ERK1/2 expression in A549 and H1299 cells expressing sh-NC or sh-FANCI. (G) Western blot analysis of IMPDH2 expression in A549 cells treated with cycloheximide (CHX) and then incubated for 1.5, 3, and 6 h. *P<0.05, **P<0.01.
Because IMPDH2 has been shown to activate the MEK/ERK (UniProt ID: Q02750 and P27361, respectively) pathway, we performed GSEA analysis of GSE62213 datasets to determine the possible involvement of FANCI and IMPDH2 in regulating this pathway. The results of showed enrichment of MEK/ERK pathway components in tissues expressing high levels of either FANCI or IMPDH2 compared with tissues expressing low levels (Figure 6D, P<0.05). To verify these data, we performed Western blot analysis of MEK/ERK pathway components in control or sh3-FANCI-expressing cells and found that expression of the phosphorylated (activated) forms of MEK1/2 and ERK1/2 was decreased significantly by FANCI knockdown (Figure 6E and F, P<0.05). Previous studies have shown that IMPDH2 expression levels are regulated in part through modulation of its degradation rate, raising the possibility that FANCI binding to IMPDH2 may affect IMPDH2 stability. To examine this, we treated control and sh3-FANCI-expressing A549 cells with cycloheximide (4-[(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl]piperidine-2,6-dione) to inhibit protein synthesis and then followed the expression levels of IMPDH2 by Western blot analysis. The results showed that the degradation rate of IMPDH2 was increased significantly in cells with FANCI knockdown (Figure 6G, P<0.05).
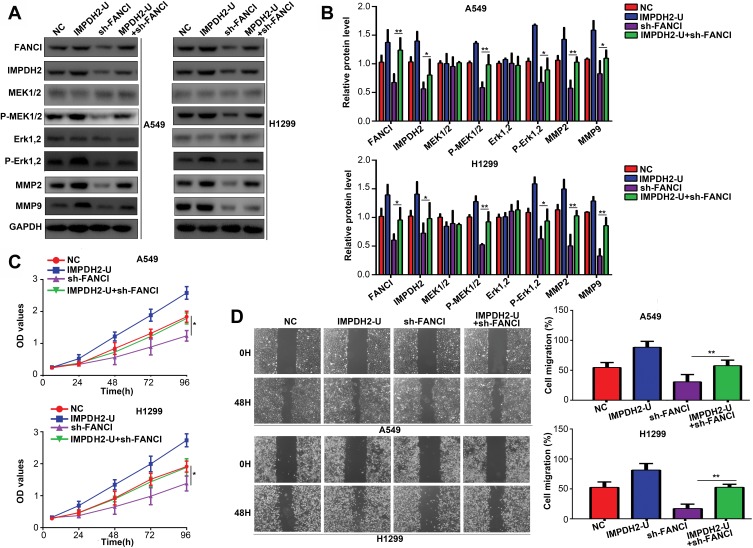
IMPDH2 Overexpression Alleviates the Inhibitory Effects of FANCI Knockdown
To determine whether IMPDH2 might mediate the biological effects of FANCI, we overexpressed IMPDH2 in FANCI knockdown cells. Notably, overexpression of IMPDH2 in FANCI knockdown A549 and H1299 cells restored the activity of MEK/ERK pathway, as well as two target genes of ERK, MMP2, and MMP9 (Figure 7A and B, P<0.05). Overexpression of IMPDH2 in FANCI knockdown A549 and H1299 cells also rescued the inhibition of LUAD cell proliferation (Figure 7C, P<0.05) and migration (Figure 7D, P<0.05). These results confirm that IMPDH2 plays an important role in mediating the functions of FANCI in LUAD.
Figure 7.
IMPDH2 overexpression decreases the inhibitory effects of FANCI knockdown in LUAD cells. (A–D) Western blot analysis of the indicated proteins (A, B), CCK-8 cell proliferation assay (C), and wound healing assay of migration (D) in cells expressing control NC shRNA, an IMPDH2 overexpression vector (IMPDH2-U), FANCI shRNA (sh-FANCI), or an IMPDH2 overexpression vector plus sh-FANCI (IMPDH2-U+sh-FANCI). *P<0.05, **P<0.01.
Discussion
The results of the current study demonstrate a key role for FANCI in LUAD pathogenesis and prognosis. We showed that expression of FANCI is upregulated in LUAD tissues compared with normal lung tissues, and high expression is positively associated with several aggressive features. Furthermore, FANCI was found to be an independent predictor of patient outcome. Knockdown of FANCI in LUAD cell lines inhibited their growth in vitro and in vivo, and reduced their migration and invasion capacity in vitro. Mechanistic studies demonstrated that the interaction between FANCI and IMPDH2 decreases the degradation of IMPDH2, promotes activation of MEK/ERK/MMPs pathway signaling. Collectively, these results suggest that FANCI may be a novel predictive biomarker and a potential therapeutic target for LUAD.
The FA family of proteins contributes to the regulation of genomic stability and the cell cycle.16 The roles and molecular mechanisms of action of many FA proteins in various pathological processes, including cancer, have been reported. FANCD2 is a modulator of AMPK and plays a key role in energy metabolism. Moreover, inhibition of FANCD2 increases the chemosensitivity of the renal cell carcinoma cell line Caki-1 and induces apoptosis via its regulation of AMPK.17 In colorectal cancer, high expression of FANCJ is positively associated with resistance to 5-fluoruracil.18 FANCD2 also inhibits the inflammatory response by decreasing TNF-α transcription.19 Mutation of both FANCD220 and FANCO (HGNC ID: 9820)22 have been shown to contribute to ovarian cancer, whereas FANCA deletions are associated with the development of familial breast cancer.21 Finally, truncation of FANCT (HGNC ID: 25009) is associated with a high-risk for breast cancer.23 In the present study, we identified a novel role for FANCI as a predicted hub gene with a key role in the development of LUAD. Similar with other family members which play as onco-genes in cancer, we demonstrated that FANCI was upregulated in LUAD tissues and cell lines and showed that its expression is positively associated with lymphatic metastasis, distant metastasis, and poor outcome. FANCI was also shown to be an independent predictor of patient outcome. In vitro cell biology assays and mouse xenograft experiments showed that FANCI promotes the proliferation and metastatic behavior of LUAD cells.
Our results are consistent with an accumulating body of evidence that IMPDH2 is involved in the development of various cancer types. In glioblastoma, IMPDH2 increases the synthesis of rRNA and tRNA, thereby promoting rapid cell proliferation.24 High expression of IMPDH2 in osteosarcoma cells was shown to be associated with radioresistance and chemoresistance.25 Conversely, pharmacological inhibition of IMPDH2 expression induced cancer cell apoptosis via regulation of multiple pathways, including the Bax–AIF–Endo G and MEK/ERK pathways.26,27 In the present study, we identified a novel mechanism of FANCI function through its direct binding to IMPDH2, thereby protecting IMPDH2 from degradation. MEK/ERK pathway genes were found in GSEA analysis to be significantly enriched in LUAD tissues with high FANCI or IMPDH2 expression. In vitro, we found that inhibition of FANCI decreased the expression of IMPDH2 and the activity of the MEK/ERK pathway, but this was reversed by concomitant overexpression of IMPDH2, confirming the functional importance of the FANCI/IMPDH2 interaction.
Conclusion
We demonstrated that FANCI is highly expressed in LUAD tissues and cell lines, and we uncovered positive associations between FANCI expression and malignant processes, leading to poor prognosis. Knockdown of FANCI suggested that these effects were mediated via modulation of IMPDH2 degradation, MEK/ERK/MMPs pathway activation. Collectively, our data identify a novel oncogenic role for FANCI and suggest that it may have utility as a prognostic biomarker and/or therapeutic target for LUAD.
Acknowledgment
We thank Anne M. O’Rourke, PhD, from Liwen Bianji, Edanz Group China, for editing the English text of a draft of this manuscript.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no proprietary or commercial interest in any product mentioned or concept discussed in this article.
References
- 1.Cui Y, Fang W, Li C, et al. Development and validation of a novel signature to predict overall survival in “driver gene-negative” lung adenocarcinoma (LUAD): results of a multicenter study. Clin Cancer Res. 2019;25(5):1546–1556. doi: 10.1158/1078-0432.CCR-18-2545 [DOI] [PubMed] [Google Scholar]
- 2.Chen M, Liu X, Du J, Wang XJ, Xia L. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget. 2017;8(1):133–144. doi: 10.18632/oncotarget.13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelaidi C, Makis A, Petrikkos L, et al. Bone marrow failure in fanconi anemia: clinical and genetic spectrum in a cohort of 20 pediatric patients. J Pediatr Hematol Oncol. 2019;41:612–617. doi: 10.1097/MPH.0000000000001549 [DOI] [PubMed] [Google Scholar]
- 4.Engel NW, Schliffke S, Schuller U, et al. Fatal myelotoxicity following palliative chemotherapy with cisplatin and gemcitabine in a patient with stage IV cholangiocarcinoma linked to post mortem diagnosis of fanconi anemia. Front Oncol. 2019;9:420. doi: 10.3389/fonc.2019.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katsuki Y, Takata M. Defects in homologous recombination repair behind the human diseases: FA and HBOC. Endocr Relat Cancer. 2016;23(10):T19–T37. doi: 10.1530/ERC-16-0221 [DOI] [PubMed] [Google Scholar]
- 6.Yao C, Du W, Chen H, Xiao S, Huang L, Chen FP. Involvement of Fanconi anemia genes FANCD2 and FANCF in the molecular basis of drug resistance in leukemia. Mol Med Rep. 2015;11(6):4605–4610. doi: 10.3892/mmr.2015.3288 [DOI] [PubMed] [Google Scholar]
- 7.Park E, Kim H, Kim JM, et al. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol Cell. 2013;50(6):908–918. doi: 10.1016/j.molcel.2013.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L, Li N, Yu JK, et al. RNAi-mediated knockdown of FANCF suppresses cell proliferation, migration, invasion, and drug resistance potential of breast cancer cells. Braz J Med Biol Res. 2014;47(1):24–34. doi: 10.1590/1414-431X20132938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai CH, Li J, Chen P, Jiang HG, Wu M, Chen YC. RNA interferences targeting the Fanconi anemia/BRCA pathway upstream genes reverse cisplatin resistance in drug-resistant lung cancer cells. J Biomed Sci. 2015;22:77. doi: 10.1186/s12929-015-0185-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah CP, Purushothaman G, Thiruvenkatam V, Kirubakaran S, Juvale K, Kharkar PS. Design, synthesis and biological evaluation of Helicobacter pylori inosine 5ʹ-monophosphate dehydrogenase (HpIMPDH) inhibitors. Further optimization of selectivity towards HpIMPDH over human IMPDH2. Bioorg Chem. 2019;87:753–764. doi: 10.1016/j.bioorg.2019.04.001 [DOI] [PubMed] [Google Scholar]
- 11.Ciliao HL, Camargo-Godoy R, Souza MF, Zanuto A, Delfino V, Colus I. Polymorphisms in IMPDH2, UGT2B7, and CES2 genes influence the risk of graft rejection in kidney transplant recipients taking mycophenolate mofetil. Mutat Res Genet Toxicol Environ Mutagen. 2018;836(Pt B):97–102. doi: 10.1016/j.mrgentox.2018.06.008 [DOI] [PubMed] [Google Scholar]
- 12.Duan S, Huang W, Liu X, et al. IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways. J Exp Clin Cancer Res. 2018;37(1):304. doi: 10.1186/s13046-018-0980-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Yang Y, Dai J, Xing D, Dong Y. IMPDH2 is highly expressed in breast cancer and predicts unfavourable prognosis. Biomarkers. 2018;1–9. doi: 10.1080/1354750X.2018.1496360 [DOI] [PubMed] [Google Scholar]
- 14.He Y, Zheng Z, Xu Y, et al. Over-expression of IMPDH2 is associated with tumor progression and poor prognosis in hepatocellular carcinoma. Am J Cancer Res. 2018;8(8):1604–1614. [PMC free article] [PubMed] [Google Scholar]
- 15.Hayward D, Kouznetsova VL, Pierson HE, et al. ANKRD9 is a metabolically-controlled regulator of IMPDH2 abundance and macro-assembly. J Biol Chem. 2019;294:14454–14466. doi: 10.1074/jbc.RA119.008231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Martinez D, Kupculak M, Yang D, et al. Phosphorylation of FANCD2 Inhibits the FANCD2/FANCI complex and suppresses the Fanconi anemia pathway in the absence of DNA damage. Cell Rep. 2019;27(10):2990–3005. doi: 10.1016/j.celrep.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun MJ, Choi H, Jun DW, et al. Fanconi anemia protein FANCD2 is activated by AICAR, a modulator of AMPK and cellular energy metabolism. FEBS Open Bio. 2017;7(2):284–292. doi: 10.1002/feb4.2017.7.issue-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakanishi R, Kitao H, Fujinaka Y, et al. FANCJ expression predicts the response to 5-fluorouracil-based chemotherapy in MLH1-proficient colorectal cancer. Ann Surg Oncol. 2012;19(11):3627–3635. doi: 10.1245/s10434-012-2349-8 [DOI] [PubMed] [Google Scholar]
- 19.Matsushita N, Endo Y, Sato K, et al. Direct inhibition of TNF-alpha promoter activity by Fanconi anemia protein FANCD2. PLoS One. 2011;6(8):e23324. doi: 10.1371/journal.pone.0023324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rockfield S, Kee Y, Nanjundan M. Chronic iron exposure and c-Myc/H-ras-mediated transformation in fallopian tube cells alter the expression of EVI1, amplified at 3q26.2 in ovarian cancer. Oncogenesis. 2019;8(9):46. doi: 10.1038/s41389-019-0154-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbasi S, Rasouli M. A rare FANCA gene variation as a breast cancer susceptibility allele in an Iranian population. Mol Med Rep. 2017;15(6):3983–3988. doi: 10.3892/mmr.2017.6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somyajit K, Subramanya S, Correction: NG. Distinct roles of FANCO/RAD51C protein in DNA damage signaling and repair: implications for Fanconi anemia and breast cancer susceptibility. J Biol Chem. 2019;294(35):13198. doi: 10.1074/jbc.AAC119.010430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson R, Senan S, Harrow S, et al. Quality of life outcomes after Stereotactic Ablative Radiotherapy (SABR) Vs. Standard of care treatments in the oligometastatic setting: a secondary analysis of the SABR-COMET Randomized Trial. Int J Radiat Oncol Biol Phys. 2019;105(5):943–947. [DOI] [PubMed] [Google Scholar]
- 24.Kofuji S, Hirayama A, Eberhardt AO, et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat Cell Biol. 2019;21(8):1003–1014. doi: 10.1038/s41556-019-0363-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li HX, Meng QP, Liu W, et al. IMPDH2 mediate radioresistance and chemoresistance in osteosarcoma cells. Eur Rev Med Pharmacol Sci. 2014;18(20):3038–3044. [PubMed] [Google Scholar]
- 26.Li M, Su BS, Chang LH, et al. Oxymatrine induces apoptosis in human cervical cancer cells through guanine nucleotide depletion. Anticancer Drugs. 2014;25(2):161–173. doi: 10.1097/CAD.0000000000000012 [DOI] [PubMed] [Google Scholar]
- 27.Karigane D, Kobayashi H, Morikawa T, et al. p38alpha activates purine metabolism to initiate hematopoietic stem/progenitor cell cycling in response to stress. Cell Stem Cell. 2016;19(2):192–204. doi: 10.1016/j.stem.2016.05.013 [DOI] [PubMed] [Google Scholar]