Abstract
OBJECTIVES:
Intestinal neuronal dysplasia (IND) is a common malformation of the enteric nervous system. Diagnosis requires a full-thickness colonic specimen and an experienced pathologist, emphasizing the need for noninvasive analytical methods. Recently, the methylation level of the Sox10 promoter has been found to be critical for enteric nervous system development. However, whether it can be used for diagnostic purposes in IND is unclear.
METHODS:
Blood and colon specimens were collected from 32 patients with IND, 60 patients with Hirschsprung disease (HD), and 60 controls. Sox10 promoter methylation in the blood and the Sox10 expression level in the colon were determined, and their correlation was analyzed. The diagnostic efficacy of blood Sox10 promoter methylation was analyzed by receiver operating characteristic curve.
RESULTS:
The blood level of Sox10 promoter methylation at the 32nd locus was 100% (90%–100%; 95% confidence interval [CI], 92.29%–96.37%) in control, 90% (80%–90%; 95% CI, 82.84%–87.83%) in HD, and 60% (50%–80%; 95% CI, 57.12%–69.76%) in IND specimens. Sox10 promoter methylation in the peripheral blood was negatively correlated with Sox10 expression in the colon, which was low in control, moderate in HD, and high in IND specimens (r = −0.89). The area under the curve of Sox10 promoter methylation in the diagnosis of IND was 0.94 (95% CI, 0.874–1.000, P = 0.000), with a cutoff value of 85% (sensitivity, 90.6%; specificity, 95.0%). By applying a cutoff value of 65%, promoter methylation was more indicative of IND than HD.
DISCUSSION:
The analysis of Sox10 promoter methylation in the peripheral blood can be used as a noninvasive method for IND diagnosis.
INTRODUCTION
Intestinal neuronal dysplasia (IND) is a common congenital malformation of the enteric nervous system (ENS) (1,2). It was first reported by Meier-Ruge in 1971 as a malformation of the enteric plexus in children who presented with clinical symptoms resembling Hirschsprung disease (HD). In 1983, Fadda et al. (3) proposed the classification of 2 clinically and histologically distinct subtypes of IND. Type A IND (IND-A), accounting for less than 5% of the cases observed, presents in the neonatal period and is due to delayed maturation of neuronal cells. Type B IND (IND-B) is a permanent illness with hyperganglionosis and giant ganglia of the submucosal and myenteric plexus and with symptoms of constipation and abdominal bloating. Because it comprises over 95% of all IND cases (4–6), many authors have considered IND almost as a synonym for IND-B (7).
Because of the overlap between the symptoms and signs of HD and IND, misdiagnosis often occurs in the initial diagnosis. Unfortunately, there is still no uniform diagnostic standard for IND. The current diagnostic criteria have been gradually revised by the Meier-Ruge et al. (8), and the disease concepts have changed over time. Muto et al. (9) have recently established the Japanese clinical practice guidelines for diagnosis, which were developed using the methodologies in the Medical Information Network Distribution System and a unique systematic review approach for the evaluation of small numbers of cases. Wu et al. (10) presented a novel diagnostic scoring system that was able to differentiate HD from IND by the area under the receiver operating characteristic (ROC) curve, which combines the 3 risk factors (meconium, age <3 years, and male sex) and the results of the barium enema radiography, anorectal manometry, and acetylcholinesterase reaction tests. Among all mentioned diagnostic systems, the histological evaluation of colon tissue is considered as the gold standard for assessing the size of enteric ganglia and the proportion of immature neurons (11). However, the collection of a full-thickness colonic biopsy requires general anesthesia and suturing. Thus, to a large extent, patients are unwilling to accept the procedure. Moreover, IND diagnosis is complex and subjective and requires experienced pathologists. Therefore, a noninvasive and objective method for IND diagnosis is strongly needed.
DNA methylation has been found to be relevant for various human diseases such as neurodevelopmental, neurodegenerative, and psychiatric disorders and is a well-recognized biomarker (12). DNA methylation plays an important role in the proliferation, migration, and differentiation of neural crest stem cells, which are all involved in the development of the ENS. DNA methylation occurs in the promoters of all the transcription factors that are associated with ENS development, such as DNMT3B, Sox10, EDNRB, PAX6, PAX7, NGFR, and NEUROD1 (13–19). Sox10, in particular, has been considered to be critical for neural progenitor differentiation during ENS development (20). It is first expressed in neural crest cells, including cells of vagal origin, as they delaminate from the neural tube, reflecting its crucial role in the development of these cells and of the ENS in particular (21,22). However, whether Sox10 promoter methylation in the peripheral blood can be used for the diagnosis of IND remains unclear. In this study, we performed Sox10 promoter methylation sequencing in the peripheral blood of patients with HD and IND to determine whether this event could be associated with ENS anomalies and could therefore be exploited to develop a noninvasive method for IND diagnosis.
PATIENTS AND METHODS
Sample collection
This study was approved by the ethics committee of China Medical University. The colon specimens and paired blood samples were collected at the Department of Pediatric Surgery in Shengjing Hospital of China Medical University, Capital Institute of Pediatrics of Capital Medical University, and Harbin Children's Hospital from 2016 to 2018. Detailed information on the patients, including age, sex, clinical symptoms, and diagnosis, was retrieved. The subject cohort included 60 controls, 60 HDs, and 32 INDs. The control subjects were children without constipation and consisted of 21 colon/blood sample pairs from colonic trauma/colostomy patients (used in all the following analyses) and blood samples from 39 patients with inguinal hernia or superficial hemangioma but no gastrointestinal diseases. The latter blood samples were only used in the analysis of the area under the curve (AUC) in the ROC curve and to evaluate the diagnostic efficacy of blood Sox10 promoter methylation. Diagnosis of IND-B and HD was made by 3 blinded pathologists, according to the most recent diagnostic criteria (7). Blood/colon sample pairs were collected from all patients with HD and IND. All the fresh samples were immediately stored in a −80 °C freezer.
Immunohistochemistry
Sections were incubated overnight at 4 °C with murine Sox10 primary antibody (1:100 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) and then incubated for 45 minutes at room temperature. Samples were incubated with Biotin-conjugated rabbit anti-mouse secondary antibody (1:200 dilution; Santa Cruz Biotechnology) for 20 minutes and then incubated with streptomycin-biotin-peroxidase solution for 20 minutes. A DAB kit was used for staining, and sections were counterstained with hematoxylin. The sections were dehydrated by gradient alcohol and rehydrated with xylene. Sections were examined under a microscope, and the density of the positively stained area was calculated at a ×400 magnification. The total number of glial cells in 5 light microscopy fields was counted.
Western blot
Protein extracts were heated to 100 °C for 6 minutes and size fractionated. After blocking with 5% fat-free milk in Tris-buffered saline (1.5 hours, room temperature), extracts were incubated with mouse anti-Sox10 (58 kDa; 1:200 dilution; Santa Cruz Biotechnology) or anti-glyceraldehyde-3-phosphate dehydrogenase (36 kDa; 1:4,000 dilution; ImmunoWay Biotechnology Company, Plano, TX) at 4 °C overnight. Protein bands were detected using a chemiluminescent substrate kit (Pierce, Rockford, IL), and GAPDH served as a control.
Real-time PCR
Total RNA was reverse transcribed into complementary DNA (cDNA; PrimeScript RT Reagent Kit with DNA Eraser, Takara Biotechnology, Shiga, Japan). Quantitative real time polymerase chain reaction (qRT-PCR) was performed using SYBR Premix Ex Taq (Takara Biotechnology, Shiga, Japan) on a 7500HT Fast Real-time PCR system (Applied Biosystems, Foster City, CA) as per the manufacturer's instructions. The qRT-PCR primers were as follows: Sox10F, 5′-CATCCAGGCCCACTACAAG-3′; Sox10R, 5′-AACTTCAGTTTCCCCATCTCG-3′; β-actinF, 5′-ACTCTTC CAGCCTTCCTTCC-3′; β-actinR, 5′-CGTCATACTCCTGCTTGCTG-3″.
Bisulfite genomic sequencing
The 24 CpG sites in the Sox10 gene fragment were analyzed (Figure 1). The DNA methylation levels were measured with an enzyme-linked immunosorbent assay–based commercial kit (MDQ1, Imprint Methylated DNA Quantification Kit, Sigma-Aldrich, St. Louis, MO). Specifically, 2 μg of DNA was diluted in the appropriate volume of lysis and binding buffers and incubated in a water bath at 42 °C for 30 minutes. PCR was performed using a Verity 96-well PCR reaction amplification instrument (ABI, Shanghai, China) under the following conditions: 98 °C for 4 minutes, 94 °C for 45 seconds, 66 °C for 45 seconds, 72 °C for 1 minute, 94 °C for 45 seconds, 56 °C for 45 seconds, 72 °C for 1 minute, 72 °C for 8 minutes, and 20 cycles under 94 °C. PCR products were examined by electrophoresis in 3% agarose. The purified PCR products were cloned and sequenced. SeqMan software 8.1 (Shanghai, China) was used to analyze the methylation level.
Figure 1.
The frame is the location of the primer, and the middle is the sequence to be tested. The Sox10 gene fragment contains 24 CpG sites, which are indicated above in the following order: 5, 9, 19, 32, 47, 54, 57, 68, 75, 96, 107, 111, 123, 127, 131, 179, 194, 225, 261, 282, 285, 294, 297, and 335.
Statistical analysis
Statistical analyses were performed using SPSS software version 13.0 (IBM, Armonk, NY). Continuous variables were expressed as medians with interquartile ranges (P25–P75), and P ≤ 0.05 was considered statistically significant. The ROC curve was constructed, and the maximal AUC was calculated to assess the optimal cutoff values of Sox10 methylation to diagnose IND and to discriminate it from HD. The cutoff values for 100% of specificity and the 100% of positive predictive value (PPV) were also determined. Both of these 2 optimal cutoff values were declared before the start of the study.
RESULTS
Methylation level of the Sox10 promoter gene in IND
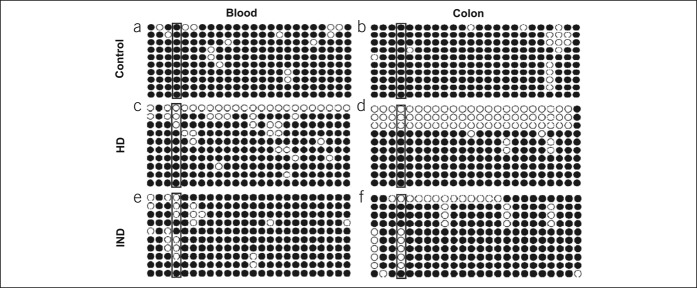
Methylation-sensitive PCR was applied to analyze the status of Sox10 promoter methylation in the peripheral blood of normal controls, patients with HD, and patients with IND at 24 CpG sites (Figure 2). In result, the 32nd locus of the Sox10 promoter was the only site exhibiting differential methylation between the control and patient groups. At this locus, the blood level of methylation of the Sox10 promoter was 100% (90%–100%; 95% confidence interval [CI], 92.29%–96.37%) in the control, 90% (80%–90%; 95% CI, 82.84%–87.83%) in HD, and 60% (50%–80%; 95% CI, 57.12%–69.76%) in IND (Figure 2a, b, e). In the colonic tissues, the 32nd locus of the Sox10 promoter was completely methylated in controls (80%; range, 80%–90%; 95% CI, 80.77%–89.70%), moderately methylated in patients with HD (80%; range, 80%–90%; 95% CI, 79.58%–84.09%), and hypomethylated in patients with IND (60%; range, 50%–70%; 95% CI, 56.50%–67.24%; Figure 2b, d, f).
Figure 2.
Promoter methylation level of the Sox10 gene in IND. In the peripheral blood, the promoter methylation level at the 32nd locus of the Sox10 promoter was (a) 100% (90%–100%) in the control, (c) 90% (80%–90%) in HD, and (e) 60% (50%–80%) in IND. In colon specimens, the 32nd locus of the Sox10 gene promoter was (b) completely methylated in the control (80%; range, 80%–90%), (d) moderately methylated in the aganglionic segment in HD (80%; range, 80%–90%), and (f) hypomethylated in IND (60%; range, 50%–70%). HD, Hirschsprung disease; IND, intestinal neuronal dysplasia.
Expression of Sox10 in the ENS
Immunohistochemistry
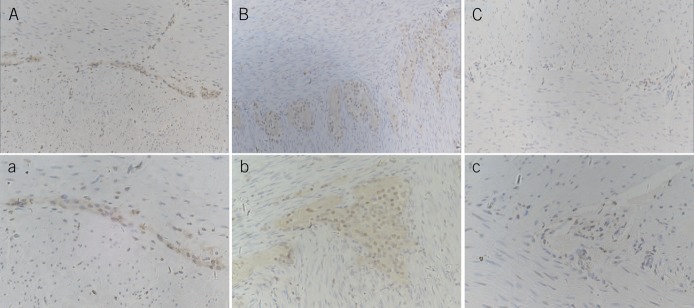
To address the expression pattern of Sox10 in the ENS, immunohistochemical staining was performed, and the result revealed that in the control group, Sox10 was mainly expressed in glial cells and in a few enteric neurons; the nerve plexus and cell morphology were homogeneous (Figure 3A,a). The number of glial cells and the plexus showed high proliferation in patients with IND (Figure 3B,b), and they moderately proliferated in the aganglionic segment of patients with HD (Figure 3C,c). The number of glial cells was 1.52 ± 0.98/cm2 in the control group, 4.27 ± 1.26/cm2 in the aganglionic segment of the HD group, and 7.54 ± 2.41/cm2 in the IND group.
Figure 3.
Immunohistochemical staining of Sox10 in the enteric nervous system. A and a are the control group: Sox10 was mostly expressed in glial cells and a few enteric neurons; the nerve plexus and cell morphology were homogeneous. B and b are the intestinal neuronal dysplasia group. The number of glial cells and the area of the nerve plexus markedly proliferated. There were more glial cells than in the control group and giant ganglia in the plexus with more than 8 nerve cells. C and c are the aganglionic segment of HD. No reactivity was seen in ganglia stained with Sox10, but more positively stained nerve fibers and glial cells were observed than the control (A–C: ×200, a–c: ×400). HD, Hirschsprung disease.
Western blot
The level of Sox10 expression was quantified by Western blot, and it was demonstrated lower in the normal colon than in the aganglionic segment of patients with HD and IND. The difference was statistically significant (P < 0.05). The difference in Sox10 expression between the controls and the patients with HD was not statistically significant (P > 0.05) (Figure 4).
Figure 4.
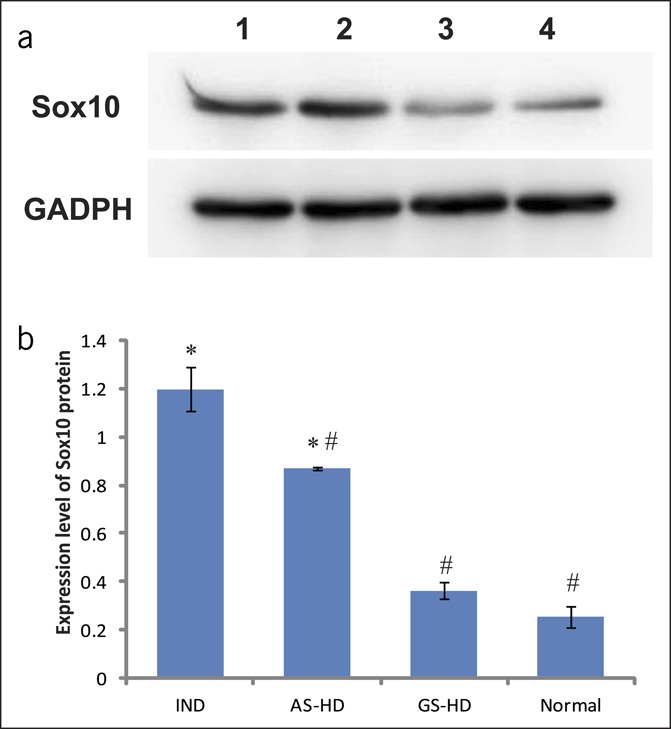
Western blot analysis of Sox10 protein in the colon. (a) Electrophoresis of Sox10 protein in the colon. Lane 1 = aganglionic segment of HD, lane 2 = IND, lane 3 = ganglionic segment of HD, and lane 4 = normal colon. (b) Expression level analysis of Sox10 protein. *Compared with the normal group, P < 0.05, #compared with IND, P < 0.05. AS-HD, aganglionic segment of HD; GS-HD, ganglionic segment of HD; HD, Hirschsprung disease; IND, intestinal neuronal dysplasia.
Messenger RNA expression
The Sox10 messenger RNA expression level was quantified by qRT-PCR; it was highest in patients with IND, moderate in patients with HD, and lowest in normal controls; qRT-PCR results were consistent with those of Western blot analysis.
Blood Sox10 promoter methylation correlates with Sox10 expression levels in the colon
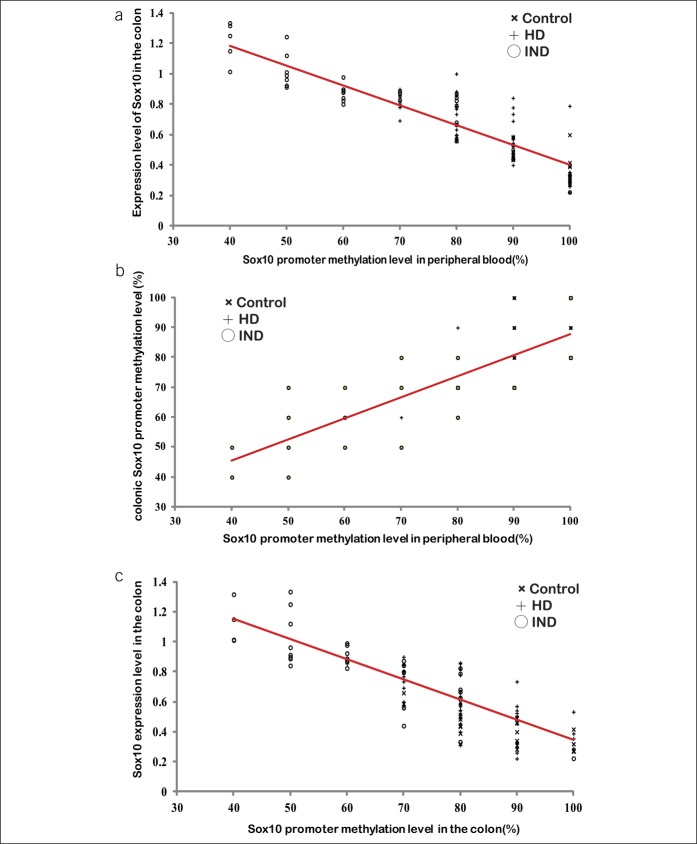
To address the relationship between the level of Sox10 promoter methylation in the blood and in the colon, blood and colon promoter methylation levels were correlated with the levels of Sox10 expression in paired blood and colon samples of 21 controls, 60 patients with HD, and 32 patients with IND. A negative correlation between the blood level of Sox10 promoter methylation at the 32nd locus and Sox10 expression level was noted in all groups (r = −0.89, Figure 5a). The levels of Sox10 promoter methylation at the 32nd locus in the blood and the colon were positively correlated (r = 0.84, Figure 5b), and colonic Sox10 promoter methylation was negatively correlated with Sox10 expression in the colon (r = −0.83, Figure 5c).
Figure 5.
Correlation analysis between the Sox10 promoter methylation level in the peripheral blood and the Sox10 expression level in the colon. (a) Correlation analysis between the Sox10 promoter methylation in the blood and its expression level in the colon (r = −0.89). (b) Correlation analysis between the levels of Sox10 promoter methylation in the peripheral blood and the colon (r = 0.84). (c) Correlation analysis between the Sox10 promoter methylation and its expression level in the colon (r = −0.83). The red lines are the correlation lines. HD, Hirschsprung disease; IND, intestinal neuronal dysplasia.
Efficacy of Sox10 promoter methylation in the blood in the diagnosis of IND
To investigate the efficacy of blood Sox10 promoter methylation in the diagnosis of IND and HD, we firstly expanded the control group from 21 to 60 cases to improve the confidence level and then calculated the AUC of the ROC curve. The AUC for the level of Sox10 promoter methylation in the diagnosis of HD was 0.776 (95% CI, 0.692–0.859; P = 0.000; Figure 6a). The cutoff value of blood Sox10 promoter methylation was determined to be 85% for HD, with a sensitivity of 46.7% and a specificity of 95.0%; the negative predictive value was 64.06%, and the PPV was 90.33%. However, with a cutoff of 95%, the sensitivity and specificity were 83.3% and 53.3%, respectively. Thus, Sox10 promoter methylation was not suitable for HD diagnosis because of low efficacy.
Figure 6.
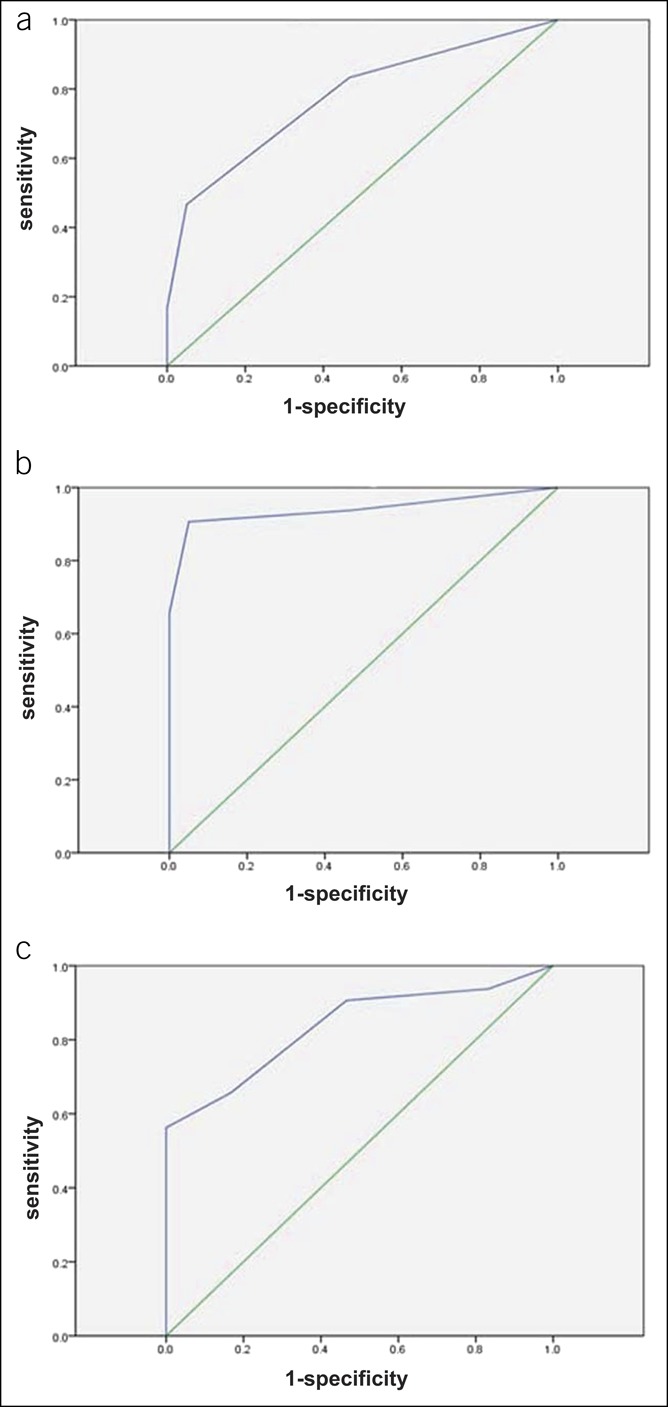
Diagnostic efficacy of blood Sox10 promoter methylation in IND. (a) ROC curve of the Sox10 promoter methylation level in the diagnosis of HD. (b) ROC curve of the Sox10 promoter methylation level in the diagnosis of IND. The AUC of Sox10 methylation in the diagnosis of IND was 0.94 (95% CI, 0.874–1.000, P = 0.000). At the cutoff value of 85%, the sensitivity is 90.6%, the specificity is 95.0%, the NPV is 90.99%, and PPV is 94.77%. (c) ROC curve of the Sox10 promoter methylation level in the differentiation of IND from HD. The optimal cutoff value of 65% was instructive for IND, with a sensitivity of 56.3%, a specificity of 100%, and a PPV of 100%. Although both sensitivity and accuracy were not satisfactory, the 100% specificity and PPV indicated that when blood Sox10 promoter methylation was lower than 65%, IND was more likely than HD. CI, confidence interval; HD, Hirschsprung disease; IND, intestinal neuronal dysplasia; NPV, negative predictive value; PPV, positive predictive value; ROC, receiver operating characteristic.
The performance of blood Sox10 promoter methylation at the 32nd locus in the diagnosis of IND was also evaluated. The AUC of Sox10 methylation in the diagnosis of IND was 0.94 (95% CI, 0.874–1.000, P = 0.000; Figure 6b). The cutoff value for IND was determined to be 85%, with a sensitivity of 90.6%, a specificity of 95.0%, and an accuracy of 92.5%, and resulted in an negative predictive value of 90.99% and in a PPV of 94.77%.
Although IND could be diagnosed at a cutoff of 85%, this was not sufficient to distinguish IND from HD because of the high degree of overlap between the symptoms and signs and Sox10 promoter methylation levels. To better distinguish between these 2 conditions, we calculated the AUC of the combined ROC curve, which demonstrated that the AUC for Sox10 promoter methylation in the diagnosis of IND was 0.835 (95% CI, 0.739–0.931; P = 0.000; Figure 6c). The optimal cutoff value was determined to be 65% for IND, with a sensitivity of 56.3%, a specificity of 100%, and a PPV of 100%. Although both sensitivity and accuracy were not satisfactory, the 100% specificity and PPV indicated that when blood Sox10 promoter methylation was lower than 65%, IND was more likely than HD.
DISCUSSION
Diagnosis of IND is still a challenge because of the high degree of overlap with HD in terms of symptoms and signs. To date, several standards for IND diagnosis have been proposed, and the identification of giant ganglia has been considered to be critical (23). To visualize the giant ganglia, many neuronal and muscular markers have been developed, such as the S-100 protein for Schwann cells, CD56 for the size of enteric ganglia (11), and Bcl-2 for immature neurons (24). In the present study, Sox10 was also shown to be a potential indicator. Immunohistochemical staining demonstrated that glial cells were highly proliferative in patients with IND but not in patients with HD. The expression level of Sox10 was correlated with the number of glial cells. Similar results were reported by Bondurand et al. and Young et al., who argued that Sox10 is mainly expressed in glial cells of the peripheral nervous system but is silenced in neurons (25,26). Sox10 was proposed to play a role in cell fate specification and glial cell differentiation (21,27,28). It can inhibit neuronal differentiation and must be downregulated during neuronal specification (29,30). Consistently, Sox10 was mainly expressed in immature ganglion and mature glial cells and could serve as a biomarker for the identification of giant ganglia in IND.
In recent years, DNA methylation has been reported to be critical for ENS development, playing important roles in the proliferation, migration, and differentiation of neural crest stem cells. In this context, the importance of Sox10 gene promoter methylation has been demonstrated (14). Hu et al. (14) have reported that the methylation of the Sox10 promoter by DNMT3B is required for the cessation of neural crest migration. The upregulation of Sox10 can also induce the demethylation of other neural crest genes, such as Snail2 and FoxD3, leading to their concomitant upregulation (14). Although the role of Sox10 methylation in the development of ENS has been verified, whether it can be used in the diagnosis of ENS anomalies remains unclear. Therefore, we applied a noninvasive genetic method based on the evaluation of the peripheral blood for the diagnosis of ENS anomalies. The results demonstrated that in the peripheral blood, the level of Sox10 promoter 32nd locus methylation was negatively correlated with Sox10 expression in the colon. Hence, we concluded that the 32nd locus of the Sox10 promoter contributed to the regulation of the number of intestinal glial cells and to the maturation of enteric neurons. Thus, Sox10 DNA methylation could be related to the expression of this gene in response to external environmental stimuli and to the development and maturation of synapses (31). Based on the present results, Sox10 methylation at the 32nd locus in the peripheral blood could be used as a diagnostic marker for IND. To further verify this possibility, we evaluated the diagnostic efficiency of this analysis. We found that the level of methylation at the 32nd locus of the Sox10 promoter was higher in controls than in HD and IND specimens. At a cutoff value of either 85% or 95%, the accuracy of the blood promoter methylation level in HD diagnosis was barely satisfactory because of low specificity or sensitivity. However, this parameter proved effective in IND diagnosis. Because in patients with IND, the blood level of Sox10 promoter methylation was significantly lower than in control children, and there was little overlap between the 2 groups, the diagnostic efficiency was evaluated in patients with IND. The results showed that at a cutoff value of 85%, the blood level of Sox10 promoter methylation at the 32nd locus displayed a 90.6% sensitivity and a 95.0% specificity, resulting in high diagnostic accuracy. Therefore, the blood level of Sox10 promoter methylation may represent a suitable alternative to the invasive examination of colon specimens, also avoiding the need of experienced pathologists. However, because of the overlap of symptoms, signs, and the Sox10 promoter methylation level between IND and HD, an 85% cutoff was not sufficient to distinguish IND from HD. Therefore, we calculated the AUC of the combined IND and HD ROC curve. The result of this analysis was that when the cutoff value was less than 65%, IND was more likely than HD because the specificity and PPV were both 100%. Notably, this means that when Sox10 promoter methylation in the blood is between 65% and 85%, additional criteria are required to distinguish IND from HD.
Although the determination of Sox10 promoter methylation in the blood implies enormous advantages over the conventional approach to IND diagnosis, this method also has a few intrinsic drawbacks. First, although it may allow for IND diagnosis, it provides no clues on the extent of the lesion, and colon specimens are still necessary to obtain this information. Second, because of the low incidence of IND, large numbers of samples are difficult to achieve in the short term. Larger studies must therefore be performed to further explore the efficacy of blood DNA testing as a noninvasive method for IND diagnosis. Finally, the level of DNA methylation in the blood can be influenced by many factors, such as viral infection, pesticide exposure, radiation, and age (32–35). On the other hand, as long as these confounding factors are properly controlled, we believe that in most cases, blood DNA methylation can be a suitable method for IND diagnosis.
In summary, blood Sox10 promoter methylation can be used as a noninvasive and efficient diagnostic method for IND, particularly when the extent of Sox10 promoter methylation is lower than 65%.
CONFLICTS OF INTEREST
Guarantor of the article: Shu-Cheng Zhang, MD.
Specific authors contributions: X.L., S.-W.Z., and S.-C.Z. participated in the design of the study and contributed to sample collection. L.-J.C. conducted the expression analysis of Sox10. Y.-R.L. conducted the BSP analysis of Sox10 and drafted the manuscript. F.B. contributed to sample collection and storage and conducted the data analysis. All authors read and approved the final manuscript. X.L. and S.-W.Z. have the equal rights with the author S.-C.Z. F.B. has the equal rights with the first author Y.-R.L.
Financial support: This study was supported by the National Natural Science Foundation of China (No.81570465, 30700917).
Potential competing interests: None to report.
Study Highlights.
WHAT IS KNOWN
✓ Diagnosis of IND is problematic due to its high overlap with HD symptoms and signs.
✓ Currently, the diagnosis of IND requires an invasive procedure based on colon biopsy.
WHAT IS NEW HERE
✓ Patients with IND exhibit lower Sox10 promoter methylation compared with patients with HD and control subjects.
✓ Sox10 promoter methylation can be measured in the peripheral blood.
✓ Peripheral Sox10 promoter methylation is inversely correlated with colonic Sox10 expression
TRANSLATIONAL IMPACT
✓ Noninvasive diagnosis of IND can be developed by determining the level of peripheral Sox10 methylation.
Supplementary Material
References
- 1.Martucciello G, Pini Prato A, Puri P, et al. Controversies concerning diagnostic guidelines for anomalies of the enteric nervous system: A report from the fourth International symposium on Hirschsprung's disease and related neurocristopathies. J Pediatr Surg 2005;40:1527–31. [DOI] [PubMed] [Google Scholar]
- 2.Sergi C. Hirschsprung's disease: Historical notes and pathological diagnosis on the occasion of the 100th anniversary of Dr. Harald Hirschsprung's death. World J Clin Pediatr 2015;4:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fadda B, Maier WA, Meier-Ruge W, et al. Neuronal intestinal dysplasia. Critical 10-years' analysis of clinical and biopsy diagnosis. Z Kinderchir 1983;38:305–11. [German.] [DOI] [PubMed] [Google Scholar]
- 4.Puri P, Gosemann JH. Variants of Hirschsprung disease. Semin Pediatr Surg 2012;21:310–8. [DOI] [PubMed] [Google Scholar]
- 5.Puri P. Variant Hirschsprung's disease. J Pediatr Surg 1997;32:149–57. [DOI] [PubMed] [Google Scholar]
- 6.Puri P, Rolle U. Variant Hirschsprung's disease. Semin Pediatr Surg 2004;13:293–9. [DOI] [PubMed] [Google Scholar]
- 7.Schäppi MG, Staiano A, Milla PJ, et al. A practical guide for the diagnosis of primary enteric nervous system disorders. J Pediatr Gastroenterol Nutr 2013;57:677–86. [DOI] [PubMed] [Google Scholar]
- 8.Meier-Ruge WA, Ammann K, Bruder E, et al. Updated results on intestinal neuronal dysplasia (IND B). Eur J Pediatr Surg 2004;14:384–91. [DOI] [PubMed] [Google Scholar]
- 9.Muto M, Matsufuji H, Taguchi T, et al. Japanese clinical practice guidelines for allied disorders of Hirschsprung's disease 2017. Pediatr Int 2018;60:400–10. [DOI] [PubMed] [Google Scholar]
- 10.Wu XJ, Zhang HY, Li N, et al. A new diagnostic scoring system to differentiate Hirschsprung's disease from Hirschsprung's disease-allied disorders in patients with suspected intestinal dysganglionosis. Int J Colorectal Dis 2013;28:689–96. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimaru K, Taguchi T, Obata S, et al. Immunostaining for Hu C/D and CD56 is useful for a definitive histopathological diagnosis of congenital and acquired isolated hypoganglionosis. Virchows Archiv 2017;470:679–85. [DOI] [PubMed] [Google Scholar]
- 12.Ai S, Shen L, Guo J, et al. DNA methylation as a biomarker for neuropsychiatric diseases. Int J Neurosci 2012;122:165–76. [DOI] [PubMed] [Google Scholar]
- 13.Enguix-Riego MV, Torroglosa A, Fernandez RM, et al. Identification of different mechanisms leading to PAX6 down-regulation as potential events contributing to the onset of Hirschsprung disease. Sci Rep 2016;6:21160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu N, Strobl-Mazzulla PH, Simoes-Costa M, et al. DNA methyltransferase 3B regulates duration of neural crest production via repression of Sox10. Proc Natl Acad Sci USA 2014;111:17911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martins-Taylor K, Schroeder DI, LaSalle JM, et al. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics 2012;7:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang W, Li B, Tang J, et al. Methylation analysis of EDNRB in human colon tissues of Hirschsprung's disease. Pediatr Surg Int 2013;29:683–8. [DOI] [PubMed] [Google Scholar]
- 17.Amiel J, Lyonnet S. Hirschsprung disease, associated syndromes, and genetics: A review. J Med Genet 2008;38:1–14. [DOI] [PubMed] [Google Scholar]
- 18.Wallace AS, Anderson RB. Genetic interactions and modifier genes in Hirschsprung's disease. World J Gastroenterol 2011;17:4937–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luzon-Toro B, Espino-Paisan L, Fernandez RM, et al. Next-generation-based targeted sequencing as an efficient tool for the study of the genetic background in Hirschsprung patients. BMC Med Genet 2015;16:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet 1998;18:60–4. [DOI] [PubMed] [Google Scholar]
- 21.Heanue TA, Pachnis V. Enteric nervous system development and Hirschsprung's disease: Advances in genetic and stem cell studies. Nat Rev Neurosci 2007;8:466–79. [DOI] [PubMed] [Google Scholar]
- 22.Herbarth B, Pingault V, Bondurand N, et al. Mutation of the Sry-related Sox10 gene in dominant megacolon, a mouse model for human Hirschsprung disease. Proc Natl Acad Sci USA 1998;95:5161–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier-Ruge WA, Schmidt PC, Stoss F. Intestinal neuronal dysplasia and its morphometric evidences. Pediatr Surg Int 1995;10:447–53. [Google Scholar]
- 24.Park SH, Min H, Chi JG, et al. Immunohistochemical studies of pediatric intestinal pseudo-obstruction: bcl2, a valuable biomarker to detect immature enteric ganglion cells. Am J Surg Pathol 2005;29:1017–24. [PubMed] [Google Scholar]
- 25.Bondurand N, Natarajan D, Thapar N, et al. Neuron and glia generating progenitors of the mammalian enteric nervous system isolated from foetal and postnatal gut cultures. Development 2003;130:6387–400. [DOI] [PubMed] [Google Scholar]
- 26.Young HM, Bergner AJ, Müller T. Acquisition of neuronal and glial markers by neural crest-derived cells in the mouse intestine. J Comp Neurol 2003;456:1–11. [DOI] [PubMed] [Google Scholar]
- 27.Bondurand N, Mai HS. The role of SOX10 during enteric nervous system development. Dev Biol 2013;382:330–43. [DOI] [PubMed] [Google Scholar]
- 28.Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Dev Biol 2012;366:64–73. [DOI] [PubMed] [Google Scholar]
- 29.Bondurand N, Natarajan D, Barlow A, et al. Maintenance of mammalian enteric nervous system progenitors by SOX10 and endothelin 3 signalling. Development 2006;133:2075–86. [DOI] [PubMed] [Google Scholar]
- 30.Sham MH, Lui VC, Fu M, et al. SOX10 is abnormally expressed in aganglionic bowel of Hirschsprung's disease infants. Gut 2001;49:220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallegos DA, Chan U, Chen LF, et al. Chromatin regulation of neuronal maturation and plasticity. Trends Neurosci 2018;41:311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito G, Yoshimura K, Momoi Y. Analysis of DNA methylation of potential age-related methylation sites in canine peripheral blood leukocytes. J Vet Med Sci 2017;79:745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuzmina NS, Lapteva NSh, Rubanovich AV. Hypermethylation of gene promoters in peripheral blood leukocytes in humans long term after radiation exposure. Environ Res 2016;146:10–7. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowska M, Reszka E, Woźniak K, et al. DNA damage and methylation induced by glyphosate in human peripheral blood mononuclear cells (in vitro study). Food Chem Toxicol 2017;105:93–8. [DOI] [PubMed] [Google Scholar]
- 35.Matsusaka K, Funata S, Fukuyo M, et al. Epstein-Barr virus infection induces genome-wide de novo DNA methylation in non-neoplastic gastric epithelial cells. J Pathol 2017;242:391–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.