Abstract
Sodium taurocholate cotransporting polypeptide (NTCP) is expressed at the surface of human hepatocytes and functions as an entry receptor of hepatitis B virus (HBV). Recently, we have reported that epidermal growth factor receptor (EGFR) is involved in NTCP-mediated viral internalization during the cell entry process. Here, we analyzed which function of EGFR is essential for mediating HBV internalization. In contrast to the reported crucial function of EGFR-downstream signaling for the entry of hepatitis C virus (HCV), blockade of EGFR-downstream signaling proteins, including mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), and signal transducer and activator of transcription (STAT), had no or only minor effects on HBV infection. Instead, deficiency of EGFR endocytosis resulting from either a deleterious mutation in EGFR or genetic knockdown of endocytosis adaptor molecules abrogated internalization of HBV via NTCP and prevented viral infection. EGFR activation triggered a time-dependent relocalization of HBV preS1 to the early and late endosomes and to lysosomes in concert with EGFR transport. Suppression of EGFR ubiquitination by site-directed mutagenesis or by knocking down two EGFR-sorting molecules, signal-transducing adaptor molecule (STAM) and lysosomal protein transmembrane 4β (LAPTM4B), suggested that EGFR transport to the late endosome is critical for efficient HBV infection. Cumulatively, these results support the idea that the EGFR endocytosis/sorting machinery drives the translocation of NTCP-bound HBV from the cell surface to the endosomal network, which eventually enables productive viral infection.
Keywords: hepatitis B virus (HBV, Hep B); epidermal growth factor receptor (EGFR); infection; endocytosis; virus entry; entry; sodium taurocholate cotransporting polypeptide (NTCP); endosomal sorting complex required for transport (ESCRT); viral translocation; receptor tyrosine kinase
Introduction
Hepatitis B virus (HBV)3 is a hepatotropic virus that specifically infects human and related species (1). The entry receptor for HBV, sodium taurocholate cotransporting polypeptide (NTCP, also known as solute carrier family 10A1 (SLC10A1)), is a liver-specific bile acid transporter expressed mainly on the cell surface (2). NTCP allows the specific attachment mediated by the preS1 region of the HBV large surface protein (LHBs) to target cells (3, 4). After HBV preS1 attachment, NTCP is thought to trigger HBV endocytosis using a yet unknown mechanism. Recently, we have reported that epidermal growth factor receptor (EGFR) initiates the internalization of NTCP-mediated viral internalization (5). Whereas NTCP supports HBV attachment to the cell surface, loss of interaction between EGFR and NTCP abrogated HBV internalization from the cell surface. However, how EGFR mediates the internalization of HBV remains to be clarified.
EGFR is a receptor tyrosine kinase that triggers signaling events essential for cellular processes, including Ras-mitogen activated protein kinase (MAPK), phosphatidylinositide 3-kinase (PI3K)-Akt, and JAK-STAT pathways (6). Upon stimulation with its ligand, epidermal growth factor (EGF), EGFR is autophosphorylated to activate these signaling pathways. In addition, activated EGFR itself is transported to the endosomal network through the endocytosis machinery, destined to be degraded in the lysosome or recycled back to the plasma membrane. To date, EGFR has been reported to play a pivotal role in the entry mechanism of different classes of viruses, such as human cytomegalovirus, hepatitis C virus (HCV), and respiratory syncytial virus (7). Typically, in such cases, virus entry essentially requires EGFR-triggered activation of the downstream signaling, including that leading to actin rearrangement necessary to the migration of internalized viral particles to the site of replication (see “Discussion”). In the present study, we analyze the function of EGFR that essentially regulates the NTCP-mediated viral entry.
Results
EGFR-downstream signaling has only a minor or no role in HBV infection
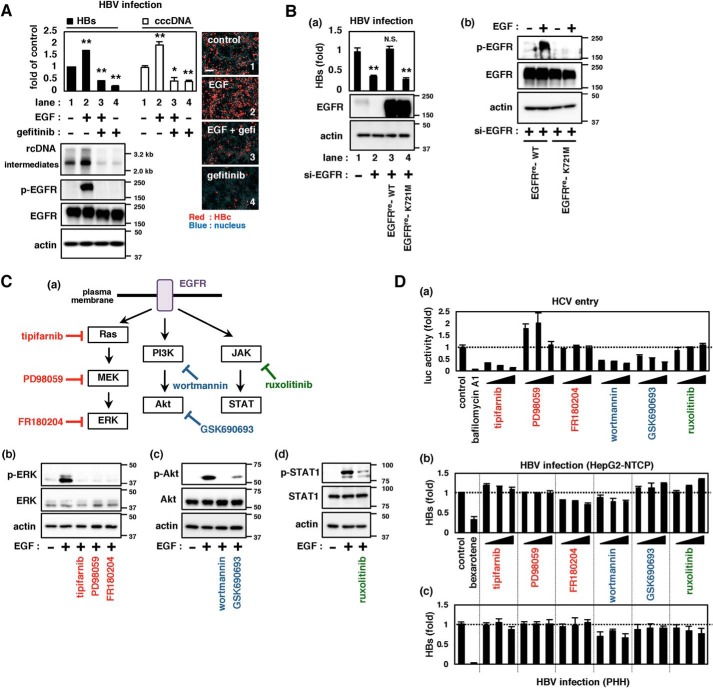
Stimulation with EGF activates EGFR through the induction of dimerization and autophosphorylation, leading to the activation of downstream signaling (6). Activation of EGFR in HepG2-NTCP cells by EGF treatment during HBV inoculation enhanced HBV infection (Fig. 1A, lane 2). In contrast, inactivation of EGFR by gefitinib, an EGFR autophosphorylation inhibitor (8), remarkably down-regulated HBV infection (Fig. 1A, lane 4) without cytotoxicity (Fig. S1A). Overexpression of WT EGFR restored HBV infection in HepG2-NTCP cells transfected with si-EGFR, whereas that with a mutation at Lys-721, an essential residue for ATP binding and EGFR phosphorylation (Fig. 1B (b)) (9, 10), had no ability to restore infection (Fig. 1B (a) and Fig. S1C, lane 2 versus lane 3 or 4). These results suggest that EGFR activation supports HBV infection.
Figure 1.
Role of EGFR-downstream signaling in supporting HBV infection. A, HBV infection upon activation or inactivation of EGFR. EGF or gefitinib was applied during HBV inoculation of HepG2-NTCP cells for 16 h, and HBV infection was evaluated by detecting HBs in the culture supernatant (top left, black bars) as well as cccDNA (top left, white bars), HBV DNAs (bottom left, top panel, relaxed circular (rc) DNA and replicative intermediates (intermediates)), and HBc (right pictures, red) in the cells at 12 days post-inoculation. Total EGFR (EGFR) and EGFR phosphorylated at Tyr-1068 (p-EGFR) as well as actin as an internal control (actin) were detected by immunoblotting (bottom left). The numbers on the right of the panels indicate the size markers of DNA (kb, top panel) and protein (kDa, second, third, and bottom panels). Scale bars, 100 μm. B, ability of EGFR and its mutant to support HBV infection. HepG2-NTCP cells transfected with or without siRNA against EGFR (si-EGFR) and transduced with either EGFRre-WT or EGFRre-K721M by lentivirus vector were evaluated for HBV infection by detecting HBs in the culture supernatant (a, top graph). Protein expressions were also detected (a (middle and bottom panels) and b). C (a), schematic representations of the EGFR-downstream signaling (Ras-MAPK, PI3K-Akt, and JAK-STAT) and inhibitors for each cascade. b–d, effect of inhibitors on the target signaling. The indicated proteins were detected by immunoblot in HepG2-NTCP (b and c) or Huh-7 cells (d) treated with or without the indicated inhibitors. D, infection of HCV pseudoparticles to Huh-7 cells (a) and of HBV to HepG2-NTCP cells (b) and primary human hepatocytes (c) were evaluated upon treatment with or without the indicated inhibitors. Precise experimental procedures, including compound concentrations and treatment times, are shown under “Experimental procedures.” Error bars, S.D. Experiments were repeated three times (n = 3 independent experiments, each with triplicate samples). *, p < 0.05; **, p < 0.01; N.S., not significant.
Activated EGFR triggers its downstream signaling, including Ras-MAPK, PI3K-Akt, and JAK-STAT pathways (11) (Fig. 1C, top). We examined the significance of these signaling pathways in the infection of HBV as well as HCV as a reference: EGFR has been reported to facilitate HCV entry through activation of Ras and PI3K-Akt (12, 13). As reported, inhibition of either Ras (tipifarnib), PI3K (wortmannin), or Akt (GSK690693) reduced HCV entry in a dose-dependent manner (Fig. 1D (a) and Fig. S2a). In contrast, pharmacological inhibition of the Ras-MAPK, PI3K-Akt, or JAK-STAT pathway had no drastic effect on HBV infection of HepG2-NTCP cells or primary cultures of human hepatocytes, with only a marginal reduction of HBV infection upon wortmannin treatment in both cell types (Fig. 1D (b and c) and Fig. S2 (b and c)). Also in the viral internalization assay, Ras inhibitor (tipifarnib) reduced the internalization of HCV, but not HBV (Fig. S3). In contrast, knockdown of EGFR and treatment with gefitinib, which inhibits not only signal transduction but also EGFR endocytosis, as mentioned below, reduced the internalization of both HCV and HBV (Fig. S3). Thus, blockade of EGFR-downstream signaling did not show any remarkable effect on HBV infection.
EGFR endocytosis machinery is essential for supporting HBV internalization
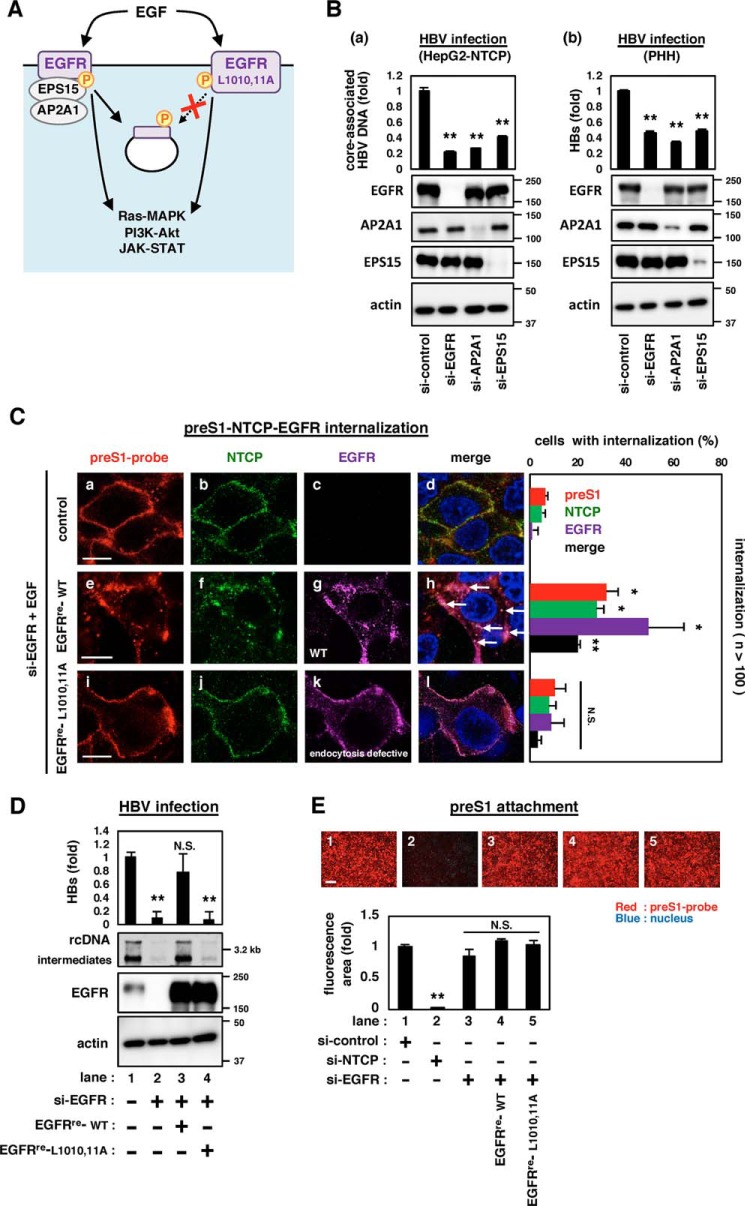
In addition to the activation of its downstream signaling, EGFR phosphorylation induces the recruitment of adaptor molecules that leads to the endocytosis of EGFR (Fig. 2A, left) (6). We then examined the contribution of the EGFR endocytosis machinery in HBV infection. As shown in Fig. 2B and Fig. S4A, knockdown of such endogenous adaptor molecules, either AP2A1 or EPS15, by multiple siRNAs significantly reduced HBV infection of HepG2-NTCP cells or primary human hepatocytes to the level seen by the knockdown of EGFR (Fig. 2B and Fig. S4A). Moreover, an EGFR variant with the L1010A/L1011A double mutation (EGFRre-L1010,11A) (Fig. 2A, right), which is deficient in endocytosis of activated EGFR (14, 15) (see Fig. 2C (k)) but still active on EGFR-downstream signaling (Fig. S4B), could not rescue HBV infection in si-EGFR–transfected HepG2-NTCP cells (Fig. 2D, lane 4). These data suggest that the active EGFR endocytosis pathway, rather than the EGFR-downstream signaling, is essential for supporting HBV infection. This EGFR regulation was not observed in the infection of hepatitis E virus (Fig. S4C). Recently, we reported that EGFR-depleted cells supported the cell attachment of fluorescence-labeled HBV preS1 peptide, a probe to visualize the HBV entry process (5, 16, 17), to a similar extent as the control cells (Fig. 2E, lane 1 versus lane 3), whereas NTCP-depleted cells completely lost the HBV preS1-cell attachment (Fig. 2E, lane 1 versus lane 2) (5). However, knockdown of EGFR attenuated the internalization of HBV preS1-NTCP complex (Fig. 2C, a–d), which was rescued by the complementation of the WT EGFR (Fig. 2C, e–h). In contrast to the WT EGFR, we found that an endocytosis-defective EGFRre-L1010,11A did not render preS1-NTCP internalization (Fig. 2C, i–l and the right graph). These results clearly suggest that the EGFR endocytosis machinery is important for mediating the HBV internalization.
Figure 2.
EGFR endocytosis was essential for mediating HBV internalization. A, schematic representation of the downstream events after activation of EGFR (left) and its mutant of Leu-Leu at 1010 and 1011 to Ala-Ala (EGFR-L1010,11A) (right). EGFR-L1010,11A is deficient in endocytosis, while possessing the intact potential to activate the EGFR-downstream signaling. B, HBV infection was evaluated by quantifying the core-associated HBV DNA in HepG2-NTCP cells (a) or the extracellular HBs of primary human hepatocytes (b) transfected with siRNAs against the indicated target gene. EGFR, AP2A1, and EPS15 as well as actin as an internal control were detected by immunoblotting (bottom panels). C, internalization of HBV preS1 (red), NTCP (green), and EGFR (purple) was evaluated by confocal microscopy. si-EGFR–transfected HepG2-NTCP cells transduced with or without either EGFRre-WT or EGFRre-L1010,11A were exposed with a fluorescence-labeled myristoylated preS1 peptide (amino acids 2–48) (preS1 probe) at 4 °C to allow attachment. After washing, the cells were then stimulated with EGF at 37 °C for 30 min to allow preS1 internalization and were observed by immunofluorescence. Scale bars, 10 μm. The percentages of cells in which preS1, NTCP, and EGFR were internalized as speckles are indicated in the right graph (n >100). D, HBV infection was examined in HepG2-NTCP cells transfected with (lanes 2–4) or without (lane 1) si-EGFR and transduced with GFP (lane 1,2), EGFR (EGFRre-WT) (lane 3), or its mutant, EGFRre-L1010,11A (lane 4) that has si-EGFR–resistant substitutions. HBV infection was evaluated with HBs by ELISA (top graph) and HBV DNA by Southern blotting (top panel). Protein expressions are also shown in the bottom panels. E, HBV preS1-cell attachment was evaluated by using the cells transfected with the indicated siRNAs (si-control, si-NTCP or si-EGFR) and transduced with or without EGFRre-WT or EGFRre-L1010,11A as shown under “Experimental procedures” (top pictures). Scale bars, 200 μm. The bottom graph shows the quantification of the fluorescence attached to the cells. Error bars, S.D. The reproducibility of the data was confirmed in three independent experiments. *, p < 0.05; **, p < 0.01; N.S., not significant.
Time-dependent transport of HBV preS1-EGFR to the endosomal network
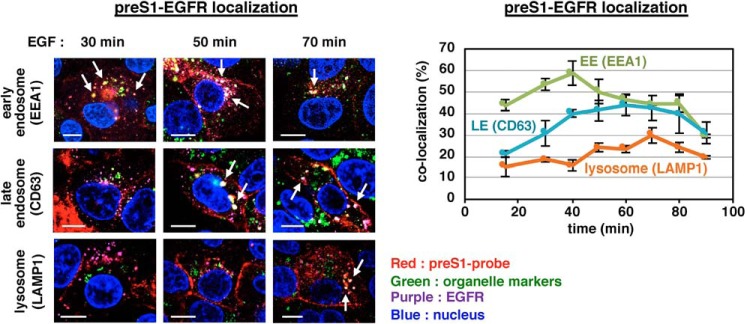
Upon endocytosis triggered by the EGFR stimulation, EGFR is sorted to the endosomal network (6). We then chased the localization of HBV preS1-EGFR after EGF stimulation by co-staining with organelle markers. As shown in Fig. 3, speckles that were positive for both preS1 and EGFR (preS1-EGFR speckles) were readily colocalized with EEA1, an early endosome marker, as early as 15–30 min after EGF stimulation and peaked at 40 min with >50% colocalization and were then gradually dissociated from the early endosome (Fig. 3, green in the right graph). Instead, colocalization of preS1-EGFR speckles with CD63 (as a late endosome marker) was markedly increased in the first 50 min after EGF stimulation and was maintained for up to 80 min (Fig. 3, blue in the right graph). The colocalization with LAMP1 (a lysosome marker) was even more gradually increased and peaked at later time points, 70–80 min (Fig. 3, orange in the right graph). A similar localization change was also observed using HBV virion along with EGFR (Fig. S5). Thus, localization of HBV-EGFR speckles was sequentially shifted from the early endosome to the late endosome and subsequently to lysosome after EGFR activation.
Figure 3.
Time-dependent transport of HBV preS1 and EGFR to the endosomal network. HepG2-NTCP cells attached with the preS1 probe on the cell surface at 4 °C were stimulated with EGF and then chased under observation by confocal microscopy up to 90 min. HBV preS1 (red), EGFR (purple), each organelle marker (green; EEA1 for early endosome, CD63 for late endosome, and LAMP1 for lysosome), and the nucleus (blue) was detected at 15, 30, 40, 50, 60, 70, 80, and 90 min after EGF stimulation. The left pictures show a representative image at 30, 50, and 70 min after EGF stimulation. (The arrows indicate the preS1-EGFR speckles colocalized with early endosome, late endosome, or lysosome). Scale bars, 10 μm. In the right graph, the number of HBV preS1 speckles was counted as the total number (n > 90), and among them, the number of those colocalized with EGFR and each organelle marker (green, early endosome; blue, late endosome; orange, lysosome) was counted to calculate the percentage of preS1-EGFR speckle localization at each organelle. This percentage (y axis) is shown against the time after EGF stimulation (x axis). Error bars, S.D. Data are based on the average of three independent experiments from at least n > 10 cells, n > 90 vesicles.
EGFR-mediated sorting of HBV preS1 to the late endosome is important for productive infection
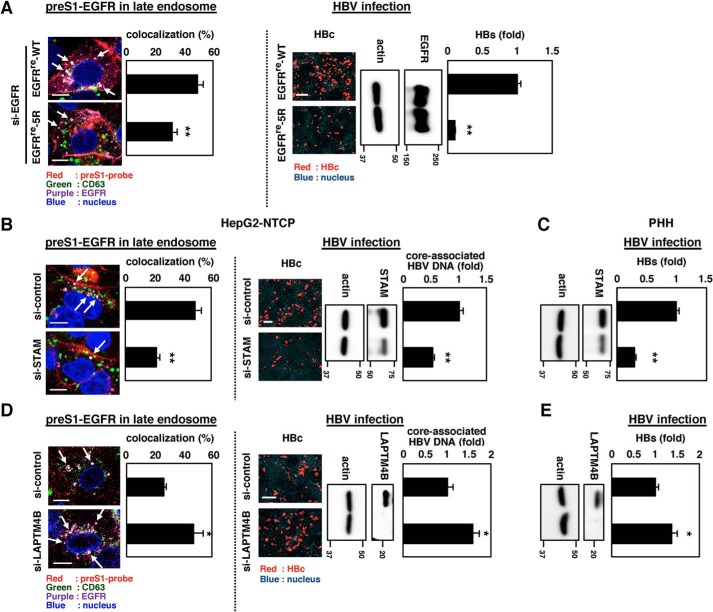
To examine the role of the observed EGFR relocalization to the endosomal network in achieving the productive infection, we employed an EGFR mutant. The EGFRre-5R mutant, having five point mutations of Lys (amino acids 692, 713, 843, 905, and 946) to Arg was reported to be less ubiquitinated to escape from the transport to the late endosome, with accumulation in the early endosome (18, 19) (Fig. 4A and Fig. S6A). Expression of this EGFRre-5R mutant in EGFR-depleted cells supported much a lower level of HBV infection compared with the WT EGFR (Fig. 4A (right panels and graph) and Fig. S6B). This observation was further supported by perturbation of the sorting pathway by knockdown of STAM and LAPTM4B; STAM is an EGFR-sorting adaptor molecule mediating the EGFR localization to the late endosome, and LAPTM4B suppresses the late endosome sorting of EGFR (see Fig. 5) (20, 21). Genetic knockdown of STAM significantly decreased the late endosome localization of HBV preS1-EGFR complex (Fig. 4B, left) and reduced the productive infection both in HepG2-NTCP cells (Fig. 4B (right) and Fig. S7) and PHH (Fig. 4C). Augmentation of late endosome localization of EGFR by knocking down LAPTM4B (Fig. 4D, left) significantly promoted the HBV infection level (Fig. 4 (D (right) and E) and Fig. S7). These results cumulatively support the important role of the EGFR-sorting machinery in the productive infection of HBV.
Figure 4.
EGFR-mediated sorting of HBV preS1 to the late endosome was important for the productive HBV infection. A, left, HepG2-NTCP cells transfected with si-EGFR and transduced with EGFR (EGFRre-WT) or that carried mutations of Lys (amino acids 692, 713, 843, 905, and 946) to Arg (EGFRre-5R) were observed by confocal microscopy as shown in Fig. 3. Arrows, preS1-EGFR speckles localized in the late endosome (left panels). Right, HBV infection was evaluated with intracellular HBc (right panels, red) and core-associated HBV DNA (right graph). Protein expressions of EGFR or its variant (EGFR) and actin as an internal control (actin) are also shown. B–E, similar analysis was performed using HepG2-NTCP cells (B and D) and primary human hepatocytes (C and E) transfected with or without siRNA against STAM (B and C) or LAPTM4B (D and E), known trafficking adaptors that facilitate and suppress EGFR sorting to the late endosome, respectively. Scale bars, 10 μm (left panels of A, B, and D) and 100 μm (right panels of A, B, and D). Error bars, S.D. Data are representative of at least n = 3 independent experiments. *, p < 0.05; **, p < 0.01.
Figure 5.
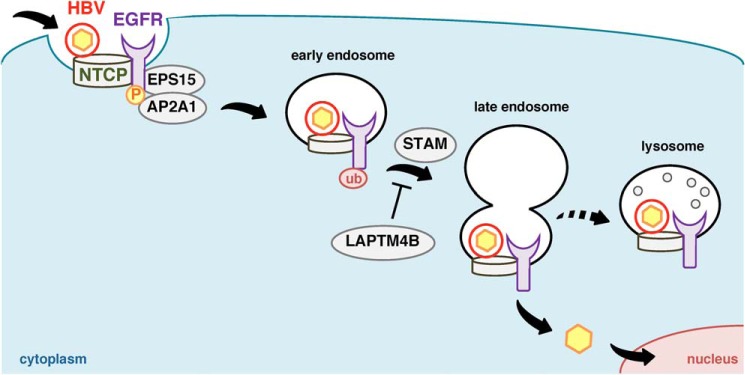
Schematic model of the EGFR-mediated HBV sorting for viral entry. After HBV attachment to the cell surface NTCP, the HBV-NTCP complex is internalized inside the cell by depending on the EGFR endocytosis machinery, which involves phosphorylation of EGFR and the following recruitment of adaptor molecules such as AP2A1 and EPS15. The EGFR-sorting machinery (involving EGFR ubiquitination, STAM, and LAPTM4B) coordinates HBV transport in the endosomal network (early and late endosome and lysosome). HBV localization, especially to the late endosome, is suggested to be important for achieving productive infection.
Discussion
EGFR has been reported to play a role in the entry mechanism of a variety of viruses. For example, human cytomegalovirus interacts with EGFR as a host receptor and activates its downstream signaling to reorganize the cytoskeletal structures and facilitate the viral particle translocation to the nucleus (22). Herpes simplex virus type 1, respiratory syncytial virus, vaccinia virus, and African swine fever virus also activate EGFR signaling, leading to actin rearrangement that enables the internalization/migration of incoming viral particles (7). HCV-induced activation of EGFR facilitates viral internalization by mechanisms that include the activation of Ras downstream signaling to induce assembly of the viral receptor complex required for viral internalization (13, 23). These examples highlight the essential role of EGFR-downstream signaling and the resulting cellular rearrangement in facilitating the viral entry process. In contrast to the above, EGFR plays a unique role in HBV internalization, in mediating the internalization of the HBV-NTCP complex through its endocytosis/sorting pathway (Fig. 5). We also suggest that the transport of incoming HBV to the endosomal network, especially to the late endosome, is key to achieving productive infection. Although the mechanisms underlying HBV entry, especially post-attachment, remain largely unknown, our results provide significant information and tools to further investigate this process, including the precise site of membrane fusion that is assumed to be engaged in the release of vesicular nucleocapsids into the cytosol.
The EGFR endocytosis/sorting pathway is regulated by multistep processes, including the dimerization and posttranslational modification of EGFR, recruitment of adaptor molecules, and complex formation of membrane trafficking components (11, 20, 24). This regulatory system is modulated by factors such as extracellular ligands, stresses, and microenvironments. This raises the possibility that cell susceptibility to HBV infection could be affected by these conditions and be dependent upon the expression level of NTCP. In addition, our demonstration that EGFR is a key factor in the HBV life cycle should open the field of investigation for antiviral discovery and prevention.
Experimental procedures
HBV infection assay
HBV infection was performed as described (25). HBV (genotype D) used as inoculum was recovered from the culture supernatant of Hep38.7-Tet cells cultured under tetracycline depletion and concentrated up to 200-fold by PEG concentration. For infection experiment, HepG2-NTCP cells or primary human hepatocytes were inoculated with HBV at 12,000 and 500 genome equivalent/cell, respectively, for 16 h in the presence of 4% PEG 8000. After washing out free HBV, the cells were cultured for an additional 12–15 days and harvested for evaluating HBV infection. HBV infection was evaluated either by detecting HBs in the culture supernatant by ELISA, HBc in the cells by immunofluorescence, HBV DNA in the cells by Southern blotting, or core-associated HBV DNA and covalently closed circular DNA (cccDNA) by real-time PCR, as described (26). In Fig. 1, the cells during HBV inoculation were treated together with the indicated ligand or compounds (EGF (10 ng/ml) or gefitinib (50 μm), tipifarnib (6.25, 12.5, or 25 μm), PD98059 (12.5, 25, or 50 μm), FR180204 (12.5, 25, or 50 μm), wortmannin (12.5, 25, or 50 μm), GSK690693 (12.5, 25, or 50 μm), or ruxolitinib (12.5, 25, or 50 μm)) to examine HBV infection. Bexarotene, a retinoid X receptor agonist, was used as a positive control that has been reported to reduce HBV infection (27).
Indirect immunofluorescence analysis
For evaluation of HBV infection, HBc in the cells after infection was detected by immunofluorescence as described (25) using anti-HBc antibody (Thermo Scientific).
For detecting the subcellular localization of preS1 probe, NTCP, EGFR, and the organelle markers, the indicated cells were incubated with the preS1 probe at 4 °C for 30 min to 2 h and then washed out. The preS1-attached cells were cultured at 37 °C to allow internalization under treatment with or without EGF (100 ng/ml) for the indicated times (30 min in Fig. 2E; 15, 30, 40, 50, 60, 70, 80, and 90 min in Fig. 3; 50 min in Fig. 4A; 60 min in Fig. 4B; 40 min in Fig. 4D). The cells were fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100, followed by incubation with anti-EGFR (Cell Signaling Technology), anti-HA (Sigma) (for detection of HA-tagged EGFRre-WT or EGFRre-5R in Fig. 4A), and anti-NTCP (28) antibodies. The immunocomplex was visualized with Alexa 488– or Alexa 647–conjugated secondary antibody (Thermo Scientific) by staining the nucleus with 4′,6-diamidino-2-phenylindole. For co-detection with organelle markers, we used anti-EEA1 (early endosome) (BD Biosciences), anti-CD63 (late endosome) (Santa Cruz Biotechnology, Inc.), and anti-LAMP1 (lysosome) (Abcam). Fluorescence signal was observed with confocal microscopy TCS SP8 (Leica).
For quantification of the localization in Fig. 2E, we observed at least 100 cells per sample as a total cell number and counted the number of cells having intracellular speckles for each signal (preS1 probe, NTCP, and EGFR) to calculate the percentage of cells having internalized signal (shown in the right graph). In Fig. 3, we counted the intracellular speckles that were positive for preS1, at least 90 speckles per sample, as a total number, and these preS1 speckles colocalized with both EGFR and each organelle marker (organelle marker–colocalized preS1-EGFR speckles). We calculated the percentage of the organelle marker–colocalized preS1-EGFR speckles over the total preS1 speckles, and this is shown in the graph.
Statistics
Statistical significance was determined with at least three samples by using Student's test (*, p < 0.05; **, p < 0.01; N.S., not significant) in Figs. 1 (A and B), 2 (B, D, and E), 4 (A–E) and Figs. S3 (A and B) and S4A.
Other experimental procedures are described in the supporting Experimental procedures.
Author contributions
M. I. and K. W. conceptualization; M. I., W. S., and K. W. data curation; M. I., W. S., K. N., H. O., R. Suzuki, H. A., M. M., T. M., S. I., T. W., and K. W. formal analysis; M. I., W. S., K. N., H. O., R. Sugiyama, and K. W. investigation; M. I., W. S., and K. W. methodology; M. I., W. S., and K. W. writing-original draft; A. R., M. O., J.-H. Y., S.-Y. P., T. O., and C. S. resources; K. W. supervision; M. I., W. S., and K. W. funding acquisition; K. W. project administration.
Supplementary Material
Acknowledgments
Plasmids for production of lentivirus were kindly provided by Dr. Hiroyuki Miyoshi at RIKEN. Plasmids for preparing hepatitis C virus pseudoparticle were generous gifts from Dr. Francois-Loic Cosset at the University de Lyon.
This study was supported in part by Japan Society for the Promotion of Science KAKENHI Grants JP17H04085, JP19K16672, and JP19J21455; the Japan Science and Technology Agency (JST) Core Research for Evolutional Science and Technology (CREST) and MIRAI program; Japan Agency for Medical Research and Development (AMED) Grants JP19fk0310114j0003, JP19fk0310101j1003, JP19fk0310103j0203, JP19fm0208019j0003, and JP19fk0210036j0002; the Takeda Science Foundation; the Yasuda Medical Foundation; the Smoking Research Foundation; the Mochida Memorial Foundation for Medical and Pharmaceutical Research; the Mitsui Life Social Welfare Foundation; and the Kyushu University Qdai-jump Research Program. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting Experimental procedures and Figs. S1–S7.
- HBV
- hepatitis B virus
- AP2A1
- adaptor-related protein complex 2 subunit α1
- cccDNA
- covalently closed circular DNA
- EEA1
- early endosome antigen 1
- EGF
- epidermal growth factor
- EGFR
- epidermal growth factor receptor
- EPS15
- epidermal growth factor receptor pathway substrate 15
- HBc
- hepatitis B virus core protein
- HCV
- hepatitis C virus
- LAMP1
- lysosome-associated membrane protein 1
- LAPTM4B
- lysosomal protein transmembrane 4β
- MAPK
- mitogen activated protein kinase
- NTCP
- sodium taurocholate cotransporting polypeptide
- PHH
- primary human hepatocytes
- PI3K
- phosphatidylinositide 3-kinase
- STAM
- signal-transducing adaptor molecule
- STAT
- signal transducers and activators of transcription
- JAK
- Janus kinase.
References
- 1. Watashi K., and Wakita T. (2015) Hepatitis B virus and hepatitis D virus entry, species specificity, and tissue tropism. Cold Spring Harb. Perspect. Med. 5, a021378 10.1101/cshperspect.a021378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., Fu L., Song M., Chen P., Gao W., Ren B., et al. (2012) Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1, e00049 10.7554/eLife.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le Seyec J., Chouteau P., Cannie I., Guguen-Guillouzo C., and Gripon P. (1999) Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 73, 2052–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gripon P., Cannie I., and Urban S. (2005) Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 79, 1613–1622 10.1128/JVI.79.3.1613-1622.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iwamoto M., Saso W., Sugiyama R., Ishii K., Ohki M., Nagamori S., Suzuki R., Aizaki H., Ryo A., Yun J. H., Park S. Y., Ohtani N., Muramatsu M., Iwami S., Tanaka Y., et al. (2019) Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc. Natl. Acad. Sci. U.S.A. 116, 8487–8492 10.1073/pnas.1811064116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tomas A., Futter C. E., and Eden E. R. (2014) EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 24, 26–34 10.1016/j.tcb.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng K., Kitazato K., and Wang Y. (2014) Viruses exploit the function of epidermal growth factor receptor. Rev. Med. Virol. 24, 274–286 10.1002/rmv.1796 [DOI] [PubMed] [Google Scholar]
- 8. Nishimura Y., Bereczky B., and Ono M. (2007) The EGFR inhibitor gefitinib suppresses ligand-stimulated endocytosis of EGFR via the early/late endocytic pathway in non-small cell lung cancer cell lines. Histochem. Cell Biol. 127, 541–553 10.1007/s00418-007-0281-y [DOI] [PubMed] [Google Scholar]
- 9. Glenney J. R. Jr, Chen W. S., Lazar C. S., Walton G. M., Zokas L. M., Rosenfeld M. G., and Gill G. N. (1988) Ligand-induced endocytosis of the EGF receptor is blocked by mutational inactivation and by microinjection of anti-phosphotyrosine antibodies. Cell 52, 675–684 10.1016/0092-8674(88)90405-9 [DOI] [PubMed] [Google Scholar]
- 10. Wheeler D. L., Dunn E. F., and Harari P. M. (2010) Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat. Rev. Clin. Oncol. 7, 493–507 10.1038/nrclinonc.2010.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Di Domenico M., and Giordano A. (2017) Signal transduction growth factors: the effective governance of transcription and cellular adhesion in cancer invasion. Oncotarget 8, 36869–36884 10.18632/oncotarget.16300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Z., Tian Y., Machida K., Lai M. M., Luo G., Foung S. K., and Ou J. H. (2012) Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J. Biol. Chem. 287, 41922–41930 10.1074/jbc.M112.414789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zona L., Lupberger J., Sidahmed-Adrar N., Thumann C., Harris H. J., Barnes A., Florentin J., Tawar R. G., Xiao F., Turek M., Durand S. C., Duong F. H., Heim M. H., Cosset F. L., Hirsch I., Samuel D., Brino L., Zeisel M. B., Le Naour F., McKeating J. A., and Baumert T. F. (2013) HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe 13, 302–313 10.1016/j.chom.2013.02.006 [DOI] [PubMed] [Google Scholar]
- 14. Wang Q., Zhu F., and Wang Z. (2007) Identification of EGF receptor C-terminal sequences 1005–1017 and di-leucine motif 1010LL1011 as essential in EGF receptor endocytosis. Exp. Cell Res. 313, 3349–3363 10.1016/j.yexcr.2007.06.014 [DOI] [PubMed] [Google Scholar]
- 15. Liu L., Shi H., Chen X., and Wang Z. (2011) Regulation of EGF-stimulated EGF receptor endocytosis during M phase. Traffic 12, 201–217 10.1111/j.1600-0854.2010.01141.x [DOI] [PubMed] [Google Scholar]
- 16. König A., Döring B., Mohr C., Geipel A., Geyer J., and Glebe D. (2014) Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J. Hepatol. 61, 867–875 10.1016/j.jhep.2014.05.018 [DOI] [PubMed] [Google Scholar]
- 17. Fukano K., Tsukuda S., Oshima M., Suzuki R., Aizaki H., Ohki M., Park S. Y., Muramatsu M., Wakita T., Sureau C., Ogasawara Y., and Watashi K. (2018) Troglitazone impedes the oligomerization of sodium taurocholate cotransporting polypeptide and entry of hepatitis B virus into hepatocytes. Front. Microbiol. 9, 3257 10.3389/fmicb.2018.03257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang F., Kirkpatrick D., Jiang X., Gygi S., and Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21, 737–748 10.1016/j.molcel.2006.02.018 [DOI] [PubMed] [Google Scholar]
- 19. Huang F., Goh L. K., and Sorkin A. (2007) EGF receptor ubiquitination is not necessary for its internalization. Proc. Natl. Acad. Sci. U.S.A. 104, 16904–16909 10.1073/pnas.0707416104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bakker J., Spits M., Neefjes J., and Berlin I. (2017) The EGFR odyssey—from activation to destruction in space and time. J. Cell Sci. 130, 4087–4096 10.1242/jcs.209197 [DOI] [PubMed] [Google Scholar]
- 21. Tan X., Sun Y., Thapa N., Liao Y., Hedman A. C., and Anderson R. A. (2015) LAPTM4B is a PtdIns(4,5)P2 effector that regulates EGFR signaling, lysosomal sorting, and degradation. EMBO J. 34, 475–490 10.15252/embj.201489425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X., Huang D. Y., Huong S. M., and Huang E. S. (2005) Integrin αvβ3 is a coreceptor for human cytomegalovirus. Nat. Med. 11, 515–521 10.1038/nm1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diao J., Pantua H., Ngu H., Komuves L., Diehl L., Schaefer G., and Kapadia S. B. (2012) Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J. Virol. 86, 10935–10949 10.1128/JVI.00750-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan X., Lambert P. F., Rapraeger A. C., and Anderson R. A. (2016) Stress-induced EGFR trafficking: mechanisms, functions, and therapeutic implications. Trends Cell Biol. 26, 352–366 10.1016/j.tcb.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watashi K., Liang G., Iwamoto M., Marusawa H., Uchida N., Daito T., Kitamura K., Muramatsu M., Ohashi H., Kiyohara T., Suzuki R., Li J., Tong S., Tanaka Y., Murata K., et al. (2013) Interleukin-1 and tumor necrosis factor-α trigger restriction of hepatitis B virus infection via a cytidine deaminase activation-induced cytidine deaminase (AID). J. Biol. Chem. 288, 31715–31727 10.1074/jbc.M113.501122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iwamoto M., Cai D., Sugiyama M., Suzuki R., Aizaki H., Ryo A., Ohtani N., Tanaka Y., Mizokami M., Wakita T., Guo H., and Watashi K. (2017) Functional association of cellular microtubules with viral capsid assembly supports efficient hepatitis B virus replication. Sci. Rep. 7, 10620 10.1038/s41598-017-11015-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song M., Sun Y., Tian J., He W., Xu G., Jing Z., and Li W. (2018) Silencing Retinoid X receptor α expression enhances early-stage hepatitis B virus infection in cell cultures. J. Virol. 92, e01771–17 10.1128/JVI.01771-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyakawa K., Matsunaga S., Yamaoka Y., Dairaku M., Fukano K., Kimura H., Chimuro T., Nishitsuji H., Watashi K., Shimotohno K., Wakita T., and Ryo A. (2018) Development of a cell-based assay to identify hepatitis B virus entry inhibitors targeting the sodium taurocholate cotransporting polypeptide. Oncotarget 9, 23681–23694 10.18632/oncotarget.25348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.