Abstract
Insoluble, hyperubiquitylated TAR DNA-binding protein of 43 kDa (TDP-43) in the central nervous system characterizes frontotemporal dementia and ALS in many individuals with these neurodegenerative diseases. The causes for neuropathological TDP-43 aggregation are unknown, but it has been suggested that stress granule (SG) formation is important in this process. Indeed, in human embryonic kidney HEK293E cells, various SG-forming conditions induced very strong TDP-43 ubiquitylation, insolubility, and reduced splicing activity. Osmotic stress–induced SG formation and TDP-43 ubiquitylation occurred rapidly and coincided with colocalization of TDP-43 and SG markers. Washout experiments confirmed the rapid dissolution of SGs, accompanied by normalization of TDP-43 ubiquitylation and solubility. Surprisingly, interference with the SG process using a protein kinase R–like endoplasmic reticulum kinase inhibitor (GSK2606414) or the translation blocker emetine did not prevent TDP-43 ubiquitylation and insolubility. Thus, parallel pathways may lead to pathological TDP-43 modifications independent of SG formation. Using a panel of kinase inhibitors targeting signaling pathways of the osmotic shock inducer sorbitol, we could largely rule out the stress-activated and extracellular signal–regulated protein kinase modules and glycogen synthase kinase 3β. For arsenite, but not for sorbitol, quenching oxidative stress with N-acetylcysteine did suppress both SG formation and TDP-43 ubiquitylation and insolubility. Thus, sodium arsenite appears to promote SG formation and TDP-43 modifications via oxidative stress, but sorbitol stimulates TDP-43 ubiquitylation and insolubility via a novel pathway(s) independent of SG formation. In conclusion, pathological TDP-43 modifications can be mediated via multiple distinct pathways for which SGs are not essential.
Keywords: TAR DNA-binding protein 43 (TDP-43) (TARDBP), ubiquitylation (ubiquitination), protein aggregation, RNA splicing, stress granule, oxidative stress, protein kinase, signal transduction, amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), frontotemporal dementia, osmotic shock
Introduction
Frontotemporal lobar degeneration (FTLD)2 and ALS are human neurodegenerative diseases causing symptoms of dementia and motoneuron degeneration, respectively. Neuropathological inclusions containing TAR DNA–binding protein of 43 kDa (TDP-43) occur in most cases, whereas a subset of these diseases is characterized by the protein fused-in-sarcoma (FUS). Moreover, mutations in the genes encoding TDP-43 and FUS are linked to ALS and rarely also to FTLD (1). It was evident from the beginning that TDP-43 is pathologically modified. Disease-associated modifications of TDP-43 include protein insolubility, ubiquitylation, phosphorylation, and proteolytic processing (2). Nevertheless, despite very much knowledge gained in the past decade about the RNA-binding proteins (RBPs) TDP-43 and FUS (3), disease etiologies and pathophysiological consequences remain uncertain.
In cells subjected to protein misfolding stress, a portion of TDP-43 and FUS (4–11) translocate from the nucleus to the cytosol to become incorporated into stress granules (SGs). Translationally stalled mRNAs are assembled into SGs as part of a cellular defense mechanism against proteotoxic stress (12). A variety of environmental stresses reduce the delivery of the initiator tRNAiMet to the 40S ribosomal subunit, thus suppressing translation initiation. Cells can achieve this by phosphorylation of the eukaryotic initiation factor eIF2α, which inhibits efficient GDP-GTP exchange and thus prevents the interaction with the initiator tRNAiMet and its delivery to the ribosome. Ser-51 phosphorylation of eIF2α can be mediated by four kinases, namely protein kinase R (PKR), PKR-like ER kinase (PERK), general control nonderepressible protein 2 (GCN2), and the heme-regulated inhibitor (HRI) (13). SG formation can also be initiated independent of eIF2α phosphorylation by interference with the cap-binding eIF4F complex either by inhibition of the RNA helicase subunit eIF4A or by alteration of mTOR-regulated phosphorylations of eIF4F components (14). The accumulation of untranslated mRNAs promotes the formation of SGs, and upon removal of the stressor, protein translation is resumed, and the SGs quickly disassemble (14).
SG composition is diverse and depends on the nature of the cellular stressor. The canonical SGs contain poly(A) mRNAs, mRNA-associated translation initiation complexes, including the eIF3 complex, as well as RBPs and 40S ribosomal subunits (14, 15). The formation of SGs involves liquid-liquid phase separation promoted by intrinsically disordered proteins, such as T-cell–restricted intracellular antigen-1 (TIA-1) and the TIA-1–related protein (TIAR). Interestingly, TDP-43 and FUS also belong to the class of intrinsically disordered RBPs containing low complexity (prion-like) domains like the TIA proteins. Thus, it was hypothesized that the formation of liquid droplets under SG-forming conditions would lead to proteopathy in FTLD/ALS (16, 17) or even neurodegeneration in general (18). Also, SG components are found at times in pathological TDP-43 and FUS inclusions in ALS and FTLD (15). Finally, a genetic link between ALS and SGs is indicated by mutations in the SG components TIA1 and ataxin-2 (19, 20).
Various cellular stresses can induce SG formation, such as oxidative, osmotic, ER, heat shock, and viral stress. More than 60% of the studies used oxidative stress mediated by sodium arsenite, and about one-third applied heat shock to induce SGs (15), whereas osmotic stress caused by sorbitol is a rather new tool (8). The exact pathways that are involved in osmotic stress-induced SG formation by sorbitol are not well-understood. In general, cells exposed to osmotic stress shrink due to water efflux. This negatively affects the cell in many ways, like decreased protein degradation and translation, altered enzymatic function as well as increased macromolecular crowding, cell cycle arrest, DNA damage, oxidative stress, and protein carbonylation (21, 22). Hyperosmotic stress also stimulates many signaling cascades, including the stress-activated c-Jun N-terminal kinase (JNK), p38 mitogen-activated protein kinase (MAPK), and extracellular signal regulated kinase (ERK) modules (23–26).
We investigated pathological modifications of TDP-43 under SG-inducing conditions. Treatment of HEK293E cells with arsenite and sorbitol caused particularly strong TDP-43 ubiquitylation and insolubility and induced functional impairment of TDP-43. Interestingly, SG suppression did not prevent TDP-43 ubiquitylation and shifts into more insoluble fractions. Arsenite affected TDP-43 via oxidative stress, whereas sorbitol apparently stimulated novel pathways leading to TDP-43 ubiquitylation and insolubility. Importantly, TDP-43 accumulation within SGs was not a prerequisite for early steps of TDP-43 pathogenesis.
Results
Proteasome inhibition, sorbitol, sodium arsenite, and heat shock promote TDP-43 ubiquitylation and insolubility
To investigate stress-induced changes of potentially pathogenic TDP-43 polyubiquitylation, insolubility, and aggregate formation, we exposed human embryonic kidney HEK293E cells to various stress conditions and analyzed these modifications of TDP-43 by His6-ubiquitin pulldown, solubility assay, and immunofluorescence.
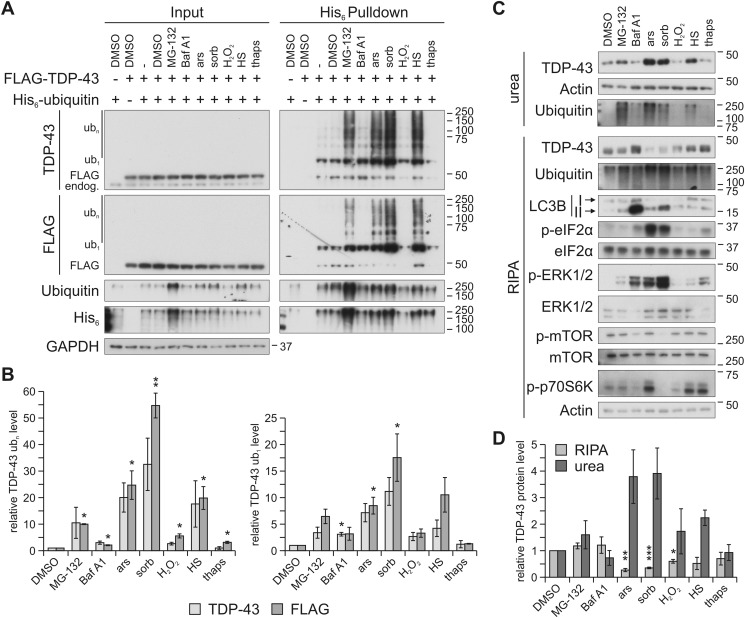
As reported before (27, 28), the proteasome inhibitor MG-132 but not the autophagy inhibitor bafilomycin (Baf) A1 promoted ubiquitylation of FLAG-tagged TDP-43 in HEK293E cells (Fig. 1, A (right) and B). We added several stressors that had been implicated with TDP-43 mismetabolism, namely hyperosmolar concentrations of sorbitol (8), heat shock, oxidative stress mediated by sodium arsenite or hydrogen peroxide (H2O2), and the ER stress inducer thapsigargin (9). These treatments can activate cellular stress responses involving the formation of SGs, which was suggested to be an important step in the development of TDP-43 and FUS inclusions in FTLD and ALS (17). We found that treatment with 0.4 m sorbitol most strongly induced ubiquitylation of TDP-43 (Fig. 1, A and B). Thermal stress mediated by 30-min temperature elevation to 43 °C also robustly caused TDP-43 ubiquitylation. Oxidative stress induced by treatment with 250 μm sodium arsenite also caused robust TDP-43 ubiquitylation, but treatment with 500 μm H2O2 failed in this experiment. Thapsigargin (1 μm) also caused no TDP-43 ubiquitylation.
Figure 1.
Osmotic, oxidative, and heat stress enhance TDP-43 ubiquitylation and insolubility. A, HEK293E cells expressing FLAG-TDP-43 and His6-ubiquitin (+) or empty control vectors (−) were exposed to MG-132, Baf A1, sodium arsenite (ars), sorbitol (sorb), H2O2, heat shock (HS), or thapsigargin (thaps) or DMSO as control. The cells were lysed with 8 m urea lysis buffer followed by affinity purification of His6-ubiquitin–conjugated proteins with Ni-NTA–agarose. Total protein (Input) and eluates (His6 Pulldown) were analyzed by Western blotting with antibodies detecting TDP-43, FLAG and His6 tag, ubiquitin, and GAPDH as loading control. Ub1, monoubiquitylated TDP-43; ubn, polyubiquitylated TDP-43; endog., endogenous TDP-43. B, quantification of mono- (ub1, right bar graph) and polyubiquitylated (ubn, left bar graph) TDP-43 band intensities (TDP-43 and FLAG signal) from four independent experiments as in A (His6 Pulldown). Relative values compared with DMSO control conditions (lane 4, His6 Pulldown) are displayed. Data represent the mean ± S.E. (error bars): *, p ≤ 0.05; **, p ≤ 0.005. C, HEK293E cells exposed to stresses as in A were sequentially extracted into RIPA-soluble and -insoluble urea fractions, followed by Western blot analysis with antibodies against TDP-43, ubiquitin, LC3B, phosphorylated and total eIF2α, ERK1/2 and mTOR, phosphorylated p70S6K, and actin as loading control. All conditions except H2O2 treatment elicited strong cellular responses as evidenced by phosphorylation of ERK1/2. Impaired autophagic flux was confirmed for the direct lysosomal inhibitor Baf A1. Accumulation of LC3B II, although to a lesser degree, was also observed after proteasome inhibition as well as sorbitol and arsenite treatment. Reduced mTOR phosphorylation was caused by lysosomal inhibition and osmotic stress, and both treatments as well as H2O2 exposure reduced phosphorylation of ribosomal S6 protein kinase (p70S6K). D, quantification of TDP-43 band strengths normalized to actin from four independent experiments as in C. Relative values compared with DMSO control conditions are displayed. Data represent the mean ± S.E.: *, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005.
As reported previously (27, 28), proteasome inhibition with MG-132 not only enhanced TDP-43 ubiquitylation, but also shifted TDP-43 into the RIPA-insoluble urea fraction. The strongest TDP-43 ubiquitylation inducers (Fig. 1, A and B), sorbitol, arsenite, and heat shock shifted a relatively large portion of endogenous TDP-43 into the insoluble (urea) fraction (Fig. 1, C and D). The effect was even stronger than for MG-132 (Fig. 1D), consistent with enhanced TDP-43 ubiquitylation (Fig. 1A). We confirmed successful treatments by detection of various downstream (phosphorylation) targets (see Fig. 1C and the figure legend for further details). Interestingly, osmotic stress reduced mTOR phosphorylation and phosphorylation of ribosomal S6 protein kinase (p70S6K), consistent with translation-blocking SG signaling. At the same time, membrane-bound LC3B II accumulated. Thus, under these conditions, reduced mTOR signaling might initiate autophagy but with impaired autophagic flux.
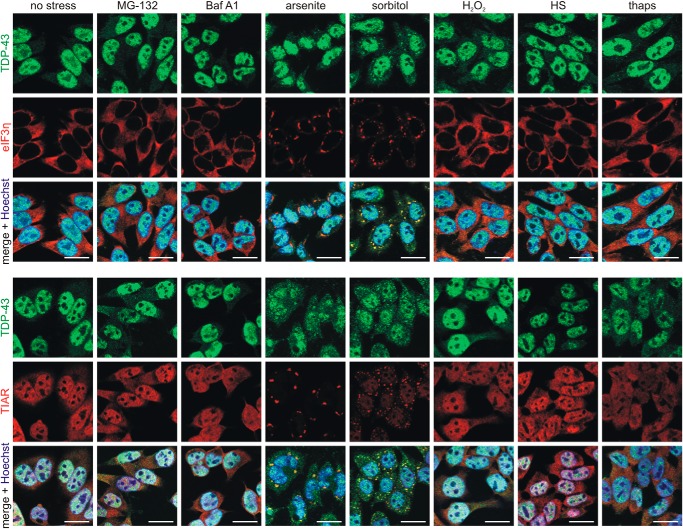
Immunofluorescence microscopy using two different SG markers (eIF3η and TIAR) did not reveal any SG formation after treatment with MG-132 and Baf A1, and TDP-43 remained mostly nuclear under these conditions (Fig. 2). H2O2 failed in our hands. Short exposure to the ER stressor thapsigargin caused cellular reactions, as evidenced by ERK1/2 and p70S6K phosphorylations (Fig. 1B), but did not effectively lead to SG formation (Fig. 2). As expected, sorbitol and arsenite treatments caused most prominent SG formation, as well as translocation of a considerable portion of TDP-43 into the cytosol, where it co-localized with SGs (Fig. 2). Strong induction of SG signaling by arsenite and sorbitol exposure was confirmed by detection of eIF2α phosphorylation (Fig. 1B). Heat shock caused quite strong TDP-43 ubiquitylation and insolubility (Fig. 1), although under these conditions no SG formation was visible (Fig. 2). SG signaling was not detected after 30-min heat shock; in fact, p70S6K phosphorylation was even enhanced (Fig. 1B). It appears that such a short heat shock does not induce global protein-misfolding stress in HEK293E cells, but the aggregation-prone TDP-43 may be affected even under limiting thermal stress conditions. Thus, although SG-forming conditions after treatment with arsenite and sorbitol cause very strong TDP-43 ubiquitylation and insolubility, TDP-43 recruitment into SGs is not a universal prerequisite for such TDP-43 modifications, which can happen after brief heat shock even in the absence of SGs.
Figure 2.
Osmotic and oxidative stress induce stress granule formation and TDP-43 translocation. HEK293E cells were stressed with MG-132, Baf A1, sodium arsenite, sorbitol, H2O2, heat shock (HS), or thapsigargin (thaps). The fixed cells were immunostained with rabbit anti-TDP-43 (green) and mouse anti-eIF3η (red) (top) or mouse anti-TDP-43 (green) and rabbit anti-TIAR (red) (bottom). The cell nuclei were counterstained with Hoechst 33342 (blue). Co-localization of TDP-43 with the SG marker eIF3η or TIAR is visible in the merged pictures (yellow). Scale bars, 20 μm.
Osmotic and oxidative stress impair splice activity of TDP-43
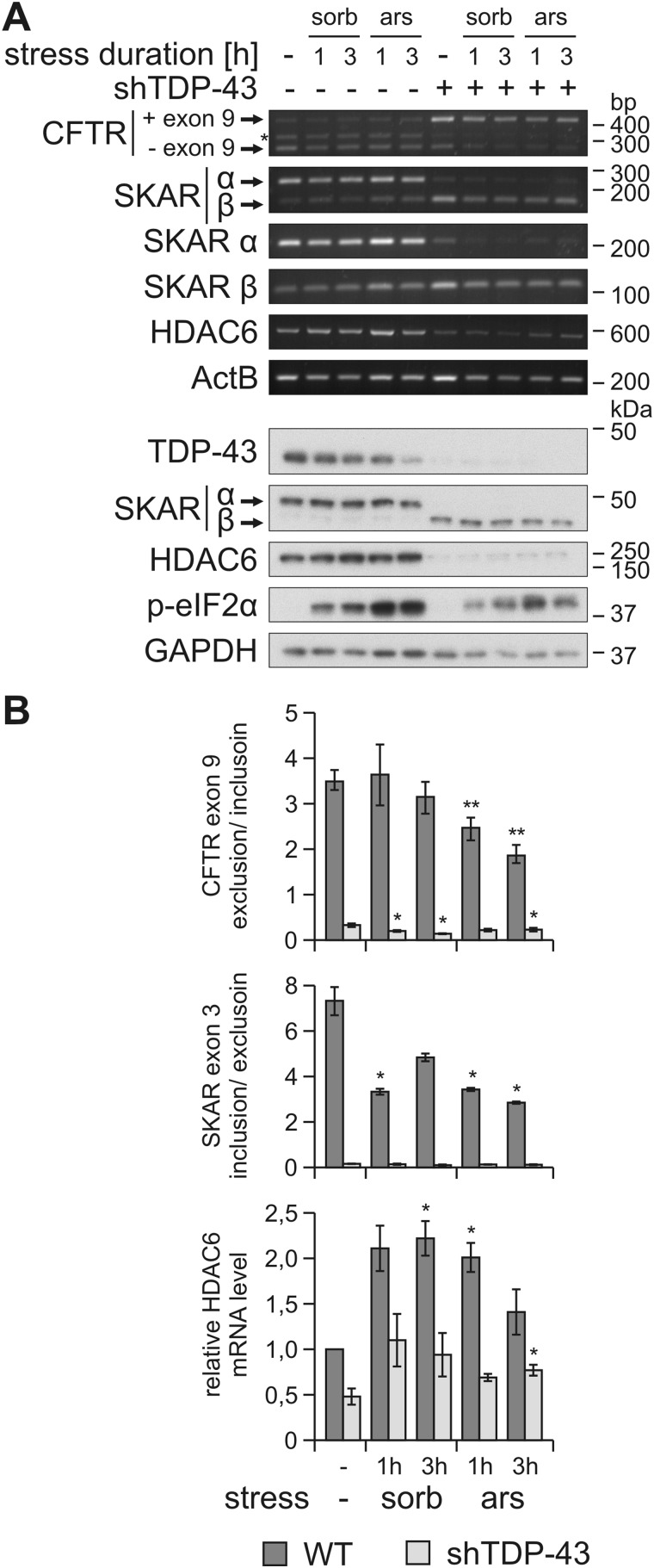
Enhanced TDP-43 ubiquitylation and insolubility could impair its functions by reducing the amount of active TDP-43 molecules. TDP-43 regulates many RNA-processing steps, including pre-mRNA splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) (29) and the ribosomal subunit protein S6 kinase 1 Aly/REF-like target (SKAR) (29, 30). TDP-43 also promotes histone deacetylase 6 (HDAC6) mRNA stability (31). We assessed whether the two strongest TDP-43–modifying stressors sorbitol and sodium arsenite could impair these TDP-43 functions. For comparison, the same stressors were applied to cells stably depleted of TDP-43 by RNAi. Cells were transfected with a CFTR splice reporter minigene (29), and as expected under control conditions, TDP-43 promoted strong exon 9 skipping (Fig. 3). Arsenite treatment for 1–3 h led to a small accumulation of the unspliced exon 9 included product and thus resulted in a significant reduction of the splicing ratio (Fig. 3). The same effect was seen for the endogenous splice target SKAR. Arsenite treatment for 1–3 h caused an accumulation of the mis-spliced exon-skipped SKAR β isoform, resulting in significant reduction of the TDP-43–dependent splice ratio (Fig. 3). For sorbitol treatments, the trends were less significant, where we only observed impaired SKAR splicing. The arsenite- and sorbitol-induced effects were not confirmed for protein levels of SKAR α and β, likely because only short time courses could be reliably followed. Prolonged stress may be necessary for mis-spliced mRNA and protein turnover to reach saturation, but the cells did not tolerate well longer exposure times to sorbitol or arsenite.
Figure 3.
Osmotic and oxidative stress impair TDP-43 splice activity. A, HEK293E WT (−)– or shTDP-43–expressing cells were transfected with a CFTR exon 9 mini gene expression vector. After exposure to sorbitol (sorb) or sodium arsenite (ars) for 1 or 3 h or to control medium (−), RNA and RIPA-soluble proteins were extracted. RNA was analyzed with RT-PCR using specific primers for CFTR mini gene, SKAR α and β isoforms, HDAC6, and the housekeeping gene ActB. CFTR exon 9–included (+ exon 9) and excluded (− exon 9) products, as well as SKAR α (exon 3 included) and β (exon 3 excluded) isoforms are labeled. *, nonspecific splice band (top). RIPA lysates were subjected to Western blot analysis with antibodies against TDP-43, SKAR, HDAC6, and phospho-eIF2α as stress induction control and GAPDH as loading control (bottom). B, quantification of CFTR exon 9 exclusion/inclusion ratio (top bar graph), SKAR exon 3 inclusion/exclusion ratio (middle bar graph), and HDAC6 mRNA level normalized to ActB and control conditions (bottom bar graph) from three independent experiments as in A. Data represent the mean ± S.E. (error bars): *, p ≤ 0.05; **, p ≤ 0.005.
The HDAC6 mRNA levels were reduced in cells with stable TDP-43 shRNA expression (Fig. 3), as expected. Surprisingly, sorbitol and arsenite treatment did not diminish HDAC6 expression, but actually enhanced the levels of HDAC6 mRNA and, to a lesser extent, also HDAC6 protein levels (Fig. 3). HDAC6 induction occurred both in WT cells and in TDP-43 knockdown cells. Thus, TDP-43–independent pathways regulating HDAC6 expression must exist in our experimental system. Therefore, HDAC6 does not serve as a surrogate marker of TDP-43 activity under these conditions. Nevertheless, the observed CFTR and SKAR mis-splicing (see above) is indicative of some loss of TDP-43 activity under stress conditions that lead to ubiquitinylation and insolubility of a fraction of TDP-43 protein.
Sorbitol and sodium arsenite do not promote ubiquitylation of FUS
The SG inducers sorbitol and arsenite very strongly promoted His6-ubiquitylation of FLAG-tagged TDP-43 in transfected HEK293E cells. In contrast, no HMW ubiquitylation was detected for 3xFLAG-tagged FUS in HEK293E cells treated with sorbitol or sodium arsenite (Fig. S1A), although endogenous FUS was efficiently recruited to SGs upon sorbitol exposure (Fig. S1B). Arsenite-induced SG translocation of FUS was barely detectable. Thus, the stress-induced ubiquitylations investigated here and our previous studies (27, 28) are unique to TDP-43 and do not seem to directly relate to FUS.
Nonconventional TDP-43 ubiquitylations induced by sorbitol are sensitive to the ubiquitin isopeptidase Y (UBPY)
Zinc finger protein 179 was recently identified as a ubiquitin ligase that mediated proteasome-targeting polyubiquitylation of TDP-43 (32). Enzymes that could affect the nonconventional TDP-43 ubiquitylations investigated here include the ubiquitin ligase parkin (33) and the deubiquitylating enzyme UBPY (also known as USP8) (27). To check whether parkin is the responsible enzyme promoting the TDP-43 ubiquitylations in response to sorbitol and arsenite, we used HeLa cells that lack parkin expression (34). Robust TDP-43 ubiquitylations were observed after treatment of HeLa cells with sorbitol and arsenite, indicating that ubiquitin ligases other than parkin can mediate stress-induced TDP-43 ubiquitylations (Fig. S2A). Similar to our previous findings for MG-132–stabilized TDP-43 ubiquitylations (27), expression of catalytically active MYC-UBPY WT but not the catalytically inactive C786S mutant or a C-terminally truncated UBPY (ΔC) could reduce the amount of polyubiquitylated TDP-43 smears formed after osmotic shock (Fig. S2B).
Seven lysine residues are present in ubiquitin that could form isopeptide bonds to assemble polyubiquitin chains, determining the fate of target proteins (35). To gain more insight into the nature of sorbitol-induced TDP-43 polyubiquitin linkages, we employed ubiquitin constructs lacking all seven lysine residues due to arginine substitution (K0) and individual lysine mutants. Lysine-less K0 was found in TDP-43–positive HMW smears, suggesting multiple monoubiquitylations (Fig. S3A). Lys-48–only ubiquitin that should form exclusively proteasome-targeting polyubiquitin chains was least incorporated into sorbitol-induced TDP-43 polyubiquitin chains. Interestingly, ubiquitin variants containing only Lys-27 and Lys-29 showed the strongest incorporation into TDP-43 HMW smears in sorbitol-treated HEK293E cells (Fig. S3A). However, arginine substitution of Lys-27 alone hardly reduced overall TDP-43 ubiquitylation, possibly indicating mixed polyubiquitin chains.
We also confirmed the linkages with specific polyubiquitin antibodies (Fig. S3B). The Lys-27 linkage was confirmed for Lys-27–ubiquitin, and importantly sorbitol treatment strongly enhanced Lys-27 linkage of WT ubiquitin (Fig. S3B, compare lanes 3 and 4 in the Lys-27–linked ubiquitin blot). Lys-63 linkages were also observed with a specific antibody and were moderately enhanced upon sorbitol treatment. In contrast, Lys-48–linked TDP-43 appeared unaffected by osmotic shock. These results indicate that sorbitol exposure induces highly complex TDP-43 ubiquitin modifications. The prominent unconventional Lys-27–linked polyubiquitylations of TDP-43 are remarkable. Potentially autophagy-targeting Lys-63 linkages occur as well, but proteasome-targeting Lys-48 linkages appear less relevant for sorbitol-induced TDP-43 polyubiquitylations.
Thus, SG-inducing conditions most strongly promote nonconventional ubiquitylations partially sensitive to UBPY and the insolubility of TDP-43. In the following, we characterize in more detail TDP-43 modifications after treatment with the SG inducers sorbitol and arsenite.
Kinetics of osmotic stress–induced SG formation and dissolution
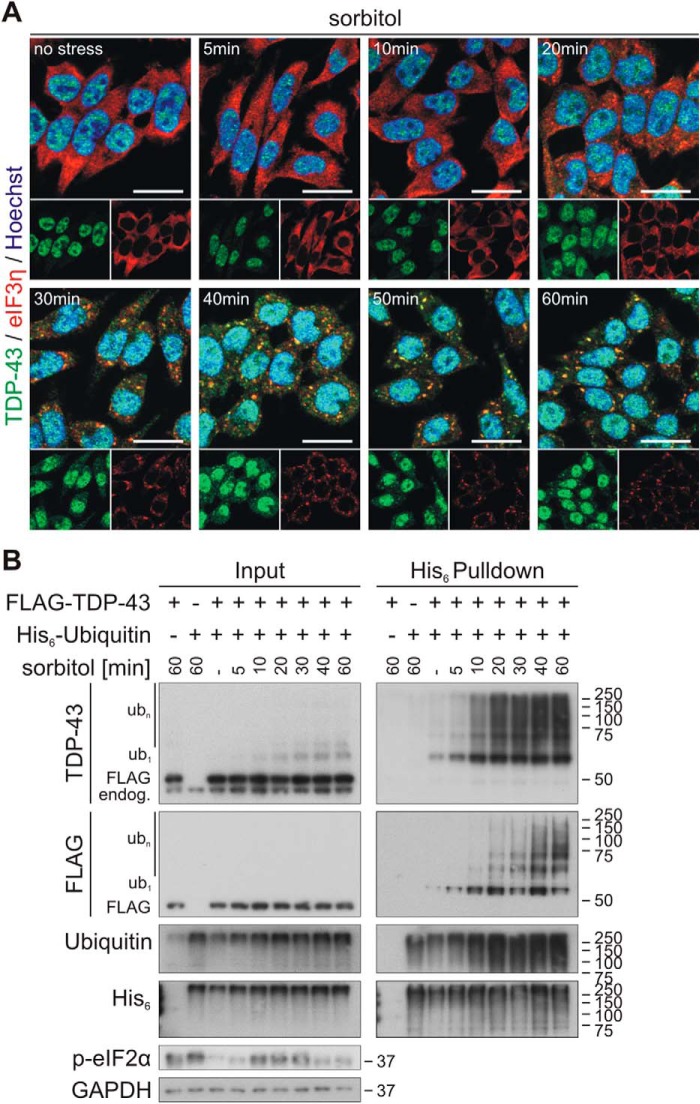
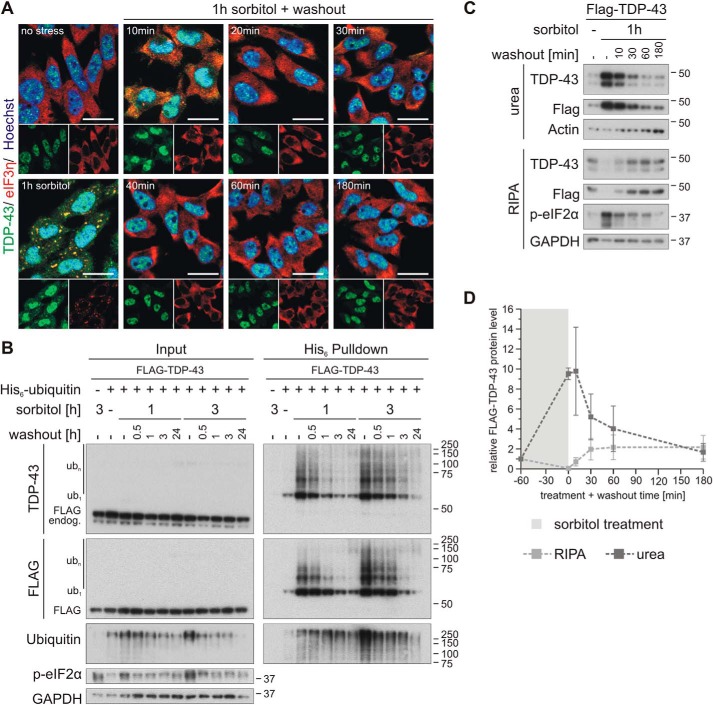
Next, we studied the kinetics of osmotic stress–induced SG formation and disassembly and TDP-43 SG translocation, ubiquitylation, and solubility. SGs formed very rapidly upon treatment with 0.4 m sorbitol. Assembly of the SG marker eIF3η into cytoplasmic granular structures started within 20 min, and clear SG formation could be observed 30–60 min after sorbitol addition (Fig. 4A). At these time points, TDP-43 co-localized with SGs. Already after 10 min, hyperosmotic shock–distinct ubiquitylation of TDP-43 was detectable (Fig. 4B) and thus occurred faster than visible SG formation and cytoplasmic TDP-43 translocation.
Figure 4.
Osmotic stress induces rapid ubiquitylation of TDP-43. A, HEK293E cells were exposed to osmotic shock with sorbitol for 5–60 min followed by fixation and immunostaining of endogenous TDP-43 (green) and the SG marker eIF3η (red) with specific antibodies. Cell nuclei were counterstained with Hoechst 33342 (blue). Scale bars, 20 μm. B, FLAG-TDP-43 and His6-ubiquitin (+) or empty control vectors (−) were expressed in HEK293E cells that were stressed with 0.4 m sorbitol for 5–60 min. Cells were lysed with 8 m urea lysis buffer, and His6-ubiquitin–conjugated proteins were isolated with Ni-NTA–affinity purification. Total protein (Input) and eluates (His6 Pulldown) were analyzed by Western blotting with antibodies against TDP-43, FLAG- and His6-tagged proteins, ubiquitin, phospho-eIF2α, and GAPDH as loading control.
SGs disassemble rapidly after removal of the cell stressor. Indeed, only a 10-min washout was sufficient to dissolve most SGs formed by 1-h treatment with 0.4 m sorbitol. After a 20-min washout, no more cytoplasmic granular structures were detectable (Fig. 5A). TDP-43 ubiquitylation normalized in a slower time course, reaching basal levels after a 3-h sorbitol washout (Fig. 5B, 1 h sorbitol). The time course was a little protracted after a 3-h SG formation incubation with 0.4 m sorbitol (Fig. 5B, 3 h sorbitol). Likewise, the solubility of TDP-43 normalized following a similar time course (Fig. 5, C and D).
Figure 5.
Osmotic stress–induced TDP-43 ubiquitylation and insolubility are rapidly reversible. A, HEK293E cells were stressed with 0.4 m sorbitol for 1 h followed by washout with fresh medium for 10–180 min, as indicated. The fixed cells were subjected to immunofluorescence analysis with specific antibodies against endogenous TDP-43 (green) and the SG marker eIF3η (red). Cell nuclei were counterstained with Hoechst 33342 (blue). Scale bars, 20 μm. B, cells transfected with FLAG-TDP-43 and His6-ubiquitin (+) or His6-control vector (−) were exposed to osmotic stress with sorbitol for 1 or 3 h or left untreated (−). After a washout for 0.5–24 h, as indicated, the cells were lysed with urea buffer, and ubiquitylated proteins were isolated by His6 pulldown with Ni-NTA–agarose. Total protein (Input) and eluates (His6 Pulldown) were analyzed by Western blotting with antibodies detecting TDP-43, FLAG, ubiquitin, phospho-eIF2α, and GAPDH. C, after treatment with sorbitol for 1 h, HEK293E cells expressing FLAG-TDP-43 were washed out for 10–180 min and were subjected to sequential extraction. The RIPA-soluble and -insoluble urea protein lysates were analyzed by Western blotting with antibodies against TDP-43, FLAG, and phospho-eIF2α and GAPDH and actin as loading controls. D, quantification of FLAG-TDP-43 protein levels normalized to actin from three independent experiments as in C. Band intensities were normalized to nonstressed control conditions (−). The data represent the mean ± S.E. (error bars).
Thus, SGs build up rapidly upon osmotic shock and dissolve quickly after sorbitol washout. TDP-43 recruitment to SGs, ubiquitylation, and solubility shifts roughly but not perfectly coincide with SG formation and dissolution.
TDP-43 ubiquitylation and insolubility in response to sorbitol and arsenite can occur independent of SG formation
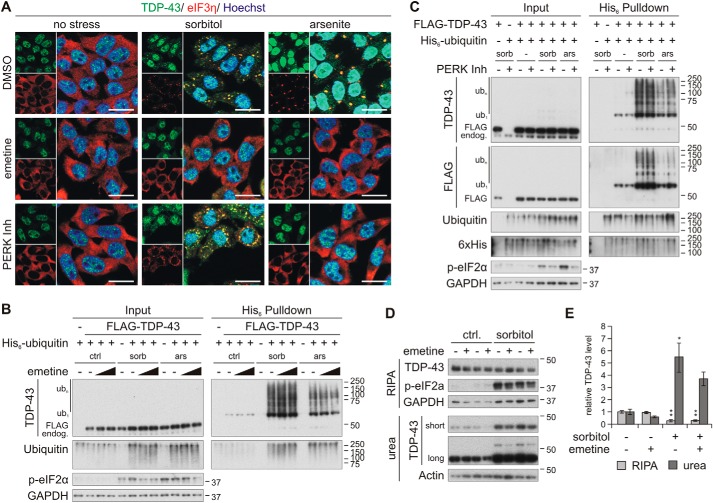
Next, we investigated whether SG formation would promote the pathological conversion of TDP-43. We employed the translation inhibitor emetine, which prevents SG formation by polysome stabilization (36), and the PERK inhibitor GSK2606414. Indeed, pretreatment with emetine completely blocked SG formation in response to sorbitol and arsenite (Fig. 6A). Interestingly, a fraction of TDP-43 was mislocalized to the cytosol in sorbitol-treated cells, although emetine prevented SG formation. In contrast, PERK inhibition reduced only arsenite-induced SG formation and had no effect on SGs formed upon osmotic stress.
Figure 6.
Osmotic stress-induced ubiquitylation and insolubility of TDP-43 are not dependent on its translocation into stress granules. A, HEK293E cells were pretreated with 25 μg/ml emetine or the PERK inhibitor GSK2606414 (PERK Inh, 10 μm) and stressed with sorbitol or arsenite. The fixed cells were immunostained with rabbit-anti-TDP-43 (green) and mouse-anti-eIF3η (red). Cell nuclei were counterstained with Hoechst 33342 (blue). Scale bars, 20 μm. B and C, FLAG-TDP-43 (+), His6-ubiquitin (+), or empty control vectors (−) were expressed in cells that were pretreated with 10 or 25 μg/ml emetine (B) or 10 μm PERK inhibitor (C), followed by exposure to sorbitol (sorb) or arsenite (ars) in the presence of the inhibitors, as indicated. His6-ubiquitin–conjugated proteins were purified from urea cell lysates with Ni-NTA–agarose and were subjected to Western blot analysis with antibodies detecting TDP-43, FLAG, ubiquitin, His6, phospho-eIF2α, and the loading control GAPDH. D, solubility analysis of emetine-pretreated HEK293E cells that were exposed to control medium (−) or stressed with sorbitol (+). RIPA-soluble and -insoluble urea fractions were analyzed with Western blotting using TDP-43, phospho-eIF2a, and GAPDH or actin antibodies. E, quantification of TDP-43 protein levels normalized to actin from three independent experiments as in D. Band intensities were normalized to control conditions. Data represent the mean ± S.E. (error bars): *, p ≤ 0.05; **, p ≤ 0.005.
Surprisingly, emetine did not abolish TDP-43 ubiquitylation mediated by sorbitol and sodium arsenite (Fig. 6B). Because emetine interferes with SG formation at a very late step, stress-induced eIF2α phosphorylation was only slightly reduced, maybe caused by its translation inhibitory function. Likewise, we observed the sorbitol-induced TDP-43 insolubility shift even in the presence of the SG-preventing agent emetine (Fig. 6, D and E). Like emetine, the PERK inhibitor did not affect stress-induced TDP-43 ubiquitylation, although reduction of eIF2α phosphorylation due to both stress conditions was achieved by GSK2606414 treatment (Fig. 6C). The PERK inhibitor did not prevent SG formation in response to sorbitol (Fig. 6A). Perhaps under these conditions, sorbitol induces eIF2α phosphorylation via one of the other three known eIF2α kinases (PKR, HRI, or GCN2), or hyperosmotic stress might induce SG formation independent of eIF2α phosphorylation (37–39).
In summary, we conclude that stress-induced ubiquitylations and insolubility shifts can occur independent of TDP-43 translocation into SGs.
Investigation of sorbitol signaling pathways leading to TDP-43 ubiquitylation
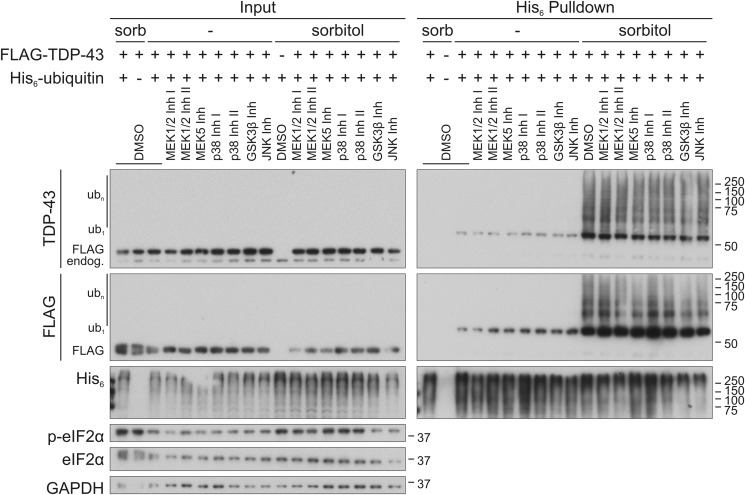
Osmotic shock induced by sorbitol triggers several MAPK signal transduction cascades, including the ERK1/2, ERK5, JNK, and p38MAPK pathways (23–26). Also, GSK3β was reported to affect TDP-43 translocation into SGs (40). Therefore, we asked whether signaling via one or several of these pathways is involved in the ubiquitylation of TDP-43. We employed a panel of kinase inhibitors and validated their performance (Fig. S4, A–E). First, we verified the sorbitol-induced activation of these pathways by detection of phosphorylated p38MAPK, JNK, and the JNK downstream target c-Jun, as well as ERK1/2 phosphorylation (Fig. S4, A, B, and D). Also, the activating GSK3β phosphorylation at Tyr-216 (41) was enhanced upon osmotic stress (Fig. S4E), in contrast to the inactivating GSK3β phosphorylation at Ser-9, which was not changed by sorbitol exposure (Fig. S4D). ERK5 activation was indirectly confirmed by increased phosphorylation of its downstream target FoxO3a (Fig. S4C).
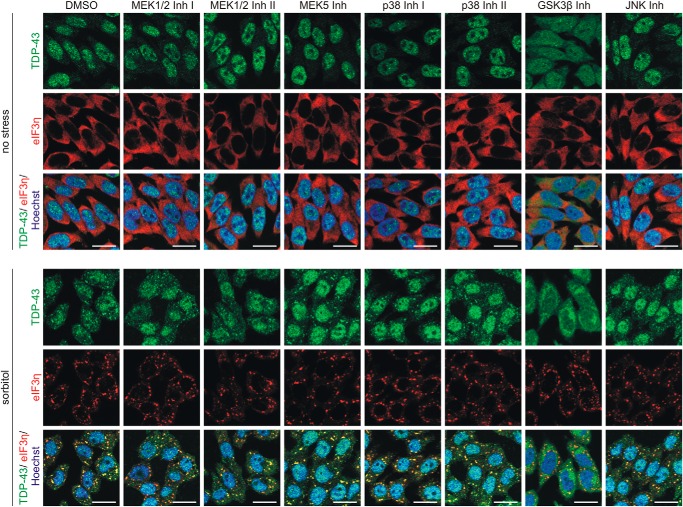
When investigating the effects of these inhibitors on the ubiquitylation of TDP-43, we detected no prominent alterations (Fig. 7). Also, sorbitol-induced SG formation and TDP-43 translocation were not clearly affected upon inhibition of most pathways (Fig. 8). Interestingly, GSK3β inhibition induced cytoplasmic translocation of TDP-43, which became even more pronounced after sorbitol treatment. Thus, cytoplasmic translocation alone is not sufficient for TDP-43 ubiquitylation, but further signals are necessary. In conclusion, sorbitol stimulates novel pathways promoting TDP-43 ubiquitylation parallel to SG formation.
Figure 7.
Osmotic stress induced ubiquitylation of TDP-43 is not dependent on classical stress signaling pathways. Stress-activated signaling pathways were inhibited with the inhibitors PD98059 (MEK1/2 Inh I), PD325901 (MEK1/2 Inh II), BIX02189 (MEK5 Inh), SB203580 (p38MAPK Inh I), SB202190 (p38MAPK Inh II), SB216763 (GSK3β Inh), and JNK-In-8 (JNK Inh) in HEK293E cells expressing FLAG-TDP-43 (+) and His6-ubiquitin (+) or empty control vector (−). After exposure to sorbitol (sorb) or control medium (−), cells were lysed in 8 m urea buffer, and His6-ubiquitin–conjugated proteins were purified with Ni-NTA–agarose. Total protein lysates (Input) and eluates (His6 Pulldown) were analyzed by Western blotting using antibodies against TDP-43, FLAG tag, and His6 tag, total and phosphorylated eIF2α, and GAPDH as loading control. Successful kinase pathway inhibition is shown in Fig. S4.
Figure 8.
MAPK pathway inhibition does not prevent sorbitol-induced SG formation and TDP-43 translocation. HEK293E cells were pretreated with the indicated kinase inhibitors before they were exposed to osmotic stress with 0.4 m sorbitol or no stress with control medium in the presence of the inhibitors. The fixed cells were immunostained with rabbit anti-TDP-43 (green) and mouse anti-eIF3η (red), and nuclei were counterstained with Hoechst 33342 (blue). Scale bars, 20 μm.
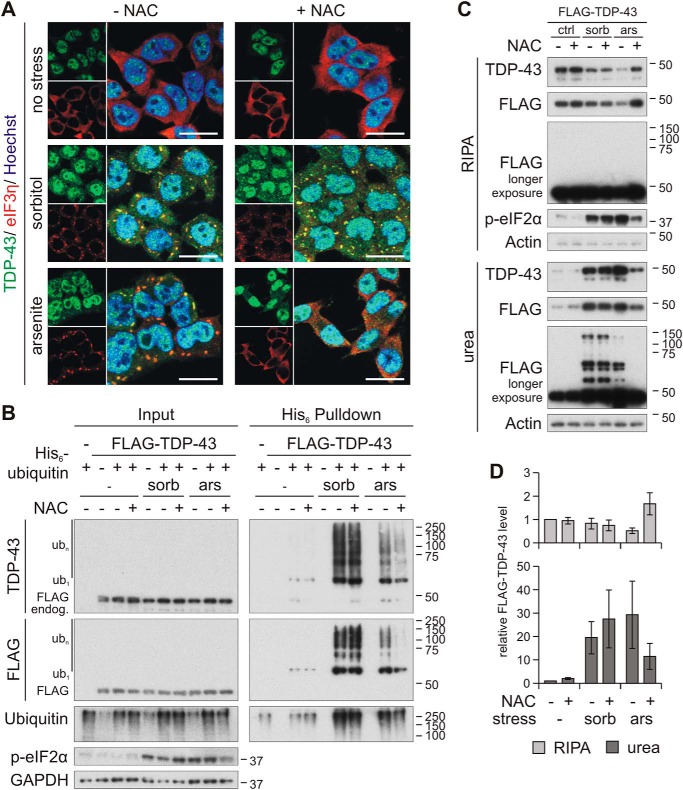
Oxidative stress is involved in arsenite- but not in sorbitol-induced ubiquitylation and insolubility of TDP-43
Sodium arsenite is known to promote oxidative stress (42), and osmotic shock is accompanied by oxidative stress as well (22). In addition, sorbitol can theoretically induce oxidative stress, because it is metabolized via the polyol pathway (8). Therefore, we utilized the antioxidant N-acetyl cysteine (NAC) and found it powerful to inhibit SG formation induced by arsenite in HEK293E cells (Fig. 9A). However, NAC did not affect SG formation and TDP-43 translocation in response to sorbitol, indicating that distinct pathways are activated by sorbitol and arsenic stress. Importantly, NAC treatment strongly reduced arsenite-induced TDP-43 ubiquitylation (Fig. 9B) and insolubility (Fig. 9, C and D) but was ineffective for sorbitol. Longer exposure of the sequentially extracted FLAG-TDP-43 also showed the complete clearing of arsenite-induced insoluble HMW FLAG-TDP-43 due to NAC treatment, whereas the antioxidant showed no effect on the sorbitol-induced HMW shift (Fig. 9C).
Figure 9.
Oxidative stress is involved in arsenite- but not in sorbitol-induced TDP-43 ubiquitylation and insolubility. A, immunofluorescence analysis of HEK293E cells that were pretreated with N-acetylcysteine (+ NAC) or control medium (− NAC), followed by exposure to sorbitol, arsenite, or control conditions (no stress), as indicated. Cells were immunolabeled with rabbit anti-TDP-43 (green) and mouse-anti-eIF3η (red), and nuclei were counterstained with Hoechst 33342 (blue). Scale bars, 20 μm. B, cells transiently expressing FLAG-TDP-43 and His6-ubiquitin (+) or control vectors (−) were treated with sorbitol (sorb), sodium arsenite (ars), or control medium (−) for 1 h in the presence (+) or absence (−) of the antioxidant NAC, as indicated. The cells were lysed in 8 m urea buffer, and ubiquitylated proteins were isolated with His6 pulldown. Total proteins (Input) and eluates (His6 Pulldown) were subjected to Western blot analysis, detecting TDP-43, FLAG, ubiquitin, phospho-eIF2α, and GAPDH. C, FLAG-TDP-43–transfected cells were exposed to stress with sorbitol or arsenite for 1 h in the presence of NAC, where indicated. The soluble RIPA and insoluble urea fractions were analyzed by Western blotting with TDP-43, FLAG, phospho-eIF2α, and actin antibodies. D, quantification of soluble (RIPA) and insoluble (urea) FLAG-TDP-43 from three independent experiments as in C. Band intensities were normalized to no stress and no NAC treatment conditions. Error bars, S.E.
In conclusion, we confirm that arsenite promotes SG formation and TDP-43 modifications involving oxidative stress. Both processes seem to be parallel but independent, as inhibition of SG formation at late steps with emetine and inhibiting the eIF2α kinase PERK did not reduce arsenite-induced TDP-43 ubiquitylation. Our sorbitol treatment study identifies that novel pathways parallel to SG formation may lead to TDP-43 pathology. In summary, multiple stress-induced pathways potentially cause TDP proteinopathy, and the direct SG formation process does not seem to be necessary for pathological TDP-43 modifications.
Discussion
The involvement of SGs in the formation of pathological TDP-43 and FUS inclusions in ALS and FTLD is controversial. A straightforward hypothesis states that SGs failing to disassemble nucleate these pathological inclusions (16, 18, 43–45). This finds support in studies that identified typical SG marker proteins like TIA-1, PABP1, eIF3, or eIF4G in TDP-43– and FUS–positive pathological inclusions in ALS and FTLD (5–7, 10). However, it is not a general finding that SG components are located in pathological deposits (15), as several studies did not find certain SG components in cytoplasmic TDP-43 inclusions (4, 6, 46). Other studies claim that TDP-43 pathology is independent of SGs (47) or does not arise directly from SGs but is either only induced by the same stress that mediates SG formation or transits through an SG intermediate (46, 48) (reviewed in Ref. 15). In addition, the formation of SGs could even protect from pathological aggregation of TDP-43 (48, 49). It is also possible that pathological TDP-43 inclusions might evolve in some cases through a SG, but in other cases their origin could be independent of SGs (50).
Here, we provide novel insights into the generation of typical pathological modifications on TDP-43 due to exposure to different stress situations that can induce the formation of SGs. We show that sorbitol and arsenite can promote pathological modifications like ubiquitylation and insolubility (28, 51). Importantly, the formation of SGs and TDP-43 incorporation due to exposure to these stressors is not required for ubiquitylation, and the solubility shift of TDP-43 as inhibiting the SG process with emetine and PERK inhibition showed no reduction of stress-induced TDP-43 ubiquitylation. Further evidence that these modifications can form independent of SGs is provided by the strong TDP-43 ubiquitylation upon heat shock for 30 min up to 1 h (data not shown), although at these time points SG formation did not occur (Fig. 2).
It may be assumed that pathological modifications impair the functional activity of TDP-43. Two studies showed that oxidative stress caused dramatic CFTR exon 9 inclusion and RNA and protein amounts of the splice-impaired SKAR β isoform were increased upon expression of an aggregation-prone TDP-43 variant (52, 53), supporting our findings. Also, HDAC6 mRNA levels were reduced upon overnight exposure with arsenite (52), as is expected when aggregating TDP-43 cannot stabilize the mRNA anymore. In contrast, we observed increased HDAC6 level after brief exposure to sorbitol or arsenite, but this was independent of TDP-43 expression. A study suggested that HDAC6 might be a critical stress regulator upon exposure to various stresses (54). Thus, our observation of increased HDAC6 level might be the result of a general stress response. Furthermore, mRNA splicing is regulated by many further factors, which also might be affected by arsenite or sorbitol. Thus, we cannot be absolutely certain that the impaired splicing of CFTR and SKAR observed in this study are entirely dependent on loss of TDP-43 activity due to increased insolubility.
We observed the strongest increase of TDP-43 ubiquitylation and heavy insolubility upon exposure to osmotic stress with sorbitol, and because SG translocation is not necessary for these modifications of TDP-43, we aimed to identify other involved pathways. In this study, sorbitol activated the ERK1/2, ERK5, JNK, and p38MAPK stress-responsive signaling pathways, as was demonstrated by others before (23, 24, 55). Furthermore, MEK5 suppression reduced TDP-43 toxicity in a recent study (56). However, using specific inhibitors, we could not identify a distinct signaling pathway that is involved in sorbitol-mediated TDP-43 ubiquitylation and/or SG formation. Although a study demonstrated that osmotic stress increases the amount of ubiquitylated proteins mediated by p38MAPK-dependent phosphorylation of the proteasomal subunit Rpn2 (57), in our hands, p38MAPK inhibition did not increase the total amount of ubiquitylated proteins (Fig. 7, Input). Hock et al. (58) also did not observe changes of sorbitol-induced cytoplasmic redistribution of FUS via the same pathways.
It is possible that failure of proteasomal or autophagosomal degradation under oxidative and osmotic stress could also contribute—at least partly—to the strong accumulation of insoluble, ubiquitylated TDP-43. Indeed, oxidative stress could impair proteasomal function, decreasing the degradation of oxidized proteins (59). However, another study suggested that acute and prolonged oxidative stress enhances the activity of the proteasome, but extensive, sublethal stress eventually impairs proteasomal degradation, which could cause development of pathology in neurodegenerative disease (60). Indeed, proteasomal inhibition led to the presence of moderate levels of ubiquitylated and insoluble TDP-43 (Fig. 1), but no SG formation was observed. Thus, MG-132 cannot induce TDP-43 pathogenesis via SG nucleation. Nevertheless, SG-forming conditions in arsenite- and sorbitol-treated cells caused stronger TDP-43 ubiquitylation and insolubility. Sorbitol treatment inactivated mTOR and p70S6K, consistent with translational repression and SG formation. At the same time, mTOR signaling should initiate autophagy, as also seen by the activation of the transcription factor FoxO3a (Fig. S4C) that contributes to autophagy induction (61). However, the accumulation of membrane-bound LC3B II (Fig. 1) suggests impaired autophagic flux. However, the autophagy inhibitor Baf A1 did not promote TDP-43 ubiquitylation and insolubility. Although impaired autophagic elimination could contribute to an accumulation of ubiquitylated and insoluble TDP-43, autophagy inhibition alone is not sufficient for the pathological TDP-43 modifications observed here. Additional upstream signaling events induced by hyperosmotic stress are required.
Interestingly, osmotic stress–induced mTOR inactivation occurs much faster than activation of the p38MAPK or JNK pathway (55). This suggests that separation of sorbitol-activated signaling pathways that lead to either TDP-43 ubiquitylation or SG formation or stress-responsive gene expression via MAPK pathways takes place very early, or these pathways are completely independent. Although arsenite induced similar effects on SG formation and TDP-43 modifications, it affected mTOR signaling differently than sorbitol. Consistent with our findings (Fig. 1), several studies showed that osmotic stress inhibits mTOR and p70S6K activity (55, 62–64), whereas arsenite can actually induce p70S6K (62). In contrast to sorbitol, arsenite induced TDP-43 ubiquitylation, and insolubility was attenuated with the antioxidant NAC. NAC has been also found to recover nuclear TDP-43 localization in neuron-like cells from patients affected by Niemann–Pick disease, type C (65). Oxidative stress induces cysteine oxidation and disulfide bond formation and was found to enhance insolubility of TDP-43 (52). Although arsenite and sorbitol both induce SGs as well as TDP-43 modifications, the pathways are different. Arsenite appears to act via oxidative stress, whereas sorbitol triggers pathological TDP-43 modifications via distinct pathways that do not rely on TDP-43 nucleation within SGs.
Our immunofluorescence studies did not reveal where the stress-induced ubiquitylated TDP-43 species are located in the cell, because we did not detect any visible accumulation beside the TDP-43 subpopulation that translocated into SGs. Upon exposure to sorbitol, cytoplasmic TDP-43 was slightly increased, but most TDP-43 was located in the nucleus as under basal conditions. We observed before that import-impaired TDP-43 NLS mutant exhibits increased ubiquitylation (28). Thus, it is conceivable that the mislocalization could be involved in the sorbitol-induced ubiquitylation. However, inhibition of GSK3β caused a dramatic TDP-43 translocation into the cytoplasm, although this did not increase the amount of ubiquitylated TDP-43 in absence or presence of sorbitol, suggesting that cytoplasmic mislocalization alone is not sufficient to induce ubiquitylation of TDP-43. Additionally, GSK3β inhibition did not increase TDP-43 engagement into SGs, indicating that cytoplasmic translocation alone is insufficient to promote SG incorporation. Although the cytoplasmic deubiquitylating enzyme UBPY reduced sorbitol-induced ubiquitylation partially, ubiquitin was not completely removed from TDP-43 as was the case for cytoplasmic TDP-43 C-terminal fragments in our previous study (27). Only 3-h recovery allowed complete clearance of ubiquitylated TDP-43. This could mean that either further DUBs or other mechanisms are involved in the reduction of ubiquitylated TDP-43 or that the mainly nuclear localized TDP-43 is spatially separated from UBPY.
In contrast to TDP-43, we could not detect polyubiquitylated FUS upon exposure to osmotic and oxidative stress. Moreover, blocking of proteasomal or autophagosomal degradation with MG-132, 3-methyladenine, and Baf A1 had no effect on the ubiquitylation of FUS.3 Although FUS-positive pathological inclusions in FTLD contain also ubiquitylated proteins and proteins of the UPS (66–68), Western blot analysis of FUSopathy brains did not show the prominent ubiquitylation HMW shifts that are typical for insoluble pathological TDP-43 (66). Thus, ubiquitylation is not an abundant pathological modification of FUS, and therefore the present findings for TDP-43 cannot be generalized to the related protein FUS.
In conclusion, we provide evidence that the typical pathological TDP-43 modifications ubiquitylation and insolubility may be induced by various stress conditions via distinct signaling pathways but—importantly—independent of SG formation and TDP-43 translocation into these cytoplasmic granules.
Experimental procedures
Constructs
The expression constructs pcDNA3.1(−)-FLAG-TDP-43, pCMV-His6-ubiquitin WT and lysine mutants, as well as pCMV-MYC-UBPY WT, C786S, and ΔC (lacking amino acids 735–1118) mutants were described before (27, 31, 69). FUS was amplified from HEK293E cDNA with specific primers introducing BglII and NotI restriction sites and cloned into the pCMV vector (Clontech) with a 5′-3xFLAG tag (Bsp120I and EcoRI). The pTB-CFTR exon 9 mini gene expression construct was a kind gift from E. Buratti (ICGEB, Trieste).
Antibodies
In this study, we used the following antibodies for Western blot analysis (WB) and immunofluorescence staining (IF): mouse anti-β-actin (WB 1:25.000, #A5441, Sigma, clone AC-15); rabbit anti-c-Jun (WB 1:1,000, #9165, Cell Signaling, clone 60A8); rabbit anti-phospho-c-Jun (Ser-73) (WB 1:1,000, #9164, Cell Signaling); rabbit anti-eIF2α (WB 1:2,000, #9722, Cell Signaling); rabbit anti-phospho-eIF2α (WB 1:2,000, #ab4837, Abcam, clone S51); mouse anti-eIF3η (IF 1:100, #sc-137214, Santa Cruz Biotechnology, Inc., clone C-5); rabbit anti-ERK1/2 (WB 1:2,000, #9102, Cell Signaling); rabbit anti-phospho-ERK1/2 (Thr-202/Tyr-204) (WB 1:500, #4370, Cell Signaling, clone D13.14.4E); rabbit anti-ERK5 (WB 1:1,000, #3372, Cell Signaling); peroxidase-conjugated anti-FLAG (WB 1:2,000–1:60,000, #A8592, Sigma, clone M2); rabbit anti-FoxO3a (WB 1:1,000, #2497, Cell Signaling, clone 75D8); rabbit anti-phospho-FoxO3a (Ser-413) (WB 1:1000, #8174, Cell Signaling, clone D77C9); mouse anti-FUS (WB 1:2,000, IF 1:250, sc-47711, Santa Cruz Biotechnology, clone 4H11); mouse anti-GAPDH (WB 1:35,000, #H86504M, Biodesign International, clone 6C5); rabbit anti-GSK3β (WB 1:2,000, #9315, Cell Signaling, clone 27C10); rabbit anti-phospho-GSK3β (Ser-9) (WB 1:1000, #9336, Cell Signaling); rabbit anti-phospho-GSK3β (Tyr-216) (WB 1:500, ab75745, Abcam); rabbit anti-HDAC6 (WB 1:16.000, sc-11420, Santa Cruz Biotechnology); mouse anti-His6 (WB 1:5,000–1:10,000, #MA1-21315, Invitrogen, clone HIS.H8); rabbit anti-JNK (WB 1:1,000, #9258, Cell Signaling, clone 56G8); rabbit anti-phospho-JNK (Thr-183/Tyr-185) (WB 1:2,000, #9251, Cell Signaling); rabbit anti-LC3B (WB 1:1,000, #2775, Cell Signaling); rabbit anti-mTOR (WB 1:1,000, #2972, Cell Signaling); rabbit anti-phospho-mTOR (Ser-2448) (WB 1:1,000, #2971, Cell Signaling); peroxidase-conjugated anti-MYC (WB 1:10,000, #11814150001, Roche Applied Science, clone 9E10); rabbit anti-p38MAPK (WB 1:1,000, #9212, Cell Signaling); rabbit anti-phospho-p38MAPK (Thr-180/Tyr-182) (WB 1:1,000, #9215, Cell Signaling); rabbit anti-phospho-p70S6K (Thr-389) (WB 1:1,000, #9234, Cell Signaling); rabbit anti-SKAR α/β (WB 1:1,000, #3794S, Cell Signaling); rabbit anti-TDP-43 (WB 1:10,000, IF 1:1,000; #10782-2-AP, ProteinTech Group); mouse anti-TDP-43 (IF 1:500, #H00023435-M01, Abnova, clone 2E2-D3); rabbit anti-TIAR (IF 1:1,000, #8509, Cell Signaling, clone D32D3); mouse anti-ubiquitin (mono- and polyubiquitin) (WB 1:10,000, #MAB1510, Millipore, clone Ubi-1); rabbit anti-ubiquitin (Lys-27–specific) (WB 1:20,000, #ab181537, Abcam, clone EPR17034); rabbit anti-ubiquitin (Lys-48–specific) (WB 1:20,000, #05-1307, Millipore, clone Apu2); rabbit anti-ubiquitin (Lys-63–specific) (WB 1:10,000, #05-1308, Millipore, clone Apu3).
The secondary HRP-conjugated antibodies for Western blot analysis were purchased from Jackson ImmunoResearch Laboratories (1:5,000–1:30,000), and secondary Alexa Fluor 488– or 568–conjugated antibodies for immunofluorescence were from Invitrogen (1:1,000–1:2,000).
Cell culture and transfection
HEK293E (WT and stably TDP-43 silenced (31)) and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum at 37 °C and 5% CO2. For the different experiments, 0.15 × 105, 0.1 × 106, 0.35 × 106, or 1 × 106 cells were seeded in 24-well, 6-well, 6-cm, or 10-cm cell culture dishes. For overexpression experiments, cells were transiently transfected 24 or 48 h after seeding with plasmid DNA using FuGENE6 (Promega), according to the manufacturer's instructions. The DNA/FuGENE6 ratio was 1:3. Further analysis of the cells was performed 48 h after transfection.
Stress induction and inhibition of SG formation and signaling cascades
For all treatments, the medium was replaced with fresh DMEM plus 10% fetal calf serum containing the various inhibitors and/or stressors and incubated for varying periods of time as indicated in the experiments. As control, cells were treated either with 1:1,000 diluted DMSO in DMEM or with medium without any compounds.
Stress was induced by exposing HEK293E cells to the following substances: proteasomal inhibition with MG-132 (10 μm for 6 h); autophagy inhibition with Baf A1 (20 nm for 24 h); osmotic shock with sorbitol (0.4 m for 1 h unless otherwise noted); ER stress with thapsigargin (1 μm for 1 h); oxidative stress with sodium arsenite (250 μm for 1 h, all from Sigma-Aldrich) or H2O2 (500 μm for 1 h in pyruvate-free medium, Roth). Heat shock was induced by cultivating the cells at 43 °C for 30 min.
SG formation was prevented by pretreatment with emetine (25 μg/ml unless otherwise noted for 30 min), GSK2606414 (10 μm for 3 h), or NAC (10 mm for 30 min). MAPK signaling was prevented by pretreatment with the following inhibitors for 1 h: PD98059 (MEK1/2 Inh I, 100 μm); PD325901 (MEK1/2 Inh II, 0.1 μm); BIX02189 (MEK5 Inh, 50 μm); SB216763 (GSK3β Inh, 40 μm); JNK-In-8 (JNK Inh, 8 μm); SB203580 (p38MAPK Inh I, 50 μm, all from Selleckchem); SB202190 (p38 InhMAPK II, 50 μm, Sigma-Aldrich). All compounds were also applied to the medium during stress induction with sorbitol or arsenite.
Washout assay
After exposure of the cells to sorbitol for 1 or 3 h, the medium was replaced with fresh DMEM without sorbitol, and washout took place for 5–60 min followed by immunofluorescence analysis or for 0.5–3 h before lysis for further pulldown or solubility experiments.
Pulldown of total ubiquitylated proteins with Ni-NTA–affinity chromatography
HEK293E cells (1 × 106 cells/dish) grown in a 10-cm cell culture dish were co-transfected with the indicated constructs and His6-ubiquitin for 48 h. The cells were exposed to the indicated treatments, followed by washing with PBS. They were lysed with 8 m urea buffer (10 mm Tris, pH 8.0, 100 mm NaH2PO4, 8 m urea) containing 10 mm imidazole, and the DNA was sheared by passing the lysate 20 times through a 23-gauge needle. The cell debris was pelleted at 14,000 × g for 15 min at 4 °C, and the protein concentration was determined with a Bradford assay (Bio-Rad protein assay). Total protein (500–750 μg) was incubated with Ni-NTA–agarose beads (Qiagen) for 3 h at room temperature. The beads were washed three times with washing buffer (10 mm Tris, pH 6.3, 100 mm NaH2PO4, 8 m urea) containing 20 mm imidazole, followed by elution of proteins in 3× Laemmli buffer + 20 μm DTT at 95 °C for 10 min. Total protein and one-fifth of eluates were subjected to analysis by Western blotting.
Sequential extraction of HEK293E proteins
HEK293E cells (0.35 × 106 cells/dish) cultivated in a 6-cm cell culture dish were lysed with RIPA lysis buffer (50 mm Tris/HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 10 mm NaPPi) containing 1× Complete protease inhibitor and PhosphoStop (both from Roche Applied Science). The RIPA-insoluble fraction was pelleted at 14,000 × g for 15 min at 4 °C, and the supernatant contained the RIPA-soluble fraction. The RIPA-insoluble pellets were washed twice with RIPA buffer and lysed in 8 m urea buffer (10 mm Tris, pH 8.0, 100 mm NaH2PO4, 8 m urea). The total protein concentration of RIPA lysates was determined with the bicinchoninic acid (BCA) protein assay kit (Pierce), and the total protein concentration of urea lysates was determined with the Bradford assay (Bio-Rad). RIPA-soluble protein was boiled in 1× Laemmli buffer including 20 μm DTT at 95 °C for 5 min, and the urea fraction was incubated with 1× Laemmli buffer + 20 μm DTT at room temperature for 10 min. 5 μg of soluble and 1–2.5 μg of insoluble protein were analyzed by Western blotting.
Western blot analysis
The denatured protein samples were separated on 7.5, 10, or 15% self-cast polyacrylamide gels or on 4–12% BisTris NuPAGE gradient gels (Invitrogen). Separated proteins were transferred onto Hybond-P polyvinylidene difluoride membranes (Millipore), which were blocked in 5% skim milk/TBST or 5% BSA/TBST, incubated with primary antibody in Western Blocking Reagent (Roche Applied Science) at 4 °C overnight. This was followed by incubation with HRP-conjugated secondary antibodies for 1–2 h at RT. The proteins were detected with the Immobilon Western chemiluminescent HRP substrate (Millipore) on Amersham Biosciences HyperfilmTM ECL (GE Healthcare).
Immunofluorescence
HEK293E cells (0.15 × 105/coverslip) were seeded on poly-d-lysine (Sigma-Aldrich)– and collagen (Millipore)–coated coverslips in 24-well plates and cultivated for 72 h. After the indicated treatments, the cells were washed once with prewarmed PBS and fixed with 4% (w/v) paraformaldehyde/PBS for 25 min. The cells were permeabilized with 1% Triton X-100/PBS for 5 min and blocked for 1 h with 10% normal goat serum/PBS at RT. The cells were incubated with primary antibodies in 1% BSA/PBS for 2 h at RT, followed by incubation with secondary Alexa Fluor–conjugated antibodies in the dark for 2 h at RT. The nuclei were counterstained with Hoechst 33342 (2 μg/ml/PBS) for 10 min at RT, and the coverslips were mounted in fluorescent mounting medium (Dako) onto microscope slides. The cells were analyzed with the ApoTome Imaging system and processed with ZEN software (both from Zeiss).
RNA extraction and semiquantitative RT-PCR
HEK293E cells (0.1 × 105) were seeded in 6-well plates, transfected with the CFTR exon 9 mini gene construct, and cultivated for an additional 48 h. RNA was isolated using the RNeasy Mini kit (Qiagen). 600 ng of total RNA was reverse-transcribed with anchored oligo(dT) primer using the Transcriptor High Fidelity cDNA Synthesis kit (Roche Applied Science). As a template for transcript amplification in a semiquantitative RT-PCR in a 25-μl reaction, 17.9 μl of cDNA (diluted 1:10 with DEPC-H2O) was used with 5 μl of 5× GoTaq buffer, 0.1 μl of GoTaq DNA polymerase (Promega), and a 2 mm concentration of the following specific primer pairs: CFTR mini gene (5′-CAACTTCAAGCTCCTAAGCCACTGC-3′ and 5′-TAGGATCCGGTCACCAGGAAGTTGGTTAAATCA-3′); HDAC6 (5′-GGTTCGCCCAGCTAACCCACCT-3′ and 5′-GGAACTCTCACGGTGCAGCTCC-3′); SKAR α/β (5′-CCTTCATAAACCCACCCATTGGGACAG-3′ (SKAR α/β for), 5′-CCAAAACCATCCAGGTTCCACAGCAG-3′ (SKAR α for), 5′-CCAAAACCATCCAGAATTTATATGACCTGG-3′ (SKAR β for), and 5′-GTGGTGGAGAAAGCCGCCTGAG-3′ (SKAR rev)); and ActB (5′-TGACCCAGATCATGTTTGAGAC-3′ and 5′-GAGGTAGTCTGTCAGGTCCC-3′). PCR products were separated by electrophoresis using a 2% (w/v) agarose/TBE gel stained with Midori Green Advance (Biozym) and photoimaged with a gel documentation system (Vilber Lourmat).
Data analysis, quantifications, and statistics
Western blotting and RT-PCR signals were quantified using the ImageJ software (version 1.52a, National Institute of Health). The statistical significance of the data was calculated with paired, two-sided Student's t test. Probability values below 0.05 (*, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005) were considered as statistically significant. Quantitative data are expressed as mean ± S.E. from at least three independent experiments.
Author contributions
F. H. and P. J. K. conceptualization; F. H. formal analysis; F. H. and P. J. K. supervision; F. H. validation; F. H. and H. G. investigation; F. H. visualization; F. H. and P. J. K. writing-original draft; P. J. K. funding acquisition.
Supplementary Material
Acknowledgments
We thank Catherine M. Thömmes for cloning of the pCMV-3xFLAG-FUS construct, Emanuelle Buratti for the pTB-CFTR exon 9 mini gene, and Manuela Neumann and Jorge García Morato for critical reading of the manuscript.
This work was supported by the NOMIS Foundation, German Research Foundation (DFG) Grant KA1675/3-2, the German Center for Neurodegenerative Diseases (DZNE) within the Helmholtz Association, and the Hertie Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
F. Hans and P. J. Kahle, unpublished data.
- FTLD
- frontotemporal lobar degeneration
- Baf
- bafilomycin
- CFTR
- cystic fibrosis transmembrane conductance regulator
- eIF
- eukaryotic initiation factor
- ERK
- extracellular signal–regulated kinase
- FUS
- fused-in-sarcoma
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- GSK3β
- glycogen synthase kinase 3β
- HDAC6
- histone deacetylase 6
- HEK
- human embryonic kidney
- HMW
- higher molecular weight smear
- HRP
- horseradish peroxidase
- JNK
- c-Jun N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- NAC
- N-acetylcysteine
- PKR
- protein kinase R
- PERK
- PKR-like endoplasmic reticulum kinase
- RIPA
- radioimmunoprecipitation assay
- SG
- stress granule
- SKAR
- ribosomal subunit protein S6 kinase 1 Aly/REF-like target
- TAR
- transactive response element
- TDP-43
- TAR DNA–binding protein of 43 kDa
- RBP
- RNA-binding protein
- ER
- endoplasmic reticulum
- HRI
- heme-regulated inhibitor
- mTOR
- mechanistic target of rapamycin
- TIA-1
- T-cell–restricted intracellular antigen-1
- TIAR
- TIA-1–related protein
- p70S6K
- S6 protein kinase
- UBPY
- ubiquitin isopeptidase Y
- WB
- Western blotting
- DMEM
- Dulbecco's modified Eagle's medium
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- Inh
- inhibitor
- Ni-NTA
- nickel-nitrilotriacetic acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Mackenzie I. R., Rademakers R., and Neumann M. (2010) TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9, 995–1007 10.1016/S1474-4422(10)70195-2 [DOI] [PubMed] [Google Scholar]
- 2. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- 3. Ratti A., and Buratti E. (2016) Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 138, 95–111 10.1111/jnc.13625 [DOI] [PubMed] [Google Scholar]
- 4. Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., and Ratti A. (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061 10.1111/j.1471-4159.2009.06383.x [DOI] [PubMed] [Google Scholar]
- 5. Volkening K., Leystra-Lantz C., Yang W., Jaffee H., and Strong M. J. (2009) Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability: implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res. 1305, 168–182 10.1016/j.brainres.2009.09.105 [DOI] [PubMed] [Google Scholar]
- 6. Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M. E., Mackenzie I. R., Capell A., Schmid B., Neumann M., and Haass C. (2010) ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29, 2841–2857 10.1038/emboj.2010.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu-Yesucevitz L., Bilgutay A., Zhang Y. J., Vanderwyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., Petrucelli L., and Wolozin B. (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE 5, e13250 10.1371/journal.pone.0013250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dewey C. M., Cenik B., Sephton C. F., Dries D. R., Mayer P. 3rd, Good S. K., Johnson B. A., Herz J., and Yu G. (2011) TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell. Biol. 31, 1098–1108 10.1128/MCB.01279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDonald K. K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G. A., and Vande Velde C. (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20, 1400–1410 10.1093/hmg/ddr021 [DOI] [PubMed] [Google Scholar]
- 10. Bentmann E., Neumann M., Tahirovic S., Rodde R., Dormann D., and Haass C. (2012) Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem. 287, 23079–23094 10.1074/jbc.M111.328757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andersson M. K., Ståhlberg A., Arvidsson Y., Olofsson A., Semb H., Stenman G., Nilsson O., and Aman P. (2008) The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 9, 37 10.1186/1471-2121-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson P., and Kedersha N. (2008) Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- 13. Donnelly N., Gorman A. M., Gupta S., and Samali A. (2013) The eIF2α kinases: their structures and functions. Cell Mol. Life Sci. 70, 3493–3511 10.1007/s00018-012-1252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panas M. D., Ivanov P., and Anderson P. (2016) Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215, 313–323 10.1083/jcb.201609081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aulas A., and Vande Velde C. (2015) Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front. Cell Neurosci. 9, 423 10.3389/fncel.2015.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y. R., King O. D., Shorter J., and Gitler A. D. (2013) Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bentmann E., Haass C., and Dormann D. (2013) Stress granules in neurodegeneration—lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J. 280, 4348–4370 10.1111/febs.12287 [DOI] [PubMed] [Google Scholar]
- 18. Wolozin B. (2012) Regulated protein aggregation: stress granules and neurodegeneration. Mol. Neurodegener. 7, 56 10.1186/1750-1326-7-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elden A. C., Kim H. J., Hart M. P., Chen-Plotkin A. S., Johnson B. S., Fang X., Armakola M., Geser F., Greene R., Lu M. M., Padmanabhan A., Clay-Falcone D., McCluskey L., Elman L., Juhr D., et al. (2010) Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 10.1038/nature09320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mackenzie I. R., Nicholson A. M., Sarkar M., Messing J., Purice M. D., Pottier C., Annu K., Baker M., Perkerson R. B., Kurti A., Matchett B. J., Mittag T., Temirov J., Hsiung G. R., Krieger C., et al. (2017) TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95, 808–816.e9 10.1016/j.neuron.2017.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brocker C., Thompson D. C., and Vasiliou V. (2012) The role of hyperosmotic stress in inflammation and disease. Biomol. Concepts 3, 345–364 10.1515/bmc-2012-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burg M. B., Ferraris J. D., and Dmitrieva N. I. (2007) Cellular response to hyperosmotic stresses. Physiol. Rev. 87, 1441–1474 10.1152/physrev.00056.2006 [DOI] [PubMed] [Google Scholar]
- 23. Zhou X., Naguro I., Ichijo H., and Watanabe K. (2016) Mitogen-activated protein kinases as key players in osmotic stress signaling. Biochim. Biophys. Acta 1860, 2037–2052 10.1016/j.bbagen.2016.05.032 [DOI] [PubMed] [Google Scholar]
- 24. Lu Z., Xu S., Joazeiro C., Cobb M. H., and Hunter T. (2002) The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9, 945–956 10.1016/S1097-2765(02)00519-1 [DOI] [PubMed] [Google Scholar]
- 25. Tatake R. J., O'Neill M. M., Kennedy C. A., Wayne A. L., Jakes S., Wu D., Kugler S. Z. Jr, Kashem M. A., Kaplita P., and Snow R. J. (2008) Identification of pharmacological inhibitors of the MEK5/ERK5 pathway. Biochem. Biophys. Res. Commun. 377, 120–125 10.1016/j.bbrc.2008.09.087 [DOI] [PubMed] [Google Scholar]
- 26. Abe J., Kusuhara M., Ulevitch R. J., Berk B. C., and Lee J. D. (1996) Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J. Biol. Chem. 271, 16586–16590 10.1074/jbc.271.28.16586 [DOI] [PubMed] [Google Scholar]
- 27. Hans F., Fiesel F. C., Strong J. C., Jäckel S., Rasse T. M., Geisler S., Springer W., Schulz J. B., Voigt A., and Kahle P. J. (2014) UBE2E ubiquitin-conjugating enzymes and ubiquitin isopeptidase Y regulate TDP-43 protein ubiquitination. J. Biol. Chem. 289, 19164–19179 10.1074/jbc.M114.561704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hans F., Eckert M., von Zweydorf F., Gloeckner C. J., and Kahle P. J. (2018) Identification and characterization of ubiquitinylation sites in TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem. 293, 16083–16099 10.1074/jbc.RA118.003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., and Baralle F. E. (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20, 1774–1784 10.1093/emboj/20.7.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fiesel F. C., Weber S. S., Supper J., Zell A., and Kahle P. J. (2012) TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 40, 2668–2682 10.1093/nar/gkr1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M. L., Kern J. V., Rasse T. M., Schmidt T., Springer W., Kirchner R., Bonin M., Neumann M., Baekelandt V., Alunni-Fabbroni M., Schulz J. B., and Kahle P. J. (2010) Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 29, 209–221 10.1038/emboj.2009.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee Y. C., Huang W. C., Lin J. H., Kao T. J., Lin H. C., Lee K. H., Lin H. C., Shen C. J., Chang W. C., and Huang C. C. (2018) Znf179 E3 ligase-mediated TDP-43 polyubiquitination is involved in TDP-43- ubiquitinated inclusions (UBI) (+)-related neurodegenerative pathology. J. Biomed Sci. 25, 76 10.1186/s12929-018-0479-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hebron M. L., Lonskaya I., Sharpe K., Weerasinghe P. P., Algarzae N. K., Shekoyan A. R., and Moussa C. E. (2013) Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6). J. Biol. Chem. 288, 4103–4115 10.1074/jbc.M112.419945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Denison S. R., Wang F., Becker N. A., Schüle B., Kock N., Phillips L. A., Klein C., and Smith D. I. (2003) Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene 22, 8370–8378 10.1038/sj.onc.1207072 [DOI] [PubMed] [Google Scholar]
- 35. Kwon Y. T., and Ciechanover A. (2017) The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 42, 873–886 10.1016/j.tibs.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 36. Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., and Anderson P. (2000) Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–1268 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kedersha N., Panas M. D., Achorn C. A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G. M., Ivanov P., and Anderson P. (2016) G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845–860 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bevilacqua E., Wang X., Majumder M., Gaccioli F., Yuan C. L., Wang C., Zhu X., Jordan L. E., Scheuner D., Kaufman R. J., Koromilas A. E., Snider M. D., Holcik M., and Hatzoglou M. (2010) eIF2α phosphorylation tips the balance to apoptosis during osmotic stress. J. Biol. Chem. 285, 17098–17111 10.1074/jbc.M110.109439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bounedjah O., Hamon L., Savarin P., Desforges B., Curmi P. A., and Pastré D. (2012) Macromolecular crowding regulates assembly of mRNA stress granules after osmotic stress: new role for compatible osmolytes. J. Biol. Chem. 287, 2446–2458 10.1074/jbc.M111.292748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moujalled D., James J. L., Parker S. J., Lidgerwood G. E., Duncan C., Meyerowitz J., Nonaka T., Hasegawa M., Kanninen K. M., Grubman A., Liddell J. R., Crouch P. J., and White A. R. (2013) Kinase inhibitor screening identifies cyclin-dependent kinases and glycogen synthase kinase 3 as potential modulators of TDP-43 cytosolic accumulation during cell stress. PLoS ONE 8, e67433 10.1371/journal.pone.0067433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hughes K., Nikolakaki E., Plyte S. E., Totty N. F., and Woodgett J. R. (1993) Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 12, 803–808 10.1002/j.1460-2075.1993.tb05715.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamann I., and Klotz L. O. (2013) Arsenite-induced stress signaling: modulation of the phosphoinositide 3′-kinase/Akt/FoxO signaling cascade. Redox Biol. 1, 104–109 10.1016/j.redox.2012.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harrison A. F., and Shorter J. (2017) RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 474, 1417–1438 10.1042/BCJ20160499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chew J., Cook C., Gendron T. F., Jansen-West K., Del Rosso G., Daughrity L. M., Castanedes-Casey M., Kurti A., Stankowski J. N., Disney M. D., Rothstein J. D., Dickson D. W., Fryer J. D., Zhang Y. J., and Petrucelli L. (2019) Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener. 14, 9 10.1186/s13024-019-0310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang P., Fan B., Yang P., Temirov J., Messing J., Kim H. J., and Taylor J. P. (2019) Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife 8, e39578 10.7554/eLife.39578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y., and Cohen T. J. (2019) Aggregation of the nucleic acid-binding protein TDP-43 occurs via distinct routes that are coordinated with stress granule formation. J. Biol. Chem. 294, 3696–3706 10.1074/jbc.RA118.006351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gasset-Rosa F., Lu S., Yu H., Chen C., Melamed Z., Guo L., Shorter J., Da Cruz S., and Cleveland D. W. (2019) Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357.e7 10.1016/j.neuron.2019.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mann J. R., Gleixner A. M., Mauna J. C., Gomes E., DeChellis-Marks M. R., Needham P. G., Copley K. E., Hurtle B., Portz B., Pyles N. J., Guo L., Calder C. B., Wills Z. P., Pandey U. B., Kofler J. K., et al. (2019) RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e8 10.1016/j.neuron.2019.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGurk L., Gomes E., Guo L., Mojsilovic-Petrovic J., Tran V., Kalb R. G., Shorter J., and Bonini N. M. (2018) Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9 10.1016/j.molcel.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolozin B. (2019) The evolution of phase-separated TDP-43 in stress. Neuron 102, 265–267 10.1016/j.neuron.2019.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wobst H. J., Delsing L., Brandon N. J., and Moss S. J. (2017) Truncation of the TAR DNA-binding protein 43 is not a prerequisite for cytoplasmic relocalization, and is suppressed by caspase inhibition and by introduction of the A90V sequence variant. PLoS ONE 12, e0177181 10.1371/journal.pone.0177181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cohen T. J., Hwang A. W., Unger T., Trojanowski J. Q., and Lee V. M. (2012) Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. EMBO J. 31, 1241–1252 10.1038/emboj.2011.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Prpar Mihevc S., Baralle M., Buratti E., and Rogelj B. (2016) TDP-43 aggregation mirrors TDP-43 knockdown, affecting the expression levels of a common set of proteins. Sci. Rep. 6, 33996 10.1038/srep33996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ryu H. W., Won H. R., Lee D. H., and Kwon S. H. (2017) HDAC6 regulates sensitivity to cell death in response to stress and post-stress recovery. Cell Stress Chaperones 22, 253–261 10.1007/s12192-017-0763-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yuan H. X., Wang Z., Yu F. X., Li F., Russell R. C., Jewell J. L., and Guan K. L. (2015) NLK phosphorylates Raptor to mediate stress-induced mTORC1 inhibition. Genes Dev. 29, 2362–2376 10.1101/gad.265116.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jo M., Lee S., Kim K., Lee S., Kim S. R., and Kim H. J. (2019) Inhibition of MEK5 suppresses TDP-43 toxicity via the mTOR-independent activation of the autophagy-lysosome pathway. Biochem. Biophys. Res. Commun. 513, 925–932 10.1016/j.bbrc.2019.04.088 [DOI] [PubMed] [Google Scholar]
- 57. Lee S. H., Park Y., Yoon S. K., and Yoon J. B. (2010) Osmotic stress inhibits proteasome by p38 MAPK-dependent phosphorylation. J. Biol. Chem. 285, 41280–41289 10.1074/jbc.M110.182188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hock E. M., Maniecka Z., Hruska-Plochan M., Reber S., Laferrière F., Sahadevan M. K. S., Ederle H., Gittings L., Pelkmans L., Dupuis L., Lashley T., Ruepp M. D., Dormann D., and Polymenidou M. (2018) Hypertonic stress causes cytoplasmic translocation of neuronal, but not astrocytic, FUS due to impaired transportin function. Cell Rep. 24, 987–1000.e7 10.1016/j.celrep.2018.06.094 [DOI] [PubMed] [Google Scholar]
- 59. Aiken C. T., Kaake R. M., Wang X., and Huang L. (2011) Oxidative stress-mediated regulation of proteasome complexes. Mol. Cell. Proteomics 10, R110.006924 10.1074/mcp.M110.006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pajares M., Jiménez-Moreno N., Dias I. H. K., Debelec B., Vucetic M., Fladmark K. E., Basaga H., Ribaric S., Milisav I., and Cuadrado A. (2015) Redox control of protein degradation. Redox Biol. 6, 409–420 10.1016/j.redox.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Webb A. E., and Brunet A. (2014) FOXO transcription factors: key regulators of cellular quality control. Trends Biochem. Sci. 39, 159–169 10.1016/j.tibs.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Patel J., McLeod L. E., Vries R. G., Flynn A., Wang X., and Proud C. G. (2002) Cellular stresses profoundly inhibit protein synthesis and modulate the states of phosphorylation of multiple translation factors. Eur. J. Biochem. 269, 3076–3085 10.1046/j.1432-1033.2002.02992.x [DOI] [PubMed] [Google Scholar]
- 63. Plescher M., Teleman A. A., and Demetriades C. (2015) TSC2 mediates hyperosmotic stress-induced inactivation of mTORC1. Sci. Rep. 5, 13828 10.1038/srep13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Peña-Oyarzun D., Troncoso R., Kretschmar C., Hernando C., Budini M., Morselli E., Lavandero S., and Criollo A. (2017) Hyperosmotic stress stimulates autophagy via polycystin-2. Oncotarget 8, 55984–55997 10.18632/oncotarget.18995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dardis A., Zampieri S., Canterini S., Newell K. L., Stuani C., Murrell J. R., Ghetti B., Fiorenza M. T., Bembi B., and Buratti E. (2016) Altered localization and functionality of TAR DNA binding protein 43 (TDP-43) in Niemann-Pick disease type C. Acta Neuropathol. Commun. 4, 52 10.1186/s40478-016-0325-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Neumann M., Rademakers R., Roeber S., Baker M., Kretzschmar H. A., and Mackenzie I. R. (2009) A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132, 2922–2931 10.1093/brain/awp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Seelaar H., Klijnsma K. Y., de Koning I., van der Lugt A., Chiu W. Z., Azmani A., Rozemuller A. J., and van Swieten J. C. (2010) Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J. Neurol. 257, 747–753 10.1007/s00415-009-5404-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mackenzie I. R., and Neumann M. (2016) Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J. Neurochem. 138, Suppl. 1, 54–70 10.1111/jnc.13588 [DOI] [PubMed] [Google Scholar]
- 69. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., and Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.