Abstract
Stressful environments accelerate the formation of isoaspartyl (isoAsp) residues in proteins, which detrimentally affect protein structure and function. The enzyme PROTEIN l-ISOASPARTYL METHYLTRANSFERASE (PIMT) repairs other proteins by reverting deleterious isoAsp residues to functional aspartyl residues. PIMT function previously has been elucidated in seeds, but its role in plant survival under stress conditions remains undefined. Herein, we used molecular, biochemical, and genetic approaches, including protein overexpression and knockdown experiments, in Arabidopsis to investigate the role of PIMTs in plant growth and survival during heat and oxidative stresses. We demonstrate that these stresses increase isoAsp accumulation in plant proteins, that PIMT activity is essential for restricting isoAsp accumulation, and that both PIMT1 and PIMT2 play an important role in this restriction and Arabidopsis growth and survival. Moreover, we show that PIMT improves stress tolerance by facilitating efficient reactive oxygen species (ROS) scavenging by protecting the functionality of antioxidant enzymes from isoAsp-mediated damage during stress. Specifically, biochemical and MS/MS analyses revealed that antioxidant enzymes acquire deleterious isoAsp residues during stress, which adversely affect their catalytic activities, and that PIMT repairs the isoAsp residues and thereby restores antioxidant enzyme function. Collectively, our results suggest that the PIMT-mediated protein repair system is an integral part of the stress-tolerance mechanism in plants, in which PIMTs protect antioxidant enzymes that maintain proper ROS homeostasis against isoAsp-mediated damage in stressful environments.
Keywords: plant physiology, protein deamidation, plant biochemistry, reactive oxygen species (ROS), Arabidopsis, antioxidant enzyme, isoAsp, PIMT, protein repair, ROS
Introduction
The survival of plants under various abiotic stress conditions is largely determined by their ability to maintain the structural and functional integrity of proteins because these stresses can have devastating effects on protein structure and biological function. Under these stressful environments, various spontaneous covalent modifications are known to occur in proteins, which often detrimentally affects their structure and function (1). One of these modifications includes the formation of abnormal isoaspartyl (isoAsp)4 residues that arise spontaneously in proteins from either the isomerization of aspartyl residues or the deamidation of asparaginyl residues (2). The formation of such isoAsp residues in the polypeptide backbone can disrupt the local secondary structure, thus compromising biological function of the proteins. Nevertheless, organisms have evolved a mechanism for the detection and repair of such isoAsp-mediated protein damage by employing an enzyme called PROTEIN l-ISOASPARTYL METHYLTRANSFERASE (PIMT, EC 2.1.1.77). PIMT repairs this damage by catalyzing the conversion of abnormal isoaspartyl residues to their normal aspartyl residues through an S-adenosyl-l-methionine (AdoMet)-dependent methyl esterification reaction (3–5). PIMT is a well-conserved enzyme in nature and is found in organisms across nearly all phylogenetic domains of life. A number of studies in microbial and mammalian systems have demonstrated its in vivo function as an essential intracellular protein-repairing enzyme and also its contribution in enhancing life span and survivability under stress conditions (6–10). Even though PIMT has been well-characterized in microbes and animals, such studies are rather limited in plants. So far, the deleterious effect of isoAsp and the physiological significance of PIMT in plants have been elucidated only in seeds. Unlike microbes and animals, PIMT enzymes are encoded by two genes (PIMT1 and PIMT2) in plants, and both the genes were shown to be involved in maintaining seed vigor and longevity (11–15). Apart from seeds, considerable PIMT activity was also reported in the vegetative organs of several plant species. Furthermore, a comparatively low level of PIMT activity in seedlings of Triticum aestivum (wheat) and Cicer arietinum (chickpea) was shown to be significantly increased in response to various abiotic stresses (16–18). Despite the demonstrated up-regulation of PIMT activity in several plant species, the involvement of PIMT in abiotic stress tolerance has not yet been properly explored. Furthermore, isoAsp-mediated protein damage and its possible adverse effects on plant growth and survival during abiotic stresses remain elusive. Thus, it is important to study how this protein-repairing enzyme PIMT contributes to the plant's survival under stressful environments.
In this study, we have elucidated the role and contribution of PIMT in plant's survival under stressful environments. We provide evidence that isoAsp accumulation in proteins is significantly increased in plants during the heat and oxidative stresses. We have shown that increased PIMT activity is important in restricting stress-induced isoAsp-mediated protein damage in plants. To further assess the role of PIMT in plant survivability under stress conditions, overexpression and RNAi lines of PIMT1 and PIMT2 in Arabidopsis were generated and characterized. We have demonstrated a strong correlation of increased PIMT activity with increased tolerance to heat and oxidative stresses, and increased isoAsp accumulation with compromised stress tolerance. Analyses further demonstrate that the PIMT-mediated protein repair system is critical in maintaining ROS homeostasis apparently by protecting the catalytic efficiency of antioxidant enzymes that are otherwise affected by stress-induced isoAsp formation. We provide clear evidence that superoxide dismutase (SOD) and catalase (CAT) enzymes are susceptible to isoAsp formation under stressful conditions impeding their function, and PIMT repairs such isoAsp residues in these antioxidant enzymes and restores their biological function. Thus, PIMT forms the first tier of protection shielding a second tier of protective proteins from dysfunction, allowing them to provide protection against diverse stresses.
Results
Heat and oxidative stresses induce isoAsp accumulation in proteins in Arabidopsis
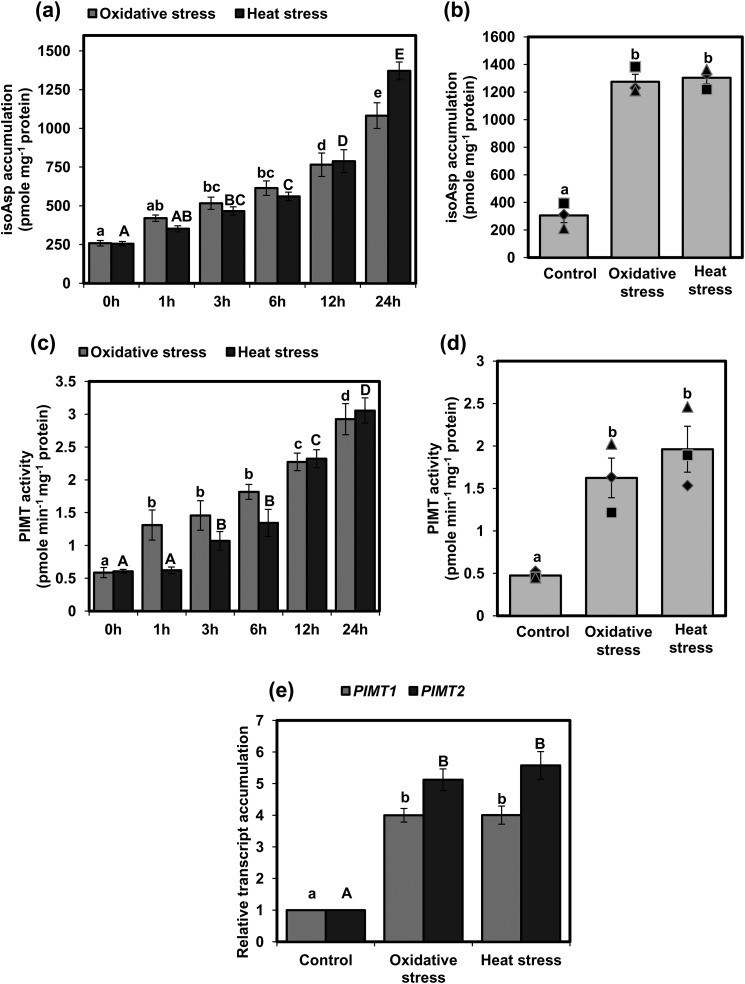
As a first step toward understanding the importance of the PIMT activity in plant stress tolerance, we surveyed isoAsp accumulation in proteins at the seedling and mature plant stage of Arabidopsis under standard growth conditions as well as under heat and oxidative stresses. Total isoAsp accumulation in proteins was determined following the standard protocol used in previous studies (12, 14, 15, 19). Under standard growth conditions, isoAsp content in 14-day-old seedlings was detected only at low levels, and under heat and oxidative stress conditions, isoAsp content progressively increased, albeit at varying levels, with increasing duration of the stress treatments (Fig. 1a). More than 2.2-fold increases in isoAsp content were observed as early as 6 h after these stress treatments (Fig. 1a). Next, to examine whether stress-induced isoAsp accumulation also occurs at the mature stage of Arabidopsis, isoAsp accumulation was also quantified in 4-week-old plants. Like seedlings, significantly increased isoAsp accumulation was also observed in mature plants following the exposure of heat and oxidative stresses (Fig. 1b). These results indicate that isoAsp formation in proteins is accelerated under heat and oxidative stresses at both the seedling stage as well as the mature stage in plants.
Figure 1.
isoAsp accumulation and PIMT activity in response to heat and oxidative stresses in Arabidopsis. a and b, isoAsp accumulation was estimated in the following: a, 14-day-old seedlings (in increasing time points (0–24 h) of stress exposure); b, 4-week-old mature plant grown under control and 24 h of heat and oxidative stress. c and d, PIMT activity was determined in 14-day-old seedlings (in increasing time points (0–24 h) of stress exposure) (c); and 4-week-old mature plants grown under control, heat, and oxidative stress (24 h) (d). Fourteen-day-old seedlings and mature plants were subjected to oxidative stress (seedlings, 1 μm paraquat; mature plants, 10 μm paraquat with 0.05% Triton X-100) and heat stress (37 °C) followed by extraction of total proteins. isoAsp content and PIMT activity were determined using 50 μg of crude protein extract from seedlings and mature plants. e, qRT-PCR analysis showing relative transcript accumulation of AtPIMT1 and AtPIMT2 in 14-day-old seedlings grown under control, heat, and oxidative stress for 12 h. The relative expression value of each gene was normalized with endogenous control actin and calculated using the ΔΔCT method (Applied Biosystems). Data are means ± S.E. of four biological replicates. Significant differences among means (α = 0.01) are denoted by the different letters. For a and c, oxidative stress is in lowercase letters, and heat stress is in uppercase letters. For e, PIMT1 is in lowercase letters and PIMT2 is in uppercase letters.
PIMT activity is up-regulated as a result of transcriptional induction of PIMT genes in response to heat and oxidative stresses in Arabidopsis
PIMT genes were previously shown to be induced by various abiotic stress conditions in several plant species, including Arabidopsis (13, 17–19). To further extend and corroborate these analyses, PIMT activity was analyzed in 14-day-old Arabidopsis seedlings following exposure to heat and oxidative stresses. Results showed that the PIMT activity, which was apparently very low in normally grown seedlings, has progressively increased following exposure to heat and oxidative stresses (Fig. 1c). Significant up-regulation of PIMT activity was seen as early as 1 h of exposure to paraquat treatment, while marked up-regulation of PIMT activity was observed only after 3 h of heat treatment. PIMT activity was also analyzed in 4-week-old mature plants following exposure to heat and oxidative stresses. As shown in Fig. 1d, PIMT activity in mature plants was also significantly increased following exposure to heat and oxidative stresses (Fig. 1d). Furthermore, to analyze whether up-regulation of PIMT activity in response to stress is due to transcriptional induction of PIMT genes, PIMT1 and PIMT2 transcript accumulation was analyzed in seedlings exposed to heat and oxidative stresses by quantitative real time PCR (qRT-PCR). Our results showed that heat and oxidative stress treatments strongly induced AtPIMT1 and AtPIMT2 transcript expression (Fig. 1e and Fig. S1). Consistent with previous studies (13, 14), AtPIMT2 was found to be more responsive toward abiotic stresses than AtPIMT1 (Fig. 1e). These data indicate that the low level of PIMT activity in seedlings or mature plants is up-regulated in response to heat and oxidative stresses, possibly to meet increased isoAsp repairing demands during stressful environments.
Arabidopsis transgenic lines constitutively overexpressing AtPIMT1 or AtPIMT2 exhibits increased tolerance to heat and oxidative stresses
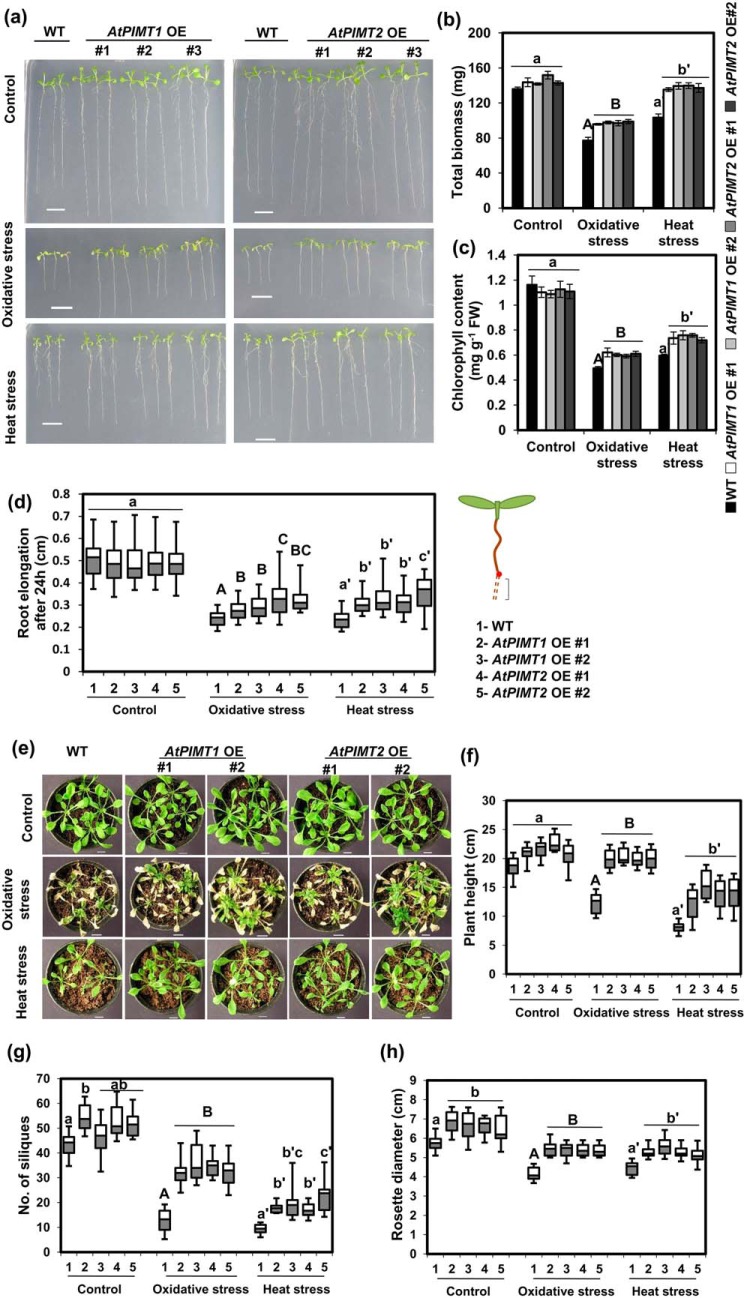
To establish the potential role of PIMTs in plant stress tolerance, we generated Arabidopsis transgenic lines overexpressing either AtPIMT1 or AtPIMT2. Overexpression of AtPIMT1 and AtPIMT2 was confirmed by qRT-PCR in several independent homozygous transgenic lines in the T3 generation (Fig. S2). Thereafter, growth responses of these PIMT-overexpressing and WT plants were assessed under standard growth conditions as well as under heat and oxidative stresses. No significant differences in seedling growth or biomass were observed among AtPIMT1- and AtPIMT2-overexpressing and WT seedlings under normal standard growth conditions (Fig. 2, a–c, and Fig. S5). However, marked differences in seedling growth and tolerance to stress were observed among PIMT overexpressing and WT seedlings under heat and oxidative stresses. AtPIMT1- and AtPIMT2- overexpressing seedlings showed enhanced tolerance to heat and oxidative stresses and were superior in mitigating the adverse effects imposed by these abiotic stress conditions than the WT seedlings. To obtain further evidence, we measured root growth, total biomass, and chlorophyll content. As shown in Fig. 2, b–d, under heat and oxidative stresses conditions, AtPIMT1- and AtPIMT2-overexpressing seedlings showed greater biomass, root growth, and chlorophyll content than the WT seedlings (Fig. 2, a–d). Overall, PIMT-overexpressing seedlings showed better growth response and increased tolerance to heat and oxidative stresses compared with the WT seedlings (Fig. 2, a–c). We have also assessed the growth pattern of soil-grown mature plants of AtPIMT1- and AtPIMT2-overexpressing and the WT plants under standard growth conditions as well as under heat and oxidative stress conditions. To get conclusive evidence, we measured several growth parameters such as plant height, rosette diameter, leaf chlorophyll content, and numbers of siliques of these plants. Under standard growth conditions, both AtPIMT1- and AtPIMT2-overexpressing plants and the WT plants showed a reasonably similar growth phenotype, except increased rosette diameter and slightly greater numbers of siliques in PIMT-overexpressing plants. However, under heat and oxidative stress conditions, striking differences in both vegetative and reproductive growth was observed between PIMT-overexpressing and WT plants. Under heat and oxidative stress conditions, both vegetative and reproductive growth of the WT plants were drastically affected. Under these stress conditions, particularly under oxidative stress, plant height and the number of siliques were drastically reduced. In contrast, the PIMT-overexpressing plants remained reasonably healthy, and both vegetative and reproductive growth were remarkably less affected (Fig. 2, e–h, and Fig. S6). PIMT-overexpressing plants retained greater chlorophyll content (Fig. S7) and produced a significantly greater number of siliques than that of the WT plants under heat and oxidative stress conditions (Fig. 2, e–h). From the results of the above studies, it was evident that transgenic plants expressing either AtPIMT1 or AtPIMT2 showed increased stress tolerance and survived better than the WT plants under heat and oxidative stress conditions.
Figure 2.
Overexpression (OE) of AtPIMT1 and AtPIMT2 improves tolerance to heat and oxidative stresses. a, phenotype of AtPIMT1 OE, AtPIMT2 OE, and WT seedlings (10 days old) grown under control, heat, and oxidative stress conditions. Three representative independent lines of AtPIMT1 (line 1–3) and AtPIMT2 (lines 1, 2, and 5) are shown here. Transgenic lines with variable phenotype were purposely chosen here. Five-day-old Arabidopsis seedlings were transferred to ½MS plates subjected to oxidative (1 μm paraquat) and heat stress (37 °C). The phenotype of WT and transgenic seedlings were monitored and photographed. b, biomass accumulation of AtPIMT1 OE, AtPIMT2 OE, and WT seedlings under control, heat, and oxidative stress conditions. Fresh weight of seedlings was measured 3 days post-treatments. The results are presented as total biomass of 20 seedlings. The values represent mean ± S.E. (n = 3, 20 seedlings each). c, chlorophyll content of AtPIMT1 OE, AtPIMT2 OE, and WT seedlings under normal, heat, and oxidative stress conditions. d, root elongation of AtPIMT1 OE, AtPIMT2 OE, and WT seedlings under normal, heat, and oxidative stress conditions. Elongation of root was determined by measuring root growth during the 24-h post stress treatments using ImageJ software. The values are shown in box plots (n = 30). e, growth pattern of Arabidopsis mature plants overexpressing AtPIMT1, AtPIMT2, and WT plants grown under normal, heat, and oxidative stress conditions. Four-week-old WT and transgenic plants were subjected to oxidative stress (10 μm paraquat and 0.05% Triton X-100) and heat stress (37 °C). Growth parameters are as follows: f, plant height; g, no. of siliques; and h, rosette diameter were measured in AtPIMT1 OE, AtPIMT2 OE, and WT plants (WT) grown under control, heat, and oxidative stress conditions. The box plots represent values obtained from 10 independent plants per line. Significant differences (α = 0.01) are denoted by different letters. Control is shown in lowercase letters, and oxidative stress is in uppercase letters, and heat stress is in lowercase letters with a prime. Scale bar = 1 cm for a and e.
Arabidopsis AtPIMT1 and AtPIMT2 RNAi lines are hypersensitive to heat and oxidative stresses and are associated with increased isoAsp accumulation
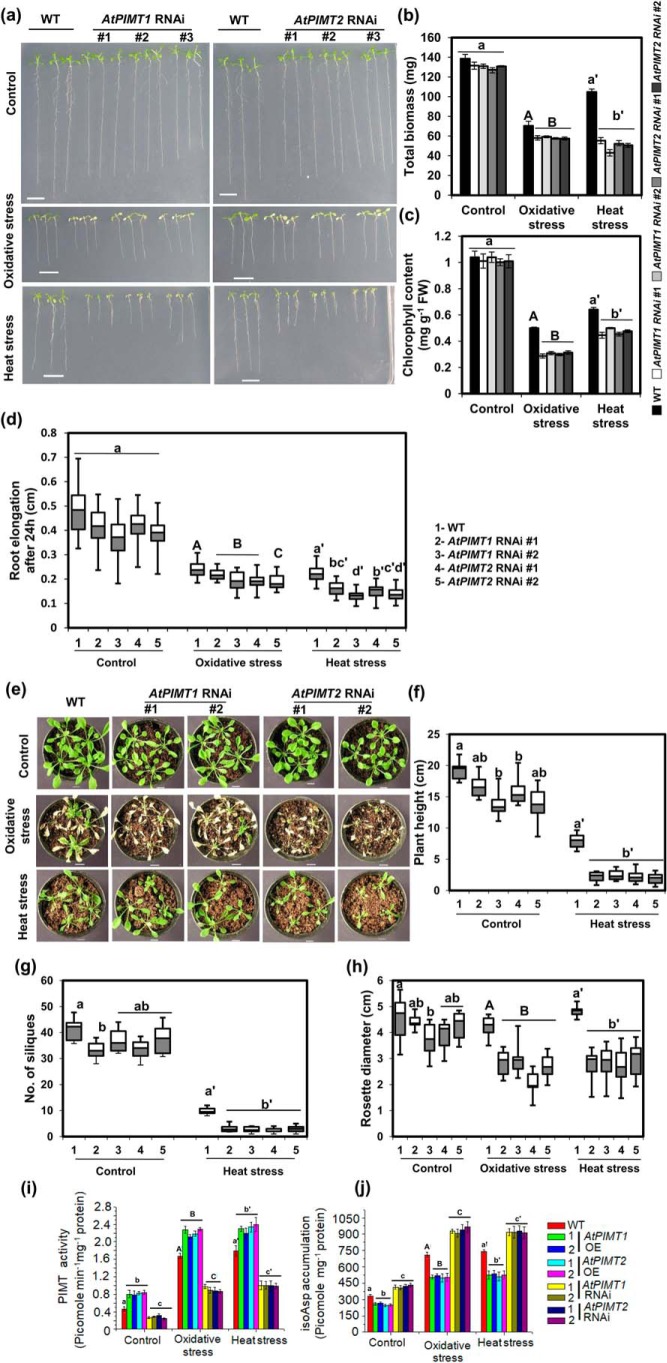
To further confirm the role of PIMTs in plant stress tolerance, we generated AtPIMT1 and AtPIMT2 intron-spliced hairpin RNA interference (RNAi) lines in Arabidopsis using unique sequences of AtPIMT1 and AtPIMT2 as the targeted interference region (Fig. S3). Several RNAi lines were successfully generated, and significantly decreased expression of AtPIMT1 or AtPIMT2 was observed in a number of respective RNAi lines as compared with the WT plants (Fig. S4). Subsequently, the growth pattern of AtPIMT1 and AtPIMT2 RNAi seedlings and WT control seedlings was evaluated under standard growth conditions as well as under heat and oxidative stress conditions. AtPIMT1 and AtPIMT2 RNAi seedlings exhibited slightly reduced growth than the WT seedlings when grown in standard growth condition (Fig. 3, a–d, and Fig. S5). However, significant differences in growth responses were observed between RNAi and WT seedlings under heat and oxidative stresses (Fig. 3, a–d). All RNAi lines exhibited greater sensitivity toward these stress treatments than the WT plants. As expected, growth responses of WT seedlings with respect to biomass, root elongation, and chlorophyll content were greatly affected by high temperature and paraquat treatment. However, these adverse effects were more pronounced in PIMT RNAi seedlings, as RNAi seedlings showed significantly reduced biomass, root growth, and chlorophyll content than the WT seedlings under these stress conditions (Fig. 3, a–d). Growth responses of soil-grown mature plants of AtPIMT1, AtPIMT2 RNAi, and the WT plants were also assessed. Under standard growth conditions, AtPIMT1 and AtPIMT2 RNAi lines and WT plants were visually somewhat similar, although RNAi lines displayed slightly reduced growth patterns than the WT plants. AtPIMT1 and AtPIMT2 RNAi lines showed a somewhat reduced plant height, rosette diameter, and silique numbers compared with WT plants under standard growth conditions. However, significant differences in growth responses were observed under heat and oxidative stress conditions between RNAi and the WT plants (Fig. 3, e–h, and Fig. S6). AtPIMT1 and AtPIMT2 RNAi lines were hypersensitive to these stress conditions and were much more severely affected. Plant height, rosette diameter, number of siliques, and chlorophyll content of these RNAi lines were significantly more compromised than that of the WT plants when grown under heat and oxidative stress conditions (Fig. 3, e–h, and Figs. S6 and S7). In the presence of oxidative stress, PIMT RNAi lines could barely grow and were not able to produce any siliques.
Figure 3.
AtPIMT1 and AtPIMT2 RNAi lines are hypersensitive to heat and oxidative stresses. a, phenotype of AtPIMT1 RNAi, AtPIMT2 RNAi, and WT seedlings (10-day-old) under control, heat, and oxidative stress conditions. Three representative independent lines of AtPIMT1 RNAi (lines 3, 5, and 6) and AtPIMT2 RNAi (lines 2, 5, and 6) are shown here. Five-day-old Arabidopsis seedlings were transferred to ½MS plates subjected to oxidative (1 μm paraquat) and heat stress (37 °C). The phenotype of WT and transgenic seedlings was monitored and photographed. Transgenic lines with variable phenotypes were purposely shown here. b, biomass accumulation of AtPIMT1 RNAi, AtPIMT2 RNAi, and WT seedlings under control, heat, and oxidative stress conditions. Fresh weight of seedlings was measured 3 days post-treatments. The results are presented as total biomass of 20 seedlings. The values represent mean ± S.E. (n = 3, 20 seedlings each). c, chlorophyll content of AtPIMT1 RNAi, AtPIMT2 RNAi, and WT seedlings grown under control, oxidative, and heat stress. d, root elongation of AtPIMT1 RNAi, AtPIMT2 RNAi, and WT seedlings grown under control, heat, and oxidative stress conditions. Elongation of root was determined by measuring root growth during 24-h post-stress treatments using ImageJ software. The values are shown in box plots (n = 30). e, growth pattern of soil-grown Arabidopsis mature plants of AtPIMT1 RNAi, AtPIMT2 RNAi, and WT under control, heat, and oxidative stress conditions. Four-week-old WT and transgenic plants were subjected to oxidative stress (10 μm paraquat and 0.05% Triton X-100) and heat stress (37 °C). Growth parameters are as follows: plant height (f), no. of siliques (g); and rosette diameter (h) were measured of AtPIMT1 RNAi, AtPIMT2 RNAi, and WT plants grown under control, heat, and oxidative stress conditions. The box plots represent values obtained from 10 independent plants per line. Significant differences (α = 0.01) are denoted by the different letters. (The RNAi lines died at rosette stage after paraquat treatment. Therefore, plant height and no. of siliques in response to oxidative stress could not be measured). i and j, quantitation of PIMT activity (i) and isoAsp accumulation (j) in proteins extracted from seedlings grown under control, heat, and oxidative stress conditions. Fifty micrograms of crude protein were used for isoAsp estimation and PIMT activity. Data are means ± S.E. of four biological replicates. Significant differences (α = 0.01) are denoted by the different letters. Control is shown in lowercase letters, oxidative stress in uppercase letters, and heat stress in lowercase letters with a prime. Scale bar = 1 cm for a and e.
To further establish the relationship among PIMT activities, isoAsp accumulation, and stress tolerance, PIMT activity and isoAsp accumulation were quantified in PIMT overexpression and RNAi and WT plants with and without exposure to these stress conditions. Upon imposing stress, elevated levels of isoAsp content were observed in all genotypes; however, the degree of elevation of isoAsp content significantly differed among PIMT overexpression, RNAi and WT plants. AtPIMT1 or AtPIMT2 RNAi seedlings, which had reduced PIMT activity, showed increased accumulation of isoAsp relative to the WT plants. In contrast, AtPIMT1- and AtPIMT2-overexpressing seedlings with increased PIMT activity had significantly reduced isoAsp content much below that of the WT plants (Fig. 3, i and j). These results provide evidence that PIMT activity is important to reduce stress-induced isoAsp accumulation, which otherwise affects plant growth and development and survivability under stress conditions.
PIMT restricts ROS accumulation and lipid peroxidation during heat and oxidative stresses, by protecting functionality of antioxidant enzymes, and improves stress tolerance
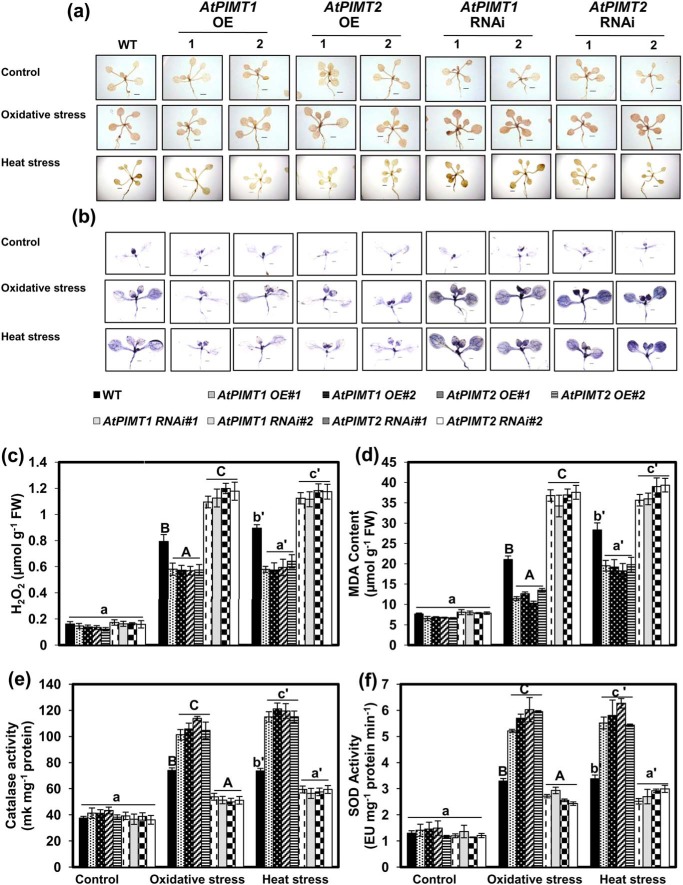
ROS play a critical role in abiotic stress tolerance. ROS can act as signaling molecules and is beneficial to plants; however, it can also be detrimental to a cell at a high concentration (20). To explore whether the stress tolerance phenotype of PIMT-overexpressing lines and stress hypersensitivity of RNAi lines are due to the alteration of ROS homeostasis, we examined ROS (hydrogen peroxide (H2O2) and superoxide (O2˙̄)) accumulation in AtPIMT-overexpressing, RNAi lines and the WT plants. To measure changes in the levels of ROS, we first analyzed H2O2 and O2˙̄ accumulation through DAB and NBT staining. The pattern of DAB and NBT staining clearly revealed that stress-induced accumulation of H2O2 and O2˙̄ was greatly reduced in AtPIMT-overexpressing lines relative to the WT plants. In contrast, RNAi lines accumulated significantly increased levels of H2O2 and O2˙̄ than the WT plants under similar stress conditions (Fig. 4, a and b, and Fig. S8a). These results were further validated by the quantitative analysis of H2O2 content in these transgenic and WT plants (Fig. 4c and Fig. S8b). Subsequently, malondialdehyde (MDA) content was also measured in PIMT overexpression, RNAi lines, and WT. Like ROS, MDA content was significantly increased in RNAi lines and markedly reduced in overexpressing lines as compared with the WT plants under heat and oxidative stress conditions (Fig. 4d and Fig. S8c). The results of these experiments suggest that PIMT somehow plays an important role in restricting ROS accumulation under stressful environments. Considering the mode of action of PIMT, it can be speculated that PIMT possibly prevents stress-induced isoAsp-mediated damage of ROS-scavenging enzymes and thereby maintains their functional competence for efficient detoxification of ROS in plants under stressful conditions. To test this hypothesis, we analyzed CAT and SOD activity in all PIMT transgenic lines as well as in the WT plants. As expected, CAT and SOD activities were up-regulated in all genotypes in response to these stress conditions; however, the level of these enzyme activities significantly differed among PIMT-overexpressing, RNAi and the WT plants. Significantly compromised activities of CAT and SOD were observed in AtPIMT1 and AtPIMT2 RNAi lines, whereas enhanced CAT and SOD activities were seen in PIMT-overexpressing lines relative to that of the WT plants, particularly under stressful environments (Fig. 4, e and f). These results suggest that PIMT plays an important role in maintaining the functionality of antioxidant enzymes, possibly by preventing them from isoAsp-mediated damage, under stressful conditions, thereby preventing ROS accumulation in plants during stressful environments. We also examined NADPH oxidase activity in these transgenic and WT seedlings, as this enzyme is considered to be the major source of ROS generation. Under the control condition, all genotypes exhibited similar NADPH oxidase activity. However, under heat stress, AtPIMT RNAi lines displayed significantly increased NADPH activity, whereas the AtPIMT OE lines showed reduced NADPH activity compared with the WT (Fig. S9). These results indicate that PIMT might be required to suppress stress-induced NADPH activity, through an unidentified mechanism, in order to maintain ROS homeostasis during stress. Overall, our results strongly suggest that PIMT plays an important role in plant stress tolerance by maintaining ROS homeostasis during stress.
Figure 4.
PIMT OE lines exhibit reduced accumulation of ROS and increased activity of antioxidant enzymes, and RNAi lines exhibit increased accumulation of ROS and reduced activity of antioxidant enzymes. a and b, comparison of temporal accumulation of H2O2 (a) and superoxide radicals (O2˙̄) (b) in WT and transgenic AtPIMT1 OE, AtPIMT2 OE, AtPIMT1 RNAi, and AtPIMT2 RNAi seedling lines treated with heat or oxidative stress. Fourteen-day-old seedlings were subjected to oxidative (1 μm PQ) and heat stress (37 °C), and then DAB staining and or NBT staining was performed for in situ localization of H2O2 and superoxide anions (O2˙̄), respectively. The presence of dark brown DAB precipitate or the blue formazan precipitate indicates higher H2O2 and O2˙̄ accumulation, respectively. Scale bar = 0.5 cm for a and b. c–f, quantitative analysis of the following: H2O2 accumulation (c); MDA accumulation (d); CAT activity (e); and SOD activity (f) of WT, AtPIMT1 OE, AtPIMT2 OE, AtPIMT1 RNAi, and AtPIMT2 RNAi seedlings grown in control conditions or treated with heat or oxidative stress. These data represent means ± S.E. of four biological replicates. Significantly deviating means are denoted by different letters (α = 0.01). Control is shown in lowercase letters, oxidative stress in uppercase letters, and heat stress in lowercase letters with a prime.
CAT and SOD are susceptible to isoAsp accumulation upon thermal insult
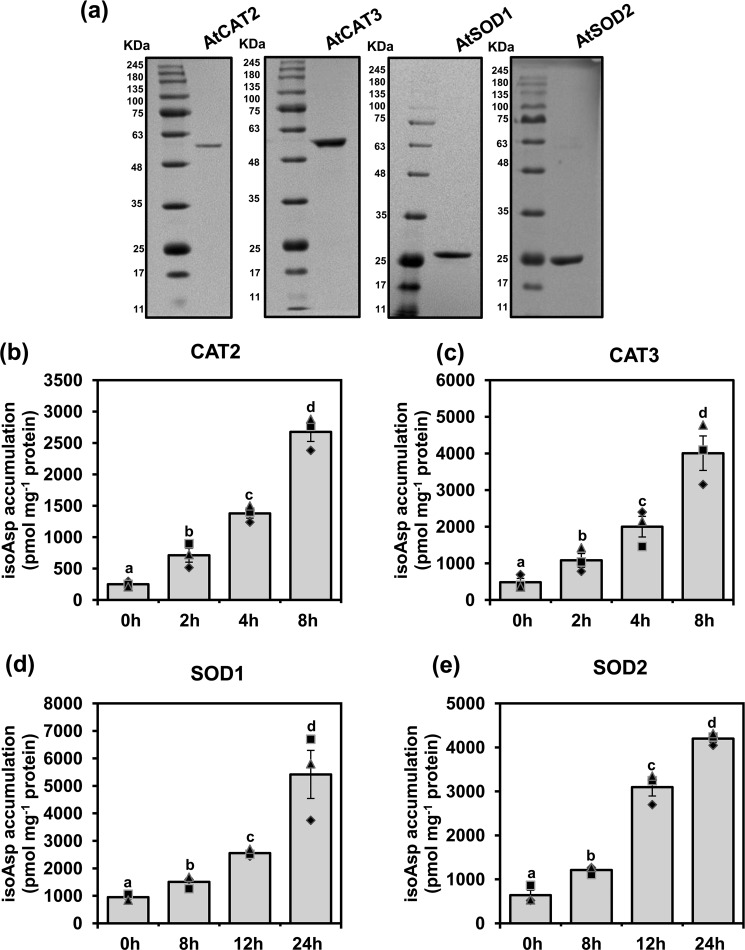
Reduction of catalytic efficiency of antioxidant enzymes in AtPIMT RNAi lines raised an intriguing possibility that antioxidant enzymes are susceptible to isoAsp modification under stressful environments. To verify this, we initially analyzed the amino acid sequences of several CAT and SOD enzymes for their potential aspartate (Asp) and asparagine (Asn) residues that are susceptible to isoAsp formation. It has previously been reported that Asp and Asn residues preceding glycine, serine, and histidine are highly susceptible to isoAsp formation (1). Sequence analysis of these enzymes revealed a few potential Asp and Asn residues highly susceptible to isoAsp formation (Fig. S10). To investigate whether these potential Asp and Asn residues indeed undergo isoAsp formation during stressful environments, representative CAT (AtCAT2 and AtCAT3) and SOD (AtSOD1 and AtSOD2) enzymes were overexpressed in Escherichia coli and subsequently purified to near-homogeneity through nickel-charged affinity chromatography (Fig. 5a). These purified proteins were then subjected to thermal insult at 37 °C for increasing time periods (21). Subsequently, isoAsp accumulation was determined in treated and untreated proteins as described under “Experimental procedures.” Our results demonstrated that isoAsp content in CAT and SOD enzymes was significantly increased upon thermal insult as compared with untreated proteins (Fig. 5, b–e). In the case of CAT, significant accumulation of isoAsp was observed as early as 2 h of treatment at 37 °C (Fig. 5, b and c), whereas SOD enzyme showed a significant accumulation at 37 °C only after 8 h of treatment (Fig. 5, d and e).
Figure 5.
CAT and SOD enzymes are susceptible to isoAsp accumulation upon thermal insult. a, 12% SDS-PAGE analysis of purified AtCAT2, AtCAT3, AtSOD1, and AtSOD2 recombinant proteins. The gel was stained with CBB R-250. b–e, isoAsp accumulation in the following: AtCAT2 (b); AtCAT3 (c); AtSOD1 (d); and AtSOD2 (e) upon thermal insult. Thirty micrograms (1 μg/μl) of purified CAT2, CAT3, SOD1, and SOD2 proteins were thermally insulted at 37 °C for increasing durations of time, and then isoAsp was quantified. Data represent means ± S.E. of four repetitions. Significantly deviating means are denoted by different letters (α = 0.01).
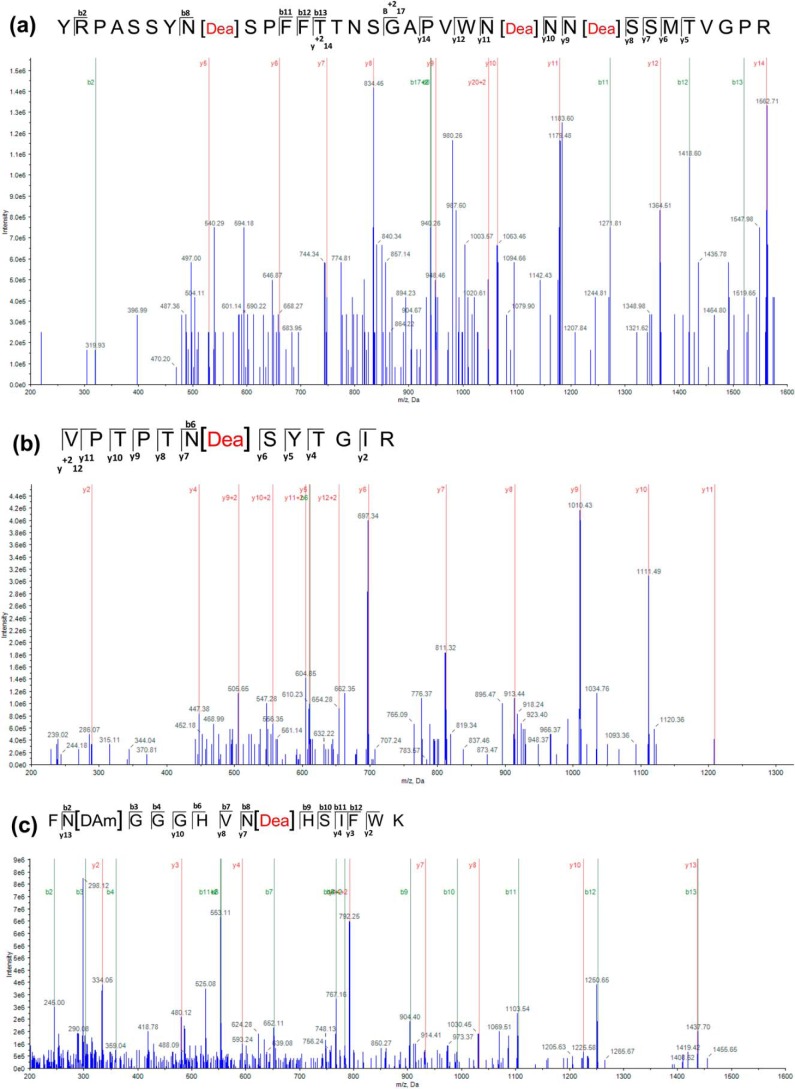
To further confirm whether deamidation of Asn or isomerization of Asp residues indeed occurs during stress to form isoAsp, MS/MS analysis of these treated and untreated CAT and SOD proteins was performed. The Asn/Asp residues susceptible to isoAsp formation were identified directly by comparing peptides from a tryptic digest of control and thermally insulted CAT and SOD enzymes using the AB Sciex QTRAP 6500 system. Only sequences that yielded more than 50% coverage with 99% confidence were considered for further analyses. All the selected peptides were 1% FDR-corrected. Subsequent analysis showed that at least three Asn residues (Asn13, Asn27, and Asn29) of the CAT2 enzyme had undergone deamidation after the thermal insult (Fig. 6), whereas only one Asn residue (Asn28) was found deamidated without pre-exposure to high temperature (Fig. S11 and Table S2). In the case of CAT3, only one Asn (Asn411) residue had undergone deamidation after thermal insult. SOD1 showed deamidation only at Asn102 after 24 h of heat treatment (Fig. 6). In the case of SOD2, the sequences produced 40–50% coverage with low confidence (81.2% for control and 45.4% for treated protein) and showed deamidation at Asn201 (Fig. S12 and Table S2). These results suggest that antioxidant enzymes are indeed susceptible to isoAsp formation during stressful environments.
Figure 6.
Identification of isoAsp formation sites in CAT and SOD enzymes. (a) ESI mass spectra of tryptic peptide YRPASSYNSPFFTTNSGAPVWNNNSSMTVGPR resulting from CAT2 incubated at 37 °C for 8 h. (b) Tryptic peptide VPTPTNSYTGIR resulting from CAT3 incubated at 37 °C for 8 h. (c) ESI mass spectra of tryptic peptide FNGGGHVNHSIFWK resulting from SOD1 incubated at 37 °C for 24 h. The proteins were PAGE-purified and in-gel digested with trypsin, and the resulting peptides were extracted and analyzed using the AB Sciex QTRAP 6500 Exion AD LC system (AB Sciex). The fragmentation occurs at peptide bonds to generate b ions (N-terminal fragments) and y ions (C-terminal fragments) at specific m/z ratios and intensities that provide information regarding amino acid sequence. Site(s) of deamidation are represented square brackets enclosing residue(s) of the peptide sequence [Dea]. Analysis of y and b ion fragmentation patterns with Protein Pilot version 5.0.1 showed that Asn13, Asn27, and Asn29 in CAT2, Asn411 in CAT3, and Asn102 in SOD1 were deamidated. Detailed information of peptides shown here is given in Table S2 and is highlighted.
Compromised catalytic activity of CAT and SOD due to isoAsp accumulation is restored by PIMT
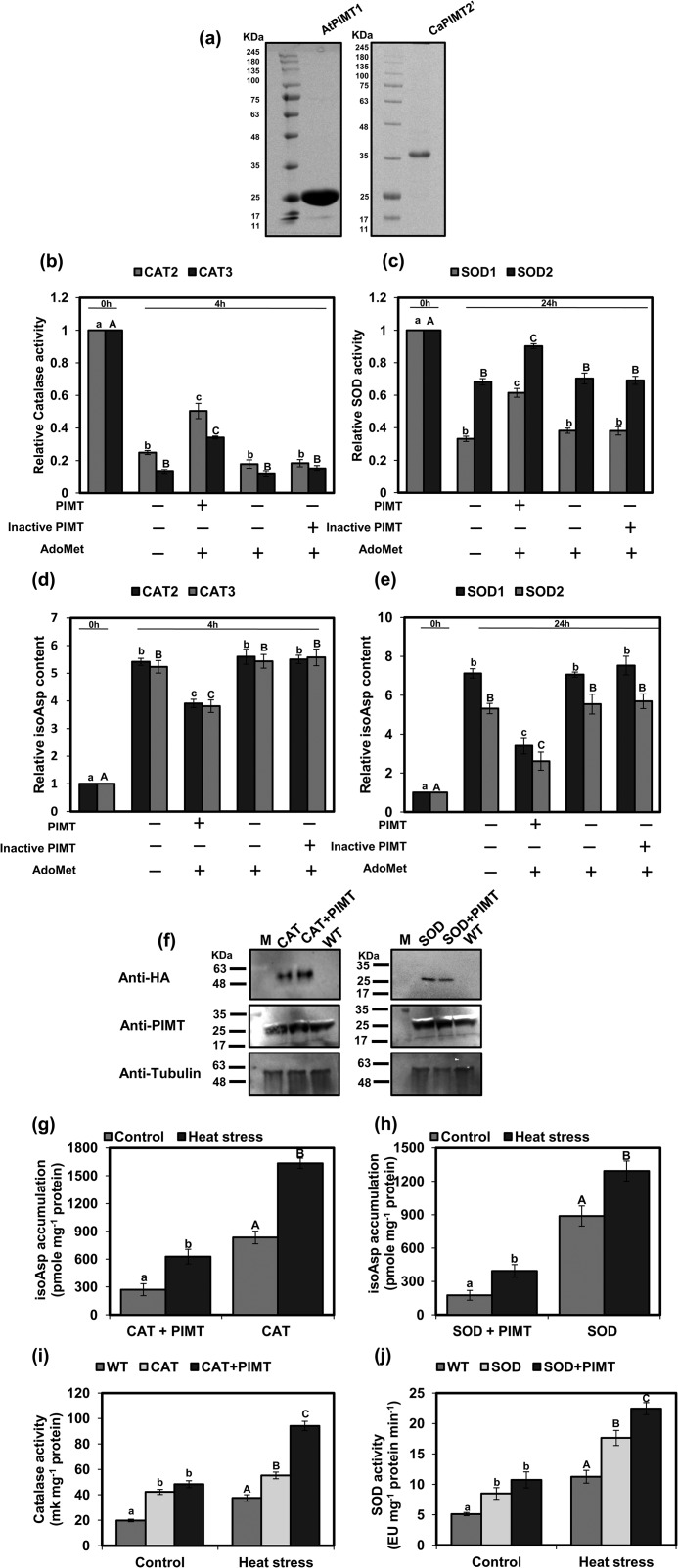
Based on the above results, it is apparent that isoAsp accumulation in these antioxidant enzymes is accelerated upon thermal insult. Next, to examine whether isoAsp formation after thermal treatment has any effect on their catalytic activity, enzymes were assessed for their activity before and after the heat treatment. As shown in Fig. 7, b and c, and Fig. S13, enzyme activities of both CATs and SODs were significantly affected after thermal insult when isoAsp residues were formed. These data suggest that the catalytic efficiency of the antioxidant enzymes is greatly and negatively affected by the formation of isoAsp upon thermal insult. To examine whether PIMT is capable of restricting isoAsp accumulation and restoring the activity of these enzymes, we performed PIMT repair assays with thermally insulted CAT and SOD proteins, and then isoAsp content and catalytic activity were determined. For this purpose, biochemically active PIMT (AtPIMT1) was overexpressed in E. coli, purified to near-homogeneity, and subsequently used for the in vitro protein repair assay (Fig. 7a). PIMT repair assay was conducted with suitable concentrations of heat-treated SOD and CAT substrates and PIMT enzyme protein. Following PIMT repair assay, isoAsp accumulation and catalytic activity of SOD and CAT enzymes were measured. Results showed that the level of heat stress–induced accumulation of isoAsp in these enzymes was significantly reduced when biochemically active PIMT and AdoMet were present in the PIMT repair reaction mix (Fig. 7, d and e). Similarly, the activity loss of CAT and SOD due to thermal insult was partially restored only when biochemically-active PIMT and AdoMet were present in the reaction mix (Fig. 7, b and c). To further ascertain whether the reduction of isoAsp content in these antioxidant proteins was only due to PIMT activity, a biochemically inactive PIMT isoform (CaPIMT2′) from chickpea (14) was similarly expressed, purified, and subsequently used in the reaction mix (Fig. 7a). Contrary to biochemically-active PIMT, isoAsp content in SOD and CAT was found unchanged in the presence of biochemically-inactive PIMT isoforms (CaPIMT2′) and AdoMet (Fig. 7, d and e). Similarly, we did not observe any recovery of the activity of antioxidant enzymes in the presence of biochemically-inactive PIMT and AdoMet (Fig. 7, b and c). These results strongly suggest that PIMT repairs isoAsp accumulation and consequently restores the catalytic activity of the antioxidant enzymes.
Figure 7.
Compromised catalytic activity of CAT and SOD enzymes due to isoAsp accumulation upon stress is restored by PIMT enzyme. a, 12% SDS-PAGE analysis of purified AtPIMT1 (biochemically active) and CaPIMT2′ (biochemically inactive) recombinant proteins. The gel was stained with CBB R-250. b and c, partial restoration of compromised activity of thermally insulted as follows: CAT2 and CAT3 (b) and SOD1 and SOD2 enzymes by PIMT (c). d and e, relative isoAsp content of thermally insulted as follows: CAT2 and CAT3 (d) and SOD1 and SOD2 enzymes with and without repair (e). All purified substrate proteins were thermally insulted at 37 °C for different time periods (CAT2 and CAT3 for 4 h, and SOD1 and SOD2 for 24 h). Following heat treatment, PIMT repair assay was carried out with heat-treated 10 μm CAT and 20 μm SOD with (+) or without (−) biochemically active PIMT (30 μm AtPIMT1), biochemically inactive PIMT (30 μm CaPIMT2′) (as negative control), and AdoMet (10 μm). Following the PIMT repair assay, enzyme activity and isoAsp accumulation in these samples were determined. f–j, in vivo analyses of PIMT repair of CAT and SOD proteins. Tobacco leaves were agroinfiltrated with CAT and SOD alone or along with PIMT. f, immunoblot analyses of HA-tagged CAT and SOD and PIMT proteins in agroinfiltrated and control leaves. Fifty micrograms of total protein of each sample were run in 12% SDS-PAGE and probed with different antibodies (anti-HA, anti-PIMT, and anti-tubulin (loading control)). g and h, analysis of isoAsp accumulation in immunoprecipitated CAT (g) and SOD (h) proteins extracted from agroinfiltrated leaves with and without heat stress. Immunoprecipitation was done using anti-HA antibody, and isoAsp accumulation was estimated in immunoprecipitated proteins. i and j, analysis of catalytic activity of CAT and SOD in control and respective agroinfiltrated leaves subjected to heat stress and without stress. Fifty micrograms of total proteins were used to determine the CAT and SOD activity. Data represent means ± S.E. of four biological repetitions. Significantly deviating means are denoted by different letters (α = 0.01) as follows: b and d, CAT2 in lowercase and CAT3 in uppercase; c and e, SOD1 in lowercase and SOD2 in uppercase; g and h, enzyme + PIMT in lowercase and enzyme only in uppercase; and i and j, control in lowercase and heat stress in uppercase.
Next, to ascertain whether isoAsp formation in these enzymes indeed occurs in planta during stressful situations and whether PIMT repairs isoAsp and restores enzymatic activities in vivo, CAT3 and SOD1 were cloned into a binary expression vector fused to the N-terminal HA tag. HA-tagged CAT3 and SOD1 were then expressed alone or in combination with AtPIMT1 through agroinfiltration in tobacco leaves. Immunoblot analyses were carried out to confirm the expression of these proteins in respective agroinfiltrated leaves (Fig. 7f). The agroinfiltrated leaves, expressing either CAT3/SOD1 alone or in combination with AtPIMT1, were subjected to heat stress at 37 °C for 24 h or were grown normally. Subsequently, HA-tagged CAT3 and SOD1 were immunoprecipitated with anti-HA antibody from total proteins extracted from the treated and untreated agroinfiltrated leaves. Subsequently, isoAsp content in immunoprecipitated protein samples was determined and compared. The isoAsp content in all immunoprecipitated CAT and SOD protein samples extracted from heat-treated agroinfiltrated leaves was significantly greater than that of the untreated leaves, indicating that isoAsp formation in CAT and SOD proteins was accelerated upon heat stress in vivo. However, the immunoprecipitated CAT and SOD proteins exhibited a significantly reduced isoAsp level when co-expressed with AtPIMT1 than when expressed alone (Fig. 7, g and h). These results strongly suggest that isoAsp indeed occurs in CAT and SOD enzymes upon thermal stress in planta, and PIMT restricts such isoAsp accumulation in these proteins (Fig. 7, g and h). In parallel, catalytic activities of these antioxidant enzymes were also attempted in these immunoprecipitated samples; however, immunoprecipitated CAT3 and SOD1 proteins were found to be insufficient to obtain reliable enzyme activity. Therefore, CAT and SOD activities were determined in total proteins from the WT leaves and agroinfiltrated leaves. As expected, total CAT or SOD activity was greater in the respective agroinfiltrated leaves than in the WT leaves. Increased SOD or CAT activity was also observed in heat-treated WT and infiltrated leaves than the corresponding untreated leaves, possibly because of stress-induced transcriptional up-regulation of endogenous SOD and CAT. However, the tobacco leaves co-expressing AtPIMT1 and CAT3 or SOD1 exhibited significantly greater CAT (Fig. 7i) or SOD activity (Fig. 7j) in the respective agroinfiltrated leaves than the leaves expressing either CAT3 or SOD1 alone. Total PIMT activity was also measured in these agroinfiltrated and WT leaves. Results showed increased PIMT activity in leaves co-expressing PIMT and CAT3/SOD1 than control leaves or leaves expressing either CAT3 or SOD1 alone (Fig. S14). These results suggest that PIMT protects antioxidant enzymes from heat stress–induced isoAsp-mediated damage in plants.
Discussion
Plants have evolved several mechanisms to cope with stressful environments, and in these tolerance mechanisms proteins play an indispensable role. Starting from stress perception to actuate various adaptive biochemical, molecular, and physiological responses, a wide range of proteins, such as stress sensors, transcription regulators, and enzymes, participate and play a pivotal role during the whole protective process (22, 23). However, proteins are often susceptible to a variety of reversible and irreversible damages, particularly in stressful environments, which negatively affect their functional competence (1). Therefore, it is essential to keep these proteins unimpaired and functionally competent under hostile stressful environments to optimally perform their biological functions, which is likely to be achieved by protein-repairing enzymes. PIMT, one of the protein-repairing enzymes, repairs isoAsp-induced protein damage in a cell. In this study, we have shown the detrimental effects of stress induced isoAsp accumulation in protein functions and the consequent adverse effect on plant growth and survivability under heat and oxidative stress environments, and the protective role of PIMT in mitigating such detrimental effects. Analyses of isoAsp accumulation and PIMT activity revealed that heat and oxidative stress induces isoAsp accumulation in proteins not only at the seedling stage but also in the mature stage of Arabidopsis, and increased PIMT activity is apparently essential to meet the isoAsp repairing demands in stressful environments. Our analyses further suggest that the PIMT activity perhaps becomes limiting during the prolonged exposure to stress, potentially due to a more rapid generation of isoAsp residues than PIMT can repair. Previous studies have also documented stress-induced up-regulation of PIMT activity in various plant species such as rice (Oryza sativa), wheat, and chickpea (13, 18, 24). Furthermore, in agreement with previous studies, our results suggest that stress-induced up-regulation of PIMT activity in plants is achieved by transcriptional induction of both the PIMT-coding genes (PIMT1 and PIMT2), although PIMT2 is more responsive toward environmental stresses (13, 14). Subsequently, by analyzing Arabidopsis PIMT1 and PIMT2 overexpression and knockdown RNAi lines, we demonstrated a correlation of increased PIMT activity with increased tolerance to heat and oxidative stresses and increased isoAsp accumulation with compromised stress tolerance. Arabidopsis plants overexpressing AtPIMT1 or AtPIMT2 exhibit reduced accumulation of isoAsp and show improved tolerance and survivability to heat and oxidative stresses. In contrast, Arabidopsis plants with significantly reduced AtPIMT1 or AtPIMT2 are hypersensitive to heat and oxidative stresses. In both AtPIMT1 and AtPIMT2 RNAi lines, isoAsp residues in proteins are over-represented following stress treatments. These data strongly suggest that isoAsp accumulation is particularly detrimental for the growth and survival of plants under stressful environments, and both PIMT1 and PIMT2 play important roles in restricting such stress-induced deleterious isoAsp accumulations in plants. The implication of both PIMT1 and PIMT2 enzymes in plant stress tolerance can be justified, as PIMT1 and PIMT2 localize in different subcellular compartments to safeguard respective subcellular proteomes from isoAsp-induced damage (13, 14, 25). Previously, increased accumulation of isoAsp was also shown to be associated with reduced survivability of PIMT-deficient prokaryotes and lower eukaryotes under stressful conditions (6–8). Additionally, overexpression of PIMT in E. coli and Drosophila melanogaster resulted in high heat-shock survival and extended life span under high-temperature conditions (9, 10). Taken together, these studies strongly suggest that the protective role of PIMT in stress adaptation is not only confined to microbes and animals but is also extended to higher plants. It has now become clearly evident that PIMT protects the stability and functionality of proteins by repairing isoAsp-induced protein damage during stressful conditions and thereby improves tolerance and survivability of organisms under stress conditions. However, to get better insight, it is necessary to identify the PIMT target proteins that are essentially affected by isoAsp modification during stressful environments. So far, only a handful of PIMT target proteins, such as HSP, LEA, and RNA helicase, important for maintaining seed germination vigor and longevity, have been identified in the seed proteome (21, 27). Here, we sought to explore mechanisms by which PIMT functions to mediate heat and oxidative stress tolerance in Arabidopsis. Contrasting patterns of ROS accumulation in PIMT-overexpressing and RNAi lines following exposure to heat and oxidative stress offers a clear indication of the role of PIMT in maintaining ROS homeostasis under stressful environments. Subsequently, compromised catalytic activities of SOD and CAT in PIMT RNAi lines under stressful environments provide important evidence to support the possibility of isoAsp-mediated damage of these antioxidant enzymes and also the protective role of PIMT for repairing such damage, during stressful environments. Our results also indicate PIMT-mediated regulation of NADPH oxidase for maintaining ROS homeostasis during stress. However, a detailed study is required to understand how PIMT suppresses stress-induced up-regulation of NADPH oxidase activity. Subsequently, following a similar approach described by Nayak et al. (27), we demonstrated that SOD and CAT activities are indeed susceptible to isoAsp modification during stressful environments. Through in vitro biochemical and MS/MS analysis we could identify the asparagine residues susceptible to isoAsp formation in CAT and SOD enzymes during heat stress. Further analysis of protein-repairing assays with SOD and CAT revealed that compromised catalytic activity of CAT and SOD, due to isoAsp accumulation upon heat stress, is not completely but is significantly restored by PIMT. Previously, Nayak et al. (27) have shown through in vitro analysis that the RNA helicase activity of PRH75, required for successful seed germination, is compromised by stress-induced isoAsp, which is subsequently repaired by PIMT (27). In addition to in vitro analyses, we established the stress-induced isoAsp formation in SOD and CAT enzymes and the role of PIMT in repairing such damage in planta through agroinfiltration of SOD/CAT alone or with PIMT in tobacco leaves. Overall, our results strongly suggest that antioxidant enzymes are susceptible to isoAsp formation in stressful environments, which compromises their catalytic activity; however, PIMT restricts such damage and maintains their functionality during stressful conditions. We suggest that PIMT-mediated repair is a first tier of protection of the cell against abiotic stress. PIMT protects the second-tier enzymes such as CAT and SOD that, when functional, detoxify ROS. We strongly believe that isoAsp-mediated protein damage is not only restricted to antioxidant enzymes, but it may also occur in several other proteins that particularly participate in stress-tolerance mechanisms, although further effort is clearly required to identify more PIMT substrates to authenticate such assumptions. In conclusion, the PIMT-mediated protein repair system is an integral part of the stress-tolerance mechanism in plants, where PIMT plays a key role in protecting the integrity and functionality of proteins, participating in ROS homeostasis, from isoAsp-mediated damage under stressful environments.
Experimental procedures
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) and Nicotiana tabacum were used in this study and were grown and maintained in a plant growth facility maintained at 22 ± 2 °C, Relative Humidity (RH) of 60% with a 16-h light (200 μmol m−2 s−1 light intensity)/8-h dark cycle (28).
Plasmid construction and Arabidopsis transformation
For the generation of AtPIMT1 and AtPIMT2 overexpression lines, the coding sequences of AtPIMT1 (At3g48330) and AtPIMT2 or PIMT2Ψ (At5g50240) were amplified and cloned into a plant expression–modified binary vector pCAMBIA2301. For the generation of AtPIMT1 and AtPIMT2 RNAi lines, a modified binary vector pPZP200lox (29) was used. Initially, an RNAi cassette was developed in pRT100-int vector with a 35S promoter (29) where a unique fragment, 272 bp for AtPIMT1 or 243 bp for AtPIMT2, was cloned in sense and antisense orientations. The hairpin RNAi cassette for AtPIMT1 and AtPIMT2 thus created was excised and finally cloned into the pPZP200lox binary vector (primers details are given in Table S1). For transient expression of SOD1 and CAT3 in tobacco leaves, the coding sequences of CAT3 (At1g20620) and SOD1 (At3g10920) were amplified and cloned into pEG201-HA tag vector. Transgenic Arabidopsis plants were obtained by Agrobacterium tumefaciens–mediated transformation using the floral dip method (30).
Stress treatments
To study the oxidative stress tolerance, 5-day-old seedlings were transferred to ½MS agar medium supplemented with/without 1 μm methyl viologen; for heat stress, 5-day-old seedlings grown vertically in ½MS agar medium were transferred to 37 °C for 12 h and then transferred back to normal growth conditions. The growth response and stress survivability of these seedlings were assessed for the next 3–6 days. Root elongation (average of 20 seedlings) of each transgenic and control lines was analyzed using image analysis software ImageJ. To study stress tolerance at the mature plant stage, 10-day-old seedlings grown in ½MS agar medium were transferred to pots and grown under the standard conditions described above. Stress treatments were administered to the plants according to Salvi et al. (31). Growth parameters like rosette diameter, plant height, and number of siliques per plant were also measured. For growth parameters, 10 plants per line were taken. All the experiments were repeated three times.
qRT-PCR
Total RNA was extracted from the Arabidopsis seedlings using TRIzol reagent (Sigma), and qRT-PCR was done according to Ref. 32.
PIMT activity and isoAsp quantification
Total crude protein was extracted from Arabidopsis seedlings or mature plants as described previously (14). PIMT activity was assayed using the vapor diffusion method (16, 18) that involves the transfer of a radiolabeled methyl group from [3H]AdoMet to an isoaspartyl-containing methyl-accepting peptide (VYP-(l-isoD)-HA) (Peptron, Korea). The isoAsp content was measured using an ISOQUANT isoAsp detection kit (Promega, Madison, WI) according to the kit's protocol with minor alterations as described previously (14, 15). Fifty micrograms of vacuum-dried protein samples were suspended in 20 μl of denaturing digestion buffer (50 mm Tris-Cl (pH 8.0), 8 m urea, and 5 mm DTT). Next, 60 μl of 50 mm Tris-Cl (pH 8.0) was added to dilute urea to 2 m, and then 10 μl of 0.025 μg μl−1 trypsin was added to the reaction mix. The reaction mix was then incubated at 37 °C for 2 h before 0.01 volume of protease inhibitor was added to stop the proteolysis. The digested sample was then used for isoAsp detection. Next, methylation reaction was performed for determining isoAsp content by setting up a reaction mixture (25 μl) containing 5 μl of 5× reaction buffer, 1 μl of radiolabeled [3H]AdoMet (370 GBq mmol−1; PerkinElmer Life Sciences), 5 μl of PIMT (supplied in kit), and 14 μl of trypsin-digested protein sample. Methylation assay was also performed with isoAsp-DSIP (delta sleep-inducing peptide) (25 pmol) as reference standard (supplied in the kit). A blank reaction was performed in parallel without a protein sample. The reaction mixture was incubated at 30 °C for 30 min. The tubes were further kept in ice for 15 min followed by 25 μl of stop solution. The reaction mixture was absorbed onto a cotton swab inserted into the tube containing the reaction mixture and then placed inverted in a scintillation vial containing 15 ml of counting fluor (Sigma). Vials were left at 37 °C for 1 h. Radioactivity was counted using a liquid scintillation counter. Subsequently, isoAsp content in the sample (mg−1 protein) was calculated by extrapolating the radioactivity of the known isoAsp content of isoDSIP reference standard.
Histochemical detection of ROS O2˙̄ and H2O2
For superoxide detection, NBT staining was carried out (33). Fourteen-day-old seedlings of WT and transgenic lines were stained in NBT (1 mg ml−1 prepared in 10 mm phosphate buffer (pH 7.8)) freshly-prepared in the dark for 18 h at 25 °C. For H2O2, DAB staining was performed (34). Fourteen-day-old seedlings or 4-week-old plant mature leaves of WT and transgenic lines were incubated in DAB solution (1 mg ml−1 DAB in Tris acetate buffer (pH 3.8)) freshly-prepared in the dark for 18 h at 25 °C. Stained samples were then bleached using a solution of ethanol/acetic acid/glycerol at a 3:1:1 ratio. Finally, pictures of stained tissues were taken in the Nikon Stereozoom microscope (Nikon, Tokyo, Japan).
Quantification of H2O2 and MDA
To quantify MDA, the method of Heath and Packer (35) was followed. One hundred milligrams of seedlings or leaf material were ground to a fine powder in the presence of liquid nitrogen and then homogenized with 2 ml of 0.25% (w/v) thiobarbituric acid prepared in 10% (w/v) TCA. Homogenate was incubated at 95 °C for 30 min and then was kept in an ice bath for 10 min. Homogenate was then centrifuged at 13,000 × g for 30 min. The absorbance of the supernatant was measured at 532 and 600 nm. The nonspecific absorbance of the supernatant at 600 nm was deducted from the absorbance of 532 nm, and the difference was used to determine the concentration of MDA using the extinction coefficient of 155 mm−1 cm−1.
H2O2 was quantified following the method described by Alexieva et al. (36). One hundred milligrams of seedlings or leaf material were ground to a fine powder in the presence of liquid nitrogen, and then the samples were homogenized in 2 ml of 0.1% TCA. Homogenate was centrifuged at 13,000 × g for 20 min at 4 °C. The supernatant was mixed with an equal volume of 10 mm potassium phosphate buffer (pH 7.0) and a double volume of 1 m potassium iodide and kept in the dark for 1 h at room temperature. The absorption of the supernatant was measured at 390 nm. The amount of H2O2 was calculated using a standard curve prepared from known concentrations of H2O2.
Chlorophyll estimation
Total chlorophyll content in leaves was estimated according to Breeze et al. (37). Chlorophyll in mature plant leaves was extracted in 80% (v/v) acetone. The extracts were kept at −20 °C for 1 h in the dark and then centrifuged at 12,800 × g for 3 min. The absorbances were measured at 663 and 646 nm.
CAT, SOD, and NADPH oxidase activity
CAT activity (38), SOD activity (33), and NADPH oxidase activity (39) were analyzed in total protein extracts (5–50 μg) from stressed and control seedlings. In the case of bacterially-expressed purified proteins, 10–20 μg of proteins were used in the assay. For catalase activity, a 3-ml reaction mix contained 50 mm potassium phosphate buffer (pH 7.8), 3 mm H2O2, and 20 μl of protein extract. The decomposition of H2O2 by CAT was monitored spectrophotometrically at 240 nm for 2 min at room temperature. Catalase activity was calculated in katals according to Weydert and Cullen (38).
Superoxide dismutase activity was assayed by the photochemical reduction of NBT. One hundred milligrams of tissues were homogenized in 1 ml of extraction buffer consisting of 50 mm phosphate (pH 7.8), 1% (w/v) PVP, 0.1% (w/v) EDTA, 1% Triton X-100. The 3-ml assay mixture contained 50 mm phosphate buffer (pH 7.8), 13 μm l-methionine, 75 μm NBT, 0.1 mm EDTA, 2 μm riboflavin, and 100 μl of protein extract. NBT was added last to initiate the reaction mix and was incubated for 5 min at 25 °C. Absorbance was measured at 560 nm. One unit of SOD activity is defined as the amount of enzyme causing 50% inhibition of the NBT photoreduction rate.
NADPH activity was measured following the protocol described by Kaundal et al. (39). Fifty milligrams of tissues were homogenized in 1 ml of protein extraction buffer (50 mm HEPES buffer (pH 7.2), 0.25 m sucrose, 3 mm EDTA, 1 mm dithiothreitol (DTT), 3.6 mm l-cysteine, 0.1 mm MgCl2, and 0.6% PVP with protease inhibitor mixture (Sigma)). The homogenate tissue was further filtered and centrifuged at 10,000 × g for 45 min at 4 °C. Next, the supernatant was centrifuged at 203,000 × g for 60 min at 4 °C. Finally, the pellet was resuspended in 150 μl of ice-cold 10 mm Tris-Cl (pH 7.4) and was used for the enzyme assay. NADPH oxidase activity was assayed through reduction of {3′-[1-[(phenylamino)-carbonyl]-3, 4-tetrazolium]-bis(4-methoxy-6-nitro)benzene-sulfonic acid} (XTT). The assay reaction mixture contained 50 mm Tris-Cl buffer (pH 7.5), 0.5 mm XTT, 0.1 mm NADPH, and 5 μg of protein extract. The linear increase in absorption at 492 nm due to the formation of a yellow formazan was followed for 60 min or until a saturation point is reached at room temperature. Calculation was done using an extinction coefficient of 21.6 mm−1 cm−1.
Bacterial expression of CAT, SOD, and PIMT
The cDNAs used for AtCAT2 (At4g35090), AtCAT3 (At1g20620), AtSOD1 (At3g10920), AtSOD2 (At2g28190), AtPIMT1 (At3g48330), and CaPIMT2′ (JQ690077) were cloned into the pET23b vector (Novagen, Madison, WI) and transformed into E. coli BL-21(DE3) for expression. Recombinant proteins were induced by 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 20 °C and were purified to homogeneity using nickel-nitrilotriacetic acid affinity chromatography (Qiagen).
Thermal stress treatment and restoration of activity by PIMT
Thirty micrograms (1 μg/μl) of purified recombinant CAT and SOD proteins were separately incubated at 37 °C for 0, 2, 4, or 8 h. In the control set, samples remained untreated. At each time point, sample aliquots were analyzed for isoAsp content as described above. As SOD enzymes did not show significant accumulation of isoAsp before 8 h, treatment was therefore increased up to 24 h. After heat treatment, for PIMT repair assay, an initial range of heat-treated protein sample amounts (substrate) (final concentration of 5–30 μm) was used to select the most suitable concentration for consistent and reproducible results. Finally, 10 μm CAT and 20 μm SOD proteins were used in the respective PIMT repair reactions. The repair reaction consisted of 30 μm PIMT, 10 μm AdoMet in 0.1 m HEPES buffer (pH 7.5). Negative control experiments were also carried out, where reaction sets received only AdoMet or inactive PIMT (CaPIMT2′) (Verma et al. (14)) and AdoMet. After a 1-h incubation at 37 °C, isoAsp content and catalytic activities of CAT and SOD were measured as described previously (33, 38).
Sample preparation and LC-MS/MS analysis
Twenty micrograms of treated and untreated purified protein samples were run on 12% SDS-PAGE. Protein bands were then excised, and in-gel digestion was performed using trypsin (Promega) as described previously (40). In short, bands were initially destained and reduced with 10 mm DTT (1 h, 65 °C) followed by alkylation with 50 mm iodoacetamide in the dark for 30 min at room temperature. Next, in-gel trypsin digestion was carried out for 16 h at 30 °C. Eluted proteins were collected in a fresh tube, and trypsin digestion was terminated using 0.1% (v/v) formic acid. Samples were then vacuum-dried and reconstituted in 0.1% formic acid before using for LC-MS/MS analysis.
LC-MS/MS was performed on AB Sciex QTRAP 6500 Exion AD LC system (AB Sciex) with an Acquity UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm, Waters) using Turbo Spray Ion Drive electrospray ionization (ESI) source. The peptide mixture was fractionated by applying a linear gradient 5% acetonitrile (0.1% formic acid) to 30% of acetonitrile (0.1% formic acid) at a flow rate of 300 μl/min in a 32-min run. Spectra were acquired at 3 Hz with a mass range of m/z 400–1200 for mass spectrum 1 (MS1). The additional top three intense peaks were selected for MS/MS analysis with a scan rate of 10,000 Da/s in positive mode.
LC-MS/MS data were processed with Analyst 1.7 (AB Sciex) using the paragon method of protein pilot version 5.0.1 (AB Sciex), and peak lists were searched against 78,742 proteins of the Uniprot Arabidopsis proteome database (UP000006548, 39,371 proteins, version May 22, 2019). Protein pilot was searched with standard tryptic peptides with up to three allowed miscleavages, which were further corrected by 1% FDR. Cys-carbamidomethylation was set as fixed modifications, biological modifications, and amino acid substitutions as variable modifications. Mass tolerance for precursor ions is set at 0.7 Da, whereas mass tolerance for fragment ions is set at 0.6 Da. The threshold score value for accepting individual spectra is set at 95% confidence level, and the unused score of 1.3 was used to accept individual spectra.
Agroinfiltration, immunoprecipitation, and immunoblotting
pEG201/CAT3-HA and pEG201/SOD1-HA constructs were agroinfiltrated to 3-week-old tobacco leaves, as described previously (26). For immunoprecipitation, total proteins were extracted, and crude proteins were incubated with pre-washed anti-HA– conjugated agarose beads for 4 h at 4 °C with gentle shaking. The beads were collected by centrifugation and washed three times with washing buffer (1× PBS and 0.05% (v/v) Tween 20 (pH 7.5)). The protein was eluted in 50 mm NaOH and neutralized in Tris-Cl (pH 9.0). The immunoprecipitated proteins were then used for further analysis.
Fifty micrograms of protein samples were separated by 12% SDS-PAGE and then transferred onto a nitrocellulose membrane. The membrane was then probed with suitable antibodies (anti-HA, anti-PIMT, and anti-tubulin (loading control) followed by horseradish peroxidase–conjugated secondary antibody (GE Healthcare).
Statistical analysis
All data presented in this study were expressed as means ± S.E. The statistical analysis was performed by one-way analysis of variance using SPSS software (SPSS, Chicago, IL). To check the statistical significance, Duncan's multiple range test (DMRT) was done. Letters on the graph show the result of DMRT (α = 0.01); different letter refers to significant differences between mean values. Additional details for the data are given in the figure legends.
Author contributions
S. G., N. U. K., P. V., and M. M. investigation; S. G., N. U. K., P. V., P. S., B. P. P., S. R., V. R., A. H., V. V., and H. K. methodology; M. M. conceptualization; M. M. resources; M. M. formal analysis; M. M. supervision; M. M. funding acquisition; M. M. writing-original draft.
Supplementary Material
Acknowledgments
We thank Dr. N. C. Bisht (NIPGR) for providing modified binary vector pPZP200lox. We thank the technicians of NIPGR central instrumentation facility. We thank technician Dr. Sudeep Ghosh of NIPGR proteomic facility for assisting with the MS/MS analysis (BT/INF/22/SP28268/2018). We are thankful to DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
This work was supported in part by Department of Biotechnology Grant BT/PR8000/BRB/10/1210/2013, the Government of India, and a core grant from the National Institute of Plant Genome Research. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S14 and Tables S1 and S2.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier accession no. PXD016240.
- isoAsp
- isoaspartyl
- PIMT
- PROTEIN l-ISOASPARTYL METHYLTRANSFERASE
- ROS
- reactive oxygen species
- SOD
- superoxide dismutase
- MDA
- malondialdehyde
- CAT
- catalase
- AdoMet
- S-adenosyl-l-methionine
- DAB
- 3,3′-diaminobenzidine
- OE
- overexpression
- NBT
- or nitro blue tetrazolium
- CBB
- Coomassie Brilliant Blue
- ESI
- electrospray ionization
- qRT-PCR
- quantitative real-time PCR
- XTT
- {3′-[1-[(phenylamino)-carbonyl]-3, 4-tetrazolium]-bis(4-methoxy-6-nitro)benzene-sulfonic acid}
- FDR
- false discovery rate
- PVP
- polyvinylpyrrolidone
- DMRT
- Duncan's multiple range test.
References
- 1. Clarke S. (2003) Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res. Rev. 2, 263–285 10.1016/S1568-1637(03)00011-4 [DOI] [PubMed] [Google Scholar]
- 2. Geiger T., and Clarke S. (1987) Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 262, 785–794 [PubMed] [Google Scholar]
- 3. Lowenson J. D., and Clarke S. (1992) Recognition of d-aspartyl residues in polypeptides by the erythrocyte l-isoaspartyl/d-aspartyl protein methyltransferase. Implications for the repair hypothesis. J. Biol. Chem. 267, 5985–5995 [PubMed] [Google Scholar]
- 4. Brennan T. V., Anderson J. W., Jia Z., Waygood E. B., and Clarke S. (1994) Repair of spontaneously deamidated HPr phosphocarrier protein catalyzed by the l-isoaspartate-(d-aspartate) O-methyltransferase. J. Biol. Chem. 269, 24586–24595 [PubMed] [Google Scholar]
- 5. Aswad D. W., Paranandi M. V., and Schurter B. T. (2000) Isoaspartate in peptides and proteins: formation, significance, and analysis. J. Pharm. Biomed. Anal. 21, 1129–1136 10.1016/S0731-7085(99)00230-7 [DOI] [PubMed] [Google Scholar]
- 6. Kim E., Lowenson J. D., MacLaren D. C., Clarke S., and Young S. G. (1997) Deficiency of a protein-repair enzyme results in the accumulation of altered proteins, retardation of growth, and fatal seizures in mice. Proc. Natl. Acad. Sci. U.S.A. 94, 6132–6137 10.1073/pnas.94.12.6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kagan R. M., Niewmierzycka A., and Clarke S. (1997) Targeted gene disruption of the Caenorhabditis elegans-isoaspartyl protein repair methyltransferase impairs survival of Dauer stage nematodes. Arch. Biochem. Biophys. 348, 320–328 10.1006/abbi.1997.0362 [DOI] [PubMed] [Google Scholar]
- 8. Visick J. E., Cai H., and Clarke S. (1998) The l-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J. Bacteriol. 180, 2623–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chavous D. A., Jackson F. R., and Connor C. M. O. (2001) Extension of the Drosophila lifespan by overexpression of a protein repair methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 98, 14814–14818 10.1073/pnas.251446498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kindrachuk J., Parent J., Davies G. F., Dinsmore M., Attah-Poku S., and Napper S. (2003) Overexpression of l-isoaspartate O-methyltransferase in Escherichia coli increases heat shock survival by a mechanism independent of methyltransferase activity. J. Biol. Chem. 278, 50880–50886 10.1074/jbc.M308423200 [DOI] [PubMed] [Google Scholar]
- 11. Mudgett M. B., Lowenson J. D., and Clarke S. (1997) Protein repair l-isoaspartyl methyltransferase in plants. Plant Physiol. 115, 1481–1489 10.1104/pp.115.4.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ogé L., Bourdais G., Bove J., Collet B., Godin B., Granier F., Boutin J.-P., Job D., Jullien M., and Grappin P. (2008) Protein repair l-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell. 20, 3022–3037 10.1105/tpc.108.058479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu Q., Belcastro M. P., Villa S. T., Dinkins R. D., Clarke S. G., and Downie A. B. (2004) A second protein l-isoaspartyl methyltransferase gene in Arabidopsis produces two transcripts whose products are sequestered in the nucleus. Plant Physiol. 136, 2652–2664 10.1104/pp.104.046094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Verma P., Kaur H., Petla B. P., Rao V., Saxena S. C., and Majee M. (2013) PROTEIN l-ISOASPARTYL METHYLTRANSFERASE2 is differentially expressed in chickpea and enhances seed vigor and longevity by reducing abnormal isoaspartyl accumulation predominantly in seed nuclear proteins. Plant Physiol. 161, 1141–1157 10.1104/pp.112.206243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petla B. P., Kamble N. U., Kumar M., Verma P., Ghosh S., Singh A., Rao V., Salvi P., Kaur H., Saxena S. C., and Majee M. (2016) Rice PROTEIN l-ISOASPARTYL METHYLTRANSFERASE isoforms differentially accumulate during seed maturation to restrict deleterious isoAsp and reactive oxygen species accumulation and are implicated in seed vigor and longevity. New Phytol. 211, 627–645 10.1111/nph.13923 [DOI] [PubMed] [Google Scholar]
- 16. Mudgett M. B., and Clarke S. (1993) Characterization of plant l-isoaspartyl methyltransferases that may be involved in seed survival: purification, cloning, and sequence analysis of the wheat germ enzyme. Biochemistry 32, 11100–11111 10.1021/bi00092a020 [DOI] [PubMed] [Google Scholar]
- 17. Mudgett M. B., and Clarke S. (1994) Hormonal and environmental responsiveness of a developmentally regulated protein repair l-isoaspartyl methyltransferase in wheat. J. Biol. Chem. 269, 25605–25612 [PubMed] [Google Scholar]
- 18. Verma P., Singh A., Kaur H., and Majee M. (2010) PROTEIN l-ISOASPARTYL METHYLTRANSFERASE1 (CaPIMT1) from chickpea mitigates oxidative stress-induced growth inhibition of Escherichia coli. Planta 231, 329–336 10.1007/s00425-009-1050-z [DOI] [PubMed] [Google Scholar]
- 19. Wei Y., Xu H., Diao L., Zhu Y., Xie H., Cai Q., Wu F., Wang Z., Zhang J., and Xie H. (2015) Protein repair l-isoaspartyl methyltransferase 1 (PIMT1) in rice improves seed longevity by preserving embryo vigor and viability. Plant Mol. Biol. 89, 475–492 10.1007/s11103-015-0383-1 [DOI] [PubMed] [Google Scholar]
- 20. Choudhury F. K., Rivero R. M., Blumwald E., and Mittler R. (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867 10.1111/tpj.13299 [DOI] [PubMed] [Google Scholar]
- 21. Chen T., Nayak N., Majee S. M., Lowenson J., Schäfermeyer K. R., Eliopoulos A. C., Lloyd T. D., Dinkins R., Perry S. E., Forsthoefel N. R., Clarke S. G., Vernon D. M., Zhou Z. S., Rejtar T., and Downie A. B. (2010) Substrates of the Arabidopsis thaliana protein isoaspartyl methyltransferase 1 identified using phage display and biopanning. J. Biol. Chem. 285, 37281–37292 10.1074/jbc.M110.157008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bohnert H. J., Nelson D. E., and Jensen R. G. (1995) Adaptations to environmental stresses. Plant Cell. 7, 1099–1111 10.2307/3870060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu J.-K. (2016) Abiotic stress signaling and responses in Plants. Cell. 167, 313–324 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thapar N., Kim A. K., and Clarke S. (2001) Distinct patterns of expression but similar biochemical properties of protein l-isoaspartyl methyltransferase in higher plants. Plant Physiol. 125, 1023–1035 10.1104/pp.125.2.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dinkins R. D., Majee S. M., Nayak N. R., Martin D., Xu Q., Belcastro M. P., Houtz R. L., Beach C. M., and Downie A. B. (2008) Changing transcriptional initiation sites and alternative 5′- and 3′-splice site selection of the first intron deploys Arabidopsis PROTEIN ISOASPARTYL METHYLTRANSFERASE2 variants to different subcellular compartments. Plant J. 55, 1–13 10.1111/j.1365-313X.2008.03471.x [DOI] [PubMed] [Google Scholar]
- 26. Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., and Xie Q. (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61, 893–903 10.1111/j.1365-313X.2009.04109.x [DOI] [PubMed] [Google Scholar]
- 27. Nayak N. R., Putnam A. A., Addepalli B., Lowenson J. D., Chen T., Jankowsky E., Perry S. E., Dinkins R. D., Limbach P. A., Clarke S. G., and Downie A. B. (2013) An Arabidopsis ATP-dependent, DEAD-box RNA helicase loses activity upon IsoAsp formation but is restored by PROTEIN ISOASPARTYL METHYLTRANSFERASE. Plant Cell. 25, 2573–2586 10.1105/tpc.113.113456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salvi P., Saxena S. C., Petla B. P., Kamble N. U., Kaur H., Verma P., Rao V., Ghosh S., and Majee M. (2016) Differentially expressed galactinol synthase(s) in chickpea are implicated in seed vigor and longevity by limiting the age induced ROS accumulation. Sci. Rep. 6, 35088 10.1038/srep35088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Augustine R., Mukhopadhyay A., and Bisht N. C. (2013) Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea. Plant Biotechnol. J. 11, 855–866 10.1111/pbi.12078 [DOI] [PubMed] [Google Scholar]
- 30. Clough S. J., and Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- 31. Salvi P., Kamble N. U., and Majee M. (2018) Stress-inducible galactinol synthase of chickpea (CaGolS) is implicated in heat and oxidative stress tolerance through reducing stress-induced excessive reactive oxygen species accumulation. Plant Cell Physiol. 59, 155–166 [DOI] [PubMed] [Google Scholar]
- 32. Kaur H., Verma P., Petla B. P., Rao V., Saxena S. C., and Majee M. (2013) Ectopic expression of the ABA-inducible dehydration-responsive chickpea l-myo-inositol 1-phosphate synthase 2 (CaMIPS2) in Arabidopsis enhances tolerance to salinity and dehydration stress. Planta 237, 321–335 10.1007/s00425-012-1781-0 [DOI] [PubMed] [Google Scholar]
- 33. Beyer W. F. Jr., and Fridovich I. (1987) Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Anal. Biochem. 161, 559–566 10.1016/0003-2697(87)90489-1 [DOI] [PubMed] [Google Scholar]
- 34. Thordal-Christensen H., Zhang Z., Wei Y., and Collinge D. B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J. 11, 1187–1194 10.1046/j.1365-313X.1997.11061187.x [DOI] [Google Scholar]
- 35. Heath R. L., and Packer L. (1968) Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198 10.1016/0003-9861(68)90654-1 [DOI] [PubMed] [Google Scholar]
- 36. Alexieva V., Sergiev I., Mapelli S., and Karanov E. (2001) The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Environ. 24, 1337–1344 10.1046/j.1365-3040.2001.00778.x [DOI] [Google Scholar]
- 37. Breeze E., Harrison E., McHattie S., Hughes L., Hickman R., Hill C., Kiddle S., Kim Y. S., Penfold C. A., Jenkins D., Zhang C., Morris K., Jenner C., Jackson S., Thomas B., et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 23, 873–894 10.1105/tpc.111.083345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weydert C. J., and Cullen J. J. (2010) Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 5, 51–66 10.1038/nprot.2009.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaundal A., Rojas C. M., and Mysore K. S. (2012) Measurement of NADPH oxidase activity in plants. Bio-Protocol 2, e278 10.21769/BioProtoc.278 [DOI] [Google Scholar]
- 40. Kaur H., Petla B. P., Kamble N. U., Singh A., Rao V., Salvi P., Ghosh S., and Majee M. (2015) Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Front. Plant Sci. 6, 713 10.3389/fpls.2015.00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.