Abstract
Cellular membranes contain many lipids, some of which, such as sphingolipids, have important structural and signaling functions. The common sphingolipid glucosylceramide (GlcCer) is present in plants, fungi, and animals. As a major plant sphingolipid, GlcCer is involved in the formation of lipid microdomains, and the regulation of GlcCer is key for acclimation to stress. Although the GlcCer biosynthetic pathway has been elucidated, little is known about GlcCer catabolism, and a plant GlcCer-degrading enzyme (glucosylceramidase (GCD)) has yet to be identified. Here, we identified AtGCD3, one of four Arabidopsis thaliana homologs of human nonlysosomal glucosylceramidase, as a plant GCD. We found that recombinant AtGCD3 has a low Km for the fluorescent lipid C6-NBD GlcCer and preferentially hydrolyzes long acyl-chain GlcCer purified from Arabidopsis leaves. Testing of inhibitors of mammalian glucosylceramidases revealed that a specific inhibitor of human β-glucosidase 2, N-butyldeoxynojirimycin, inhibits AtGCD3 more effectively than does a specific inhibitor of human β-glucosidase 1, conduritol β-epoxide. We also found that Glu-499 and Asp-647 in AtGCD3 are vital for GCD activity. GFP-AtGCD3 fusion proteins mainly localized to the plasma membrane or the endoplasmic reticulum membrane. No obvious growth defects or changes in sphingolipid contents were observed in gcd3 mutants. Our results indicate that AtGCD3 is a plant glucosylceramidase that participates in GlcCer catabolism by preferentially hydrolyzing long-acyl-chain GlcCers.
Keywords: Arabidopsis thaliana, sphingolipid, ceramide, fatty acid, lipid metabolism, cell signaling, glucosylceramidase, plant biology, stress adaption
Introduction
Sphingolipids are important structural components of membranes and function as signaling molecules in eukaryotic cells (1–3). In plants, sphingolipids are major components of the plasma membrane, tonoplast, and endomembranes (4–6). Glucosylceramides (GlcCers)2 are a major group of plant sphingolipids (7). Plants produce a large variety of different GlcCers, and the GlcCer sphingoid bases can have different modifications, including hydroxylation or desaturation (7, 8). These sphingoid bases are linked to more than 10 different α-hydroxy fatty acids of varying chain lengths and n-9 desaturation levels, resulting in a variety of species of GlcCers (7, 8). Unlike animals, plants do not contain galactosylceramide, lactosylceramide, and sphingomyelin (8).
Although extensive studies have examined GlcCer structures in animals, plants, and fungi, data about their biological functions remain limited, and recent work has mainly focused on animals (9–12). For example, Nagata et al. (10) found that β-GlcCer induced immune responses via macrophage inducible lectin. Moreover, Brenna et al. (11) reported that C24:1 GlcCer, but not other GlcCers, acts as a self-antigen that can effectively activate invariant natural killer T cells. This research shows that GlcCers with different fatty acid chain lengths may have specific and unique biological functions, as proposed by Hannun and Obeid (12). However, fewer studies have examined the biological function of plant GlcCers. GlcCers are the major class of sphingolipid in plasma membranes of plants and appear to play important roles in protein sorting and organogenesis (13, 14). Inhibition of plant GlcCer biosynthesis led to strong morphological changes in the Golgi bodies and decreased protein secretion (13). Knockout of glucosylceramide synthase leads to seedling lethality, revealing that GlcCers are essential for plant development and growth (14). Therefore, understanding GlcCer metabolism in plants will likely provide insight into general and plant-specific membrane functions with key implications for plant development, defense, and other responses.
Here we report the cloning, heterologous expression, and enzymatic characterization of an Arabidopsis thaliana glucosylceramidase (encoded by At4g10060), which displays amino acid sequence similarity to the human nonlysosomal glucosylceramidase known as β-glucosidase 2 (GBA2), an enzyme involved in glucosylceramide catabolism (15, 16). We characterized the enzymatic activity of the protein encoded by At4g10060 and showed its optimal pH and substrate preference for long-acyl-chain GlcCers (C16-C18). Identification of this enzyme uncovers a missing link in our understanding of GlcCer metabolism in plants.
Results
Phylogenetic analysis and characterization of recombinant AtGCD3
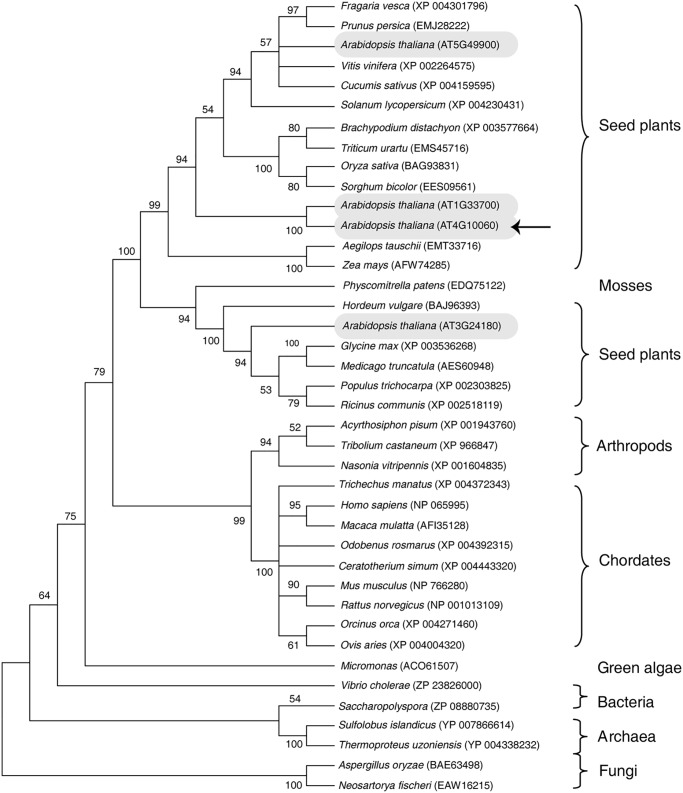
The A. thaliana genome encodes four putative GBA2-type glucosylceramidases (AT5G49900, AT1G33700, AT4G10060, and AT3G24180) on different chromosomes (Fig. 1); these proteins share 40, 37, 41, and 41% sequence identity with the human GBA2-type glucosylceramidase, respectively. To begin to investigate plant glucosylceramidases, we cloned and characterized A. thaliana At4g10060.
Figure 1.
Phylogenic analysis of AtGCD3. The phylogenetic tree of GBA2-type proteins was constructed by the maximum likelihood method using MEGA 6. The leaves of the tree are labeled with the organism genus and species and a database accession number. GBA2-type glucosylceramidases occur across Archaea, plants, and chordates. Four glucosylceramidase homologs (AT5G49900, AT1G33700, AT4G10060, and AT3G24180) in A. thaliana are shown in gray ovals. The arrow indicates AtGCD3, which we investigated in this study.
To examine the enzymatic activity of this protein, At4g10060 (designated AtGCD3) was expressed in Escherichia coli BL21 (DE3) as a His-tagged fusion protein. Recombinant AtGCD3 was successfully expressed in the soluble fraction and purified with nickel-nitrilotriacetic acid beads (Fig. 2A). After ultrafiltration, we obtained about 3 mg of purified protein per liter of E. coli culture.
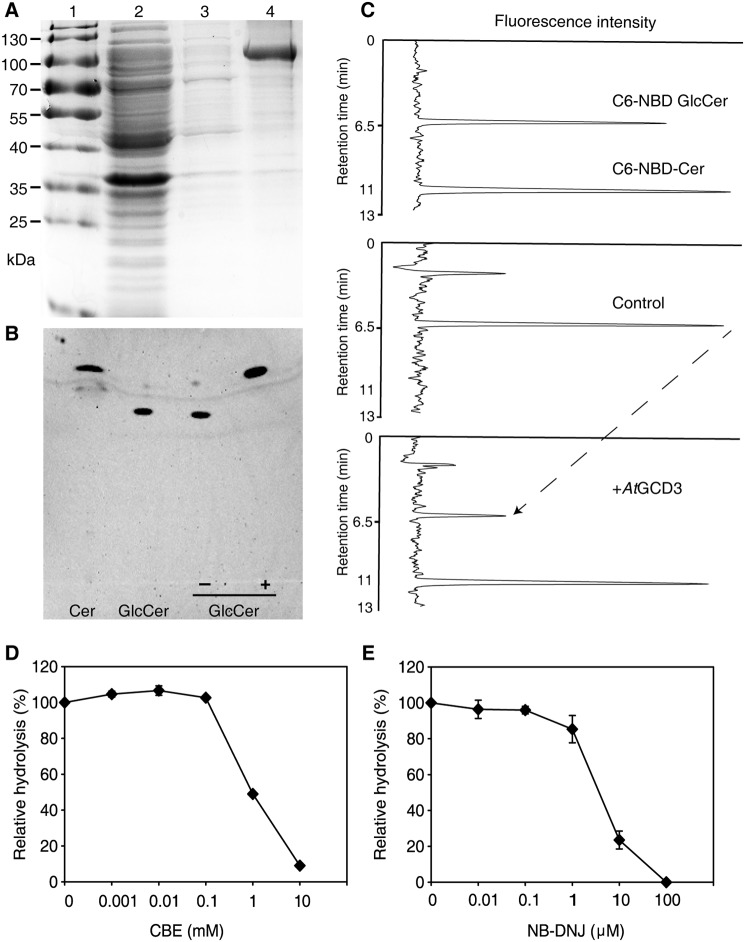
Figure 2.
Glucosylceramidase activity of purified, recombinant AtGCD3. A, purification of recombinant AtGCD3. Each fraction was subjected to SDS-PAGE and stained with Coomassie Brilliant Blue. Lane 1, protein marker; lane 2, flow-through fraction; lane 3, wash-through fraction; lane 4, purified AtGCD3. B, TLC demonstrating that the fluorescent ceramide was released from C6-NBD GlcCer by AtGCD3. In each reaction, 100 pmol of C6-NBD GlcCer was incubated with (+) or without (−) recombinant AtGCD3 for 60 min and dissolved in MeOH as described under “Experimental procedures.” C, HPLC showing the C6-NBD-Cer released from C6-NBD GlcCer by AtGCD3. Enzyme assays were conducted as described under “Experimental procedures.” The reaction was stopped by adding 4 volumes of acetonitrile, and the retention times of standard C6-NBD GlcCer and C6-NBD Ceramide were 6.5 and 11 min, respectively. Note that when AtGCD3 was added, the peak at 11-min retention time increased, and the peak at 6.5 min (the dashed line) decreased. D and E, inhibitory effects of CBE and NB-DNJ on AtGCD3. Aliquots of 100 pmol of C6-NBD GlcCer were incubated with 1 μg of AtGCD3 and various concentrations of CBE (D) and NB-DNJ (E). After 1 h, samples were loaded onto TLC plates. Fluorescence was detected with a Typhoon Trio+ scanner. Values represent the mean from at least three independent experiments, with S.D. (error bars).
To examine whether AtGCD3 has glucosylceramidase activity, we used the fluorescent lipid C6-NBD-GlcCer as a substrate. When incubated with recombinant AtGCD3, C6-NBD GlcCer was completely hydrolyzed to C6-NBD ceramide, as shown by thin-layer chromatography (TLC; Fig. 2B). We also confirmed the reaction by HPLC, where the retention times of the standards C6-NBD GlcCer and C6-NBD ceramide were 6.5 and 11 min, respectively, in an acetonitrile/H2O mobile phase (Fig. 2C, top chart). Consistent with the TLC analysis, the HPLC analysis confirmed the release of C6-NBD ceramide from C6-NBD GlcCer by recombinant AtGCD3 (Fig. 2C, middle and bottom charts). These results suggested that AtGCD3 has glucosylceramidase activity.
The activity of the mammalian nonlysosomal glucosylceramidase GBA2 can be reversibly inhibited by alkylated imino sugars (15, 17, 18); for example, N-butyldeoxynojirimycin (NB-DNJ) exclusively inhibits human GBA2 (18), and conduritol B-epoxide (CBE) inhibits GBA and affects GBA2 activity (18–20). We found that both NB-DNJ and CBE inhibited the glucosylceramidase activity of AtGCD3 (Fig. 2, D and E). CBE showed an IC50 of 1 mm, and NB-DNJ showed an IC50 of 5 μm. This result indicated that, in contrast to GBA2, AtGCD3 was relatively insensitive to CBE.
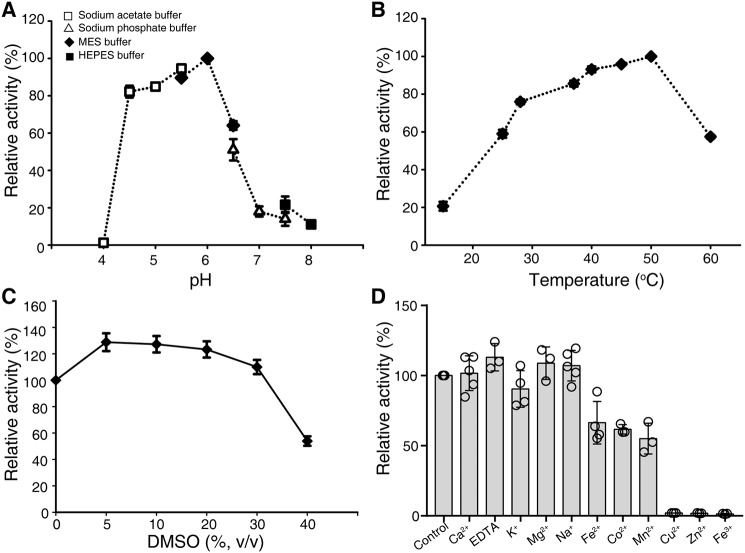
AtGCD3 exhibited low Michaelis constant (Km) values with C6-NBD GlcCer as a substrate, when compared with human neutral β-glycosylceramidase (KLrP) and fungal glucocerebrosidase (EGCrP1) (Table 1). AtGCD3 has a relatively higher enzyme activity at pH 4.5–6.0 and an optimal pH of 6.0, indicating that AtGCD3 is an acid glycosidase (Fig. 3A). The optimal temperature for AtGCD3 enzymatic activity was 40–50 °C (Fig. 3B). We found that 5–30% DMSO increased AtGCD3 activity (Fig. 3C). AtGCD3 did not need a metal ion for enzyme activity, and Cu2+, Zn2+, or Fe3+ inhibited its enzymatic activity (Fig. 3D). Taken together, these data demonstrated that the product of At4g10060 shows glucosylceramidase activity.
Table 1.
Comparison of kinetic parameters for glucosylceramidase using C6-NBD-GlcCer as a substrate
The reaction mixtures containing various concentrations of C6-NBD-GlcCer were incubated in 50 mm MES buffer, pH 6.0, at 40 °C for 5, 10, 15, 20, 30, 40, 60, 100, 150, and 240 s. The concentration of AtGCD3 was 1.3 μg/μl. The kinetic parameters were calculated using Hanes–Woolf plots. The values for human neutral β-glycosylceramidase (KLrP) and fungus endoglycoceramidase-related protein 1 (EGCrP1) are from Hayashi et al. (38) and Ishibashi et al. (26), respectively.
| Substrate | Enzyme | Km | Kcat | Kcat/Km |
|---|---|---|---|---|
| μm | s−1 | m−1 s−1 | ||
| C6-NBD-GlcCer | GCD3 | 1.52 ± 0.1 | (8.0 ± 0.9) ± 10−3 | (5.3 ± 0.6) ± 103 |
| KLrP | 4.6 ± 0.2 | (121.0 ± 5.5) ± 10−3 | (26.2 ± 0.5) ± 103 | |
| EGCrP1 | 5.8 ± 0.3 | (38.3 ± 0.2) ± 10−3 | (6.6 ± 0.4) ± 103 |
Figure 3.
General properties of the recombinant AtGCD3. A, pH dependence of glucosylceramidase activity of purified AtGCD3. To vary the pH, 50 mm sodium acetate buffer (pH 4.0–5.5), MES (pH 5.5–6.5), sodium phosphate buffer (pH 6.5–7.5), or HEPES buffer (pH 7.5–8.0) were used. B, optimal temperature. Assay mixtures were incubated at 15–60 °C. C, effects of DMSO on AtGCD3 activity. D, metal ion dependence of AtGCD3 activity. Assay mixtures were incubated with a 1 mm concentration of the indicated metal ions. Data represent the mean ± S.D. (error bars) (n ≥ 3) of independent experiments.
AtGCD3 preferentially hydrolyzes long-acyl-chain GlcCers
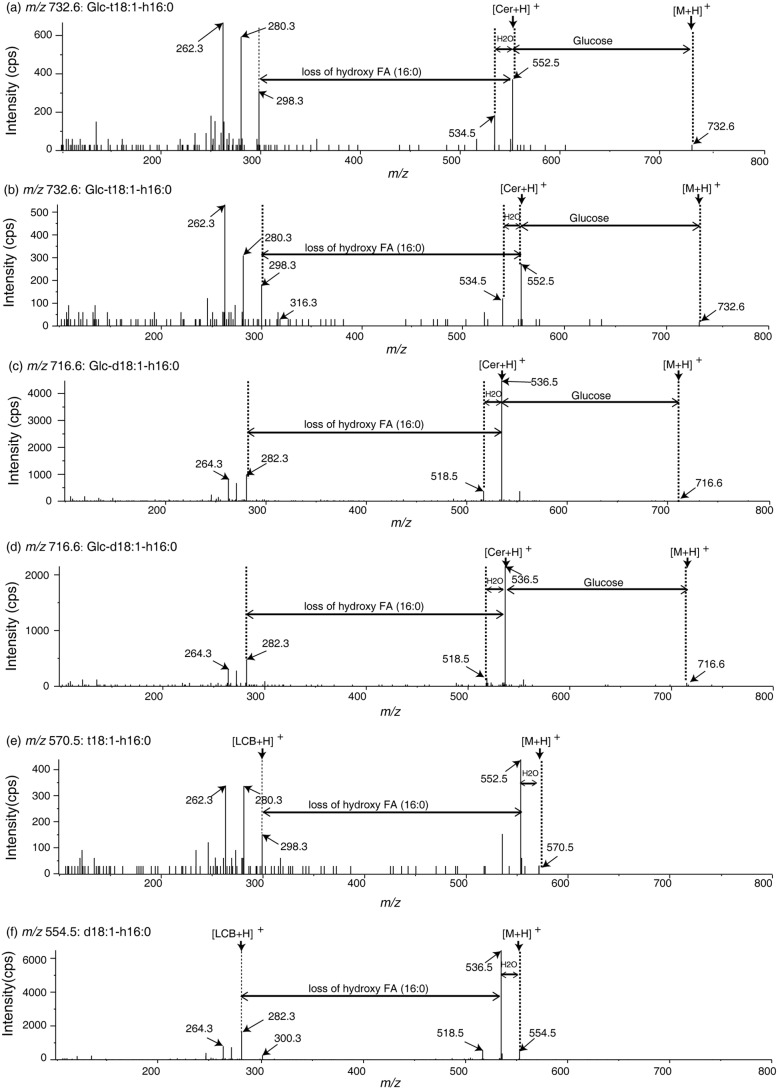
To determine the substrate specificity of AtGCD3, we used silica cartridges to purify naturally occurring GlcCers from WT Arabidopsis leaves. The fraction of naturally occurring GlcCers mainly contained t18:1 and d18:1 long-chain base GlcCers and also had d18:0 and t18:0 GlcCers (Fig. 4). We assayed the enzyme activity of AtGCD3 by incubating appropriate amounts of naturally occurring GlcCers with the enzyme for 24 h in the dark and then subjecting the reaction to LC-MS/MS analysis. As shown in Fig. 4 (A and B), peaks a and b were subsequently identified by MS/MS as Glc-t18:1-h16:0 (m/z 732.6) and Glc-d18:1-h16:0 (m/z 716.6) (Fig. 5). After incubation with AtGCD3, Glc-t18:1-h16:0 (peak a) and Glc-d18:1-h16:0 (peak b) decreased compared with corresponding peak in the control (Figs. 4 and 5). Along with the observed reduction of GlcCers, we observed two new peaks, peaks g and h, that were absent in solutions without AtGCD3; these were subsequently identified by MS/MS to be t18:1-h16:0 (m/z 570.5) and d18:1-h16:0 (m/z 554.5), respectively (Fig. 5). The fragmentation of m/z 716.6 to m/z 554.5 represents a neutral loss of glucose (m/z 162.1; Fig. 5). Statistical analysis showed that the two hydroxyceramides (ceramides with α-hydroxylated fatty acids) were produced from hydrolysis of the corresponding Glc-t18:1-h16:0 and Glc-d18:1-h16:0, respectively (Fig. 4, C and D). In addition to the two major long-acyl-chain GlcCers, Glc-d18:0-h16:0 is a minor Arabidopsis sphingolipid, and we observed hydrolysis of Glc-d18:0-h16:0 by AtGCD3 and an increase in the corresponding d18:0-h16:0 (top right corner in Fig. 4D). By contrast, very long-acyl-chain (fatty acids ≥ 20, VLACFA) GlcCers showed no significant changes when incubated with AtGCD3, and their corresponding hydroxyceramides also did not significantly increase (Fig. 4, C and D).
Figure 4.
Substrate specificity of recombinant AtGCD3. A and B, the recombinant AtGCD3 hydrolyzes C16 long-chain fatty acid GlcCer to C16 long-chain fatty acid hydroxyceramide in naturally occurring lipid substrates extracted from Arabidopsis leaves. Natural glucosylceramides were extracted from Arabidopsis, incubated with (B) or without (A) recombinant AtGCD3 at 30 °C for 24 h, and then subjected to HPLC ESI-MS/MS analysis. Each peak (a, b, c, d, e, and f) was further subjected to MS/MS, as shown in Fig. 5. C and D, quantitative analysis of hydrolysis using Arabidopsis leaf lipids. Naturally occurring glucosylceramides (C) were hydrolyzed to hydroxyceramides (D). The inset in D indicates that Glc-d18:0-h16:0 was hydrolyzed to d18:0-h16:0. E and F, the recombinant AtGCD3 preferentially hydrolyzes long-acyl-chain GlcCer. 500 nm commercially available GlcCers with different fatty acid chain length were incubated with (+) or without (−) AtGCD3 at 30 °C for 13 h, respectively, and analyzed by HPLC ESI-MS/MS. Commercial glucosylceramides (E) were hydrolyzed to ceramides (F). G, the recombinant AtGCD3 has no activity on galactosylceramide and MGDG. 500 nm Gal-d18:1–16:0 or MGDG was incubated with (+) or without (−) AtGCD3 at 30 °C for 13 h and analyzed by HPLC ESI-MS/MS. Data represent the mean ± S.D. (error bars) (n = 3) of independent experiments. The statistical significance of differences was determined with (+) or without (−) AtGCD3 in each GlcCer by Student's t test; p values are indicated where p < 0.05.
Figure 5.
Mass spectra of glucosylceramides and the product hydroxyceramides. a–d, mass spectra of purified natural GlcCer containing Glc-t18:1-h16:0 (a and b: m/z 732.6) and Glc-d18:1-h16:0 (c and d: m/z 716.6) with or without recombinant AtGCD3, as shown in Fig. 4. e and f, MS/MS spectra showing that C16 hydroxyceramide was generated through hydrolysis of relevant GlcCers with recombinant AtGCD3. t18:1-h16:0 (e, m/z 570.5) and d18:1-h16:0 (f, m/z 554.5) were hydrolyzed products of Glc-t18:1-h16:0 (b) and Glc-d18:1-h16:0 (d), respectively. The two-way arrows indicate neutral loss of sphingolipids, and one-way arrows show the product ion.
To confirm the substrate preference of AtGCD3, we used commercially available GlcCer with different acyl chain lengths as a substrate. As shown in Fig. 4E, Glc-d18:1-c12:0 and Glc-d18:1-c16:0 showed a significant decrease when incubated with AtGCD3. For Glc-d18:1-c18:0 and Glc-d18:1-c24:1, no significant change was detected in the presence of AtGCD3 (Fig. 4E); however, d18:1-c18:0 and a small amount of d18:1-c24:1 were detected (Fig. 4F). The small quantity of d18:1-c24:1 indicates that AtGCD3 could hydrolyze very long-acyl-chain GlcCers, although the glucosylceramidase activity was very low. Taken together, the results revealed that AtGCD3 preferentially hydrolyzed long-acyl-chain GlcCers.
To analyze whether the sugar headgroup is important for AtGCD3 activity, galactosylceramides and monogalactosyldiacylglycerol (MGDG) were used to evaluate the substrate specificity of AtGCD3. We used Gal-d18:1-c16:0 or MGDG as substrates and found no significant changes in the presence or absence of AtGCD3, indicating that galactosylceramide and MGDG cannot be hydrolyzed by AtGCD3 (Fig. 4G). These data suggest that the glucose headgroup of sphingolipids is important for AtGCD3 hydrolysis.
Determination of the catalytic amino acid residues of AtGCD3
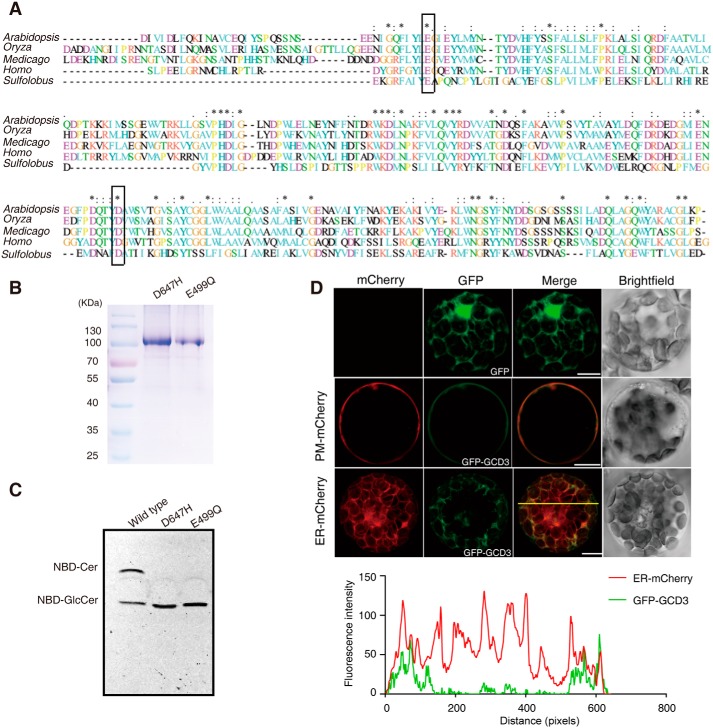
Glycosidases generally use a double displacement mechanism catalyzed by two enzymatic carboxylates; this mechanism involves formation and hydrolysis of a glycosyl intermediate (21). Conserved aspartic/glutamic acid residues generally serve as nucleophilic amino acids in acid-base catalysis of glycoside hydrolase (22). Alignment of the deduced amino acid sequences of GBA2-type glycosidases revealed that 6 Asp/Glu residues were highly conserved (Fig. 6A). Based on previous reports (22), we speculated that Glu-499 and Asp-647 might be the acid-base catalyst and nucleophile.
Figure 6.
Alignment of the AtGCD3 amino acid sequence with nonlysosomal glucosylceramidase and analysis of catalytic residues of AtGCD3. A, multiprotein alignment of the AtGCD3 amino acid sequence with other glucosylceramidases. Invariant residues are indicated with an asterisk; increased levels of conservation are indicated with a colon and a dot. The residues corresponding to the nucleophile Glu-499 and acid/base Asp-647 of AtGCD3 are boxed. Arabidopsis, gi|332657434 (AT4g10060) from A. thaliana; Oryza, gi|215704397 from Oryza sativa; Medicago, gi|355479745 from Medicago truncatula; Homo, NP_065995.1 from Homo sapiens; Sulfolobus, NC_002754.1 from S. solfataricus P2. B, SDS-PAGE of the purified E499Q and D647H mutant proteins. The two mutant enzymes were produced by site-directed mutagenesis, as shown in A (boxes), and were expressed and purified as described under “Experimental procedures.” C, TLC showing the hydrolysis of NBD-GlcCer by the WT and mutant AtGCD3. Note that AtGCD3 mutant proteins E499Q and D647H were completely inactive. D, subcellular localization of GFP-AtGCD3 fusion protein. GFP-AtGCD3 was co-transformed with organelle markers (mCherry) in WT Arabidopsis protoplasts. After incubation at 23 °C for 16 h under the light, fluorescence was observed by laser confocal microscopy. Row 3 panels are shown in three-dimensional maximum projection (eight sections at 0.84-μm step size), and others are single-plane. Free GFP was used as control (row 1 panels). The graph below shows the overlap of fluorescence intensity peaks along profiles as indicated in the merged micrograph. Bars, 10 μm.
To confirm whether these two amino acids function as catalytic residues, we used site-directed mutagenesis to produce the mutants E499Q and D647H and then expressed and purified the two mutant enzymes, which gave similar concentrations and degrees of purification (Fig. 6B). We found that the mutants assayed at 30 °C on C6-NBD GlcCer in 50 mm MES, pH 6.0, were completely inactive (Fig. 6C), suggesting that Glu-499 and Asp-647 function as an acid-base catalyst and nucleophile in AtGCD3. These invariant amino acids at the position preceding the catalytic glutamic and aspartic acids are conserved in the GH116 family (22).
Subcellular localization of AtGCD3
Subcellular location often indicates protein function, and earlier subcellular fractionation experiments using density gradient centrifugation and free-flow electrophoresis indicated that GBA2 localizes close to the cell surface (9, 15, 22). Glucosylceramide occurs in the plasma membrane in plants (4–6). To investigate whether AtGCD3 is linked to the distribution of GlcCers, AtGCD3 with enhanced GFP fused to the N-terminal was co-transformed with plasma membrane (PM), and endoplasmic reticulum (ER) markers into Arabidopsis protoplasts. As shown in Fig. 6D, AtGCD3 mainly localized to the PM and partially to the ER.
Characterization of T-DNA insertion alleles of AtGCD3
To test the function of AtGCD3 in planta, we identified two T-DNA insertion mutants, SALK_099838 (gcd3-1) in the first intron of AtGCD3 and SALK_019663 (gcd3-2) with a T-DNA insertion in the promoter region (Fig. 7A). The positions of the T-DNA insertions were verified by sequencing. The gcd3-1 and gcd3-2 alleles were both identified as homozygotes. RT-PCR experiments detected no AtGCD3 transcripts in gcd3-1 plants but showed that gcd3-2 mutants retained some AtGCD3 transcripts (Fig. 7A). Therefore, gcd3-1 was used in subsequent experiments. We found that the gcd3-1 mutants showed no obvious phenotype compared with WT (Fig. 7B).
Figure 7.
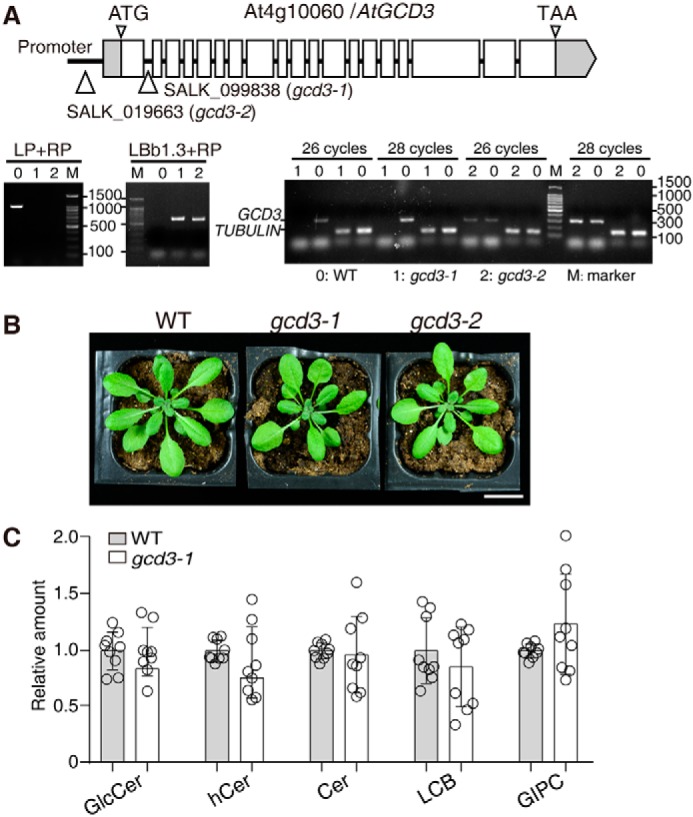
Characterization of T-DNA insertion mutants of AtGCD3. A, schematic diagram of the GCD3 locus. Gray boxes, UTR; white boxes, exons. Thin lines between exons represent introns. T-DNA was inserted in the promoter region of AtGCD3 in SALK_019663 (gcd3-2) and was inserted in the first intron of AtGCD3 in SALK_099838 (gcd3-1). The triangles indicate the insertion sites. The T-DNA insertion mutants gcd3-1 and gcd3-2 were homozygotes, as verified by two-round PCR. Primer LBb1.3 and RP indicated the existence of the T-DNA insertion, and primer LP and RP showed that both are homozygotes. RT-PCR confirmed that gcd3-1 was a genuine null mutant. TUBULIN was used as an internal control. B, normal phenotype of a 32-day-old gcd3-1 plant. Bar, 2 cm. C, sphingolipid profiles of 3-week-old rosette leaves of WT and gcd3-1 plants. The contents of sphingolipid in WT and gcd3-1 plants were quantified after extraction, separation, and identification by HPLC ESI-MS/MS as described under “Experimental procedures.” All data are available in Table S1. The relative values indicate the level of sphingolipids in gcd3-1 relative to that found in WT (set as 1). The values represent the mean ± S.D. (error bars) (n = 9 independent experiments). Significance was determined using Student's t test.
We next performed comprehensive analyses of sphingolipids in the gcd3-1 mutants. Electrospray ionization-MS/MS (ESI-MS/MS)-based profiling was used to assess long-chain bases, GlcCers, ceramides, hydroxyceramides, and glycosyl inositolphosphoceramides (GIPCs). Analyses were performed on extracts from rosettes of 3-week-old plants. We found that sphingolipids in the gcd3-1 mutants showed no significant changes compared with WT (Fig. 7C), indicating that other redundant enzymes might hydrolyze long-acyl-chain GlcCers in Arabidopsis.
Discussion
The A. thaliana genome encodes four putative glucosylceramidases (AT1g33700, AT3g24180, AT5g49900, and AT4g10060), which are annotated as GBA2-type proteins. Microarray analysis indicated that these loci are expressed in different tissues in developing embryos, dry seed, mature pollen, and elsewhere (23). AT5g49900 is predicted to be expressed exclusively in pollen (23), and the contents of glucosylceramides in leaves and pollen are strikingly different (24). AT3g24180 differs slightly from the others, as the predicted protein localized in the plasma membrane and may participate in stress responses. For this first characterization of a plant glucosylceramidase, we focused on one of these, AtGCD3 (AT4g10060), to take advantage of the available mutant line. We found that the gcd3 loss-of-function mutants showed no obvious defect in growth or sphingolipid contents. As our phylogenetic analysis showed that AT1g33700 is closely related to AtGCD3, we therefore speculate that it might have redundant functions with AtGCD3. Further research will elucidate the functions of these four proteins in sphingolipid metabolism.
As a first characterization of a plant glucosylceramidase, this study presents the characterization of the AtGCD3 glucosylceramidase from A. thaliana. Our molecular analysis using lipids purified from Arabidopsis leaves revealed that it acts mainly on long-acyl-chain glucosylceramides (fewer than 20 fatty acids). Additionally, site-directed mutagenesis of conserved glutamic/aspartic amino acids identified Glu-499 and Asp-647 as the likely acid/base and the nucleophile of the reaction, respectively. Amino acid sequence analysis showed that AtGCD3 belongs to the nonlysosome glucosylceramidase family GH116. Subcellular localization analysis revealed that AtGCD3 mainly localizes in the plasma membrane and near the ER. Our results demonstrate that this plant glucosylceramidase can hydrolyze naturally occurring long-acyl-chain glucosylceramides.
AtGCD3 displays most of the typical features of GBA2 (similar subcellular localization, pH optimum, kinetic parameters, etc.), and the Arabidopsis genome has no predicted GBA-like protein. We detected some differences between GBA2 and AtGCD3. In particular, AtGCD3 was relatively insensitive to CBE, which is an irreversible inhibitor of GBA and reduces GBA2 activity (18–20, 25). However, another inhibitor revealed the similarities between GBA2 and AtGCD3. NB-DNJ inhibits GBA2 but not GBA (19), and our results showed that NB-DNJ inhibits AtGCD3 more effectively than CBE. These observations confirmed that AtGCD3 is a GBA2-like glucosylceramidase, and not GBA-like.
AtGCD3 hydrolyzes glucosylceramides, with a substrate preference for long-acyl-chain glucosylceramides. The recombinant AtGCD3 hydrolyzed long chain fatty acid GlcCer in naturally occurring lipid substrates extracted from Arabidopsis leaves (Fig. 4, C and D). Using commercially available GlcCers with different fatty acid chain lengths as a substrate revealed that AtGCD3 has a strong preference for long-acyl-chain GlcCer substrates and very low enzymatic activity on very long-acyl-chain GlcCer substrates (Fig. 4). Previous reports indicate that the method we used for testing substrate specificity works well for VLCFA-containing GlcCers (26). Therefore, the observed substrate specificity was unlikely to be caused by the low solubility of VLCFA-containing GlcCer in the reaction buffer.
The substrate glucose headgroup is specific for enzymatic activity. The purified GlcCer fraction from leaves contains large quantities of MGDG. However, AtGCD3 cannot hydrolyze MGDG and failed to hydrolyze galactosylceramide (Fig. 4G). These results indicate that the sugar headgroup might be important for AtGCD3 activity in plants.
AtGCD3 mainly localizes in the plasma membrane and in close association with the ER membrane. Bioinformatics analysis showed that AtGCD3 has neither a transmembrane domain nor a signal peptide. Given the absence of a signal peptide, it remains unclear how AtGCD3 reaches its final destination. Homology modeling with β-glucosidase from Thermoanaerobacterium xylanolyticum TxGH116 (27) produced no additional information about the subcellular localization. Human GBA2 is also a nonintegral membrane protein that localizes at the cytosolic surface of the ER and Golgi, where it closely associates with membrane phospholipids (28). PM localization can be achieved by specific lipid-protein interactions. Therefore, we speculate that the lipid composition of the PM directly influences and drives the insertion of the AtGCD3 protein within the membrane bilayer (29); however, this remains an intriguing prospect for future research.
GlcCers and GIPCs are two of the glycosphingolipids found in Arabidopsis leaves (7). Ceramides are synthesized in the ER, and certain species (ceramides containing VLCFA bound to t18:0) are preferentially trafficked to the Golgi for GIPC biosynthesis. Ceramides remaining in the ER are available for incorporation into GlcCers (30). As shown in Fig. 7, sphingolipid profiling showed that the GIPCs show no significant change in the gcd3 mutants compared with WT. We speculate that knockout of AtGCD3 results in no perturbation in the flux of ceramide to GIPC, or another AtGCD3 homolog (AT1g33700) may have a redundant function in the gcd3 mutants.
Experimental procedures
Materials
A. thaliana WT plants of the Columbia (Col-0) ecotype were used in this study. T-DNA insertion mutants (SALK_099838, SALK_019663) were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Plant seeds were sown on soil after 2 days of stratification at 4 °C and then grown under 16-h light/8-h dark at 22 °C.
N-[6-[(7-Nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl]-d-erythro-sphingosine (C6-NBD-ceramide), C6-NBD-GlcCer, d-glucosyl-β-1,1′-N-lauroyl-d-erythro-sphingosine (C12 glucosyl(β) ceramide d18:1/12:0), d-glucosyl-β-1,1′-N-palmitoyl-d-erythro-sphingosine (C16 glucosyl(β) ceramide d18:1/16:0), d-glucosyl-β-1,1′-N-oleoyl-d-erythro-sphingosine (C18:1 glucosyl(β) ceramide d18:1/18:1(9Z)), d-glucosyl-β-1,1′-N-nervonoyl-d-erythro-sphingosine (C24:1 glucosyl(β) ceramide d18:1/24:1(15Z)), d-galactosyl-β-1,1′ N-palmitoyl-d-erythro-sphingosine (C16 glucosyl(β) ceramide d18:1/16:0), MGDG, and the sphingolipid internal standard (C17 base d-erythro-sphingosine, C12-ceramide, and C12-glucosylceramide) were purchased from Avanti Polar Lipids (Alabaster, AL). NB-DNJ and CBE were from Santa Cruz Biotechnology, Inc. (Dallas, TX). The Easy Mutagenesis System was purchased from TransGen Biotech (Beijing, China).
Extraction of natural glucosylceramides
Arabidopsis GlcCer was extracted from leaves according to the method of Bligh and Dyer (31). The total lipid extract was separated into neutral lipid (primarily free sterols) and glycolipid fractions using silica SepPak cartridges (Agela Technologies, Inc.) (32). The crude lipid extract dissolved in 2–3 ml of chloroform was applied to an equilibrated silica cartridge attached to the barrel of a 10-ml glass syringe. Residual lipids were washed into the column with 20 ml of chloroform, and neutral lipids were eluted with an additional 15 ml of chloroform/acetic acid (100:1, v/v). Ceramides and hydroxyceramides were eluted by adding 10 ml of chloroform/acetone (80:20, v/v). Glucosylceramides were eluted by adding 10 ml of chloroform/acetone (20:80, v/v). The glucosylceramide fraction was dried under nitrogen and resuspended in methanol. Natural GlcCer was identified by LC-MS/MS (AB SCIEX 5600 plus triple TOF MS) using acetonitrile as a solvent.
Sequence analysis
The alignment of AtGCD3 with related proteins was conducted with ClustalX. The phylogenetic tree was constructed, and bootstrap tests were calculated using the maximum likelihood method in MEGA 6.0.
Construction of vectors
An Arabidopsis cDNA clone with a full-length ORF of At4g10060 (stock name U21964) was ordered from the Arabidopsis Biological Resources Center. The AtGCD3 ORF was amplified using primers 10060ET-F (5′-CGGGATCCATGGAGAAGAATGGTCACACGGAAA-3′) and 10060ET-R (5′-GCGTCGACTTACAAGCGGAGAGTCTTGAGAACG-3′), which introduced BamHI and SalI restriction sites, respectively. The full-length coding sequence of AtGCD3 was subcloned into the pET28a plasmid (Novagen). The ORF was also amplified using primers with an AscI restriction site and then subcloned into the dephosphorylated GFP transient expression vector pX-DG and DsRed transient expression vector pX-DR for subcellular localization (33).
Site-directed mutagenesis
The mutants of AtGCD3 (E499Q and D647H) were generated by site-directed mutagenesis from the pET10060 plasmid, following the manufacturer's instructions. The mutagenic primers were as follows (mismatches underlined): E499Q-F, 5′-GGCCAATTCATCTACCTACAAGGAATTGAG-3′; E499Q-R, 5′-GTAGGTAGATGAATTGGCCAATGTTCTCTT-3′; D647H-F, 5′-TTTCCGGATCAGACATATCATGCTTGGAGT-3′; D647H-R, 5′-GATATGTCTGATCCGGAAACCCTTCGTTTTCG-3′. The mutant genes were confirmed by complete sequencing.
Expression and purification of recombinant AtGCD3
AtGCD3 (At4g10060) was expressed in E. coli BL21(DE3) transformed with the AtGCD3 ORF cloned in pET28a. After incubating the cells at 37 °C with shaking in 100 ml of Luria–Bertani medium supplemented with 100 μg/ml kanamycin and 30 μg/ml chloramphenicol until the A600 reached ∼0.7, isopropyl-d-thiogalactopyranoside was added to 0.5 mm to induce transcription. After being kept at 18 °C for 12 h, the cells were harvested by centrifugation (8,000 × g for 10 min) and suspended in lysis buffer (50 mm potassium phosphate buffer, pH 8.0, 300 mm KCl, 10 mm imidazole, 10% glycerol, 0.1% Triton X-100) and then incubated for 30 min on ice with 1 mg/ml lysozyme and sonicated for 15 × 10 s. Cell debris was removed by centrifugation at 12,000 × g for 30 min at 4 °C, and the supernatant was applied to a column filled with nickel-nitrilotriacetic acid His-bind beads (Novagen), which was equilibrated with lysis buffer. Then the column was washed twice with wash buffer (50 mm potassium phosphate, pH 8.0, 300 mm KCl, 20 mm imidazole, 10% glycerol) and eluted four times with elution buffer (50 mm potassium phosphate, pH 8.0, 300 mm KCl, 250 mm imidazole, 10% glycerol). The purified enzyme went through an ultrafiltration concentration step Amicon Ultra-0.5 (Merck Millipore) and was stored in enzyme storage buffer (50 mm potassium phosphate buffer, pH 7.0, 150 mm KCl, 1 mm DTT, 1 mm EDTA, 50% glycerol). The protein concentration was determined with a Quick Start Bradford Protein Assay Kit (Bio-Rad).
Enzyme activity
An aliquot of 100 pmol of C6-NBD GlcCer was incubated at 30 °C for 1 h with a suitable amount of enzyme in 20 μl of 50 mm MES buffer, pH 6.0, containing 5% DMSO. The reaction mixture was then dried using a SpeedVac concentrator, dissolved in 10 μl of MeOH, and centrifuged (12,000 × g for 5 min), and then the supernatant was applied to a TLC silica gel plate (Merck), which was developed with chloroform/methanol/water (65:25:4, v/v/v). The plate was scanned in a Typhoon Trio+ scanner (GE Healthcare) with the fluorescence mode set for excitation at 475 nm for C6-NBD GlcCer. The extent of hydrolysis was calculated as follows: hydrolysis (%) = (band area for NBD-ceramide) × 100/(band area for NBD-ceramide + band area for remaining NBD-GlcCer). For HPLC analysis, the enzyme reaction was stopped by adding a 4-fold volume of acetonitrile and centrifuged at 12,000 × g for 5 min at room temperature. The solution was then subjected to HPLC (DIONEX UltiMate 3000) analysis using acetonitrile/H2O (80:20, v/v) as the mobile phase. The extent of hydrolysis was calculated as follows: hydrolysis (%) = (peak area for NBD-ceramide) × 100/(peak area for NBD-ceramide + peak area for remaining NBD-GlcCer).
Activity on GlcCer from Arabidopsis leaves
An appropriate amount (5.8 μmol) of GlcCer purified from Arabidopsis leaves was incubated at 30 °C for 24 h with 1 μg of enzyme in 20 μl of 50 mm MES buffer, pH 6.0, containing 5% DMSO. After incubation, samples were diluted 4-fold with acetonitrile and centrifuged at 12,000 × g for 5 min at room temperature, and then sphingolipid internal standards were added. Samples were injected on the Shimadzu UFLC XR/Triple TOF 5600 LC/MS system and gradient-eluted from the Phenomenex Luna C8 column (150 × 2.0 mm, 3 μm). Peaks corresponding to the target analytes and internal standards were integrated and processed using the AB SCIEX MultiQuantTM 2.1 software. The sphingolipid composition was determined as described previously (34).
Characterization of the recombinant AtGCD3
The pH dependence of AtGCD3 activity was tested in the pH range of 4.0–8.0 using the following buffers (50 mm): sodium acetate (pH 4.0–5.5), MES (pH 5.5–6.5), sodium phosphate (pH 6.5–7.5), and HEPES (pH 7.5–8.0). Optimum temperature was determined in the range of 15–60 °C. The effect of organic solvents was determined using DMSO at 0–40%. The effects of metal ions were examined by adding CaCl2, KCl, NaCl, MgCl2, FeCl2, ZnCl2, CoCl2, FeCl3, MnCl2, CuCl2, or EDTA to the reaction at a final concentration of 1 mm. Kinetic constants of AtGCD3 were measured using C6-NBD GlcCer as a substrate ranging from 1 to 20 μm.
For inhibition of the recombinant AtGCD3, NB-DNJ and CBE were used (15, 17, 25). One microgram of purified AtGCD3 was incubated in the presence of increasing concentrations of the inhibitors (0.001–10 mm CBE and 0.01–100 μm NB-DNJ) in 50 mm MES buffer, pH 6.0, at 30 °C for 1 h and assayed by TLC as described above.
Subcellular localization of AtGCD3
The transient expression vector used was pX-DG with GFP in the N-terminal to produce GFP-AtGCD3. The CaMV 35S promoter was used to drive expression. Arabidopsis protoplasts were harvested using a sandwich method (35), and 10 μg of each plasmid, GFP-AtGCD3, and the respective organelle marker were co-transformed into the protoplasts, as described (36). After a 16-h incubation under light conditions, GFP and mCherry fluorescence were observed using a confocal laser-scanning microscope (LSM-780, Zeiss). GFP was excited with a 488-nm laser and mCherry with a 561-nm laser. Emission fluorescence signals were collected as follows: 500 to 540 nm for GFP, 590 to 640 nm for mCherry. The fluorescence of GFP and mCherry emissions was collected sequentially. Each single-fluorescence experiment showed no detectable crossover between green and red channels at the settings used for data collection.
T-DNA mutant characterization
T-DNA insertion was confirmed by PCR genotyping using the T-DNA right border primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′) and the gene-specific primers, as follows: SALK_099838 RP, 5′-TCTCTCGAACAAAATGGATGG-3′; LP primer, 5′-AGTTCAGTTTCCCTTTGACCG-3′; SALK_019663 RP primer, 5′-GTGGAGAAACAACTCCTGCAG-3′; LP primer, 5′-GAAAGATGTTGGCTTTGTTCG-3′, followed by sequencing.
RNA isolation and RT-PCR
Total RNA was isolated using the E.Z.N.A. Plant RNA Kit (R6827-01) according to the manufacturer's instructions (Omega Bio-tek). For each sample, 1 μg of RNA was reverse-transcribed into cDNA using the PrimeScript RT reagent kit with gDNA Eraser (TAKARA, DRR047A). Tissue-specific expression was assayed by RT-PCR using total RNA isolated from 3-week-old WT plants, and the relative expression of target genes was assessed using skip-intron primers (forward primer, 5′-GTCCCATTAGGTGGAATTGGTG-3′; reverse primer, 5′-CAGGCTCGTTGTATACGGTCCAAGA-3′), and TUBULIN was used as an internal control.
Sphingolipid analysis
Measurement of sphingolipids was performed by electrospray ionization/MS/MS analysis on a Shimadzu UFLC XR/Triple TOF 5600+ LC/MS system operating in a product ion mode as described previously (3, 37). Briefly, 30 mg of each lyophilized samples was homogenized. The internal standards were added, and the lipids from the leaf, together with the internal standard, were extracted with isopropyl alcohol/hexane/water (55:20:25, v/v/v). After incubation at 60 °C for 15 min, the supernatants were dried with nitrogen. The dried extract was de-esterified by dissolving in 1 ml of 33% methylamine solution in ethanol/water (7:3, v/v) and incubated at 50 °C for 1 h. After being dried with nitrogen, the sample was dissolved and injected on the Shimadzu UFLC XR/Triple TOF 5600+ LC/MS system: GS1, 50 p.s.i.; GS2, 40 p.s.i.; Cur gas, 20 p.s.i.; source temperature, 450 °C; voltage, 5000 V. Samples were gradient-eluted from Phenomenex Luna C8 column (150 × 2.0 mm, 3 μm) at a flow rate of 0.3 ml/min, and data were processed using the AB SCIEX MultiQuant 2.1 software.
Author contributions
G.-Y. D. and N. Y. conceived and designed research; G.-Y. D., J. Y., K.-E. L., D.-K. C., Z. L., F.-C. B., and C. R. conducted experiments; G.-Y. D. and N. Y. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the Arabidopsis T-DNA insertion line for AtGCD3 and organelle markers (CD3–960, CD3–1007, and CD3–975).
This work was supported by National Natural Science Foundation of China Grants 31570255 and 31771357, Natural Science Foundation of Guangdong Province Grant 2017A030311005, and Sun Yat-sen University Project 33000-31143406. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1.
- GlcCer
- glucosylceramide
- GCD
- glucosylceramidase
- GBA2
- human non-lysosomal glucosylceramidase 2
- NB-DNJ
- N-butyldeoxynojirimycin
- CBE
- conduritol B epoxide
- d18:0
- dihydrosphingosine
- t18:0
- trihydrosphingosine
- Glc-d18:0-h16:0
- glucosylceramide d18:0-h16:0
- Gal- d18:0-c16:0
- galactosylceramide d18:0-c16:0
- d18:0-h16:0
- hydroxyceramide d18:0–16:0
- d18:0-c16:0
- ceramide d18:0–16:0
- TLC
- thin-layer chromatography
- C6-NBD-ceramide
- N-[6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl]-d-erythro-sphingosine
- hydroxyceramide
- ceramide with α-hydroxylated fatty acid
- MGDG
- monogalactosyldiacylglycerol
- PM
- plasma membrane
- ER
- endoplasmic reticulum
- ESI
- electrospray ionization
- GIPC
- glycosyl inositolphosphoceramide.
References
- 1. Markham J. E., Lynch D. V., Napier J. A., Dunn T. M., and Cahoon E. B. (2013) Plant sphingolipids: function follows form. Curr. Opin Plant Biol. 16, 350–357 10.1016/j.pbi.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 2. Pata M. O., Hannun Y. A., and Ng C. K. Y. (2010) Plant sphingolipids: decoding the enigma of the Sphinx. New Phytol. 185, 611–630 10.1111/j.1469-8137.2009.03123.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bi F.-C., Liu Z., Wu J.-X., Liang H., Xi X.-L., Fang C., Sun T.-J., Yin J., Dai G.-Y., Rong C., Greenberg J. T., Su W.-W., and Yao N. (2014) Loss of ceramide kinase in Arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 26, 3449–3467 10.1105/tpc.114.127050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoshida S., and Uemura M. (1986) Lipid composition of plasma membranes and tonoplasts isolated from etiolated seedlings of mung bean (Vigna radiata L.). Plant Physiol. 82, 807–812 10.1104/pp.82.3.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sperling P., Franke S., Lüthje S., and Heinz E. (2005) Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 43, 1031–1038 10.1016/j.plaphy.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 6. Tjellström H., Hellgren L. I., Wieslander A., and Sandelius A. S. (2010) Lipid asymmetry in plant plasma membranes: phosphate deficiency-induced phospholipid replacement is restricted to the cytosolic leaflet. FASEB J. 24, 1128–1138 10.1096/fj.09-139410 [DOI] [PubMed] [Google Scholar]
- 7. Markham J. E., Li J., Cahoon E. B., and Jaworski J. G. (2006) Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281, 22684–22694 10.1074/jbc.M604050200 [DOI] [PubMed] [Google Scholar]
- 8. Warnecke D., and Heinz E. (2003) Recently discovered functions of glucosylceramides in plants and fungi. Cell Mol. Life Sci. 60, 919–941 10.1007/s00018-003-2243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ishibashi Y., Kohyama-Koganeya A., and Hirabayashi Y. (2013) New insights on glucosylated lipids: metabolism and functions. Biochim. Biophys. Acta 1831, 1475–1485 10.1016/j.bbalip.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 10. Nagata M., Izumi Y., Ishikawa E., Kiyotake R., Doi R., Iwai S., Omahdi Z., Yamaji T., Miyamoto T., Bamba T., and Yamasaki S. (2017) Intracellular metabolite β-glucosylceramide is an endogenous Mincle ligand possessing immunostimulatory activity. Proc. Natl. Acad. Sci. U.S.A. 114, E3285–E3294 10.1073/pnas.1618133114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brennan P. J., Tatituri R. V. V., Brigl M., Kim E. Y., Tuli A., Sanderson J. P., Gadola S. D., Hsu F.-F., Besra G. S., and Brenner M. B. (2011) Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat. Immunol. 12, 1202–1211 10.1038/ni.2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hannun Y. A., and Obeid L. M. (2011) Many ceramides. J. Biol. Chem. 286, 27855–27862 10.1074/jbc.R111.254359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melser S., Batailler B., Peypelut M., Poujol C., Bellec Y., Wattelet-Boyer V., Maneta-Peyret L., Faure J. D., and Moreau P. (2010) Glucosylceramide biosynthesis is involved in Golgi morphology and protein secretion in plant cells. Traffic 11, 479–490 10.1111/j.1600-0854.2009.01030.x [DOI] [PubMed] [Google Scholar]
- 14. Msanne J., Chen M., Luttgeharm K. D., Bradley A. M., Mays E. S., Paper J. M., Boyle D. L., Cahoon R. E., Schrick K., and Cahoon E. B. (2015) Glucosylceramides are critical for cell-type differentiation and organogenesis, but not for cell viability in Arabidopsis. Plant J. 84, 188–201 10.1111/tpj.13000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boot R. G., Verhoek M., Donker-Koopman W., Strijland A., van Marle J., Overkleeft H. S., Wennekes T., and Aerts J. M. (2007) Identification of the non-lysosomal glucosylceramidase as β-glucosidase 2. J. Biol. Chem. 282, 1305–1312 10.1074/jbc.M610544200 [DOI] [PubMed] [Google Scholar]
- 16. van Weely S., Brandsma M., Strijland A., Tager J. M., and Aerts J. M. (1993) Demonstration of the existence of a second, non-lysosomal glucocerebrosidase that is not deficient in Gaucher disease. Biochim. Biophys. Acta 1181, 55–62 10.1016/0925-4439(93)90090-N [DOI] [PubMed] [Google Scholar]
- 17. Overkleeft H. S., Renkema G. H., Neele J., Vianello P., Hung I. O., Strijland A., van der Burg A. M., Koomen G. J., Pandit U. K., and Aerts J. M. (1998) Generation of specific deoxynojirimycin-type inhibitors of the non-lysosomal glucosylceramidase. J. Biol. Chem. 273, 26522–26527 10.1074/jbc.273.41.26522 [DOI] [PubMed] [Google Scholar]
- 18. Walden C. M., Sandhoff R., Chuang C. C., Yildiz Y., Butters T. D., Dwek R. A., Platt F. M., and van der Spoel A. C. (2007) Accumulation of glucosylceramide in murine testis, caused by inhibition of β-glucosidase 2: implications for spermatogenesis. J. Biol. Chem. 282, 32655–32664 10.1074/jbc.M702387200 [DOI] [PubMed] [Google Scholar]
- 19. Ridley C. M., Thur K. E., Shanahan J., Thillaiappan N. B., Shen A., Uhl K., Walden C. M., Rahim A. A., Waddington S. N., Platt F. M., and van der Spoel A. C. (2013) β-Glucosidase 2 (GBA2) activity and imino sugar pharmacology. J. Biol. Chem. 288, 26052–26066 10.1074/jbc.M113.463562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daniels L. B., Glew R. H., Diven W. F., Lee R. E., and Radin N. S. (1981) An improved fluorometric leukocyte β-glucosidase assay for Gaucher's disease. Clin. Chim. Acta 115, 369–375 10.1016/0009-8981(81)90251-5 [DOI] [PubMed] [Google Scholar]
- 21. Cobucci-Ponzano B., Aurilia V., Riccio G., Henrissat B., Coutinho P. M., Strazzulli A., Padula A., Corsaro M. M., Pieretti G., Pocsfalvi G., Fiume I., Cannio R., Rossi M., and Moracci M. (2010) A new archaeal β-glycosidase from Sulfolobus solfataricus: seeding a novel retaining β-glycan-specific glycoside hydrolase family along with the human non-lysosomal glucosylceramidase GBA2. J. Biol. Chem. 285, 20691–20703 10.1074/jbc.M109.086470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferrara M. C., Cobucci-Ponzano B., Carpentieri A., Henrissat B., Rossi M., Amoresano A., and Moracci M. (2014) The identification and molecular characterization of the first archaeal bifunctional exo-β-glucosidase/N-acetyl-β-glucosaminidase demonstrate that family GH116 is made of three functionally distinct subfamilies. Biochim. Biophys. Acta 1840, 367–377 10.1016/j.bbagen.2013.09.022 [DOI] [PubMed] [Google Scholar]
- 23. Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., and Provart N. J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luttgeharm K. D., Kimberlin A. N., Cahoon R. E., Cerny R. L., Napier J. A., Markham J. E., and Cahoon E. B. (2015) Sphingolipid metabolism is strikingly different between pollen and leaf in Arabidopsis as revealed by compositional and gene expression profiling. Phytochemistry 115, 121–129 10.1016/j.phytochem.2015.02.019 [DOI] [PubMed] [Google Scholar]
- 25. Premkumar L., Sawkar A. R., Boldin-Adamsky S., Toker L., Silman I., Kelly J. W., Futerman A. H., and Sussman J. L. (2005) X-ray structure of human acid-β-glucosidase covalently bound to conduritol-B-epoxide. Implications for Gaucher disease. J. Biol. Chem. 280, 23815–23819 10.1074/jbc.M502799200 [DOI] [PubMed] [Google Scholar]
- 26. Ishibashi Y., Ikeda K., Sakaguchi K., Okino N., Taguchi R., and Ito M. (2012) Quality control of fungus-specific glucosylceramide in Cryptococcus neoformans by endoglycoceramidase-related protein 1 (EGCrP1). J. Biol. Chem. 287, 368–381 10.1074/jbc.M111.311340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Charoenwattanasatien R., Pengthaisong S., Breen I., Mutoh R., Sansenya S., Hua Y., Tankrathok A., Wu L., Songsiriritthigul C., Tanaka H., Williams S. J., Davies G. J., Kurisu G., and Cairns J. R. (2016) Bacterial β-glucosidase reveals the structural and functional basis of genetic defects in human glucocerebrosidase 2 (GBA2). ACS Chem. Biol. 11, 1891–1900 10.1021/acschembio.6b00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Körschen H. G., Yildiz Y., Raju D. N., Schonauer S., Bönigk W., Jansen V., Kremmer E., Kaupp U. B., and Wachten D. (2013) The non-lysosomal β-glucosidase GBA2 is a non-integral membrane-associated protein at the endoplasmic reticulum (ER) and Golgi. J. Biol. Chem. 288, 3381–3393 10.1074/jbc.M112.414714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Massimo A., Maura S., Nicoletta L., Giulia M., Valentina M., Elena C., Alessandro P., Rosaria B., and Sandro S. (2016) Current and novel aspects on the non-lysosomalβ-glucosylceramidase GBA2. Neurochem Res. 41, 210–220 10.1007/s11064-015-1763-2 [DOI] [PubMed] [Google Scholar]
- 30. Chen M., Markham J. E., and Cahoon E. B. (2012) Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 69, 769–781 10.1111/j.1365-313X.2011.04829.x [DOI] [PubMed] [Google Scholar]
- 31. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 32. Cahoon E. B., and Lynch D. V. (1991) Analysis of glucocerebrosides of Rye (Secale cereale L. cv. Puma) leaf and plasma membrane. Plant Physiol. 95, 58–68 10.1104/pp.95.1.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen S., Songkumarn P., Liu J., and Wang G. L. (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121 10.1104/pp.109.137125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Markham J. E., and Jaworski J. G. (2007) Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21, 1304–1314 10.1002/rcm.2962 [DOI] [PubMed] [Google Scholar]
- 35. Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., and Lin C.-S. (2009) Tape-Arabidopsis sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 10.1186/1746-4811-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson B. K., Cai X., and Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- 37. Wu J. X., Li J., Liu Z., Yin J., Chang Z. Y., Rong C., Wu J. L., Bi F. C., and Yao N. (2015) The Arabidopsis ceramidase AtACER functions in disease resistance and salt tolerance. Plant J. 81, 767–780 10.1111/tpj.12769 [DOI] [PubMed] [Google Scholar]
- 38. Hayashi Y., Okino N., Kakuta Y., Shikanai T., Tani M., Narimatsu H., and Ito M. (2007) Klotho-related protein is a novel cytosolic neutral β-glycosylceramidase. J. Biol. Chem. 282, 30889–30900 10.1074/jbc.M700832200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.