Abstract
The enigmatic and poorly studied sturgeon genus Pseudoscaphirhynchus (Scaphirhynchinae: Acipenseridae) comprises three species: the Amu Darya shovelnose sturgeon (Pseudoscaphirhynchus kaufmanni (Bogdanow)), dwarf Amu Darya shovelnose sturgeon P. hermanni (Kessler), and Syr Darya shovelnose sturgeon (P. fedtschenkoi (Bogdanow). Two species – P. hermanni and P. kaufmanni – are critically endangered due to the Aral Sea area ecological disaster, caused by massive water use for irrigation to support cotton agriculture, subsequent pesticide pollution and habitat degradation. For another species – P. fedtschenkoi – no sightings have been reported since 1960-s and it is believed to be extinct, both in nature and in captivity. In this study, complete mitochondrial (mt) genomes of these three species of Pseudoscaphirhynchus were characterized using Illumina and Sanger sequencing platforms. Phylogenetic analyses showed the significant divergence between Amu Darya and Syr Darya freshwater sturgeons and supported the monophyletic origin of the Pseudoscaphirhynchus species. We confirmed that two sympatric Amu Darya species P. kaufmanni and P. hermanni form a single genetic cluster, which may require further morphological and genetic study to assess possible hybridization, intraspecific variation and taxonomic status and to develop conservation measures to protect these unique fishes.
Subject terms: Molecular evolution, Phylogenetics, Evolutionary biology
Introduction
The animal diversity and classification remain complex with new species being continuously discovered and described. Meanwhile, an increase in human activity has led to the degradation of ecosystems, the destruction of native habitats and the direct extinction of many animal species1.
Museum specimens have become increasingly more valuable in evolutionary and conservation biology studies solving fundamental and applied problems using a novel arsenal of molecular genetic techniques. DNA isolation from historical samples and high-capacity DNA sequencing open up new opportunities associated with the investigation of evolution and phylogeny of extinct or endangered species2–6.
The order Acipenseriformes is an ancient fish group which is represented by two families: Polyodontidae and Acipenseridae7. All of the extant species from the Acipenseridae family that inhabit North America and Eurasia are endangered and listed in CITES appendices8,9.
The Aral Sea basin in Central Asia (Kazakhstan, Tajikistan, Turkmenistan and Uzbekistan) has historically had an extraordinary endemic fauna, including four acipenserid species: the Amu Darya shovelnose sturgeon (Pseudoscaphirhynchus kaufmanni Bogdanov, 1874), dwarf Amu Darya shovelnose sturgeon (P. hermanni Kessler, 1877), Syr Darya shovelnose sturgeon (P. fedtschenkoi Kessler, 1872), and Aral ship sturgeon (Acipenser nudiventris Lovetsky, 1828), with the two latter species possibly extinct in their native habitat, and, unlike A. nudiventris, P. fedtschenkoi also does not exist anywhere in aquaculture10–12. The species’ extinction in this region over the recent decades is mostly associated with the Aral Sea ecological disaster that was initially caused by anthropogenic activity through intensive water use from the Amu Darya and the Syr Darya rivers for irrigation and cotton agriculture development as well as wide application of pesticides and mineral fertilizers13.
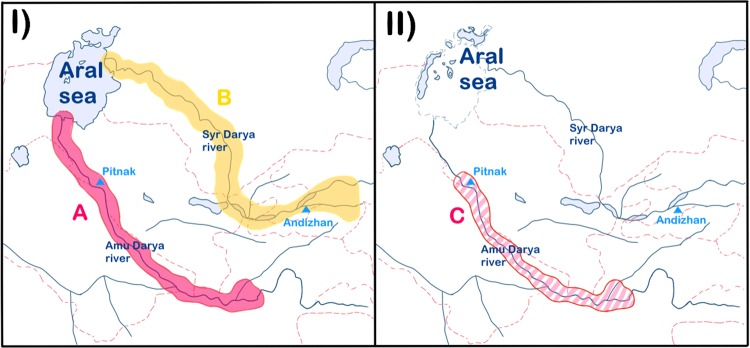
The Pseudoscaphirhynchus genus (Scaphirhynchinae: Acipenseridae) is represented by three freshwater sturgeon species that have limited distribution in Central Asia. One of them – Syr Darya shovelnose sturgeon (P. fedtschenkoi) – has not been observed since the 1960s, while the other two – P. kaufmanni and P. hermanni – are endemic and critically endangered species that live in the muddy waters of the Amu Darya and Panj river basins and are poorly studied because of their rarity14,15 (Fig. 1).
Figure 1.
Distribution map of Pseudoscaphirhynchus species investigated in our study (I – 1960s years; II – 2010s years). A – Former distribution of Amu Darya shovelnose sturgeons P. kaufmanni and P. hermanni; B – Former distribution of the extinct Syr Darya shovelnose sturgeon P. fedtschenkoi. C – Presumable current distribution of Amu Darya shovelnose sturgeons P. kaufmanni and P. hermanni.
Traditionally, Central Asian shovelnoses were placed in the Scaphirhynchinae subfamily together with three Pseudoscaphirhynchus species from North America. Based on recent morphology and molecular genetics studies, Scaphirhynchinae were reclassified as paraphyletic taxa, and Asian species were transferred to Asipenserinae subfamily, placing these species as a sister group to the stellate sturgeon (Acipenser stellatus)14–16.
Previously, the information on the phylogenetic status of P. hermanni and P. kaufmanni was based on morphological traits17 and on the sequences of the mitochondrial cytB gene, showing a monophyletic origin for both of them10. At the same time, molecular phylogenetic status of extinct P. fedtschenkoi remained unclear because it had not been caught for more than fifty years in the Syr Darya basin and can only be found in few museum collections12.
In this study, we present a comparative analysis of four mitochondrial genomes, which were assembled from DNA extracted from museum specimen tissue of extinct P. fedtschenkoi and ethanol-fixed tissues of extant P. kaufmanni and P. hermanni. Our results show that the Central Asian shovelnose species form a monophyletic group and Amu Darya sturgeons demonstrate a significant divergence from the locally extinct Syr Darya sturgeon.
Results and Discussion
We obtained four complete mitochondrial genomes of shovelnose sturgeon specimens, belonging to three Pseudoscaphirhynchus species. The Next Generation sequencing (NGS) statistics on generated and mapped Illumina reads is presented in Table 1. For all NGS-studied specimens, including the extinct Syr Darya shovelnose sturgeon, the average sequence coverage is high (from 7028X to 10859X). The read length distribution for NGS-libraries (including the museum sample) is presented in Supplementary Figs. S1–S3. Assembled mitogenomes of Pseudoscaphirhynchus species were deposited to the GenBank (BioProject PRJNA472690).
Table 1.
Illumina generated reads and the mitochondrial genome size of the Pseudoscaphirhynchus species.
| Species name | Abbreviation | No. Illumina reads generated | Mitogenome size, bp | NCBI number |
|---|---|---|---|---|
| P. fedtschenkoi | FED01 | 116,758,094 | 16,613 | SAMN09240668 |
| P. kaufmanni | KAU03 | 161,089,296 | 16,615 | SAMN09240696 |
| P. kaufmanni | KAU02 | Sanger sequencing | 15,715 | SAMN09829629 |
| P. hermanni | HER01 | 180,707,170 (Sanger sequencing was also conducted) | 16,640 | SAMN09240697 |
Assembled mitogenomes of P. fedtschenkoi, P. kaufmanni and P. hermanni consist of 16,613 bp, 16,615 bp and 16,640 bp, respectively (Table 1), and contain 13 protein coding genes (PCGs), 2 rRNA genes, and 22 tRNA genes as was shown previously for other sturgeon mitogenomes18.
To evaluate genetic diversity among species of the Pseudoscaphirhynchus genus, we conducted phylogenetic analyses of the mitochondrial cytochrome B (cytB) gene using our project specimens and the ones published in the previous Amu Darya sturgeon study10. The maximum likelihood (ML) tree is shown in Fig. 2. Unlike Amu Darya species (P. hermanni, P. kaufmanni), located in the same cluster, Syr Darya P. fedtschenkoi species is clustered apart from them.
Figure 2.
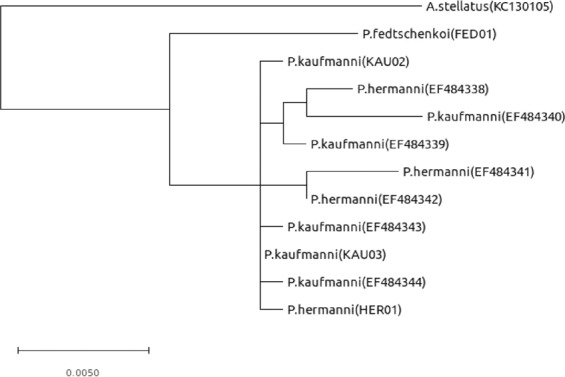
Maximum likelihood phylogenetic tree reconstruction of the Pseudoscaphirhynchus species, including an extinct Syr Darya shovelnose sturgeon based on nucleotide variability of cytB gene.
The mixed clustering of the two Amu Darya species corresponds to the data from10 and can be explained as either incomplete lineage sorting (ILS) or recent hybridization. The haplotype network on Fig. 3 also suggests a mixed nature of Amu Daria Pseudoscaphirhynchus mitochondrial haplotypes and a larger genetic distance for the Syr Daria P. fedtschenkoi specimen.
Figure 3.
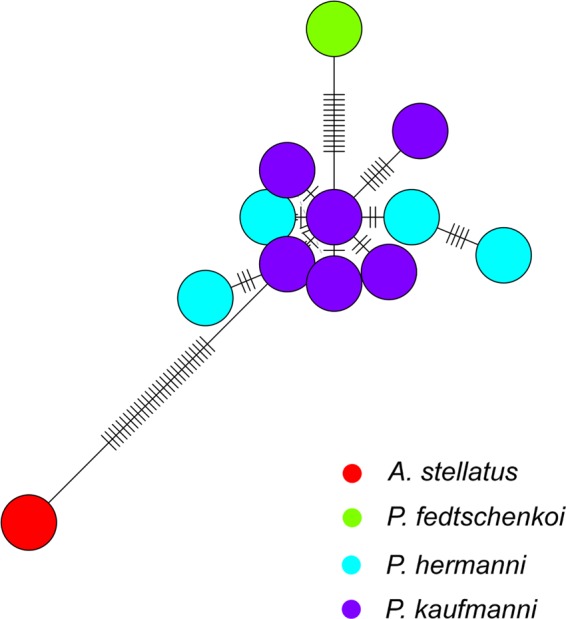
Mitochondrial DNA cytochrome B haplotype network of Pseudoscaphirhynchus species with Acipenser stellatus as an outgroup. Dashed lines indicate a number of distinguishing substitutions between the haplotypes.
Dillman et al.10 found that based on cytB gene analysis, Amu-Darya shovelnose and dwarf shovelnose sturgeons do not form distinct clusters, but the possible explanation proposed by the authors was the low rate of mitochondrial evolution. They suggested that complete genome sequencing of the sturgeons should be conducted in order to resolve this issue. Our complete mtDNA data also supports close similarity between two sympatric Amu Darya species, showing that samples KAU02 and HER01 are different by only 45 substitutions (0.27%). When the cytB region of these samples was extracted and compared with the sequences published by Dillman et al.10, it also supported lack of segregation between these two sympatric species.
The mitogenomic phylogeny of Pseudoscaphirhynchus and other Ponto Caspian sturgeons was reconstructed based on our and previously published mitochondrial datasets. The control region as well as the NADH4L gene were discarded from the alignments as they produced phylogenetic noise and made the phylogenetic tree unstable (Fig. 4).
Figure 4.
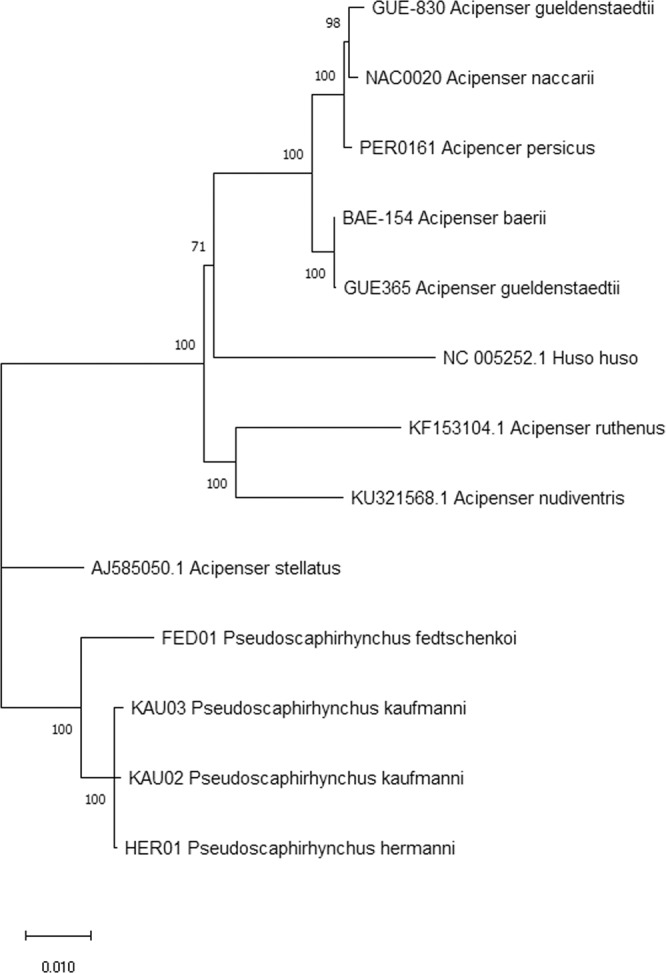
Maximum likelihood phylogenetic tree reconstruction of the Ponto Caspian sturgeon species, including the Amu Darya shovelnose sturgeon, dwarf Amu Darya shovelnose sturgeon, and extinct Syr Darya shovelnose sturgeon, based on their mitochondrial coding sequences (excluding NADH4L gene).
Mitochondrial DNA sequence analysis of the third species of this genus indicates that P. fedtschenkoi from the Syr Daria basin is clearly distinct from both Amu Darya species. Therefore, there is no support for previously suggested hypothesis of the slow evolutionary rate for mtDNA10. This finding raises the question about the taxonomic status of two Amu Darya species - P. hermanni, P. kaufmanni. The admixture of haplotypes can be explained by incomplete lineage sorting; although it is unlikely as the third species forms a distinct clade with high phylogenetic support. Another explanation could be a possible hybridization or mtDNA substitution, which would explain the absence of mtDNA lineage specific to P. hermanni. There is a third plausible explanation: the two Amu Darya species of shovelnose sturgeons, even though being morphologically very distinct17,19 (also see Table 2), are phenotypic morphs within a single species. Both of these currently recognized species are now present at the same time of year in the same part of Amu Darya river near Bukhara City (Uzbekistan) (Alexey Chernyak, pers. comm), and are also sympatric throughout the Amu Darya river basin. Large morphological variations were described for P. kaufmanni in the past, including large and dwarf morphs19. Two other forms - proposed as intermediate forms of P. kaufmanni and P. hermanni - were reported by Nikolsky (1938, cited in Berg 1948)17.
Table 2.
Morphological variation (min-max (average)) of three Central Asia shovelnose sturgeon species (after Berg 1948) and one museum specimen (Andizhan, Fergana valley of the Syr Darya basin) which we have identified as a P. fedtschenkoi.
| P. kaufmanni (after Berg 1968) | P. hermanni (after Berg 1968) | P. fedtschenkoi (after Berg 1968) | Specimen at the Andizhan museum (FED 01) | |
|---|---|---|---|---|
| Dorsal scutes | 10–14 (12) | 9–13 (10–11) | 15–22 | 19 |
| Lateral scutes | 30–38 (34.5) | 31–39 (35) | 37–46 | 36 visible plus 3–5 (?) on the caudal peduncle which is not present on the museum specimen) |
| Abdominal scutes | 6–19 (7–8) | 6–9 (7) | ? | 8 |
| Rostrum spines | present | absent | absent | absent |
| Caudal filament | present | absent | absent or present | lost or absent |
| Skin fold at the anterial edge of pectoral fin | absent | present | present | present |
Striking morphological variations are also observed in P. fedtschenkoi, where Berg described three morphs: 1) morpha typical with long rostrum and absent (or almost absent) caudal filament, 2) morpha brevirostrum with short rostrum and long caudal filament, and 3) morpha intermedia with longer rostrum and well-developed filament. According to Leo Berg, all three morphs are often found by fishermen in one catch17. These morphological variations are almost as drastic as the ones between dwarf and Amu Darya sturgeons, which makes it plausible to suggest that the latter two forms may belong to one polymorphic species. To test this hypothesis, microsatellite or genotype-by-sequencing analysis should be performed, to clarify the taxonomic status of these imperiled fishes.
The results of our study demonstrate that extinct species can be assessed using DNA sequencing of archived and museum samples and subsequent DNA sequence analysis making possible phylogenetic, population and other kinds of investigations. In our work, we showed that the extinct Syr Darya sturgeon species – P. fedtschenkoi – was quite distant genetically from the living species P. hermanni and P. kaufmanni, and its extinction was a great loss for the biological diversity of the fish fauna of the Ponto-Caspian region.
Material and Methods
Samples
DNA samples from the Amu Darya shovelnose sturgeon P. kaufmanni (KAU02 and KAU03) and dwarf Amu Darya shovelnose sturgeon P. hermanni (HER01) were obtained from specimens that are stored in 96% ethanol in the Russian reference depository of sturgeon genetic samples (Russian Federal Research Institute of Fisheries and Oceanography, Moscow, Russia).
A specimen of the Syr Darya shovelnose was obtained as a dried museum specimen, found in peculiar circumstances. During two years, a joint expedition of the Kazakh Fishery Institute (KazNIIRKh, Almaty) and the Russian Institute of Fishery and Oceanography (VNIRO, Moscow) conducted a field study throughout the Syr Darya river and its tributaries in order to assess the current status of P. fedtschenkoi. Fish protection authorities, fishermen and merchants at local fish markets were all surveyed about their encounters with this species. In 2015, the Syr Darya basin was explored from the Aral Sea to the Chardara Dam on the border between Kazakhstan and Uzbekistan, and in 2016 the exploration was continued to the Uzbekistan and Kyrgyzstan parts of the Syr Darya basin. There was no live sturgeon fish found, and no recent sightings have been recorded by local fishermen. This data leads us to conclude that this species can be assumed as extinct. During the field expedition, we found a damaged museum specimen at the Museum of Natural history in Andizhan (Fergana valley, Uzbekistan), which was labeled as asp, Aspius aspius (Linnaeus, 1758). The museum specimen register book indicates that this item has been on display since the 1960-s. Although this specimen was labeled as and A. aspius, it resembled a shovelnose, so we received permission to photograph it (Fig. 5) and to collect a small, inconspicuous fragment of fin tissue for genetic verification.
Figure 5.

P. fedtschenkoi specimen displayed in the Museum of Natural History, in Andizhan (Fergana valley, Uzbekistan).
Further evaluation of these photographs revealed that this specimen is indeed a P. fedtschenkoi (number of dorsal, lateral and abdominal scutes corresponds with the description given by Berg and clearly separates it from two other species of Pseudoscaphirhynchus genus (Table 2). This specimen was assigned as FED01.
In total, four samples of Pseudoscaphirhynchus were used for whole mitogenome sequencing in this study, which were used to ascertain the correct phylogenetic placement of the Central Asian freshwater endemic sturgeons.
DNA extraction and sequencing
For Sanger sequencing, DNA from P. kaufmanii (KAU02) and P. hermanii (HER01) was extracted using standard phenol-chloroform technique, and complete mtDNA was sequenced using Sanger sequencing. Thirty pairs of primers were designed based on previously published sturgeon genomes to amplify a complete genome by overlapping regions. We performed Sanger sequencing with BigDye v3.0 chemistry from both directions by using the same primers. Primers and PCR conditions were published previously11. However, due to the highly degraded DNA quality of P. fedtschenkoi, PCR amplification of mtDNA with this set of primers mostly failed. To obtain a complete mitochondrial genome of P. fedtschenkoi sample, we attempted to use Illumina sequencing technology which requires much shorter DNA fragments. For consistency, Illumina sequencing was also performed for P. kaufmanii and P. hermanii samples.
For Illumina sequencing, DNA from P. fedtschenkoi (FED01) was extracted from fin tissue of the museum specimen in the Ancient DNA Facilities of the National Research Center “Kurchatov Institute” (Moscow, Russia), following the previously described methodology20. Contemporary DNA samples P. kaufmanii (KAU03) and P. hermanii (HER01) were isolated from fin clips preserved in ethanol using a standard phenol-chloroform method of DNA extraction from animal tissue.
Multiplexed DNA-libraries were prepared using an Ovation® Ultralow Library System V2 (NuGEN, USA). Amplified DNA libraries were quantified using a high-sensitivity chip on a 2100 Bioanalyser instrument (Agilent Technologies, USA). The S2 flow cell of Illumina Novaseq6000 genome analyzer (Illumina, USA) was used for DNA-libraries sequencing with 150 bp paired-end reads.
The quality of the NGS data was assessed using FastQC. Data trimming was performed with default parameters using the AdapterRemoval2 tool (version 2.2.2)21. K-mer size and Depth-cutoff were set automatically by Norgal22. E-value cut-off for BLAST-search was 1e-5. Illumina paired-end reads from FED01, KAU03 and HER01 samples were used for building mtDNA sequence de-novo by a Norgal software package22.
The resulting mtDNA consensus sequences were annotated using the MitoAnnotator 3.2523. The obtained annotation was then used to define partitions in the subsequent phylogenetic analysis.
Phylogenetic analysis
The phylogenetic analysis for cytB sequences was performed for the P. hermanni, P. kaufmanni and P. fedtschenkoi species using our and previously published data10. The sequence of cytB from starry sturgeon (A. stellatus, KC130105) was used as an outgroup.
The phylogenetic analyses of the whole mitogenome sequences (excluding control region and NADH4L gene) were performed for the P. hermanni, P. kaufmanni, and P. fedtschenkoi species. As an outgroup, we used complete mitochondrial genomes of starry sturgeon – A. stellatus (NC_005795.1)16,24, sterlet sturgeon – A. ruthenus (KF153104.1), ship sturgeon – A. nudiventris (KU321568.1), beluga sturgeon – Huso huso (NC_005252.1), and mtDNA genomes of other Ponto Caspian sturgeon species, that had been previously been generated25.
The phylogenetic relationships for cytB gene sequences were reconstructed using the maximum likelihood (ML) method in the MEGA X26. The phylogenetic analysis for PCGs (excluding NADH4L gene and stop codons) was conducted in IQ-TREE software27. The consensus sequence from a multiple alignment and its annotation were generated using NCBI BLAST. Then, a partition file with the coordinates of the PCGs was used in IQ-TREE software (ML method with auto substitution model; number of bootstrap alignments – 1000).
The haplotype network of the cytB gene was built by R package Pegas. The alignment of the cytB gene of eleven Pseudoscaphirhynchus specimens and one A. stellatus specimen (1140 nucleotides sequences, 51 variable, and 10 informative positions) was loaded in ape R package. Genetic distances of the sequences were estimated by the dist.dna function using Kimura 80 (K80) evolutionary model. The haplotypes were extracted from DNA-alignment using the haplotype function of the Pegas package, and plotted with the haploNet() Pegas function28. The genetic distances were estimated by the dist.dna function of the ape R package29. The evolutionary history was inferred by using the Maximum Likelihood (ML) method based on the Hasegawa-Kishino-Yano model with gamma distribution HKY + G model30, as was estimated by the “model selection” algorithm of the MEGA X software26.
Supplementary information
Acknowledgements
This work was. was partially supported by RSF grant #16-14-00221 (Sanger mitogenome sequencing conducted by N.S.M. and A.E.B.) and RSF grant# 19-74-20189 additional research added at the major revision stage. This work was performed using high-performance computing resources of federal center for collective usage at NRC “Kurchatov Institute”, http://computing.kiae.ru/.
Author contributions
A.V.N., S.M.R. and N.S.M. conceived and designed experiments, N.S.M. performed sampling, S.V.T., E.S.B. and A.E.B. performed library preparation and sequencing, F.S.S. and K.A.A. analyzed the data, A.V.N., N.M.G., A.S.I. and N.S.M. wrote the manuscript.
Data availability
All mitogenome assemblies are publicly available at the NCBI BioProject: PRJNA472690.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sergey M. Rastorguev and Nikolai S. Mugue.
Supplementary information
is available for this paper at 10.1038/s41598-020-57581-y.
References
- 1.Ceballos Gerardo, Ehrlich Paul R., Dirzo Rodolfo. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proceedings of the National Academy of Sciences. 2017;114(30):E6089–E6096. doi: 10.1073/pnas.1704949114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Der Sarkissian Clio, Ermini Luca, Schubert Mikkel, Yang Melinda A., Librado Pablo, Fumagalli Matteo, Jónsson Hákon, Bar-Gal Gila Kahila, Albrechtsen Anders, Vieira Filipe G., Petersen Bent, Ginolhac Aurélien, Seguin-Orlando Andaine, Magnussen Kim, Fages Antoine, Gamba Cristina, Lorente-Galdos Belen, Polani Sagi, Steiner Cynthia, Neuditschko Markus, Jagannathan Vidhya, Feh Claudia, Greenblatt Charles L., Ludwig Arne, Abramson Natalia I., Zimmermann Waltraut, Schafberg Renate, Tikhonov Alexei, Sicheritz-Ponten Thomas, Willerslev Eske, Marques-Bonet Tomas, Ryder Oliver A., McCue Molly, Rieder Stefan, Leeb Tosso, Slatkin Montgomery, Orlando Ludovic. Evolutionary Genomics and Conservation of the Endangered Przewalski’s Horse. Current Biology. 2015;25(19):2577–2583. doi: 10.1016/j.cub.2015.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diez-Del-Molino, D., Sanchez-Barreiro, F., Barnes, I., Gilbert, M. T. P. & Dalen, L. Quantifying Temporal Genomic Erosion in Endangered Species. Trends Ecol Evol33, 176–185, doi:S0169-5347(17)30305-1 [pii]. [DOI] [PubMed]
- 4.Levin B, et al. High-throughput sequencing of the mitochondrial genomes from archived fish scales: an example of the endangered putative species flock of Sevan trout Salmo ischchan. Hydrobiologia. 2018;822:217–228. doi: 10.1007/s10750-018-3688-7. [DOI] [Google Scholar]
- 5.Sharko FS, et al. Phylogenetic position of the presumably extinct slender-billed curlew, Numenius tenuirostris. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2019;30:626–631. doi: 10.1080/24701394.2019.1597862. [DOI] [PubMed] [Google Scholar]
- 6.Sharko FS, et al. Molecular phylogeny of the extinct Steller’s sea cow and other Sirenia species based on their complete mitochondrial genomes. Genomics. 2019;111:1543–1546. doi: 10.1016/j.ygeno.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Nelson, J. S., Grande, T. C. & Wilson, M. V. H. Fishes of the World. 5th edn, 752 (John Wiley & Sons, 2016).
- 8.Raymakers C. CITES, the Convention on International Trade in Endangered Species of Wild Fauna and Flora: its role in the conservation of Acipenseriformes. J. Appl. Ichthyol. 2006;22:53–65. doi: 10.1111/j.1439-0426.2007.00929.x. [DOI] [Google Scholar]
- 9.Rastorguev SM, et al. High-throughput SNP-genotyping analysis of the relationships among Ponto-Caspian sturgeon species. Ecol. Evol. 2013;3:2612–2618. doi: 10.1002/ece3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillman CB, et al. Molecular systematics of Scaphirhynchinae: an assessment of North American and Central Asian freshwater sturgeon species. J. Appl. Ichthyol. 2007;23:290–296. doi: 10.1111/j.1439-0426.2007.00919.x. [DOI] [Google Scholar]
- 11.Mugue N, Barmintseva A, Schepetov D, Shalgimbayeva G, Isbekov K. Complete mitochondrial genomes of the critically endangered Ship sturgeon Acipenser nudiventris from two seas. Mitochondrial DNA B. 2016;1:195–197. doi: 10.1080/23802359.2016.1144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zholdasova I. Sturgeons and the Aral Sea ecological catastrophe. Env. Biol. Fish. 1997;48:373–380. doi: 10.1023/A:1007329401494. [DOI] [Google Scholar]
- 13.Micklin P. The Aral Sea disaster. Annu. Rev. Earth Pl. Sc. 2007;35:47–72. doi: 10.1146/annurev.earth.35.031306.140120. [DOI] [Google Scholar]
- 14.Birstein, V. J., Doukakis, P. & DeSalle, R. Molecular phylogeny of Acipenseridae: Nonmonophyly of Scaphirhynchinae. Copeia, 287–301, doi:10.1643/0045-8511(2002)002[0287:Mpoans]2.0.Co;2 (2002).
- 15.Birstein VJ, Hanner R, DeSalle R. Phylogeny of the Acipenseriformes: Cytogenetic and molecular approaches. Env. Biol. Fish. 1997;48:127–156. doi: 10.1023/A:1007366100353. [DOI] [Google Scholar]
- 16.Hilton EJ. Observations on the skulls of sturgeons (Acipenseridae): shared similarities of Pseudoscaphirhynchus kaufmanni and juvenile specimens of Acipenser stellatus. Env. Biol. Fish. 2005;72:135–144. doi: 10.1007/s10641-004-6578-y. [DOI] [Google Scholar]
- 17.Berg, L. S. Freshwater fishes of the U.S.S.R. and adjacent countries. Vol. 1 (Academy of Science of USSR Zoological institute, 1948).
- 18.Peng ZG, et al. Age and biogeography of major clades in sturgeons and paddlefishes (Pisces: Acipenseriformes) Mol. Phylogenetics Evolution. 2007;42:854–862. doi: 10.1016/j.ympev.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Sagitov NL. On the dwarf form of the large Amu-Darya shovelnose sturgeon. Nauchnye Doklady Vysshei Shkoly. 1969;6:12–15. [Google Scholar]
- 20.Orlando L, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 21.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Nakeeb K, Petersen TN, Sicheritz-Ponten T. Norgal: extraction and de novo assembly of mitochondrial DNA from whole-genome sequencing data. Bmc Bioinforma. 2017;18:510. doi: 10.1186/s12859-017-1927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwasaki W., Fukunaga T., Isagozawa R., Yamada K., Maeda Y., Satoh T. P., Sado T., Mabuchi K., Takeshima H., Miya M., Nishida M. MitoFish and MitoAnnotator: A Mitochondrial Genome Database of Fish with an Accurate and Automatic Annotation Pipeline. Molecular Biology and Evolution. 2013;30(11):2531–2540. doi: 10.1093/molbev/mst141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnason U, Gullberg A, Janke A, Joss J, Elmerot C. Mitogenomic analyses of deep gnathostome divergences: a fish is a fish. Gene. 2004;333:61–70. doi: 10.1016/j.gene.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Rastorguev S, Mugue N, Volkov A, Barmintsev V. Complete mitochondrial DNA sequence analysis of Ponto-Caspian sturgeon species. J. Appl. Ichthyol. 2008;24:46–49. doi: 10.1111/j.1439-0426.2008.01089.x. [DOI] [Google Scholar]
- 26.Kumar S, et al. Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evolution. 2018;35:1547–1549. doi: 10.1093/molbey/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26(3):419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 29.Paradis Emmanuel, Schliep Klaus. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2018;35(3):526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa M, Kishino H, Yano TA. Dating of the Human Ape Splitting by a Molecular Clock of Mitochondrial-DNA. J. Mol. Evolution. 1985;22:160–174. doi: 10.1007/Bf02101694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mitogenome assemblies are publicly available at the NCBI BioProject: PRJNA472690.