Abstract
Cytochrome P450 enzymes (P450s) are broadly distributed among living organisms and play crucial roles in natural product biosynthesis, degradation of xenobiotics, steroid biosynthesis, and drug metabolism. P450s are considered as the most versatile biocatalysts in nature because of the vast variety of substrate structures and the types of reactions they catalyze. In particular, P450s can catalyze regio- and stereoselective oxidations of nonactivated C–H bonds in complex organic molecules under mild conditions, making P450s useful biocatalysts in the production of commodity pharmaceuticals, fine or bulk chemicals, bioremediation agents, flavors, and fragrances. Major efforts have been made in engineering improved P450 systems that overcome the inherent limitations of the native enzymes. In this review, we focus on recent progress of different strategies, including protein engineering, redox-partner engineering, substrate engineering, electron source engineering, and P450-mediated metabolic engineering, in efforts to more efficiently produce pharmaceuticals and other chemicals. We also discuss future opportunities for engineering and applications of the P450 systems.
Keywords: cytochrome P450, enzyme mechanism, protein engineering, metabolic engineering, substrate specificity, directed evolution, electron source engineering, ferredoxin, light activation, redox partner proteins, substrate engineering
Introduction
Cytochrome P450 enzymes (P450s)3 are a superfamily of heme-thiolate–containing proteins named for the characteristic state of the reduced, carbon monoxide (CO)-bound complex displaying a maximum UV-visible absorption band at 450 nm, due to the heme iron group being linked to the apoprotein via an axial conserved cysteine (1, 2).
Since the first discovery of P450 as a pigment (the P denoting “pigment”) in rat liver microsomes in 1958 (3), more than 370,000 P450 sequences have been released (UniProt), which are found in human, animals, plants, microbes, and even viruses, demonstrating their incredible and significant diversity in nature (4). P450s play important roles in biosynthetic pathways for natural products, degradation of xenobiotics, biosynthesis of steroid hormones, and drug metabolism (5, 6). P450s are considered to be the most versatile biocatalysts in nature (7) and are involved in more than 20 different types of chemical oxidation reactions, including hydroxylation, epoxidation, decarboxylation, N- and O-dealkylation, nitration, and C–C bond coupling or cleavage, to name a few (5, 8) (plus some reductions). Furthermore, the substrate diversity of P450s covers almost all classes of organic structures found in nature (e.g. terpenoids, polyketides, fatty acids, alkaloids, and polypeptides) (5, 9, 10). The ubiquitous distribution and the multiplicity of reactions and substrates demonstrate the plasticity of P450 enzyme systems, providing a limitless space for mining, engineering, and designing P450 systems for practical catalysis.
Among diverse functionalities, the most important is that P450s are capable of catalyzing the regio- and stereoselective oxidation of inert C–H bonds in complex molecular scaffolds under mild conditions, making them superior to many chemical catalysts and of great interest for pharmaceutical, chemical, and biotechnological applications. However, the narrow substrate scope of some P450s, low catalytic efficiency, low stability, dependence on redox partners, high cost of cofactors, and electron uncoupling have limited the industrial applications of P450s (11, 12). More recently, innovative P450 systems have been developed to fuel industrial projects with the use of a number of new engineering strategies (e.g. interactions of essential elements, including P450 itself, redox partner, substrate, and cofactor). These include the powerful directed evolution approach pioneered by the Nobel Laureate Frances H. Arnold, used to build unnatural but more robust P450 systems (13).
Several excellent reviews have covered the diversity, functions, novel chemistry, and applications of P450s (5, 10, 14–17). For more insight into intriguing P450-related mechanisms and to deeply understand the strategies related to the practical application of P450 catalysis, we will focus on recent advances in P450 protein engineering, particularly engineering strategies for optimization of the interaction between P450s and redox partners. We will also consider substrate engineering, cofactor (NAD(P)H) regeneration, and several atypical strategies for engineering the electron transport system. Finally, a brief summary of P450-related metabolic engineering will be provided.
P450 catalytic system
In general, a P450 catalytic system includes four components: the substrate, a P450 enzyme for substrate binding and oxidative catalysis, the redox partner(s) that functions as an electron transfer shuttle, and the cofactor (NAD(P)H), which provides the reducing equivalents.
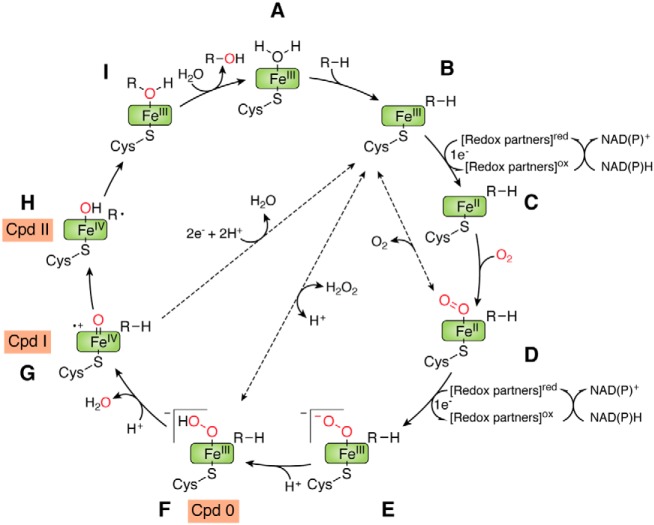
Most P450s share a common sophisticated catalytic cycle (Fig. 1) (2, 5, 18), using the typical hydroxylation reaction as a paradigm, as shown in Fig. 1. The ferric resting state (generally) of a P450 (A) first accepts a substrate (RH), which displaces an active-site water molecule but does not bond directly to the iron. The ferric iron (FeIII) of the high-spin, substrate-bound complex (B) is then reduced to ferrous iron (FeII) (C) by one electron, transferred via a redox partner. Next, binding of dioxygen to FeII results in the [FeII O2] complex (D). The complex D is reduced by the second electron to form complex E, which uses a proton from solvent to generate a ferric hydroperoxo species [FeIII–OOH] (F), referred as to Compound 0 (Cpd 0). The O–O bond of Cpd 0 is cleaved upon the addition of the second proton and releases a molecule of water to generate the high-valent porphyrin π radical cation tetravalent iron [FeIV=O] (i.e. Compound I (Cpd I; G)). This highly reactive complex abstracts a hydrogen atom from the substrate, leading to the formation of the ferryl-hydroxo compound II (Cpd II; H). Subsequently, the hydroxylated product (R-OH) is formed by the reaction of the substrate radical with the hydroxyl group of Cpd II and released from the active site of complex I. Finally, a molecule of water returns to coordinate with FeIII, restoring the resting state A. The same catalytic cycle is initiated repeatedly as substrate molecules bind to the heme-centered active site of P450.
Figure 1.
The catalytic cycle of P450s (dashed arrows indicate the peroxide shunt pathway and P450 uncoupling).
It is worth noting that some P450s are capable of directly utilizing H2O2 as the sole electron and proton donor to form Cpd 0 and do catalysis via the so-called peroxide shunt pathway (Fig. 1, dashed arrows). However, this shunt pathway is greatly limited by the low efficiency and the low H2O2 tolerance of most P450s, except P450 peroxygenases (e.g. CYP152 subfamily) (19). The well-studied and established catalytic cycle provides a theoretical basis and roadmap to understand and manipulate this P450 peroxygenase subfamily by protein and substrate engineering.
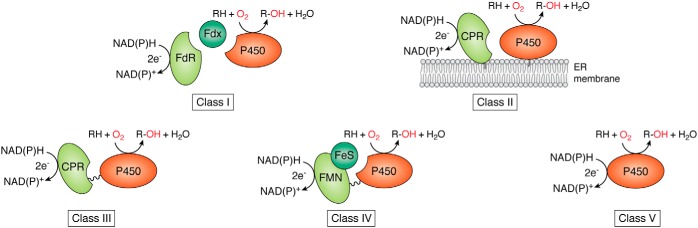
Maintenance of the P450 catalytic cycle relies on continuous electron transport to the heme-iron by redox partners, which are complicated electron-transfer systems. Based on the types of redox partners and the P450-redox partner interaction relationships, P450 systems can be divided into five main classes (10, 11, 15) (Fig. 2). The Class I P450 system present in most bacterial and mitochondrial P450s has a two-component redox partner system, comprised of an FAD-containing ferredoxin reductase (FdR) and a small iron-sulfur-containing ferredoxin (Fdx) (20, 21). The Class II P450 system employed by eukaryotic organisms has a single-component redox partner, which is a membrane-bound protein containing both an FAD and an FMN domain, termed cytochrome P450 reductase (CPR). Class III P450 systems have a eukaryotic-like CPR naturally fused to the C terminus of the P450 domain through a flexible linker, represented by Bacillus megaterium P450BM3 (CYP102A1) (22). Class IV P450 systems are exemplified by P450 RhF from Rhodococcus sp. NCIMB 9784, whose FMN/Fe2S2-containing reductase domain forms a natural fusion with the P450 domain (23). Interestingly, a few P450s can directly interact with their electron donors and are independent of additional redox partner proteins to accomplish the catalytic reactions; these Class V P450s include P450Nor (24) and P450 TxA (25). Class III–V P450s are independent of redox partner proteins and are often called self-sufficient P450s. Notably, these single-component P450 systems provide very desirable scaffolds for engineering P450 systems, due to their self-sufficiency and hence the significantly increased electron transport efficiency. It is worth noting that other classification systems also exist: Munro et al. (26) have categorized five other novel P450-fused redox partner systems in addition to the classical Class I and Class II types, and Bernhardt et al. (27) classified 10 types for P450s based on the topology of protein components involved in the electron transfer chains of P450 enzymes.
Figure 2.
Classification of the P450 systems based on redox partner proteins.
Successful applications of P450 catalytic systems
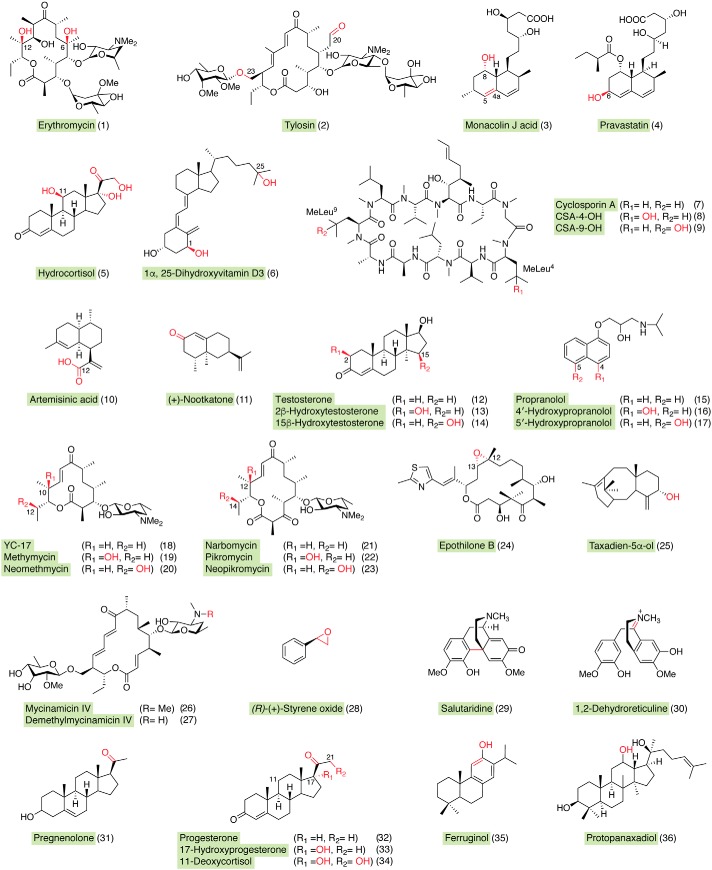
The incomparable diversity of P450s regarding substrates and reaction types provides nearly limitless application potential for production of chemicals and pharmaceuticals (28, 29), biosensor-based analysis (30), chemoenzymatic synthesis (31), and pollutant biodegradation (32). For instance, the Saccharopolyspora erythraea EryF and EryK P450s are involved in the production of the antibacterial agent erythromycin (Fig. 3, compound 1) (33); Streptomyces fradiae TylI and TylHI P450s are involved in the biosynthesis of the antimycoplasma drug tylosin (Fig. 3, compound 2) (34); and Aspergillus terreus LovA is responsible for biosynthesizing monacolin J acid (Fig. 3, compound 3), the precursor of a series of cholesterol-lowering statin drugs (35, 36). The production of high value-added chemical intermediates from phenolic environmental pollutants has been achieved with P450 biodegradation systems (32, 37, 38), and soluble P450s have been used in bacterial cell libraries to mimic human P450 drug metabolic profiles (39, 40).
Figure 3.
Structures involved in practical catalysis of diverse P450 systems. Red-colored groups are introduced by P450s.
Genome mining and high-throughput screening of P450s have proven to be effective and successful strategies for seeking suitable and robust biocatalysts in industry. P450sca-2 (CYP105A3), screened from Streptomyces carbophilus, is able to catalyze the 6β-hydroxylation of compactin produced by Penicillium citrinum, generating the cholesterol-lowering drug pravastatin (Fig. 3, compound 4) (41), considered to be one of the most successful instances of practical P450 catalysis in industry (11, 42). The bioconversion of 11-deoxycortisol into hydrocortisol by the P450lun-containing fungus Curvularia lunata has been launched by Bayer on an industrial scale (11, 43) (Table 1 and Fig. 3, compound 5). The industrially relevant P450 VD25 (CYP105A2) from Amycolata autotrophica (later renamed as Pseudonocardia autotrophica) is capable of transforming vitamin D3 into its most bioactive form, 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3) (44) (Table 1 and Fig. 3, compound 6). The P450 hydroxylase CYP-sb21 from the rare actinomycetes Sebekia benihana and CYP-pa1 from P. autotrophica are candidate biocatalysts for site-selective hydroxylation of the immunosuppressive drug cyclosporin A to two hair-stimulating agents with significantly decreased immunosuppressant activity, γ-hydroxy-N-methyl-l-Leu4-cyclosporin A and γ-hydroxy-N-methyl-l-Leu9-cyclosporin A, respectively (45–48) (Table 1 and Fig. 3, compounds 7–9).
Table 1.
Selected P450s involved in production of pharmaceuticals and chemical intermediates
| P450 | PDB code | Origin | WT/mutant | Function | Substrate | Reference |
|---|---|---|---|---|---|---|
| EryF (CYP107A1) | 1OXA | Saccharopolyspora erythraea | WT | 6-Hydroxylation | 6-Deoxyerythronolide B | 33 |
| EryK (CYP113A1) | 2JJN | S. erythraea | WT | 12-Hydroxylation | Erythromycin D | 33 |
| TylI | Streptomyces fradiae | WT | C20 hydroxylation/dehydrogenation | 5-Mycaminosyl tylacone | 34 | |
| TylHI | 6B11 | S. fradiae | WT | C23 Hydroxylation | 23-Deoxy-5-omycaminosyl-tylonolide | 34 |
| LovA | Aspergillus terreus | WT | 4a/5-Dehydrogenation, 8-hydroxylation | Dihydromonacolin L | 35 | |
| CYP71AV1 | Artemisia annua | WT | 12-Carboxylation | Amorphadiene | 49 | |
| P450sca-2 (CYP105A3) | Streptomyces carbophilus | WT | 6β-Hydroxylation | Compactin | 41 | |
| Semi-rational design | R8–5C/T85F/ T119S/V194N/N363Y | 6β-Hydroxylation | Compactin | 69 | ||
| P450lun | Curvularia lunata | WT | 11-Hydroxylation | 11-Deoxycortisol | 11, 43 | |
| P450VD25 (CYP105A2) | Pseudonocardia autotrophica | WT | 25-Hydroxylation | Vitamin D3 | 44 | |
| CYP-sb21 (CYP107Z14) | Sebekia benihana | WT | Hydroxylation at the 4th N-methyl leucine | Cyclosporin A | 45 | |
| CYP-pa1 | P. autotrophica | WT | Hydroxylation at the 9th N-methyl leucine | Cyclosporin A | 47 | |
| P450BM3 (CYP102A1) | 1JPZ | Bacillus megaterium | WT | Hydroxylation | Fatty acids | 22 |
| 2X7Y | Site-directed mutagenesis | F87A | 2β-/15β-Hydroxylation (1:1) | Testosterone | 54 | |
| Directed evolution | R47Y/T49F/V78L/A82M/F87A | 15β-Hydroxylation (96%) | Testosterone | 54 | ||
| Directed evolution | A330W/F87A | 2β-Hydroxylation (97%) | Testosterone | 54 | ||
| Heme domain of P450BM3 | Directed evolution | 9C1 A74V | 4′-Hydroxypropranolol and 5′-hydroxypropranolol | Propranolol | 39 | |
| P450 Vdh (CYP107BR1) | 3A4G | P. autotrophica | WT | 1α-/25-Hydroxylation | Vitamin D3 | 55 |
| Directed evolution | T70R/V156L/E216M/E384R | 1α-/25-Hydroxylation | Vitamin D3 | 56 | ||
| CYP105A1 | 2ZBX 2ZBZ |
Streptomyces griseolus | WT | 1α-/25-Hydroxylation | Vitamin D3 | 63 |
| Rational design | R73A/R84A | 1α-/25-Hydroxylation | Vitamin D3 | 64 | ||
| CYP105AS1 | 4OQR | Amycolatopsis orientalis | WT | 6-epi-Hydroxylation (97%) | Compactin | 29 |
| P450Prava | Directed evolution | I95T/Q127R/A180V/L263I/A265N | 6β-Hydroxylation (100%) | Compactin | 29 | |
| PikC | 2C6H 2C7X |
Streptomyces venezuelae ATCC 15439 | C10/C12 Hydroxylation, C12/C14 hydroxylation | YC-17/Narbomycin | 65 | |
| Rational design | D50N | C10/C12 Hydroxylation, C12/C14 hydroxylation | YC-17/Narbomycin | 65 | ||
| 2C6H 2C7X |
Redox parent engineering | WT | C10/C12 Hydroxylation, C12/C14 hydroxylation | YC-17/Narbomycin | 87 | |
| Substrate engineering | D50N | Regio-selective hydroxylation | Unnatural substrates | 105 | ||
| MycG | 5UHU | Micromonospora griseorubida | WT | C12/C13 Epoxidation, C14 hydroxylation | Mycinamicin IV | 102 |
| Redox parent engineering | N-Demethylation | Mycinamicin IV | 101 | |||
| CYP725A4 | Taxus cuspidata | WT | 5α-Hydroxylation | Taxadiene | 100 | |
| Redox parent engineering | N-terminal hydrophilic modifications | 5α-Hydroxylation | Taxadiene | 100 | ||
| CYP2C9 | 1OG2 | Human | Self-sufficient | N-Demethylation | Erythromycin | 89 |
| CY2C19 | Human | Self-sufficient | 4-Hydroxylation | Diclofenac | 89 | |
| CYP3A4 | 6DA2 | Human | Self-sufficient | 5-Hydroxylation | Omeprazole | 89 |
| 1,2-Dehydroreticuline synthase (CYP82Y2-like) | Papaver bracteatum | PbDRS-DRR | Dehydrogenation | (S)-Reticuline | 159 | |
| SalSyn | Papaver somniferum | yPbSalSyn92–504 | C–C Coupling | (R)-Reticuline | 159 | |
| CYP76AH1 | Salvia miltiorrhiza | WT | Hydroxylation and dehydrogenation | Miltiradiene | 161 | |
| P450 protopanaxadiol synthase | Panax ginseng | WT | Hydroxylation | Dammarenediol-II | 161 | |
| CYP11A1 | 3N9Y | Human | WT | C–C Cleavage | Egrosta-5-eneol/ergosta-5,22-dieneol | 160 |
| CYP17A1 | 3RUK | Human | WT | 17α-Hydroxylation | Progesterone | 160 |
| CYP21A1 | Human | WT | 21-Hydroxylation | 17-Hydroxyprogesterone | 160 | |
| CYP11B1 | Human | WT | 11β-Hydroxylation | 11-Deoxycortisol | 160 | |
| P450 box A | Streptomyces sp. TM-7 | WT | 6β-Hydroxylation | Compactin | 163 | |
| CYP106A2 | 4YT3 | B. megaterium ATCC 13368 | WT | 15β-Hydroxylation | Progesterone, testosterone | 167 |
P450 protein engineering has been playing a vital role in developing biocatalysts for industrial applications, as exemplified by the heterologous production of artemisinic acid (Fig. 3, compound 10), an important synthetic precursor for the potent antimalarial drug artemisinin (49). Traditional production of the anti-malarial drug artemisinin from the Chinese medicinal plant Artemisia annua L. is low-yield, unsustainable, and too expensive for millions of individuals suffering from malaria. A recombinant Saccharomyces cerevisiae strain with a heavily engineered mevalonate pathway, an amorphadiene synthase, and a key CYP71AV1 (from A. annua) produced 100 mg of artemisinic acid per liter (49) (Table 1 and Fig. 3, compound 10). By applying different synthetic biology strategies, including the introduction of the cognate reductase CPR1 of CYP71AV1 and a cytochrome b5 protein (CYB5, an electron transfer component for CYP71AV1 from A. annua), the titer of artemisinic acid was dramatically improved to 25 g/liter on an industrial scale, which successfully reduced the price and provided a stable artemisinin supply for the market (28).
The P450 systems have also been engineered for the fragrance production. For instance, the oxidation of sesquiterpene (+)-valencene to high value-added flavor (+)-nootkatone with P450 enzymes was first accomplished in the Wong group by rationally designed mutants of P450BM3 and P450cam (50) (Fig. 3, compound 11).
Strategies and progress of engineering P450 enzyme systems
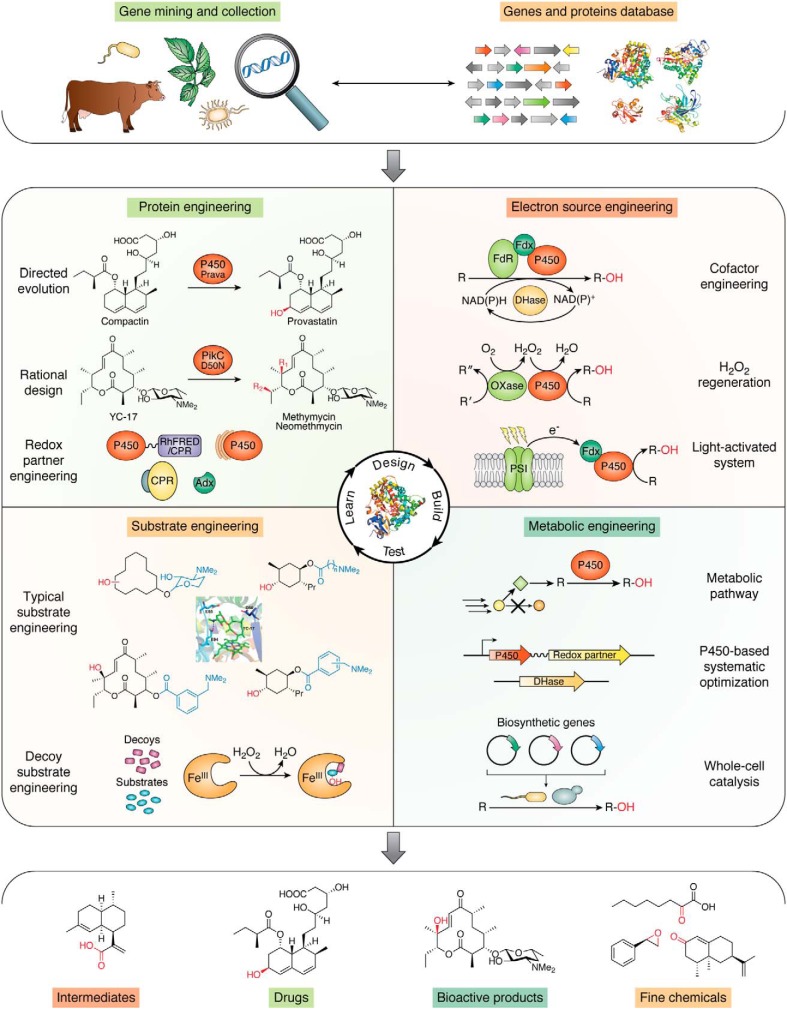
Although P450s have demonstrated amazing catalytic diversities and great prospects for application, the aforementioned limitations in industrial applications of P450s are also significant. To overcome these limitations, versatile engineering strategies have been proposed and developed to satisfy different application requirements, including protein engineering of P450s and redox partners, substrate engineering, cofactor regeneration, and P450-related metabolic engineering (Fig. 4).
Figure 4.
Engineering strategies for P450 systems discussed in the current review. DHase and OXase, dehydrogenase and oxidase, respectively.
Protein engineering
Protein engineering involves modification of the residues based on the folding principles and molecular structure of proteins, with the goal of obtaining the desired mutated proteins with enhanced properties to compensate for the poor stability, low selectivity, slow catalytic rates, and limited application space of the native proteins (51). Directed evolution and rational and semi-rational design are routinely used methods in P450 engineering and play very important roles in the development of pharmaceutical catalysts (15, 52).
Directed evolution
Directed evolution has been widely applied to engineer P450s, the structures of which are often unknown, for desired properties under artificial selective pressure, including random mutagenesis and screening (13). To obtain the desired mutant proteins, a protein library with a large number of mutants covering sufficient molecular diversity is usually generated by error-prone PCR (epPCR), combinatorial saturation mutagenesis, or DNA-shuffling methods (52). In addition, directed evolution is also an effective tool for understanding relationships between key amino acid residues surrounding catalytic pockets and catalytic abilities toward different unnatural substrates for well-characterized P450s (e.g. P450BM3 and P450cam) (53).
Reetz and co-workers (54) found that the simple P450BM3 mutant F87A, with reduced steric hindrance for substrate, was able to catalyze the nonselective 2β- and 15β-hydroxylation of testosterone to generate a 1:1 mixture of products. To alter the regioselectivity, iterative saturation mutagenesis of 20 selected residues lining the substrate-binding pocket was done, leading to two effective mutants (A330W/F87A and R47Y/T49F/V78L/A82M/F87A) that achieved specific regio-selective production of the 15β (96%) or 2β products (97%), respectively (54) (Table 1 and Fig. 3, compounds 12–14). P450 Vdh (CYP107BR1, Protein Data Bank (PDB) entry 3A4G) from P. autotrophica was also reported to produce 1α,25(OH)2D3 from vitamin D3 (55) (Table 1 and Fig. 3, compound 6). A Vdh-K1 mutant (T70R/V156L/E216M/E384R) was generated with 6-fold higher specific activity than WT P450 through high-throughput screening of a site-saturated mutagenesis library (56) (Table 1 and Fig. 3, compound 6).
Directed evolution of P450s has also been applied in the generation of drug metabolites as an effective strategy for further pharmaceutical studies. Here, the strategy is to use the bacterial P450s to generate larger amounts of drug metabolites to facilitate structural analysis of the small quantities of drug metabolites, which is a regulatory requirement for further drug development. Arnold and associates (39) constructed a mutant library of P450BM3 using epPCR and combinatorial saturation mutagenesis of seven active-site residues surrounding the heme domain. The mutants selectively oxidized the antiarrhythmic drug propranolol to its active human metabolites, including 4′-hydroxypropranolol and 5′-hydroxypropranolol, via the “H2O2” shunt pathway (39) (Table 1 and Fig. 3, compounds 15–17). Subsequently, a small panel of P450BM3 variants was further subjected to site-directed mutagenesis of the active-site residues, leading to a set of metabolites of the antihypertensive drug verapamil and the antiallergic astemizole that are the same as those metabolized by mammalian P450s (57).
The Guengerich laboratory screened a series of CYP1A2 mutants generated by random mutagenesis at six substrate recognition sites (SRSs), and the obtained variants had 2–4-fold increases in kcat/Km (specificity constant) toward the analgesic and antipyretic drug phenacetin compared with the parent enzyme (58). Similarly, human CYP2A6 mutants were screened based on the production of indole oxidation products, which could find application in production of dyestuffs or as protein kinase inhibitors (59).
The protein stability of P450s, another important factor for practical applications to enhance the total turnover numbers, can also be improved via directed evolution, as exemplified by solvent tolerance optimization of the P450BM3 variant F87A/T235A/R471A/E494K/S1024E, which was obtained from libraries constructed by saturation mutagenesis and random mutagenesis (60) (the substrates are generally hydrophobic and dissolved in organic solvents). The conversion of p-nitrophenoxydodecanoic acid to p-nitrophenol was enhanced 5.5-fold in the presence of 25% (v/v) DMSO and 10-fold in 2% THF (v/v) compared with the parental P450BM3 F87A mutant (60).
Rational and semi-rational design
Major disadvantages of directed evolution include the dependence on a high-throughput screening method, which is not always available, the requirement of automated instruments, and high cost. Rational or semi-rational design, based on the well-characterized protein tertiary structure and the mechanistic understanding of structure-activity relationships, is regarded as an effective alternate strategy. Generally (but not always), “hot spot” residues for rational and semi-rational design are usually located within the SRS, the substrate access channel, and the P450-redox partner interaction sites (15, 61).
CYP105AS1 (from Amycolatopsis orientalis) catalyzes the conversion of compactin to the cholesterol-lowering drug pravastatin plus its ineffective epimer 6-epi-pravastatin, in a ratio of 3:97. Based on the crystal structure of CYP105AS1 (PDB entry 4OQR), a single round of epPCR mutagenesis of selected residues led to a mutant with a ratio of pravastatin/6-epi-pravastatin of 48:52. A further two rounds of site-saturated mutagenesis produced a mutant P450Prava (I95T/Q127R/A180V/L263I/A265N), in which the stereoselectivity was completely inverted into a P450 pravastatin synthase with a 21-fold lower Km value for compactin (29) (Table 1).
CYP105A1 (S. griseolus) hydroxylates vitamin D3 to form 1α,25(OH)2 vitamin D3. Site-directed mutagenesis of CYP105A1 based on its three-dimensional structure (PDB entries 2ZBX and 2ZBZ) was performed. Three arginine residues (Arg-73, Arg-84, and Arg-193) located along the substrate access channel of CYP105A1 were mutated to nonpolar alanines on the basis of their important roles in substrate binding and catalysis, delineated from a crystal structure of CYP105A1 with its enzymatic product 1α,25-OH vitamin D3 (62). As hypothesized, the double mutant R73A/R84A exhibited ∼400- and 100-fold increased activity for 25-hydroxylation and 1α-hydroxylation of vitamin D3 compared with the WT enzyme (63, 64) (Table 1).
The co-crystal structures of multifunctional P450 PikC (PDB entries 2C6H and 2C7X) bound to its native substrates (narbomycin and YC-17) suggested that Asp-50, Glu-85, and Glu-94 (located in the catalytic pocket) might be critical for substrate binding and catalytic activity (Table 1 and Fig. 3, compounds 18–23). Accordingly, a series of mutants was constructed, and PikC D50N displayed significantly higher hydroxylation activities toward both narbomycin and YC-17 than did the WT enzyme (65) (Table 1 and Fig. 3, compounds 18–23).
The availability of a tertiary protein structure is considered to be a major limitation in rational and semi-rational design, in that experimental structural information is not often available for the >370,000 P450 sequences. Homology modeling has often been used to bridge the “structure knowledge gap,” based on the general observation that proteins with homologous sequences share similar structures (66, 67). A well-known example of P450 semi-rational design for industrial application is the development of a highly efficient mutant of P450sca-2 involved in the production of pravastatin (68). Based on homology modeling analysis of an active mutant (R8-5C) of P450sca-2 generated from directed evolution, five sites located in the substrate binding pocket (Arg-77/Thr-85), the substrate access channel (Val-194), and the redox partner interface (Asn-363/Thr-119) were selected for systematic site-directed saturation mutagenesis and three rounds of mutagenesis (69). As a result, a more active mutant (R8-5C/T85F/T119S/V194N/N363Y) was obtained, with 7-fold higher whole-cell biotransformation activity and a 10-fold higher kcat value than that of R8-5C (69) (Table 1). Based on the X-ray crystal structure of the high-activity progesterone hydroxylase rabbit CYP2C5 (PDB entry 1DT6), the low progesterone hydroxylase activity of CYP2B1 was re-engineered by changing active-site residues in the three-dimensional structural model of CYP2B1 to the corresponding residues of CYP2C5 (70, 71). Finally, a CYP2B1 mutant (I114A/F206V/F297G/V363L/V130I/S294D/I477F) exhibited a 3-fold higher kcat value than that of CYP2C5 for progesterone 21-hydroxylation, with 80% regioselectivity (71).
Redox partner engineering
Most P450s require redox partner proteins to sequentially transfer two electrons from NAD(P)H to the heme-iron reactive center to activate O2 for substrate oxygenation (6), which can often be the rate-limiting step of the P450 catalytic cycle (Fig. 1). However, the reconstitution of a P450 catalytic system in vitro or in a recombinant host is often hampered by the lack of information about its cognate redox partners or inaccessibility of optimal surrogate redox partners. Therefore, protein-protein interactions between P450 and surrogate redox partners have been optimized to enhance the electron transfer efficiency of P450 systems, which we will term “redox partner engineering.”
A comprehensive screening of redox partners to identify the best electron transport pathway for supporting the CYP105D5 activity was done in the Guengerich laboratory (72). Briefly, all four FdRs and six Fdxs encoded by the genome of Streptomyces coelicolor A3(2) were heterogeneously expressed and purified. A total of 24 native redox partner combinations were assembled and screened with a specific S. coelicolor P450, CYP105D5, which had been shown to hydroxylate free fatty acids (72). The results showed that the pair Fdx4/FdR1 functioned as the preferred redox partner system for this bacterial P450 enzyme in vitro (72).
Adrenodoxin and adrenodoxin reductase (Adx/AdR) were characterized as optimal redox partners in supporting the in vitro hydroxylation of lauric acid by CYP109D5 from Sorangium cellulosum So ce56, the catalytic efficiency of which was 3–4-fold higher than that of CYP109D5 supported by endogenous redox partners (Fdx2/FdR_B and Fdx8/FdR_B) (73). Interestingly, the combination of Fdx8/FdR_B was reported to be a much better pair of redox partners of P450 EpoK in the bioconversion of epothilone D to epothilone B compared with the spinach Fdx/FdR redox pair (74) (Table 1 and Fig. 3, compound 24). Thus, a certain P450 enzyme may have a differentially preferred combination of Fdx and FdR among multiple combinations, although alternative redox partners could be functionally complementary (75). It is also worth noting that surrogate redox partners may be superior to the cognate ones; thus, it can be helpful to apply a redox partner interchange approach to determine optimal electron transfer pathways, particularly in bacterial systems, to fully exploit P450 applications.
To determine whether there are any principles for guiding the screening of optimal redox partners for a given Class I bacterial P450, Zhang et al. (76) constituted a reaction matrix network based on 16 Fdxs, eight FdRs, and six P450s toward seven substrates. By analyzing the reactivity profiles of 896 reactions, plastidic-type FdR and Fe2S2 Fdx were found to be the favored types of redox partners by Class I P450 systems. Based on the empirically derived rules, the optimal cognate Fdx of PikC from Streptomyces venezuelae ATCC 15439 was predicted and confirmed in vitro to be SveFdx1948 (76). This work has provided information about the P450-preferred redox partners, and we envision that the findings will benefit future practical applications of P450 enzymes.
Notably, the protein pair SelFdx1499 (Fe2S2)/SelFdR0978 (plastidic-type FdR) from the cyanobacterial strain Synechococcus elongatus PCC 7942 has been shown to be an optimal combination for supporting in vitro reactions of prokaryotic P450s, including MycG, PikC, P450sca-2 and others (76). Besides the above-mentioned P450 reaction matrix network, the protein pair SelFdx1499/SelFdR0978 has also been shown to be optimal for the site-selective hydroxylation of CsA by CYP-sb21 and CYP-pa1 (45, 47), the uncommon ester-to-ether transformation catalyzed by Rif16 in rifamycin biosynthesis (77), the tandem ether installation and hydroxylation by AmbV involved in neoabyssomicin/abyssomicin biosynthesis (78), and the biosynthesis of phenylserine (β-OH-Phe) unit in atratumycin by Atr27 (79).
In addition to the screening and prediction of optimal redox partners, optimization of interaction modes between P450s and redox partners through redox partner engineering provides another effective strategy for P450 activity improvement. The residues located at the P450-Fdx (Fdx directly interacts with P450) interaction interface play important roles in affecting the catalytic activity of a P450. Screening of Adx derivatives modified at N-terminal or C-terminal polypeptide sequences led to the finding that Adx(4–108) truncated at N-terminal amino acids 1–3 and C-terminal amino acids 109–128 supported the 11β-hydroxylation of 11-deoxycortisol to cortisol by CYP11B1 with a higher electron transfer rate, and the specificity constant (kcat/Km) was increased 21-fold relative to that of WT Adx (80–82). The availability of co-crystal structures of P450s and their redox partners will facilitate engineering of the P450/Fdx interface, as exemplified by the artificial fusion CYP11A1-Adx (83) and the cross-linked CYP101A1-Pdx complex (84). Based on the interaction analysis, the amino acids of ferredoxin PuxB interacting with P450 were swapped (site-directed mutagenesis) to mimic the biogenic ferredoxin Pux of CYP199A2 from Rhodopseudomonas palustris CGA009. A PuxB variant with seven mutations was generated, and the rate of demethylation of 4-methoxybenzoic acid by CYP199A2 was increased 12-fold compared with WT PuxB (85). However, the semi-rational engineering approach remains challenging due to the lack of comprehensive understanding of the dynamic mechanisms for protein-protein recognition and intermolecular electron transfer. Thus, further work is needed for understanding P450-Fdx complex structures at the molecular level to address this challenge.
Inspired by the paradigm of self-sufficient P450 enzymes (Class III and IV) that contain both P450 and redox partner domains in one polypeptide chain, the construction of “unnatural” self-sufficient enzymes by making variant versions of P450-redox partner fusion proteins has been pursued (86). A self-sufficient PikC-RhFRED fusion was generated, and its catalytic activity toward YC-17 was increased ∼4-fold compared with that of a three-component system (PikC + spinach Fdx/FdR) in vitro, likely due to enhanced intramolecular electron transfer efficiency compared with the intermolecular reaction (87). Another striking example is the construction of self-sufficient P450Prava-RhFRED. The introduction of P450Prava-RhFRED into the compactin-producing P. chrysogenum delivered more than 6 g of pravastatin per liter in a one-step fermentation (29). High-throughput generation of self-sufficient P450 libraries by fusing P450 heme domains to RhFRED via a ligation-independent cloning vector, “LICRED,” was developed (88). Self-sufficient mammalian P450-reductase fusion enzymes have been prepared, mimicking the precedent of the efficient P450BM3, including CYP2C9, CY2C19, and CYP3A4 for drug metabolism studies (89). However, establishing the optimal design and length of the linker has not been trivial (90). Among seven fused P450cam-RhFRED (L1–L7) enzymes with varying linker regions, L4 was the most optimal, with 100% conversion of 3 mm (+)-camphor under the conditions tested (90).
Other chimeras have been made with diverse P450s from mammals, plants, and bacteria, including P450cam (91), P450 TxtE (92), CYP257A1 (93), OleTJE (94, 95), P450 isoflavone synthase (96), and CYP2E1 (97). In principle these fusion proteins can improve catalytic activity, coupling efficiency, and other electron transfer properties by simplifying the overall P450 redox system and process suitability (75, 98). More challenging are engineering and expression of eukaryotic P450s in prokaryotic systems. First, compartmentalization is one consideration for Class II P450 systems, in that interaction between P450 and CPR typically occurs in the endoplasmic reticulum. Second, the molar ratio of P450 and its redox partner in a chimeric system is fixed at 1:1, instead of 15:1 with membrane-bounded P450s and CPR in the liver (99). The construction of a chimeric protein will hamper the flexibility of modulating P450/CPR ratios. These shortcomings were circumvented during the heterologous production of oxygenated taxanes with engineered Taxus cuspidata P450 CYP725A4 and its native CPR in Escherichia coli. By optimizing the relative expression level of the CPR, physically unlinked to CYP725A4, the optimal ratio of P450 to CPR was shown to be ∼12 (100) (Table 1 and Fig. 3, compound 25). This information may be useful in further studies on the efficient redox partner engineering system of eukaryotic P450s in E. coli in vivo.
A change of redox partners may not only influence catalytic efficiency and product distribution (12) but also affect the type and selectivity of a P450 reaction (101). For example, the multifunctional P450 MycG interacted with a free form of the reductase domain RhFRED or the engineered Rhodococcus-spinach hybrid reductase RhFRED-Fdx, supporting unnatural reactions leading to the production of seven novel demethylated mycinamicin products (in addition to the physiological hydroxylation/epoxidation reactions), which were not observed with either the chimeric fusion MycG-RhFRED or the spinach Fdx/FdR- supported reaction (101, 102) (Table 1 and Fig. 3, compounds 26 and 27). Of particular importance, these findings highlight the potential role of redox partners in modulating the function of P450 enzymes and also suggest that P450 enzymes could be made even more versatile through interaction with a variety of redox partners to gain alternative functionalities.
Substrate engineering
Limited substrate scope is a general problem with biocatalysts. The eukaryotic Class II P450s, with high substrate promiscuity, are generally not particularly suitable for synthetic and biotechnological applications due to their membrane-bound nature. To expand the substrate repertoire of prokaryotic soluble P450s, the strategy of “substrate engineering” has been practiced more often in recent years (14, 103, 104).
Typical substrate engineering is aimed toward modification of a nonnative substrate by covalently linking an anchoring/directing group to enable the productive binding of the engineered substrate. Some pioneering work on P450 substrate engineering involved PikC, based on extensive structural studies (65). The hydrogen bond network and strong ionic interactions between the desosamine moiety (a common 2-deoxy sugar in the two native PikC substrates YC-17 and narbomycin) and several residues in the P450 BC loop and FG helices were identified as key determinants in substrate recognition (80). Thus, a series of substrates was chemically engineered to contain the desosamine anchoring group, and selective C–H bond hydroxylation of a series of unnatural carbolide substrates was achieved and mechanistically interpreted (105). The regio-selectivity of PikC hydroxylation was further probed by testing the chemically modified YC-17 analogs with varied synthetic anchoring groups. As a result, the regioselectivity of PikC could be changed significantly (106). Furthermore, PikCD50NRhFRED (a superior self-sufficient PikC mutant) was utilized to catalyze oxidation of nonactivated methylene C–H bonds of small nonnative substrates with further simplified synthetic anchors containing a dimethyl amino group (e.g. menthol and several bicyclic and bridged bicyclic compounds) (107). A substrate engineering approach was also successfully applied to the major drug-metabolizing human P450 CYP3A4 toward theobromine analogues (108), CYP2E1 toward nicotinate esters (109), and P450BM3 on mono- and polysaccharides, with predictable control of the regio- and stereoselectivity (110).
Recently, based on the understanding of the structural basis for substrate recognition in 4-cresol biodegradation by Corynebacterium glutamicum P450 CreJ, the biocatalytically installed phosphate group (attached by a ATP-dependent two-subunit phosphatase CreHI) was harnessed as an anchoring/directing group to deliver a group of p- and m-alkyphenols into the active site of P450 CreJ, achieving the highly challenging selective oxidation of the aliphatic C–H bonds of the tested alkylphenols in a controlled manner (32). This biosynthetic approach, without any chemical modification steps, may find useful applications in the pharmaceutical, biomanufacturing, and environmental remediation industries.
Distinct from typical substrate engineering using chemically or biologically modified substrates, Watanabe and his associates have systematically developed an atypical substrate engineering strategy, “decoy” substrate engineering, in which an inactive “dummy” substrate (decoy molecule) is used to trigger the P450-catalyzed reaction on the real nonnative substrate (103, 104, 111). Notably, there is no covalent linkage between the decoy and real substrates. A decoy molecule has a similar chemical structure to native substrate, so that it can be recognized and accommodated by the P450 enzyme, and its binding can reshape the substrate-binding pocket for the binding of a nonnative substrate, which can then be oxidized more efficiently. The first generation of decoy substrates for P450BSβ (CYP152A1) were short-chain fatty acids (112), followed by different types of perfluorinated fatty acids bearing shorter alkyl chains (113), N-perfluoroacyl amino acids (114), and nonfluorinated N-acyl amino acids (115) for P450BM3. Four generations of decoy molecules have been developed, not only for expanding the substrate capabilities of P450s but also for exploring the stereoselectivity and enantioselectivity toward various substrates (e.g. styrene and ethylbenzene), leading to diverse chemical scaffolds that can be applied in the pharmaceutical industry (116).
Recently, a class of dual-functional small molecules containing an anchoring group for binding to the P450 and a basic group for H2O2 activation was elegantly designed and successfully transformed the P450BM3 monooxygenase into a peroxygenase. N-(ω-Imidazolyl)-hexanoyl-l-phenylalanine (Im-C6-Phe) was the optimal co-catalyst supporting the P450BM3-H2O2 system (117). The rate of epoxidation of (R)-(+)-styrene and the enantiomeric specificity (ee value) of the product were dramatically increased (to ee 91%) by this innovative substrate engineering approach. This engineered peroxide-driven P450BM3 system was further utilized to hydroxylate small alkanes with the assistance of Im-C6-Phe (118) (Fig. 3, compound 28).
As an alternative strategy to protein engineering, the observed exquisite specificity and selectivity introduced by substrate engineering of P450 enzymes has highlighted the profound influence of the substrate-anchoring groups on the functional plasticity of P450s (103). Thus, this strategy has the potential to improve the synthetic utility of P450s. For example, it could be used for building a library of chemical structures that bear hydroxyl groups at various positions as functional group handles for further synthetic transformations (e.g. attachment of sugars).
Electron source engineering
Almost all natural P450s are cofactor-dependent enzymes, which are often expensive and must be recycled or circumvented from a process engineering perspective. To resolve this problem, several methods have been established on a laboratory scale over several decades, including cofactor regeneration systems, peroxide replacement, electrochemical approaches, and light-activated systems (14, 119, 120).
Cofactor engineering
NAD(P)H regeneration is a popular method in cell-free biocatalysis and biotransformation, in which constant supply of reducing equivalents is achieved by introducing a second reaction system to reduce NAD(P)+. Many cost-effective approaches have been widely developed in industry not only for P450s but also for many NAD(P)H-dependent oxidoreductases, including glucose dehydrogenase/glucose (121), glucose-6-phosphate dehydrogenase/glucose 6-phosphate (122), isocitrate dehydrogenase/isocitrate (123), formate dehydrogenase/formate (124), ethanol dehydrogenase/ethanol (125), and engineered phosphite dehydrogenase/phosphite (126).
The “peroxide shunt pathway” (Fig. 1) has also been successfully engineered through directed evolution because it could be industrially relevant in making P450s use the cheaper peroxides (e.g. H2O2) rather than NAD(P)H as the electron donor (38). For instance, an efficient H2O2 regeneration system was recently applied to the catalytic reaction of a P450 peroxygenase by coupling with an oxidase, as demonstrated by an enzyme cascade comprised of the P450 peroxygenase P450CLA or P450Spα and the enantioselective α-hydroxyacid oxidase (S)-α-HAO from Aerococcus viridans or d-lactate oxidase GO-LOX from Gluconobacter oxydans. This enzyme cascade efficiently converted fatty acids of various chain length (C6:0 to C10:0) into the chemical intermediate α-ketoacids in the presence of an internal H2O2-recycling system (127). Moreover, a novel P450 monooxygenase-peroxygenase cascade consisting of P450BM3 and OleTJE was recently developed for asymmetric catalysis in the conversion of 3-phenylpropionic acid to (R)-phenyl glycol without an external supply of H2O2 (128).
Electrochemical approaches
Electrochemical reductions have been used to circumvent the requirement for redox partners in shuttling electrons from NADPH to P450, with the electrode being the source of reducing equivalents. Progress with electrode-adsorbed/immobilization of P450 enzymes on various electrodes has been accomplished by engineering of both electrodes and enzymes, including layer-by-layer films with polyions (129, 130), a cobalt(III) sepulchrate (Zn/CoIIIsep) mediator (131, 132), covalent immobilization to a gold (Au) self-assembled monolayer (133), and nanomaterial-modified electrodes (134, 135). Due to the limitations of applying purified soluble P450s on various electrodes, protein film electrochemistry has been considered in electrocatalytic studies. Some of the studies include engineered membrane-bound human P450s with the reductase protein CPR added to a modified gold (Au) electrode (136, 137), (membrane-bound) liver microsomes with rat and human P450s immobilized on carbon electrodes and carbon nanostructures (138–140), and purified P450s assembled with membrane-bound CPR on pyrolytic graphite electrodes (141).
Light-activated systems
Systems have been developed by utilizing energy from light to drive the P450 catalytic cycle. Three main pathways have been designed based on the catalytic nature of P450 enzymes. The first takes advantage of the peroxide shunt pathway, with controlled generation of the reactive oxygen species in situ, mainly limited to the CYP152 P450 family with peroxygenase activity (e.g. P450BSβ, CYPC1a, and OleTJE) (142, 143). The second approach mimics the native electron transfer pathway by employment of redox partners to transfer electrons from a photosensitizer instead of cofactor, exemplified by a deazaflavin-dependent photoregeneration system (144) and photosystem I with ferredoxin as an electron mediator (145–148). The third simply involves direct shuttling of electrons to the heme active site and circumvention of redox partners by the employment of a fluorescent dye, eosin Y (149, 150), and covalently attached Ru(II)-diimine complexes (151, 152). However, there are still many challenges for photobased strategies in practical P450 catalysis, including the low efficiency of light conversion, weak coupling efficiency, low protein stability and activity, and technical difficulties.
Readers are referred to details in the individual research and reviewed publications regarding enzymatic regeneration systems (153), reactive oxygen species (154), electrochemical reduction (155), and light-activated approaches (156, 157). It is fairly important to find alternative economical electron sources for development of sustainable P450 catalytic systems to reduce the production costs. Except for the NAD(P)H- and H2O2-regenerating systems that have found practical applications in industry, other strategies are still in the developmental phase due to several critical problems (e.g. low coupling efficiency and redox potential management). Advances in biotechnology, discovery, and design of novel catalytic units and more interdisciplinary approaches may help overcome these challenges.
P450-related metabolic engineering
Rapid development of synthetic biology has led to more and more P450-related metabolic engineering work that has integrated protein, substrate, and cofactor engineering of P450 systems. These efforts have enabled cost-effective bioproduction of many commercial compounds as “natural” products for variant purposes.
As some of the most important enzymes in natural product biosynthesis, numerous natural and engineered P450s have already been included in the metabolic engineering toolbox. For example, the aforementioned CYP71AV1 has been used for bioproduction of the antimalarial drug precursor artemisinic acid (49), and CYP75 enzymes have been used for the hydroxylation on the B-ring of anthocyanidins to produce commercial blue roses and carnations (158). However, in both cases P450-related metabolic engineering has not been a straightforward process. In addition to a robust P450 with the desired activity, other requirements include high-level heterologous expression, optimization of metabolic fluxes, choice of a suitable heterologous host, and the deletion or silencing of competing pathways.
P450 SalSyn from Papaver somniferum is a key element in the complete biosynthesis of two opioid drugs in S. cerevisiae. Expression of an engineered P450 SalSyn with increased activity in generating the pro-morphinan scaffold (salutaridine) (Table 1 and Fig. 3, compound 29), together with co-expression of 19 of 21 heterologous enzymes and two native enzymes and deletion of one native yeast gene, resulted in the microbial production of thebaine/hydrocodone (159). The critical bioconversion of (S)-reticuline to (R)-reticuline was mediated by a CYP82Y2-like P450 (1,2-dehydroreticuline synthase) from Papaver bracteatum fused with 1,2-dehydroreticuline reductase, which was achieved by complementary approaches including gene mining, protein mutagenesis, codon optimization, and heterologous expression in yeast (159) (Table 1 and Fig. 3, compound 30).
Szczebara et al. (160) fully designed a de novo biosynthetic pathway involving 13 engineered enzymes in recombinant S. cerevisiae strains, in which the total biosynthesis of hydrocortisol and several steroids was achieved. First, recombinant S. cerevisiae was engineered to overproduce egrosta-5-eneol and ergosta-5,22-dieneol, which was further converted into pregnenolone by CYP11A1 (Table 1, Fig. 3, compound 31). Finally, the oxidation steps that are sequentially catalyzed by 3β-hydroxysteroid dehydrogenase/isomerase, CYP17A1, CYP21A1, and CYP11B1 were reconstituted, giving rise to the production of progesterone, 17-hydroxyprogesterone, 11-deoxycortisol, and the final product hydrocortisol (160) (Table 1, compounds 32–34).
Optimization of redox partners in vivo is also important in P450-related metabolic engineering. Huang and co-workers (161) reconstituted the catalytic activity of CYP76AH1 in the bioconversion of miltiradiene to ferruginol, a key bioactive component of the Chinese medicinal plant Salvia miltiorrhiza, in a miltiradiene-overproducing yeast strain. The production of 10.5 mg of ferruginol per liter was enabled with a surrogate redox partner protein, smCPR1, from Salvia miltiorrhiza Bungefor (Table 1 and Fig. 3, compound 35). Zhao et al. (162) designed an artificial biosynthetic pathway of protopanaxadiol (Table 1 and Fig. 3, compound 36), the precursor of bioactive ginsenosides of Panax ginseng, in an engineered S. cerevisiae strain. The self-sufficient P450 protopanaxadiol synthase was constructed by fusing it with an AtCPR from Arabidopsis thaliana, which resulted in a 71% increase in protopanaxadiol production (>1400 mg/liter) compared with co-expression of the two stand-alone components, protopanaxadiol synthase and AtCPR (162) (Table 1 and Fig. 3, compound 36).
Distinct from the de novo biosynthesis of high value-added compounds in recombinant cells from sugar sources, engineering a P450 system into a robust whole-cell biocatalyst is also a useful strategy. For instance, when P450 boxA from Streptomyces sp. TM-7 was introduced into an efflux pump inactivation mutant of E. coli, the production of 1.7 g of pravastatin per liter (from compactin) was achieved, which was 7-fold higher than that using WT E. coli (163) (Table 1). When this system was expressed in the pravastatin-tolerant actinomycetes strain P. autotrophica, accumulation of pravastatin reached a level of 14 g/liter, 8-fold higher than in its E. coli counterpart and 3-fold higher than in the original Streptomyces sp. TM-7 (164). These results indicate the importance of a suitable heterologous host for construction of robust whole-cell biocatalysts.
Cofactor regeneration and cofactor-free P450 systems have also found applications in whole-cell biocatalysts. Watanabe and associates developed E. coli as a whole-cell biocatalyst vehicle to mediate the hydroxylation of benzene into phenol by WT P450BM3 in the presence of decoy molecules (165). A novel whole-cell P450 photobiocatalysis system driven by the electrons from eosin Y instead of redox partners and cofactors was used for the bioconversion of pharmaceuticals with engineered bacterial P450s and human P450s (150). Different cofactor regeneration systems were also applied in many cases of whole-cell biotransformation, such as CYPsb-21 (45), P450 SMO from Rhodococcus sp. (166), and CYP106A2 (PDB entry 4YT3) from B. megaterium ATCC 13368 (167) (Table 1).
Conclusions and future prospects
Compared with some robust and widely applied commercial enzymes (e.g. hydrolases and ligases), P450 biocatalysts are still very limited by practical disadvantages, including low activity, poor stability, narrow substrate scope, and cofactor and redox partner dependence for most P450s. However, the irresistible regio- and stereoselectivity inherent in P450s continues to attract extensive efforts to deliver more P450 systems for industrial applications in production of pharmaceuticals, fine chemicals, flavors, and fragrances.
Exciting new biotechnology approaches have contributed to breakthroughs in P450 system engineering for practical catalysis in the past decade (14, 16, 42, 103). The multiple engineering strategies mentioned in this review have significantly improved the substrate scope, stability (60), catalytic efficiency (29), and reaction specificity (54) of P450s (Fig. 4). Moreover, P450-related metabolic engineering has opened a door for industrial application of the low-stability P450 systems (14, 61). A very recent development is the application of mammalian P450s selected by mining sequences of their relatives and prediction of primordial precursors, which unexpectedly has yielded more thermostable catalysts. These have broad specificity and can be used to generate useful products at much higher temperatures, increasing their efficiency (168). Lately, the integration of P450 catalytic systems into multienzyme cascades has been shown to be useful (128).
Versatile P450s are vital elements in the enzyme toolbox gifted from nature, and they will become much more powerful in the era of synthetic biology. In the future, we envision that functional mining of new P450s, construction of systematic libraries of P450s and redox partners, design of new electron-sourcing systems, the development of stable and highly efficient redox partner–independent P450 systems, and perhaps even the de novo design of P450s on demand will be the frontiers of P450 system engineering. Close collaboration between biologists, chemists, physicists, engineers, computer scientists, and mathematicians will be needed for engineering future new-concept P450 systems, which can create new exciting opportunities in practical catalysis for this most versatile superfamily of enzymes.
Acknowledgment
We thank Dr. Stella A. Child for comments on the manuscript.
This work was supported by National Key R&D Program of China Grant 2019YFA0905400-04 (to W. Z.), National Natural Science Foundation of China Grants 31570030 (to W. Z.), 31600045 (to L. M.), and 31872729 (to S. L.); National Institutes of Health Grant R01 GM118122 (to F. P. G.); a Qilu Youth Scholar Startup Funding of Shandong University grant (to W. Z.); and the State Key Laboratory of Bio-organic and Natural Products Chemistry, CAS, Grant SKLBNPC18242 (to W. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- P450 or CYP
- cytochrome P450
- FdR
- ferredoxin reductase
- Fdx
- ferredoxin
- Adx
- adrenodoxin
- AdR
- adrenodoxin reductase
- CPR
- cytochrome P450 reductase
- Cpd 0
- I, and II, compound 0, I, and II
- epPCR
- error-prone polymerase chain reaction
- 1α,25(OH)2D3
- 1α,25-dihydroxyvitamin D3
- PDB
- Protein Data Bank
- SRS
- substrate recognition site.
References
- 1. Guengerich F. P. (2018) Mechanisms of cytochrome P450-catalyzed oxidations. ACS Catal. 8, 10964–10976 10.1021/acscatal.8b03401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meunier B., de Visser S. P., and Shaik S. (2004) Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem. Rev. 104, 3947–3980 10.1021/cr020443g [DOI] [PubMed] [Google Scholar]
- 3. Klingenberg M. (1958) Pigments of rat liver microsomes. Arch. Biochem. Biophys. 75, 376–386 10.1016/0003-9861(58)90436-3 [DOI] [PubMed] [Google Scholar]
- 4. Nelson D. R. (2018) Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 1866, 141–154 10.1016/j.bbapap.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang X., and Li S. (2017) Expansion of chemical space for natural products by uncommon P450 reactions. Nat. Prod. Rep. 34, 1061–1089 10.1039/C7NP00028F [DOI] [PubMed] [Google Scholar]
- 6. Guengerich F. P. (2001) Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 14, 611–650 10.1021/tx0002583 [DOI] [PubMed] [Google Scholar]
- 7. Coon M. J. (2005) Cytochrome P450: nature's most versatile biological catalyst. Annu. Rev. Pharmacol. Toxicol. 45, 1–25 10.1146/annurev.pharmtox.45.120403.100030 [DOI] [PubMed] [Google Scholar]
- 8. Guengerich F. P., and Munro A. W. (2013) Unusual cytochrome P450 enzymes and reactions. J. Biol. Chem. 288, 17065–17073 10.1074/jbc.R113.462275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Podust L. M., and Sherman D. H. (2012) Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 29, 1251–1266 10.1039/c2np20020a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rudolf J. D., Chang C. Y., Ma M., and Shen B. (2017) Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function. Nat. Prod. Rep. 34, 1141–1172 10.1039/C7NP00034K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sakaki T. (2012) Practical application of cytochrome P450. Biol. Pharm. Bull. 35, 844–849 10.1248/bpb.35.844 [DOI] [PubMed] [Google Scholar]
- 12. Bernhardt R., and Urlacher V. B. (2014) Cytochromes P450 as promising catalysts for biotechnological application: chances and limitations. Appl. Microbiol. Biotechnol. 98, 6185–6203 10.1007/s00253-014-5767-7 [DOI] [PubMed] [Google Scholar]
- 13. Arnold F. H. (1998) Design by directed evolution. Acc. Chem. Res. 31, 125–131 10.1021/ar960017f [DOI] [Google Scholar]
- 14. Urlacher V. B., and Girhard M. (2019) Cytochrome P450 monooxygenases in biotechnology and synthetic biology. Trends Biotechnol. 37, 882–897 10.1016/j.tibtech.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 15. Xu L. H., and Du Y. L. (2018) Rational and semi-rational engineering of cytochrome P450s for biotechnological applications. Synth. Syst. Biotechnol. 3, 283–290 10.1016/j.synbio.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei Y., Ang E. L., and Zhao H. (2018) Recent developments in the application of P450 based biocatalysts. Curr. Opin. Chem. Biol. 43, 1–7 10.1016/j.cbpa.2017.08.006 [DOI] [PubMed] [Google Scholar]
- 17. Schmitz L. M., Rosenthal K., and Lütz S. (2019) Recent advances in heme biocatalysis engineering. Biotechnol. Bioeng. 116, 3469–3475 10.1002/bit.27156 [DOI] [PubMed] [Google Scholar]
- 18. Jiang Y., and Li S. (2018) Catalytic function and application of cytochrome P450 enzymes in biosynthesis and organic synthesis. Chinese J. Org. Chem. 38, 2307–2323 10.6023/cjoc201805055 [DOI] [Google Scholar]
- 19. Matthews S., Belcher J. D., Tee K. L., Girvan H. M., McLean K. J., Rigby S. E., Levy C. W., Leys D., Parker D. A., Blankley R. T., and Munro A. W. (2017) Catalytic determinants of alkene production by the cytochrome P450 peroxygenase OleTJE. J. Biol. Chem. 292, 5128–5143 10.1074/jbc.M116.762336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sevrioukova I. F., and Poulos T. L. (2011) Structural biology of redox partner interactions in P450cam monooxygenase: a fresh look at an old system. Arch. Biochem. Biophys. 507, 66–74 10.1016/j.abb.2010.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kido T., and Kimura T. (1979) The formation of binary and ternary complexes of cytochrome P-450scc with adrenodoxin and adrenodoxin reductase·adrenodoxin complex: the implication in ACTH function. J. Biol. Chem. 254, 11806–11815 [PubMed] [Google Scholar]
- 22. Whitehouse C. J. C., Bell S. G., and Wong L.-L. (2012) P450BM3(CYP102A1): connecting the dots. Chem. Soc. Rev. 41, 1218–1260 10.1039/C1CS15192D [DOI] [PubMed] [Google Scholar]
- 23. Roberts G. A., Celik A., Hunter D. J., Ost T. W., White J. H., Chapman S. K., Turner N. J., and Flitsch S. L. (2003) A self-sufficient cytochrome P450 with a primary structural organization that includes a flavin domain and a [2Fe-2S] redox center. J. Biol. Chem. 278, 48914–48920 10.1074/jbc.M309630200 [DOI] [PubMed] [Google Scholar]
- 24. Daiber A., Shoun H., and Ullrich V. (2005) Nitric oxide reductase (P450nor) from Fusarium oxysporum. J. Inorg. Biochem. 99, 185–193 10.1016/j.jinorgbio.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 25. Hsu P.-Y., Tsai A.-L., Kulmacz R. J., and Wang L.-H. (1999) Expression, purification, and spectroscopic characterization of human thromboxane synthase. J. Biol. Chem. 274, 762–769 10.1074/jbc.274.2.762 [DOI] [PubMed] [Google Scholar]
- 26. Munro A. W., Girvan H. M., and McLean K. J. (2007) Cytochrome P450-redox partner fusion enzymes. Biochim. Biophys. Acta 1770, 345–359 10.1016/j.bbagen.2006.08.018 [DOI] [PubMed] [Google Scholar]
- 27. Hannemann F., Bichet A., Ewen K. M., and Bernhardt R. (2007) Cytochrome P450 systems—biological variations of electron transport chains. Biochim. Biophys. Acta 1770, 330–344 10.1016/j.bbagen.2006.07.017 [DOI] [PubMed] [Google Scholar]
- 28. Paddon C. J., Westfall P. J., Pitera D. J., Benjamin K., Fisher K., McPhee D., Leavell M. D., Tai A., Main A., Eng D., Polichuk D. R., Teoh K. H., Reed D. W., Treynor T., Lenihan J., et al. (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532 10.1038/nature12051 [DOI] [PubMed] [Google Scholar]
- 29. McLean K. J., Hans M., Meijrink B., van Scheppingen W. B., Vollebregt A., Tee K. L., van der Laan J. M., Leys D., Munro A. W., and van den Berg M. A. (2015) Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum. Proc. Natl. Acad. Sci. U.S.A. 112, 2847–2852 10.1073/pnas.1419028112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bistolas N., Wollenberger U., Jung C., and Scheller F. W. (2005) Cytochrome P450 biosensors—a review. Biosens. Bioelectron. 20, 2408–2423 10.1016/j.bios.2004.11.023 [DOI] [PubMed] [Google Scholar]
- 31. Lowell A. N., DeMars M. D. 2nd, Slocum S. T., Yu F., Anand K., Chemler J. A., Korakavi N., Priessnitz J. K., Park S. R., Koch A. A., Schultz P. J., and Sherman D. H. (2017) Chemoenzymatic total synthesis and structural diversification of tylactone-based macrolide antibiotics through late-stage polyketide assembly, tailoring, and C–H functionalization. J. Am. Chem. Soc. 139, 7913–7920 10.1021/jacs.7b02875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Du L., Dong S., Zhang X., Jiang C., Chen J., Yao L., Wang X., Wan X., Liu X., Wang X., Huang S., Cui Q., Feng Y., Liu S. J., and Li S. (2017) Selective oxidation of aliphatic C–H bonds in alkylphenols by a chemomimetic biocatalytic system. Proc. Natl. Acad. Sci. U.S.A. 114, E5129–E5137 10.1073/pnas.1702317114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minas W., Brünker P., Kallio P. T., and Bailey J. E. (1998) Improved erythromycin production in a genetically engineered industrial strain of Saccharopolyspora erythraea. Biotechnol. Prog. 14, 561–566 10.1021/bp980055t [DOI] [PubMed] [Google Scholar]
- 34. Bate N., and Cundliffe E. (1999) The mycinose-biosynthetic genes of Streptomyces fradiae, producer of tylosin. J. Ind. Microbiol. Biotechnol. 23, 118–122 10.1038/sj.jim.2900707 [DOI] [PubMed] [Google Scholar]
- 35. Barriuso J., Nguyen D. T., Li J. W., Roberts J. N., MacNevin G., Chaytor J. L., Marcus S. L., Vederas J. C., and Ro D. K. (2011) Double oxidation of the cyclic nonaketide dihydromonacolin L to monacolin J by a single cytochrome P450 monooxygenase, LovA. J. Am. Chem. Soc. 133, 8078–8081 10.1021/ja201138v [DOI] [PubMed] [Google Scholar]
- 36. Huang X., Tang S., Zheng L., Teng Y., Yang Y., Zhu J., and Lu X. (2019) Construction of an efficient and robust Aspergillus terreus cell factory for monacolin J production. ACS Synth. Biol. 8, 818–825 10.1021/acssynbio.8b00489 [DOI] [PubMed] [Google Scholar]
- 37. Sulistyaningdyah W. T., Ogawa J., Li Q. S., Maeda C., Yano Y., Schmid R. D., and Shimizu S. (2005) Hydroxylation activity of P450 BM-3 mutant F87V towards aromatic compounds and its application to the synthesis of hydroquinone derivatives from phenolic compounds. Appl. Microbiol. Biotechnol. 67, 556–562 10.1007/s00253-004-1761-9 [DOI] [PubMed] [Google Scholar]
- 38. Du L., Ma L., Qi F., Zheng X., Jiang C., Li A., Wan X., Liu S. J., and Li S. (2016) Characterization of a unique pathway for 4-cresol catabolism initiated by phosphorylation in Corynebacterium glutamicum. J. Biol. Chem. 291, 6583–6594 10.1074/jbc.M115.695320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Otey C. R., Bandara G., Lalonde J., Takahashi K., and Arnold F. H. (2006) Preparation of human metabolites of propranolol using laboratory-evolved bacterial cytochromes P450. Biotechnol. Bioeng. 93, 494–499 10.1002/bit.20744 [DOI] [PubMed] [Google Scholar]
- 40. Parikh A., Gillam E. M., and Guengerich F. P. (1997) Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat. Biotechnol. 15, 784–788 10.1038/nbt0897-784 [DOI] [PubMed] [Google Scholar]
- 41. Hosobuchi M., Kurosawa K., and Yoshikawa H. (1993) Application of computer to monitoring and control of fermentation process: microbial conversion of ML-236B Na to pravastatin. Biotechnol. Bioeng. 42, 815–820 10.1002/bit.260420705 [DOI] [PubMed] [Google Scholar]
- 42. Yasuda K., Sugimoto H., Hayashi K., Takita T., Yasukawa K., Ohta M., Kamakura M., Ikushiro S., Shiro Y., and Sakaki T. (2018) Protein engineering of CYP105s for their industrial uses. Biochim. Biophys. Acta Proteins Proteom. 1866, 23–31 10.1016/j.bbapap.2017.05.014 [DOI] [PubMed] [Google Scholar]
- 43. Suzuki K., Sanga K., Chikaoka Y., and Itagaki E. (1993) Purification and properties of cytochrome P-450 (P-450lun) catalyzing steroid 11β-hydroxylation in Curvularia lunata. Biochim. Biophys. Acta 1203, 215–223 10.1016/0167-4838(93)90086-7 [DOI] [PubMed] [Google Scholar]
- 44. Kawauchi H., Sasaki J., Adachi T., Hanada K., Beppu T., and Horinouchi S. (1994) Cloning and nucleotide sequence of a bacterial cytochrome P-450VD25 gene encoding vitamin D-3 25-hydroxylase. Biochim. Biophys. Acta 1219, 179–183 10.1016/0167-4781(94)90266-6 [DOI] [PubMed] [Google Scholar]
- 45. Ma L., Du L., Chen H., Sun Y., Huang S., Zheng X., Kim E. S., and Li S. (2015) Reconstitution of the in vitro activity of the cyclosporine-specific P450 hydroxylase from Sebekia benihana and development of a heterologous whole-cell biotransformation system. Appl. Environ. Microbiol. 81, 6268–6275 10.1128/AEM.01353-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ban J. G., Woo M. W., Lee B. R., Lee M. J., Choi S. S., and Kim E. S. (2014) A novel regiospecific cyclosporin hydroxylase gene revealed through the genome mining of Pseudonocardia autotrophica. J. Ind. Microbiol. Biotechnol. 41, 879–886 10.1007/s10295-014-1432-5 [DOI] [PubMed] [Google Scholar]
- 47. Sun Y., Ma L., Han D., Du L., Qi F., Zhang W., Sun J., Huang S., Kim E. S., and Li S. (2017) In vitro reconstitution of the cyclosporine specific P450 hydroxylases using heterologous redox partner proteins. J. Ind. Microbiol. Biotechnol. 44, 161–166 10.1007/s10295-016-1875-y [DOI] [PubMed] [Google Scholar]
- 48. Lee M.-J., Kim H.-B., Yoon Y. J., Han K., and Kim E.-S. (2013) Identification of a cyclosporine-specific P450 hydroxylase gene through targeted cytochrome P450 complement (CYPome) disruption in Sebekia benihana. Appl. Environ. Microbiol. 79, 2253–2262 10.1128/AEM.03722-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ro D. K., Paradise E. M., Ouellet M., Fisher K. J., Newman K. L., Ndungu J. M., Ho K. A., Eachus R. A., Ham T. S., Kirby J., Chang M. C., Withers S. T., Shiba Y., Sarpong R., and Keasling J. D. (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943 10.1038/nature04640 [DOI] [PubMed] [Google Scholar]
- 50. Sowden R. J., Yasmin S., Rees N. H., Bell S. G., and Wong L.-L. (2005) Biotransformation of the sesquiterpene (+)-valencene by cytochrome P450cam and P450BM-3. Org. Biomol. Chem. 3, 57–64 10.1039/b413068e [DOI] [PubMed] [Google Scholar]
- 51. Woodley J. M. (2013) Protein engineering of enzymes for process applications. Curr. Opin. Chem. Biol. 17, 310–316 10.1016/j.cbpa.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 52. Turner N. J. (2009) Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 5, 567–573 10.1038/nchembio.203 [DOI] [PubMed] [Google Scholar]
- 53. Poulos T. L., Finzel B. C., and Howard A. J. (1987) High-resolution crystal structure of cytochrome P450cam. J. Mol. Biol. 195, 687–700 10.1016/0022-2836(87)90190-2 [DOI] [PubMed] [Google Scholar]
- 54. Kille S., Zilly F. E., Acevedo J. P., and Reetz M. T. (2011) Regio- and stereoselectivity of P450-catalysed hydroxylation of steroids controlled by laboratory evolution. Nat. Chem. 3, 738–743 10.1038/nchem.1113 [DOI] [PubMed] [Google Scholar]
- 55. Fujii Y., Kabumoto H., Nishimura K., Fujii T., Yanai S., Takeda K., Tamura N., Arisawa A., and Tamura T. (2009) Purification, characterization, and directed evolution study of a vitamin D3 hydroxylase from Pseudonocardia autotrophica. Biochem. Biophys. Res. Commun. 385, 170–175 10.1016/j.bbrc.2009.05.033 [DOI] [PubMed] [Google Scholar]
- 56. Yasutake Y., Fujii Y., Nishioka T., Cheon W.-K., Arisawa A., and Tamura T. (2010) Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase. J. Biol. Chem. 285, 31193–31201 10.1074/jbc.M110.147009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sawayama A. M., Chen M. M., Kulanthaivel P., Kuo M. S., Hemmerle H., and Arnold F. H. (2009) A panel of cytochrome P450 BM3 variants to produce drug metabolites and diversify lead compounds. Chem. Eur. J. 15, 11723–11729 10.1002/chem.200900643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parikh A., Josephy P. D., and Guengerich F. P. (1999) Selection and characterization of human cytochrome P450 1A2 mutants with altered catalytic properties. Biochemistry 38, 5283–5289 10.1021/bi990142+ [DOI] [PubMed] [Google Scholar]
- 59. Nakamura K., Martin M. V., and Guengerich F. P. (2001) Random mutagenesis of human cytochrome P450 2A6 and screening with indole oxidation products. Arch. Biochem. Biophys. 395, 25–31 10.1006/abbi.2001.2569 [DOI] [PubMed] [Google Scholar]
- 60. Wong T. S., Arnold F. H., and Schwaneberg U. (2004) Laboratory evolution of cytochrome P450 BM-3 monooxygenase for organic cosolvents. Biotechnol. Bioeng. 85, 351–358 10.1002/bit.10896 [DOI] [PubMed] [Google Scholar]
- 61. Urlacher V. B., and Girhard M. (2012) Cytochrome P450 monooxygenases: an update on perspectives for synthetic application. Trends Biotechnol. 30, 26–36 10.1016/j.tibtech.2011.06.012 [DOI] [PubMed] [Google Scholar]
- 62. Sugimoto H., Shinkyo R., Hayashi K., Yoneda S., Yamada M., Kamakura M., Ikushiro S.-I., Shiro Y., and Sakaki T. (2008) Crystal structure of CYP105A1 (P450SU-1) in complex with 1α,25-dihydroxyvitamin D3. Biochemistry 47, 4017–4027 10.1021/bi7023767 [DOI] [PubMed] [Google Scholar]
- 63. Hayashi K., Sugimoto H., Shinkyo R., Yamada M., Ikeda S., Ikushiro S., Kamakura M., Shiro Y., and Sakaki T. (2008) Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1. Biochemistry 47, 11964–11972 10.1021/bi801222d [DOI] [PubMed] [Google Scholar]
- 64. Hayashi K., Yasuda K., Sugimoto H., Ikushiro S., Kamakura M., Kittaka A., Horst R. L., Chen T. C., Ohta M., Shiro Y., and Sakaki T. (2010) Three-step hydroxylation of vitamin D3 by a genetically engineered CYP105A1: enzymes and catalysis. FEBS J. 277, 3999–4009 10.1111/j.1742-4658.2010.07791.x [DOI] [PubMed] [Google Scholar]
- 65. Sherman D. H., Li S., Yermalitskaya L. V., Kim Y., Smith J. A., Waterman M. R., and Podust L. M. (2006) The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J. Biol. Chem. 281, 26289–26297 10.1074/jbc.M605478200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Muhammed M. T., and Aki-Yalcin E. (2019) Homology modeling in drug discovery: overview, current applications, and future perspectives. Chem. Biol. Drug Des. 93, 12–20 10.1111/cbdd.13388 [DOI] [PubMed] [Google Scholar]
- 67. Schwede T. (2013) Protein modeling: what happened to the “protein structure gap”? Structure 21, 1531–1540 10.1016/j.str.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ito S., Matsuoka T., Watanabe I., Kagasaki T., Serizawa N., and Hata T. (1999) Crystallization and preliminary X-ray diffraction analysis of cytochrome P450sca-2 from Streptomyces carbophilus involved in production of pravastatin sodium, a tissue-selective inhibitor of HMG-CoA reductase. Acta Crystallogr. D Biol. Crystallogr. 55, 1209–1211 10.1107/S0907444999003716 [DOI] [PubMed] [Google Scholar]
- 69. Ba L., Li P., Zhang H., Duan Y., and Lin Z. (2013) Semi-rational engineering of cytochrome P450sca-2 in a hybrid system for enhanced catalytic activity: insights into the important role of electron transfer. Biotechnol. Bioeng. 110, 2815–2825 10.1002/bit.24960 [DOI] [PubMed] [Google Scholar]
- 70. Williams P. A., Cosme J., Sridhar V., Johnson E. F., and McRee D. E. (2000) Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell 5, 121–131 10.1016/S1097-2765(00)80408-6 [DOI] [PubMed] [Google Scholar]
- 71. Kumar S., Scott E. E., Liu H., and Halpert J. R. (2003) A rational approach to re-engineer cytochrome P450 2B1 regioselectivity based on the crystal structure of cytochrome P450 2C5. J. Biol. Chem. 278, 17178–17184 10.1074/jbc.M212515200 [DOI] [PubMed] [Google Scholar]
- 72. Chun Y. J., Shimada T., Sanchez-Ponce R., Martin M. V., Lei L., Zhao B., Kelly S. L., Waterman M. R., Lamb D. C., and Guengerich F. P. (2007) Electron transport pathway for a Streptomyces cytochrome P450: cytochrome P450 105D5-catalyzed fatty acid hydroxylation in Streptomyces coelicolor A3(2). J. Biol. Chem. 282, 17486–17500 10.1074/jbc.M700863200 [DOI] [PubMed] [Google Scholar]
- 73. Khatri Y., Hannemann F., Ewen K. M., Pistorius D., Perlova O., Kagawa N., Brachmann A. O., Müller R., and Bernhardt R. (2010) The CYPome of Sorangium cellulosum So ce56 and identification of CYP109D1 as a new fatty acid hydroxylase. Chem. Biol. 17, 1295–1305 10.1016/j.chembiol.2010.10.010 [DOI] [PubMed] [Google Scholar]
- 74. Kern F., Dier T. K., Khatri Y., Ewen K. M., Jacquot J. P., Volmer D. A., and Bernhardt R. (2015) Highly efficient CYP167A1 (EpoK) dependent epothilone B formation and production of 7-ketone epothilone D as a new epothilone derivative. Sci. Rep. 5, 14881 10.1038/srep14881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McLean K. J., Luciakova D., Belcher J., Tee K. L., and Munro A. W. (2015) Biological diversity of cytochrome P450 redox partner systems. in Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome P450, pp. 299–317, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 76. Zhang W., Du L., Li F., Zhang X., Qu Z., Han L., Li Z., Sun J., Qi F., Yao Q., Sun Y., Geng C., and Li S. (2018) Mechanistic insights into interactions between bacterial class I P450 enzymes and redox partners. ACS Catal. 8, 9992–10003 10.1021/acscatal.8b02913 [DOI] [Google Scholar]
- 77. Qi F., Lei C., Li F., Zhang X., Wang J., Zhang W., Fan Z., Li W., Tang G. L., Xiao Y., Zhao G., and Li S. (2018) Deciphering the late steps of rifamycin biosynthesis. Nat. Commun. 9, 2342 10.1038/s41467-018-04772-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li Q., Ding W., Yao Z., Tu J., Wang L., Huang H., Li S., and Ju J. (2018) AbmV catalyzes tandem ether installation and hydroxylation during neoabyssomicin/abyssomicin biosynthesis. Org. Lett. 20, 4854–4857 10.1021/acs.orglett.8b01997 [DOI] [PubMed] [Google Scholar]
- 79. Sun C., Yang Z., Zhang C., Liu Z., He J., Liu Q., Zhang T., Ju J., and Ma J. (2019) Genome mining of Streptomyces atratus SCSIO ZH16: discovery of atratumycin and identification of its biosynthetic gene cluster. Org. Lett. 21, 1453–1457 10.1021/acs.orglett.9b00208 [DOI] [PubMed] [Google Scholar]
- 80. Uhlmann H., Kraft R., and Bernhardt R. (1994) C-terminal region of adrenodoxin affects its structural integrity and determines differences in its electron transfer function to cytochrome P450. J. Biol. Chem. 269, 22557–22564 [PubMed] [Google Scholar]
- 81. Müller A., Müller J. J., Muller Y. A., Uhlmann H., Bernhardt R., and Heinemann U. (1998) New aspects of electron transfer revealed by the crystal structure of a truncated bovine adrenodoxin, Adx(4–108). Structure 6, 269–280 10.1016/S0969-2126(98)00031-8 [DOI] [PubMed] [Google Scholar]
- 82. Ewen K. M., Kleser M., and Bernhardt R. (2011) Adrenodoxin: the archetype of vertebrate-type [2Fe-2S] cluster ferredoxins. Biochim. Biophys. Acta 1814, 111–125 10.1016/j.bbapap.2010.06.003 [DOI] [PubMed] [Google Scholar]
- 83. Strushkevich N., MacKenzie F., Cherkesova T., Grabovec I., Usanov S., and Park H.-W. (2011) Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc. Natl. Acad. Sci. U.S.A. 108, 10139–10143 10.1073/pnas.1019441108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tripathi S., Li H., and Poulos T. L. (2013) Structural basis for effector control and redox partner recognition in cytochrome P450. Science 340, 1227–1230 10.1126/science.1235797 [DOI] [PubMed] [Google Scholar]
- 85. Bell S. G., McMillan J. H., Yorke J. A., Kavanagh E., Johnson E. O., and Wong L. L. (2012) Tailoring an alien ferredoxin to support native-like P450 monooxygenase activity. Chem. Commun. (Camb.) 48, 11692–11694 10.1039/c2cc35968e [DOI] [PubMed] [Google Scholar]
- 86. Sadeghi S. J., and Gilardi G. (2013) Chimeric P450 enzymes: activity of artificial redox fusions driven by different reductases for biotechnological applications. Biotechnol. Appl. Biochem. 60, 102–110 10.1002/bab.1086 [DOI] [PubMed] [Google Scholar]
- 87. Li S., Podust L. M., and Sherman D. H. (2007) Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. J. Am. Chem. Soc. 129, 12940–12941 10.1021/ja075842d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sabbadin F., Hyde R., Robin A., Hilgarth E. M., Delenne M., Flitsch S., Turner N., Grogan G., and Bruce N. C. (2010) LICRED: a versatile drop-in vector for rapid generation of redox-self-sufficient cytochrome P450s. Chembiochem 11, 987–994 10.1002/cbic.201000104 [DOI] [PubMed] [Google Scholar]
- 89. Dodhia V. R., Fantuzzi A., and Gilardi G. (2006) Engineering human cytochrome P450 enzymes into catalytically self-sufficient chimeras using molecular lego. J. Biol. Inorg. Chem. 11, 903–916 10.1007/s00775-006-0144-3 [DOI] [PubMed] [Google Scholar]
- 90. Robin A., Roberts G. A., Kisch J., Sabbadin F., Grogan G., Bruce N., Turner N. J., and Flitsch S. L. (2009) Engineering and improvement of the efficiency of a chimeric [P450cam-RhFRed reductase domain] enzyme. Chem. Commun. (Camb.) 14, 2478–2480 10.1039/b901716j [DOI] [PubMed] [Google Scholar]
- 91. Robin A., Köhler V., Jones A., Ali A., Kelly P. P., O'Reilly E., Turner N. J., and Flitsch S. L. (2011) Chimeric self-sufficient P450cam-RhFRed biocatalysts with broad substrate scope. Beilstein J. Org. Chem. 7, 1494–1498 10.3762/bjoc.7.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zuo R., Zhang Y., Huguet-Tapia J. C., Mehta M., Dedic E., Bruner S. D., Loria R., and Ding Y. (2016) An artificial self-sufficient cytochrome P450 directly nitrates fluorinated tryptophan analogs with a different regio-selectivity. Biotechnol. J. 11, 624–632 10.1002/biot.201500416 [DOI] [PubMed] [Google Scholar]
- 93. Kulig J. K., Spandolf C., Hyde R., Ruzzini A. C., Eltis L. D., Grönberg G., Hayes M. A., and Grogan G. (2015) A P450 fusion library of heme domains from Rhodococcus jostii RHA1 and its evaluation for the biotransformation of drug molecules. Bioorg. Med. Chem. 23, 5603–5609 10.1016/j.bmc.2015.07.025 [DOI] [PubMed] [Google Scholar]
- 94. Liu Y., Wang C., Yan J., Zhang W., Guan W., Lu X., and Li S. (2014) Hydrogen peroxide-independent production of α-alkenes by OleTJE P450 fatty acid decarboxylase. Biotechnol. Biofuels 7, 28 10.1186/1754-6834-7-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lu C., Shen F., Wang S., Wang Y., Liu J., Bai W.-J., and Wang X. (2018) An engineered self-sufficient biocatalyst enables scalable production of linear α-olefins from carboxylic acids. ACS Catal. 8, 5794–5798 10.1021/acscatal.8b01313 [DOI] [Google Scholar]
- 96. Schückel J., Rylott E. L., Grogan G., and Bruce N. C. (2012) A gene-fusion approach to enabling plant cytochromes P450 for biocatalysis. Chembiochem 13, 2758–2763 10.1002/cbic.201200572 [DOI] [PubMed] [Google Scholar]
- 97. Fairhead M., Giannini S., Gillam E. M., and Gilardi G. (2005) Functional characterisation of an engineered multidomain human P450 2E1 by molecular lego. J. Biol. Inorg. Chem. 10, 842–853 10.1007/s00775-005-0033-1 [DOI] [PubMed] [Google Scholar]
- 98. Ciaramella A., Minerdi D., and Gilardi G. (2016) Catalytically self-sufficient cytochromes P450 for green production of fine chemicals. Rend. Lincei 28, 169–181 10.1007/s12210-016-0581-z [DOI] [Google Scholar]
- 99. Shephard E. A., Phillips I. R., Bayney R. M., Pike S. F., and Rabin B. R. (1983) Quantification of NADPH cytochrome P-450 reductase in liver microsomes by a specific radioimmunoassay technique. Biochem. J. 211, 333–340 10.1042/bj2110333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Biggs B. W., Lim C. G., Sagliani K., Shankar S., Stephanopoulos G., De Mey M., and Ajikumar P. K. (2016) Overcoming heterologous protein interdependency to optimize P450-mediated taxol precursor synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 113, 3209–3214 10.1073/pnas.1515826113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang W., Liu Y., Yan J., Cao S., Bai F., Yang Y., Huang S., Yao L., Anzai Y., Kato F., Podust L. M., Sherman D. H., and Li S. (2014) New reactions and products resulting from alternative interactions between the P450 enzyme and redox partners. J. Am. Chem. Soc. 136, 3640–3646 10.1021/ja4130302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Anzai Y., Li S., Chaulagain M. R., Kinoshita K., Kato F., Montgomery J., and Sherman D. H. (2008) Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics. Chem. Biol. 15, 950–959 10.1016/j.chembiol.2008.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xu J., Wang C., and Cong Z. (2019) Strategies for substrate-regulated P450 catalysis: from substrate engineering to co-catalysis. Chem. Eur. J. 25, 6853–6863 10.1002/chem.201806383 [DOI] [PubMed] [Google Scholar]
- 104. Shoji O., and Watanabe Y. (2015) Bringing out the potential of wild-type cytochrome P450s using decoy molecules: oxygenation of nonnative substrates by bacterial cytochrome P450s. Isr. J. Chem. 55, 32–39 10.1002/ijch.201400096 [DOI] [Google Scholar]
- 105. Li S., Chaulagain M. R., Knauff A. R., Podust L. M., Montgomery J., and Sherman D. H. (2009) Selective oxidation of carbolide C–H bonds by an engineered macrolide P450 mono-oxygenase. Proc. Natl. Acad. Sci. U.S.A. 106, 18463–18468 10.1073/pnas.0907203106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Negretti S., Narayan A. R., Chiou K. C., Kells P. M., Stachowski J. L., Hansen D. A., Podust L. M., Montgomery J., and Sherman D. H. (2014) Directing group-controlled regioselectivity in an enzymatic C–H bond oxygenation. J. Am. Chem. Soc. 136, 4901–4904 10.1021/ja5016052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Narayan A. R., Jiménez-Osés G., Liu P., Negretti S., Zhao W., Gilbert M. M., Ramabhadran R. O., Yang Y. F., Furan L. R., Li Z., Podust L. M., Montgomery J., Houk K. N., and Sherman D. H. (2015) Enzymatic hydroxylation of an unactivated methylene C–H bond guided by molecular dynamics simulations. Nat. Chem. 7, 653–660 10.1038/nchem.2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Polic V., Cheong K. J., Hammerer F., and Auclair K. (2017) Regioselective epoxidations by cytochrome P450 3A4 using a theobromine chemical auxiliary to predictably produce N-protected β- or γ-amino epoxides. Adv. Synth. Catal. 359, 3983–3989 10.1002/adsc.201700637 [DOI] [Google Scholar]
- 109. Ménard A., Fabra C., Huang Y., and Auclair K. (2012) Type II ligands as chemical auxiliaries to favor enzymatic transformations by P450 2E1. Chembiochem 13, 2527–2536 10.1002/cbic.201200524 [DOI] [PubMed] [Google Scholar]
- 110. Lewis J. C., Bastian S., Bennett C. S., Fu Y., Mitsuda Y., Chen M. M., Greenberg W. A., Wong C. H., and Arnold F. H. (2009) Chemoenzymatic elaboration of monosaccharides using engineered cytochrome P450BM3 demethylases. Proc. Natl. Acad. Sci. U.S.A. 106, 16550–16555 10.1073/pnas.0908954106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Shoji O., and Watanabe Y. (2017) Monooxygenation of nonnative substrates catalyzed by bacterial cytochrome P450s facilitated by decoy molecules. Chem. Lett. 46, 278–288 10.1246/cl.160963 [DOI] [Google Scholar]
- 112. Lee D.-S., Yamada A., Sugimoto H., Matsunaga I., Ogura H., Ichihara K., Adachi S., Park S.-Y., and Shiro Y. (2003) Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis crystallographic, spectroscopic, and mutational studies. J. Biol. Chem. 278, 9761–9767 10.1074/jbc.M211575200 [DOI] [PubMed] [Google Scholar]
- 113. Kawakami N., Shoji O., and Watanabe Y. (2011) Use of perfluorocarboxylic acids to trick cytochrome P450BM3 into initiating the hydroxylation of gaseous alkanes. Angew. Chem. Int. Ed. Engl. 50, 5315–5318 10.1002/anie.201007975 [DOI] [PubMed] [Google Scholar]
- 114. Cong Z., Shoji O., Kasai C., Kawakami N., Sugimoto H., Shiro Y., and Watanabe Y. (2014) Activation of wild-type cytochrome P450BM3 by the next generation of decoy molecules: enhanced hydroxylation of gaseous alkanes and crystallographic evidence. ACS Catal. 5, 150–156 10.1021/cs501592f [DOI] [Google Scholar]
- 115. Shoji O., Yanagisawa S., Stanfield J. K., Suzuki K., Cong Z., Sugimoto H., Shiro Y., and Watanabe Y. (2017) Direct hydroxylation of benzene to phenol by cytochrome P450BM3 triggered by amino acid derivatives. Angew. Chem. Int. Ed. Engl. 56, 10324–10329 10.1002/anie.201703461 [DOI] [PubMed] [Google Scholar]
- 116. Shoji O., Fujishiro T., Nakajima H., Kim M., Nagano S., Shiro Y., and Watanabe Y. (2007) Hydrogen peroxide dependent monooxygenations by tricking the substrate recognition of cytochrome P450BSβ. Angew. Chem. Int. Ed. Engl. 119, 3730–3733 10.1002/ange.200700068 [DOI] [PubMed] [Google Scholar]
- 117. Ma N., Chen Z., Chen J., Chen J., Wang C., Zhou H., Yao L., Shoji O., Watanabe Y., and Cong Z. (2018) Dual-functional small molecules for generating an efficient cytochrome P450BM3 peroxygenase. Angew. Chem. Int. Ed. Engl. 57, 7628–7633 10.1002/anie.201801592 [DOI] [PubMed] [Google Scholar]
- 118. Chen J., Kong F., Ma N., Zhao P., Liu C., Wang X., and Cong Z. (2019) Peroxide-driven hydroxylation of small alkanes catalyzed by an artificial P450BM3 peroxygenase system. ACS Catal. 9, 7350–7355 10.1021/acscatal.9b02507 [DOI] [Google Scholar]
- 119. Urlacher V. B., Lutz-Wahl S., and Schmid R. D. (2004) Microbial P450 enzymes in biotechnology. Appl. Microbiol. Biotechnol. 64, 317–325 10.1007/s00253-003-1514-1 [DOI] [PubMed] [Google Scholar]
- 120. Shumyantseva V. V., Bulko T. V., and Archakov A. I. (2005) Electrochemical reduction of cytochrome P450 as an approach to the construction of biosensors and bioreactors. J. Inorg. Biochem. 99, 1051–1063 10.1016/j.jinorgbio.2005.01.014 [DOI] [PubMed] [Google Scholar]
- 121. Lu Y., and Mei L. (2007) Co-expression of P450 BM3 and glucose dehydrogenase by recombinant Escherichia coli and its application in an NADPH-dependent indigo production system. J. Ind. Microbiol. Biotechnol. 34, 247–253 10.1007/s10295-006-0193-1 [DOI] [PubMed] [Google Scholar]
- 122. Ding Y., Seufert W. H., Beck Z. Q., and Sherman D. H. (2008) Analysis of the cryptophycin P450 epoxidase reveals substrate tolerance and cooperativity. J. Am. Chem. Soc. 130, 5492–5498 10.1021/ja710520q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Meinhold P., Peters M. W., Hartwick A., Hernandez A. R., and Arnold F. H. (2006) Engineering cytochrome P450 BM3 for terminal alkane hydroxylation. Adv. Synth. Catal. 348, 763–772 10.1002/adsc.200505465 [DOI] [Google Scholar]
- 124. Taylor M., Lamb D. C., Cannell R. J., Dawson M. J., and Kelly S. L. (2000) Cofactor recycling with immobilized heterologous cytochrome P450 105D1 (CYP105D1). Biochem. Biophys. Res. Commun. 279, 708–711 10.1006/bbrc.2000.4002 [DOI] [PubMed] [Google Scholar]
- 125. Sandberg M., Yasar Ü., Strömberg P., Höög J. O., and Eliasson E. (2002) Oxidation of celecoxib by polymorphic cytochrome P450 2C9 and alcohol dehydrogenase. Br. J. Clin. Pharmacol. 54, 423–429 10.1046/j.1365-2125.2002.01660.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Johannes T. W., Woodyer R. D., and Zhao H. (2007) Efficient regeneration of NADPH using an engineered phosphite dehydrogenase. Biotechnol. Bioeng. 96, 18–26 10.1002/bit.21168 [DOI] [PubMed] [Google Scholar]
- 127. Gandomkar S., Dennig A., Dordic A., Hammerer L., Pickl M., Haas T., Hall M., and Faber K. (2018) Biocatalytic oxidative cascade for the conversion of fatty acids into α-ketoacids via internal H2O2 recycling. Angew. Chem. Int. Ed. Engl. 57, 427–430 10.1002/anie.201710227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Yu D., Wang J. B., and Reetz M. T. (2019) Exploiting designed oxidase-peroxygenase mutual benefit system for asymmetric cascade reactions. J. Am. Chem. Soc. 141, 5655–5658 10.1021/jacs.9b01939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Krishnan S., Abeykoon A., Schenkman J. B., and Rusling J. F. (2009) Control of electrochemical and ferryloxy formation kinetics of cyt P450s in polyion films by heme iron spin state and secondary structure. J. Am. Chem. Soc. 131, 16215–16224 10.1021/ja9065317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Krishnan S., Schenkman J. B., and Rusling J. F. (2011) Bioelectronic delivery of electrons to cytochrome P450 enzymes. J. Phys. Chem. B 115, 8371–8380 10.1021/jp201235m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Belsare K. D., Horn T., Ruff A. J., Martinez R., Magnusson A., Holtmann D., Schrader J., and Schwaneberg U. (2017) Directed evolution of P450cin for mediated electron transfer. Protein Eng. Des. Sel. 30, 119–127 10.1093/protein/gzw072 [DOI] [PubMed] [Google Scholar]
- 132. Tosstorff A., Dennig A., Ruff A. J., Schwaneberg U., Sieber V., Mangold K.-M., Schrader J., and Holtmann D. (2014) Mediated electron transfer with monooxygenases—insight in interactions between reduced mediators and the co-substrate oxygen. J. Mol. Catal. B Enzym. 108, 51–58 10.1016/j.molcatb.2014.06.011 [DOI] [Google Scholar]
- 133. Mak L. H., Sadeghi S. J., Fantuzzi A., and Gilardi G. (2010) Control of human cytochrome P450 2E1 electrocatalytic response as a result of unique orientation on gold electrodes. Anal. Chem. 82, 5357–5362 10.1021/ac101072h [DOI] [PubMed] [Google Scholar]
- 134. Lu J., Zhang Y., Li H., Yu J., and Liu S. (2014) Electrochemically driven drug metabolism via a CYP1A2-UGT1A10 bienzyme confined in a graphene nano-cage. Chem. Commun. (Camb.) 50, 13896–13899 10.1039/C4CC06200K [DOI] [PubMed] [Google Scholar]
- 135. Lu J., Cui D., Li H., Zhang Y., and Liu S. (2015) Cytochrome P450 bienzymes assembled on Au/chitosan/reduced graphene oxide nanosheets for electrochemically-driven drug cascade metabolism. Electrochim. Acta 165, 36–44 10.1016/j.electacta.2015.02.183 [DOI] [Google Scholar]
- 136. Mie Y., Suzuki M., and Komatsu Y. (2009) Electrochemically driven drug metabolism by membranes containing human cytochrome P450. J. Am. Chem. Soc. 131, 6646–6647 10.1021/ja809364r [DOI] [PubMed] [Google Scholar]
- 137. Nerimetla R., Walgama C., Singh V., Hartson S. D., and Krishnan S. (2017) Mechanistic insights into voltage-driven biocatalysis of a cytochrome P450 bactosomal film on a self-assembled monolayer. ACS Catal. 7, 3446–3453 10.1021/acscatal.6b03588 [DOI] [Google Scholar]
- 138. Walgama C., Nerimetla R., Materer N. F., Schildkraut D., Elman J. F., and Krishnan S. (2015) A simple construction of electrochemical liver microsomal bioreactor for rapid drug metabolism and inhibition assays. Anal. Chem. 87, 4712–4718 10.1021/ac5044362 [DOI] [PubMed] [Google Scholar]
- 139. Wasalathanthri D. P., Malla S., Faria R. C., and Rusling J. F. (2012) Electrochemical activation of the natural catalytic cycle of cytochrome P450s in human liver microsomes. Electroanalysis 24, 2049–2052 10.1002/elan.201200373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Nerimetla R., and Krishnan S. (2015) Electrocatalysis by subcellular liver fractions bound to carbon nanostructures for stereoselective green drug metabolite synthesis. Chem. Commun. (Camb.) 51, 11681–11684 10.1039/C5CC03364K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Krishnan S., Wasalathanthri D., Zhao L., Schenkman J. B., and Rusling J. F. (2011) Efficient bioelectronic actuation of the natural catalytic pathway of human metabolic cytochrome P450s. J. Am. Chem. Soc. 133, 1459–1465 10.1021/ja108637s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Girhard M., Kunigk E., Tihovsky S., Shumyantseva V. V., and Urlacher V. B. (2013) Light-driven biocatalysis with cytochrome P450 peroxygenases. Biotechnol. Appl. Biochem. 60, 111–118 10.1002/bab.1063 [DOI] [PubMed] [Google Scholar]
- 143. Zachos I., Gaβmeyer S. K., Bauer D., Sieber V., Hollmann F., and Kourist R. (2015) Photobiocatalytic decarboxylation for olefin synthesis. Chem. Commun. (Camb.) 51, 1918–1921 10.1039/c4cc07276f [DOI] [PubMed] [Google Scholar]
- 144. Zilly F. E., Taglieber A., Schulz F., Hollmann F., and Reetz M. T. (2009) Deazaflavins as mediators in light-driven cytochrome P450 catalyzed hydroxylations. Chem. Commun. (Camb.), 7152–7154 10.1039/b913863c [DOI] [PubMed] [Google Scholar]
- 145. Wlodarczyk A., Gnanasekaran T., Nielsen A. Z., Zulu N. N., Mellor S. B., Luckner M., Thøfner J. F. B., Olsen C. E., Mottawie M. S., Burow M., Pribil M., Feussner I., Møller B. L., and Jensen P. E. (2016) Metabolic engineering of light-driven cytochrome P450 dependent pathways into Synechocystis sp. PCC 6803. Metab. Eng. 33, 1–11 10.1016/j.ymben.2015.10.009 [DOI] [PubMed] [Google Scholar]
- 146. Nielsen A. Z., Ziersen B., Jensen K., Lassen L. M., Olsen C. E., Møller B. L., and Jensen P. E. (2013) Redirecting photosynthetic reducing power toward bioactive natural product synthesis. ACS Synth. Biol. 2, 308–315 10.1021/sb300128r [DOI] [PubMed] [Google Scholar]
- 147. Mellor S. B., Nielsen A. Z., Burow M., Motawia M. S., Jakubauskas D., Møller B. L., and Jensen P. E. (2016) Fusion of ferredoxin and cytochrome P450 enables direct light-driven biosynthesis. ACS Chem. Biol. 11, 1862–1869 10.1021/acschembio.6b00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Jensen K., Jensen P. E., and Møller B. L. (2011) Light-driven cytochrome P450 hydroxylations. ACS Chem. Biol. 6, 533–539 10.1021/cb100393j [DOI] [PubMed] [Google Scholar]
- 149. Di Nardo G., and Gilardi G. (2012) Optimization of the bacterial cytochrome P450 BM3 system for the production of human drug metabolites. Int. J. Mol. Sci. 13, 15901–15924 10.3390/ijms131215901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Park J. H., Lee S. H., Cha G. S., Choi D. S., Nam D. H., Lee J. H., Lee J. K., Yun C. H., Jeong K. J., and Park C. B. (2015) Cofactor-free light-driven whole-cell cytochrome P450 catalysis. Angew. Chem. Int. Ed. Engl. 54, 969–973 10.1002/anie.201410059 [DOI] [PubMed] [Google Scholar]
- 151. Lam Q., Kato M., and Cheruzel L. (2016) Ru(II)-diimine functionalized metalloproteins: from electron transfer studies to light-driven biocatalysis. Biochim. Biophys. Acta 1857, 589–597 10.1016/j.bbabio.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Tran N. H., Huynh N., Chavez G., Nguyen A., Dwaraknath S., Nguyen T. A., Nguyen M., and Cheruzel L. (2012) A series of hybrid P450 BM3 enzymes with different catalytic activity in the light-initiated hydroxylation of lauric acid. J. Inorg. Biochem. 115, 50–56 10.1016/j.jinorgbio.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Liu W., and Wang P. (2007) Cofactor regeneration for sustainable enzymatic biosynthesis. Biotechnol. Adv. 25, 369–384 10.1016/j.biotechadv.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 154. Shoji O., and Watanabe Y. (2014) Peroxygenase reactions catalyzed by cytochromes P450. J. Biol. Inorg. Chem. 19, 529–539 10.1007/s00775-014-1106-9 [DOI] [PubMed] [Google Scholar]
- 155. Sadeghi S. J., Fantuzzi A., and Gilardi G. (2011) Breakthrough in P450 bioelectrochemistry and future perspectives. Biochim. Biophys. Acta 1814, 237–248 10.1016/j.bbapap.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 156. Shalan H., Kato M., and Cheruzel L. (2018) Keeping the spotlight on cytochrome P450. Biochim. Biophys. Acta Proteins Proteom. 1866, 80–87 10.1016/j.bbapap.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Mellor S. B., Vavitsas K., Nielsen A. Z., and Jensen P. E. (2017) Photosynthetic fuel for heterologous enzymes: the role of electron carrier proteins. Photosynth. Res. 134, 329–342 10.1007/s11120-017-0364-0 [DOI] [PubMed] [Google Scholar]
- 158. Tanaka Y., and Brugliera F. (2013) Flower colour and cytochromes P450. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120432 10.1098/rstb.2012.0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Galanie S., Thodey K., Trenchard I. J., Filsinger Interrante M., and Smolke C. D. (2015) Complete biosynthesis of opioids in yeast. Science 349, 1095–1100 10.1126/science.aac9373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Szczebara F. M., Chandelier C., Villeret C., Masurel A., Bourot S., Duport C., Blanchard S., Groisillier A., Testet E., and Costaglioli P., Cauet G., Degryse E., Balbuena D., Winter J., Achstetter T., et al. (2003) Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 21, 143–149 10.1038/nbt775 [DOI] [PubMed] [Google Scholar]
- 161. Guo J., Zhou Y. J., Hillwig M. L., Shen Y., Yang L., Wang Y., Zhang X., Liu W., Peters R. J., Chen X., Zhao Z. K., and Huang L. (2013) CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. U.S.A. 110, 12108–12113 10.1073/pnas.1218061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Zhao F., Bai P., Liu T., Li D., Zhang X., Lu W., and Yuan Y. (2016) Optimization of a cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae. Biotechnol. Bioeng. 113, 1787–1795 10.1002/bit.25934 [DOI] [PubMed] [Google Scholar]
- 163. Fujii T., Fujii Y., Machida K., Ochiai A., and Ito M. (2009) Efficient biotransformations using Escherichia coli with tolC acrAB mutations expressing cytochrome P450 genes. Biosci. Biotechnol. Biochem. 73, 805–810 10.1271/bbb.80627 [DOI] [PubMed] [Google Scholar]
- 164. Fujii Y., Norihisa K., Fujii T., Aritoku Y., Kagawa Y., Sallam K. I., Johdo O., Arisawa A., and Tamura T. (2011) Construction of a novel expression vector in Pseudonocardia autotrophica and its application to efficient biotransformation of compactin to pravastatin, a specific HMG-CoA reductase inhibitor. Biochem. Biophys. Res. Commun. 404, 511–516 10.1016/j.bbrc.2010.12.013 [DOI] [PubMed] [Google Scholar]
- 165. Karasawa M., Stanfield J. K., Yanagisawa S., Shoji O., and Watanabe Y. (2018) Whole-cell biotransformation of benzene to phenol catalysed by intracellular cytochrome P450BM3 activated by external additives. Angew. Chem. Int. Ed. Engl. 57, 12264–12269 10.1002/anie.201804924 [DOI] [PubMed] [Google Scholar]
- 166. Zhang J. D., Li A. T., Yu H. L., Imanaka T., and Xu J. H. (2011) Synthesis of optically pure S-sulfoxide by Escherichia coli transformant cells coexpressing the P450 monooxygenase and glucose dehydrogenase genes. J. Ind. Microbiol. Biotechnol. 38, 633–641 10.1007/s10295-010-0809-3 [DOI] [PubMed] [Google Scholar]
- 167. Janocha S., Carius Y., Hutter M., Lancaster C. R., and Bernhardt R. (2016) Crystal structure of CYP106A2 in substrate-free and substrate-bound form. Chembiochem 17, 852–860 10.1002/cbic.201500524 [DOI] [PubMed] [Google Scholar]
- 168. Gumulya Y., Baek J.-M., Wun S.-J., Thomson R. E. S., Harris K. L., Hunter D. J. B., Behrendorff J. B. Y. H., Kulig J., Zheng S., Wu X., Wu B., Stok J. E., De Voss J. J., Schenk G., Jurva U., et al. (2018) Engineering highly functional thermostable proteins using ancestral sequence reconstruction. Nat. Catal. 1, 878–888 10.1038/s41929-018-0159-5 [DOI] [Google Scholar]