Abstract
Swine leukocyte antigens play indispensable roles in immune responses by recognizing a large number of foreign antigens and thus, their genetic diversity plays a critical role in their functions. In this study, we developed a new high-resolution typing method for pig SLA-1 and successfully typed 307 individuals from diverse genetic backgrounds including 11 pure breeds, 1 cross bred, and 12 cell lines. We identified a total of 52 alleles including 18 novel alleles and 9 SLA-1 duplication haplotypes, including 4 new haplotypes. We observed significant differences in the distribution of SLA-1 alleles among the different pig breeds, including the breed specific alleles. SLA-1 duplication was observed in 33% of the chromosomes and was especially high in the biomedical model breeds such as SNU (100%) and NIH (76%) miniature pigs. Our analysis showed that SLA-1 duplication is associated with the increased level of SLA-1 mRNA expression in porcine cells compared to that of the single copy haplotype. Therefore, we provide here the results of the most extensive genetic analysis on pig SLA-1.
Subject terms: Agricultural genetics, Immunogenetics
Introduction
Pigs are invaluable as a species for meat production and as experimental models in biomedical research1–3. In addition, the increase in the occurrence of infectious diseases has become a big concern for the pig production industry4. The swine major histocompatibility complex (MHC) namely swine leukocyte antigen (SLA), has been associated with the porcine immune response to various infections and vaccinations5–7. Several QTLs also have been mapped to the SLA region including antibody response to porcine reproductive and respiratory syndrome (PRRS)8.
The major histocompatibility complex (MHC) recognizes antigens and activates the immune reactions9,10. MHC polymorphisms play an essential role in determining the functional specificity of the molecules to antigens7,11. Therefore, comprehensive identification and characterization of the alleles of major MHC genes are important to predict adaptive immune responses of an individual. In the humans, about 12,000 alleles of MHC genes have been reported12. Currently, only 227 and 192 alleles have been reported for SLA class I and II genes in IPD-MHC database (https://www.ebi.ac.uk/ipd/mhc/group/SLA/). Therefore, further efforts are necessary to characterize the allelic diversity of major SLA genes.
Several methods have been used to investigate the genetic diversity of SLA genes, including the polymerase chain reaction sequence-specific primer methods (PCR-SSP)13–15, PCR-restriction fragment length polymorphism (PCR-RFLP)13,16, and cDNA based typing17. However, these DNA based typing methods still require further improvement in the resolution of the typing results, comprehensiveness in allele coverage, and usability for large-scale typing. To improve these, we previously developed the genomic sequence-based high-resolution typing (GSBT) methods for SLA-2, -DQA, -DQB1, and -DRB1, and presented the results of new allele identification using a large number of field samples and population genetic analysis on diverse breeds18–22
However, the precise typing of SLA-1 has been particularly difficult because of the presence of a large number of novel alleles and copy number variations (CNVs) of the locus. Thus, the development of a robust typing method for SLA-1 is necessary. Currently, 89 alleles for SLA-1 have been reported in the IPD database including the results from this study23.
SLA-1, SLA-2, and SLA-3 are constitutively expressed classical MHC class I genes, but their expression may vary depending on the genetic differences. For example, SLA-1 is duplicated in the haplotypes Hp-2.0, Hp-8.0, Hp-11.0, Hp-12.0, Hp-19.0, Hp-20.0, and Hp-27.014,24–26. In addition, SLA-1, 3, and 6 were not expressed in the haplotypes Hp-3.0, Hp-2.0, and Hp-5.0, respectively24. Recently, a method was reported to estimate the copy number of SLA-1 and to facilitate our understanding on the functional aspect of SLA-1 duplication27. The frequency of SLA-1 duplication could be abundant, but the detailed functional analysis is not been available.
Therefore, we developed a genomic DNA based high resolution SLA-1 typing method with high accuracy regardless of CNVs and present the extensive analysis results of SLA-1 diversity including new alleles and haplotypes, and allelic distribution among different breeds. We also analyzed the level of SLA-1 expression in pig cells according to their copy numbers which could affect MHC class I-specific immune responses. The information presented in this study should contribute to improving our understanding on the genetic polymorphisms of SLA-1 in diverse pig breeds.
Results
Determination of the SLA-1 specific region
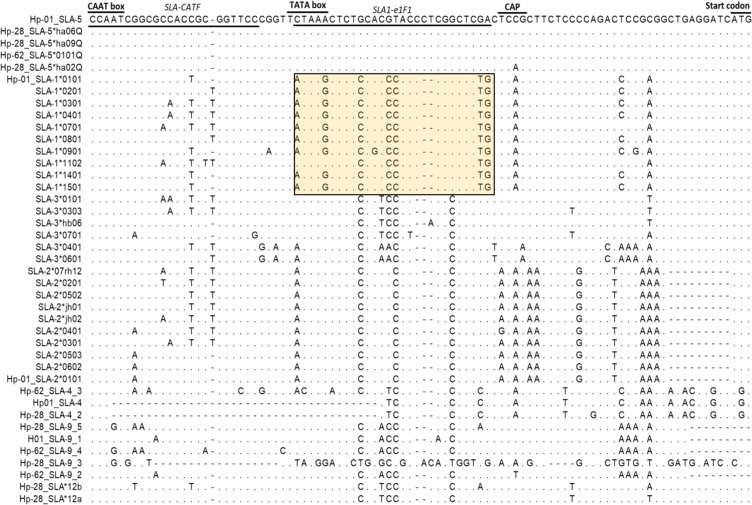
To develop a genomic DNA-based typing method of SLA-1, the determination of conserved locus specific region to design SLA-1 specific primers is required. We previously reported the locus specific nucleotide sequence variations at the downstream promoter region from six classical SLA class I-related genes including SLA-1, -2, -3, -4, -5, and -921. Here, we extended the results by incorporating genomic sequences from additional cloning and sequence analysis. As a result, we identified a SLA-1 specific motif between the TATA box and the CAP site in the 5ʹ UTR and designed a SLA-1-specific forward direction primer, SLA1-e1F1 (Table 1), from an alignment with 41 unique sequences of SLA classical class I-like genes consisting of 10 SLA-1, 10 SLA-2, 6 SLA-3, 3 SLA-4, 5 SLA-5, 5 SLA-9, and 2 SLA-12 (Fig. 1). By combining SLA1-e1F1 together with SLA-e4R4, which is the previously developed classical SLA class I gene specific reverse primer21, we successfully obtained 1844-bp SLA-1 amplicons from 9 selected samples consisting of different breeds and cell lines showing high genetic diversity (Fig. S1).
Table 1.
Primer sequences used in this study.
| Primer sequence (5′-3′) | Target region | Use | Annealing temperature (oC) | Product size (bp) | |
|---|---|---|---|---|---|
| SLA1-e1F1 | MTAARCTCTCCRCCCASCCGGCTCTG | 5′ UTR | Typing PCR | 65 | 1844 |
| SLA-e4R4 | cGGGTCACATGTGTCyTTGGAGG | Exon 4 | |||
| SLA1-e1F1 | MTAARCTCTCCRCCCASCCGGCTCTG | 5′ UTR | Real time PCR | 65 | 137 |
| SLA-e12R | AGGGAGTGGGGACCCGCCT | Exon 1–2 | |||
| GAPDH-F2 | CCTGGCCAAGGTCATCCA | Exon 6 | Real time PCR | 53 | 123 |
| GAPDH-R2 | CGGCCATCACGCCACAG | Exon 7 | |||
| SLA-CATF | CCAATCRGCGMMACYGCTGGTTCC | 5′ UTR | Cloning for primer design | 65 | 1939 |
| SLA-R |
GATCTCCTTAGGGTAGAAGCCCAaatta CAGCACCTCA |
Exon 4 | |||
| LSPE2F | CSTGTCCCGGCCCGAC | Exon 2 | Cloning sequencing | 50 | |
| SLA-CQ2R | TTCCTGGGGATGGGGATG | Intron 2 | |||
| LSPE3F | GCGGGGTCAGGGTCTC | Exon 3 | |||
| SLA-i3R2 | GAGGGGAGATGGTGGAG | Intron 3 | |||
| SLA1-Seq. 2-F | tgctatgctgtgCGCCGARAGGAGGGT | Intron 1 | Typing sequencing | ||
| SLA1-Seq. 2-R | ACCCGGAGGTCGGGGT | Intron 2 | |||
| SLA1-Seq. 3-F | atgctgattatcgCCCKGGTTGGWCGCG | Intron 2 | |||
| SLA1-Seq. 3-R | TCCTCCCTCTCAGGACAG | Intron 3 | |||
| T7 | TAATACGACTCACTATAGGG | cloning vector | Cloning sequencing | ||
| T3 | ATTAACCCTCACTAAAG | ||||
| SP6 | ATTTAGGTGACACTATAG |
Note: Nucleotides in bold indicate degenerate bases. Sequences in lower case indicate the non-template-based regions to improve PCR and sequencing efficiency.
Figure 1.
Comparison of the nucleotide sequence variations in the 5′-UTR region among the seven swine leukocyte antigen (SLA) classical class I-related genes. An alignment using 41 sequences of the 92-bp region containing CAT box, TATA box, CAP region from SLA-1, -2, -3, -4, -5, -9 and -12 is shown. The primer positions of SLA-CATF, the forward primers used for the co-amplification of SLA classical class I gene, and SLA1-e1F1, SLA-1-specific amplification, are underlined. SLA-1 specific regions are indicated in a rectangle. The identical and missing nucleotides are indicated by dots (.) and dashes (–), respectively.
Development of a genomic DNA-based high-resolution SLA-1 typing
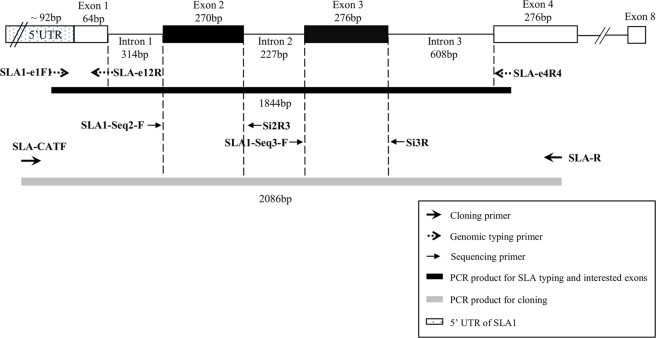
The complete coverage of SLA-1 exons 2 and 3 sequence information is the minimum requirement to officially assign alleles of SLA class I genes28. It has been proven that combining the locus-specific PCR and subsequent direct sequencing using independent primers is a successful method for the comprehensive typing of hyper polymorphic MHC genes18,20–22. Taking the advantage of the small sizes of the introns surrounding the exons 2 and 3, we succeeded in comprehensively amplifying the 1844-bp SLA-1-specific amplicons from all the tested samples (Fig. 2). However, the direct sequencing of heterozygous amplicons carrying the nucleotide deletion and non-deletion alleles (namely deletion heterozygotes) at the intronic regions often resulted in unresolvable chromatograms. To address the problem, we characterized indels at introns 1, 2, and 3 of diverse SLA-1 alleles including SLA-1*01:01, 02:01, 03:01, 04:01, 07:01, 08:01, 09:01, 14:01, and 15:01 by cloning and sequence analysis (Fig. S2). From the analysis, we identified three deletions located at −41 and −42 bp upstream (intron 1) of exon 2, +13 bp downstream (intron 2) of exon 2, and +24 bp downstream (intron 3) of exon 3, respectively.
Figure 2.
General overview of the genomic sequence-based SLA-1 typing method. The diagram shows the location and directions of primers used for polymerase chain reaction (PCR) amplification and sequencing. The sizes (bp) of introns, exons, and PCR products are indicated. Because the upstream boundary of the 5′ untranslated region of SLA-1 was unknown, only the minimum size (>92 bp) is indicated.
Through reiterative primer design and sequencing, we finally developed the sequencing primers, SLA1-seq2-F and SLA1-seq3-F, which generated clear sequencing results of SLA-1 exons 2 and 3 even from the deletion heterozygotes (Table 1, Fig. 2, Fig. S2). For sequencing from the reverse direction to confirm new alleles, we also developed the primers Si2R3 and Si3R, suitable for the direct sequencing of the homozygotes. However, cloning was required for the deletion heterozygotes for successful sequencing (Fig. S2). Consequently, we obtained high resolution typing results of SLA-1 from the genomic DNA of 307 individuals from 14 sample sets without any failure (Table S1), demonstrating the successful development of genomic DNA-based comprehensive high-resolution SLA-1 typing.
Confirmation of the accuracy of new SLA-1 genomic sequence-based typing (GSBT)
To validate the accuracy of our typing results using SLA-1 GSBT, we firstly investigated the presence of conflicts in Mendelian segregation from the typing results of nine KNP families (39 pigs) and two KNP x Landrace cross families (10 pigs) (Table S2). Nine SLA-1 alleles from 13 homozygotes and 36 heterozygotes were observed with complete agreement with Mendelian segregation. Secondly, typing eight ATCC pig cell lines identified 15 alleles which cover 9 subgroups and the results were consistent with those previously reported14,29. In addition, the primers SLA1-e1F1 and SLA-e4R4 also generated SLA-1 specific 727-bp amplicons from the cDNA, in addition to genomic PCR because they are located on exonal regions. Therefore, reverse transcription PCR using the primers and the subsequent direct sequencing results in the successful typing for SLA-1, showing a complete agreement between the SLA-1 GSBT and cDNA typing (Table S3). Lastly, we carried out blind sample testing (n = 40) in collaboration with the ISAG nomenclature committee. The results concorded with the expected results, excepting the new alleles additionally identified from our typing results (data not shown), supporting the accuracy and comprehensiveness of our new SLA-1 typing method.
Identification of novel alleles of SLA-1
A total of 52 SLA-1 alleles corresponding to 34 IPD-SLA curated, 8 NCBI noncurated, and 10 novel sequences were detected from the typing of 307 samples (Tables 1–3). The new alleles were confirmed by cloning and bidirectional sequencing. Among the new alleles, SLA-1*19:02 and *23:03 were observed only once in the Lanyu and Ossabaw pigs probably because of the sample size limit. The remainder were observed from at least two individuals (Table S4). The allele names for new and non-curated NCBI alleles were assigned by the ISAG-SLA nomenclature committee and submitted to IPD (Table S4). The new alleles were clustered into 7 existing subgroups (SLA-1*07, 08, 15, 16, 18, 19, and 20) and formed two new subgroups (SLA-1* 21 and 23) (Fig. S3).
Table 3.
Identified allelic linkages by SLA-1 duplication.
| Haplotypes* | Number of pigs | Breeds | Haplotypes | References |
|---|---|---|---|---|
| 23:02–21:02 | 10 | Berk | New | |
| 12:01–13:01 | 3 | York, Land, ATCC | Hp-35.23 | Gao et al., 2017 |
| 02:02–18:01 | 5 | York, ATCC | New | |
| 11:03–21:01 | 8 | York, KNP | New | |
| 02:01–07:01 | 32 | SNU, NIH, ATCC | HP-2.0 | Ho et al., 2009 |
| 06:01–18:02 | 8 | SNU | New | |
| 10:02–17:01 | 2 | Meishan | Hp-20.18 | Ho et al., 2006 |
| 08:13–13:02 | 1 | Meishan | Hp-19.15 | Ho et al., 2006 |
| 15:01–09:01 | 3 | Land, Land x KNP | Hp-28.8b | Gao et al., 2017 |
*Haplotypes indicate allelic linkages between two duplicated SLA-1 alleles on the same chromosome. The linkage is indicated with a dash (–).
Identification of new SLA-1 duplication haplotypes
We identified 123 cases of SLA-1 duplication-bearing typing results from the typing of 307 samples, and they were classified into 9 groups either belonging to previously reported haplotypes (Hp-2, 19, 20, 28, and 35)28,30 or new SLA-1 haplotypes (SLA-1*21:02–23:02 11:03–21:01, 06:01–18:02 and 02:02–18:01 in which two linked SLA-1 loci are indicated by a dash ‘–’) detected for the first time in this study (Table 3, Table S1). SLA-1*02:02–18:01 was a new combination of previously reported alleles and the remainder were associated with new alleles (Table 3). We also observed 9 additional cases of SLA-1 duplication, but were unable to separate them into individual haplotypes because the numbers of cases were not enough to determine their haplotypic phases (Table S1).
Copy number variation of SLA-1 among different pig breeds
We observed SLA-1 duplication in at least one chromosome from 40.06% (123 pig out of 307) of our typing results (Table S1). In detail, the frequencies of the duplication heterozygotes (4 alleles or duplication in both chromosomes), duplication hemizygotes (3 alleles or duplication in a single chromosome), and duplication homozygotes (2 alleles from duplication in both chromosomes but homozygotes) were 2.93% (n = 9), 13.68% (n = 42), and 23.45% (n = 72), respectively (Table S1). To estimate the frequency of chromosomes with SLA-1 duplication, we assigned values 1.0, 1.5, and 2.0 to each typing result for no duplication, duplication in only one chromosome, and duplication in both chromosomes, respectively. Then, the average value was 1.33 (204/614), indicating that 33% of chromosomes contains SLA-1 duplication (Table 4, Table S1).
Table 4.
Duplication, heterozygosity and diversity of SLA-1 for various breeds of pigs.
| Breeds | Number of samples |
Number of duplicated chromosomes |
aDuplication rate |
bObserved heterozygosity |
cExpected heterozygosity |
Shannon Index |
Evenness |
|---|---|---|---|---|---|---|---|
| KNP | 114 | 13 | 0.06 | 0.64 | 0.73 | 1.57 | 0.78 |
| SNU | 52 | 104 | 1 | 1 | 0.62 | 1.57 | 0.78 |
| NIH | 29 | 44 | 0.76 | 0.83 | 0.61 | 1.44 | 0.91 |
| Duroc | 21 | 0 | 0 | 0.33 | 0.42 | 1.22 | 0.53 |
| Berkshire | 19 | 15 | 0.39 | 0.89 | 0.85 | 3.02 | 0.87 |
| Yorkshire | 19 | 11 | 0.29 | 0.89 | 0.92 | 3.81 | 0.93 |
| Landrace | 19 | 5 | 0.13 | 0.84 | 0.91 | 3.89 | 0.92 |
| ATCC | 8 | 3 | —d | — | — | — | — |
| Land x KNP | 6 | 2 | — | — | — | — | — |
| Lanyu | 5 | 0 | — | — | — | — | — |
| Ossabaw | 5 | 2 | — | — | — | — | — |
| Meishan | 4 | 5 | — | — | — | — | — |
| Local PAM cells | 4 | 0 | — | — | — | — | — |
| AGH | 2 | 0 | — | — | — | — | — |
| All | 307 | 204 | 0.33 | 0.75 | 0.91 |
aDuplication rate = number of chromosomes containing SLA-1 duplication/total number of chromosomes.
bObserved heterozygosity = number of heterozygotes/total number of individuals.
cExpected heterozygosity = 1 − ∑ pi2, where pi is the allele frequency of the i-th allele
dValues were not calculated due to low numbers (<10) of individuals in the population.
SLA-1 duplication was observed from most of the breeds except for Duroc, Lanyu, and AGH in this study. The result could be affected by the sample size limit in those breeds. Indeed, SLA-1 duplication was reported from Duroc, previously28. When the frequency of SLA-1 duplication was compared among breeds, the highest was in Berkshire (39%), followed by Yorkshire (29%), Landrace (13%), and KNP (6%) (Table 4). We also observed the presence of breed-specific duplication haplotypes in which SLA-1*21:02–23:02, 06:01–16:02 and 10:02–17:01 and 08:13–13:02 were only observed in Berkshire, SNU, and Meishan, respectively. The remainder of the haplotypes were shared among the breeds (Table 3).
High SLA-1 heterozygosity in inbred pigs for biomedical study due to locus duplication
SLA-1 duplication prevents simple interpretation of typing results in determining allelic combinations. Therefore, our definition for SLA-1 heterozygosity was simply based on the presence of more than one allele in the typing results regardless of the locus duplication. The average observed heterozygosity of SLA-1 from our typing results was 51% (n = 158, out of 307) which is affected by a high frequency (54.07%) of inbred pigs such as SNU and NIH in our samples (Table 4, Table S1). However, the values were much higher in the commercial breeds such as Berkshire (89%), Yorkshire (89%), and Landrace (84%) except for Duroc (33%), than the inbred pigs. Interestingly, the observed heterozygosity of the SNU miniature pigs was 100% (n = 52). Further analysis showed that all the SLA-1 alleles in the SNU miniature pigs were associated with SLA-1 duplication (Table S1). The result indicates that SLA-1 duplication can contribute to maintaining the functional heterozygosity in inbred animals (Table 4, Table S1).
Comparison of SLA-1 genetic diversity among different pig breeds
When we compared the distribution of SLA-1 alleles across different breed groups with ≥15 individuals in our typing, the allelic constitution differs significantly among the breeds (Table 4). The allelic diversity was the lowest in the NIH miniature pigs with only three, following by the SNU, Duroc, and KNP pigs with 4, 5, and 7 alleles, respectively. Landrace had the highest with 19 alleles, followed by Yorkshire (n = 17), and Berkshire (n = 11), indicating a high genetic diversity of SLA-1 within these breeds, although the number of analyzed animals were much smaller than the inbred pigs. Interestingly, we observed 14 SLA-1 alleles from the 8 ATCC pig cell lines, indicating their diverse origins.
In regard to the allelic dominance or prevalence, SLA-1*07:01 and 02:01 were the most abundant with 16.5% and 16.38%, respectively, followed by 11:02 (12.84%), 08:01 (8.92%), and 04:01 (8.31%). However, their frequencies were influenced by the large sample size of SNU (n = 52) and KNP (n = 114), and their allelic dominance in the breeds. In contrast, 11 alleles including 07:04, 07:05, 08:05, 08:07, 08:13, 13:02, 14:02, 20:02, 07:03, 23:03, and 19:02 were detected only once from the typing results of local breeds with limited sample sizes including Lanyu, Ossabaw, Meishan, and AGH (Table 2, Table S1).
Table 2.
The distribution of allele frequencies of SLA-1 among different pig breeds.
| Allele | Accession | KNP (114)d |
SNU (52) |
NIH (29) |
Duroc (21) |
Berkshire (19) |
Yorkshire (19) |
Landrace (19) |
ATCC (8) |
Land x KNP (6) | Lanyu (5) |
Ossabaw (5) |
Meishan (4) |
Local PAM cells (4) | AGH (2) |
All (307) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01:01 | AK236893 | 0e | 0 | 0 | 0 | 0 | 10.2 | 4.65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.86 |
| 02:01 | AY135592 | 0 | 43.27 | 43.14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16.38 |
| 02:02 | EU440334 | 0 | 0 | 0 | 0 | 0 | 8.16 | 0 | 10.53 | 0 | 0 | 0 | 15.38 | 0 | 0 | 0.98 |
| 04:01 | AK396599 | 0 | 0 | 13.73 | 73.81 | 1.89 | 4.08 | 4.65 | 21.05 | 0 | 90 | 0 | 30.77 | 0 | 25 | 8.31 |
| 04:02 | AK398003 | 0 | 0 | 0 | 0 | 3.77 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 0 | 0.49 |
| 06:01 | AK398228 | 0 | 6.73 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.71 |
| 07:01 | EU440339 | 0 | 43.27 | 43.14 | 0 | 0 | 0 | 0 | 5.26 | 0 | 0 | 0 | 0 | 0 | 0 | 16.5 |
| 07:02 | AY135587 | 0 | 0 | 0 | 2.38 | 1.89 | 0 | 2.33 | 5.26 | 0 | 0 | 0 | 0 | 12.5 | 0 | 0.61 |
| 07:03a | KU754555 | 0 | 0 | 0 | 0 | 0 | 2.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 |
| 07:04 | KU754554 | 0 | 0 | 0 | 0 | 0 | 2.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 |
| 07:05 | EU440331 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.26 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 |
| 07:07b | MF871653 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 0.24 |
| 07:08b | MF871650 | 0 | 0 | 0 | 0 | 0 | 8.16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.49 |
| 08:01 | NM001246245 | 28.22 | 0 | 0 | 0 | 0 | 0 | 4.65 | 0 | 14.29 | 0 | 0 | 0 | 12.5 | 0 | 8.92 |
| 08:03 | AF464043 | 0 | 0 | 0 | 0 | 0 | 0 | 4.65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.24 |
| 08:04 | AK396218 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 33.33 | 0 | 0 | 0 | 0.49 |
| 08:05 | AF464015 | 0 | 0 | 0 | 0 | 0 | 0 | 2.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 |
| 08:07 | KU953375 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.26 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 |
| 08:08 | EU440332 | 0 | 0 | 0 | 0 | 0 | 8.16 | 0 | 5.26 | 0 | 0 | 0 | 0 | 0 | 0 | 0.61 |
| 08:10 | AK231553 | 0 | 0 | 0 | 0 | 0 | 16.33 | 2.33 | 0 | 0 | 0 | 0 | 0 | 12.5 | 0 | 1.22 |
| 08:11 | AK351685 | 0 | 0 | 0 | 0 | 0 | 2.04 | 6.98 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.49 |
| 08:13 | AY459299 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.69 | 0 | 0 | 0.12 |
| 08:15a | KJ555020 | 0 | 0 | 0 | 0 | 11.32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.73 |
| 08:16b | MF871654 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 0.37 |
| 08:17b | MF871647 | 0 | 0 | 0 | 16.67 | 0 | 0 | 2.33 | 0 | 0 | 0 | 0 | 0 | 12.5 | 0 | 1.1 |
| 09:01 | AP009556 | 0 | 0 | 0 | 0 | 0 | 0 | 4.65 | 0 | 7.14 | 0 | 0 | 0 | 0 | 0 | 0.37 |
| 10:02 | DQ303230 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15.38 | 0 | 0 | 0.24 |
| 11:01(01|02)c | AK396545 | 0 | 0 | 0 | 0 | 5.66 | 2.04 | 0 | 10.53 | 0 | 0 | 0 | 0 | 0 | 0 | 0.73 |
| 11:02 | DQ992488 | 40.66 | 0 | 0 | 4.76 | 0 | 0 | 0 | 0 | 21.43 | 0 | 0 | 0 | 25 | 0 | 12.84 |
| 11:03 | DQ883209 | 5.39 | 0 | 0 | 0 | 0 | 8.16 | 0 | 0 | 7.14 | 0 | 0 | 0 | 0 | 0 | 2.2 |
| 11:04 | EU440338 | 0 | 0 | 0 | 0 | 7.55 | 0 | 4.65 | 5.26 | 0 | 0 | 25 | 0 | 0 | 0 | 1.22 |
| 12:01 | KC510996 | 6.64 | 0 | 0 | 0 | 0 | 4.08 | 2.33 | 5.26 | 0 | 0 | 0 | 0 | 0 | 0 | 2.44 |
| 13:01 | AK237395 | 0 | 0 | 0 | 0 | 0 | 4.08 | 2.33 | 5.26 | 0 | 0 | 0 | 0 | 0 | 0 | 0.49 |
| 13:02 | AY459297 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.69 | 0 | 0 | 0.12 |
| 14:01 | EU440343 | 0 | 0 | 0 | 0 | 1.89 | 0 | 20.93 | 5.26 | 21.43 | 0 | 0 | 0 | 0 | 0 | 1.71 |
| 14:02 | EU440342 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.26 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 |
| 15:01 | AK398067 | 1.24 | 0 | 0 | 0 | 0 | 0 | 9.3 | 0 | 7.14 | 0 | 0 | 0 | 0 | 0 | 0.98 |
| 15:02 | AK346419 | 12.45 | 0 | 0 | 0 | 0 | 0 | 6.98 | 0 | 14.29 | 0 | 0 | 0 | 0 | 0 | 4.28 |
| 15:03a | MF498783 | 0 | 0 | 0 | 0 | 9.43 | 0 | 0 | 0 | 0 | 0 | 0 | 7.69 | 0 | 0 | 0.73 |
| 16:01 | MF871646 | 0 | 0 | 0 | 2.38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 0.24 |
| 17:01 | DQ303229 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15.38 | 0 | 0 | 0.24 |
| 18:01 | EU440333 | 0 | 0 | 0 | 0 | 0 | 8.16 | 0 | 5.26 | 0 | 0 | 0 | 0 | 0 | 0 | 0.61 |
| 18:02a | AB845314 | 0 | 6.73 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.71 |
| 19:02b | MF871652 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0.12 |
| 20:02b | MF871649 | 0 | 0 | 0 | 0 | 0 | 0 | 2.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 |
| 21:01a | KU754556 | 5.39 | 0 | 0 | 0 | 0 | 6.12 | 0 | 0 | 7.14 | 0 | 0 | 0 | 0 | 0 | 2.08 |
| 21:02b | MF871648 | 0 | 0 | 0 | 0 | 22.64 | 0 | 0 | 0 | 0 | 0 | 8.33 | 0 | 0 | 0 | 1.59 |
| 21:03a | AK394788 | 0 | 0 | 0 | 0 | 0 | 4.08 | 2.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.37 |
| 23:01a | KF026021 | 0 | 0 | 0 | 0 | 0 | 0 | 9.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.49 |
| 23:02b | MF871651 | 0 | 0 | 0 | 0 | 22.64 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.47 |
| 23:03b | MF871655 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.33 | 0 | 0 | 0 | 0.12 |
| 23:04a | KJ555027 | 0 | 0 | 0 | 0 | 11.32 | 2.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.86 |
aAlleles identified by other labs previously, but not reported to IPD.
bNovel alleles identified in this study.
cThe sequences of exons 2 and 3 are identical between the alleles 11:01:01 and 11:01:02. Six-digit allele naming is used to indicate nucleotide difference at exon 4.
dThe number of animals for each breed.
eAllele frequencies.
The allelic distribution pattern of SLA-1 across breeds was divided into two groups, breeds showing either dominant or balanced allelic distributions (Fig. 3). Among the 11 breeds with sample sizes ≥5, Duroc, NIH, and Lanyu showed allelic dominance with 04:01 (73.81%), 02:01–07:01 (86.28%), and 04:01 (90%), respectively. In contrast, the rest of the breeds showed a more balanced distribution of alleles in their frequencies than the former. Consistently, the genotypic richness which is the number of genotypes that would be expected was low for NIH (1.44), KNP (1.57), and SNU (1.57) while the values were much higher for Landrace (3.89), Yorkshire (3.81), and Berkshire (3.02) (Table 4). The high genotypic richness may indicate the sign of balancing of selection which results in maintaining the gene pool diversity. Consistently, genotypic evenness in which equally abundant genotypes yields a value equal to 1 was high for Landrace (0.92), Yorkshire (0.93), and Berkshire (0.87). NIH miniature pigs with only three alleles also showed high evenness (0.91) due to the allelic frequency balance among three alleles (Table 4). The allelic diversity of SLA-1 was low in Duroc with the genetic richness of 1.22 and evenness of 0.53 (Table 4), which is consistent to the results of other SLA genes in previous studies including SLA-2, -DQB1, and -DRB118,20,21.
Figure 3.
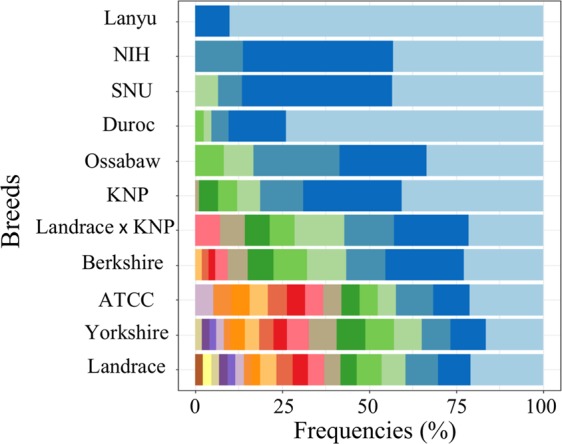
A diagram showing the patterns of allelic diversity and evenness of SLA-1 among the diverse breeds used in this study. The X-axis shows the frequencies of each allele in the horizontally stacked bar chart, within each breed (Y-axis). The results from the breeds with >5 individuals were used. Note that the color does not indicate the same allele.
Low allele sharing in SLA-1 across pig breeds
Pair-wise genetic identity analysis (Nei’s) for SLA-1 among breeds with sample sizes >10 showed distant relationships among them, except between the SNU and NIH miniature pigs (Table S5) which share the same ancestral origin as the Minnesota miniature pigs31. Interestingly, the result of allele sharing analysis using 38 SLA-1 alleles from six selected breeds (KNP, NIH, Duroc, Berkshire, Yorkshire, Landrace) also showed the presence of no common allele across all six breeds (Fig. S4). SLA-1*04:01 was the most common allele shared across 5 breeds (Berkshire, Duroc, Yorkshire, Landrace, and NIH) (Fig. S4, Table S6). This finding is consistent with previous studies which reported the presence of 04:01 in various other rare breeds including the Micromini pigs, Clawn, Yucatan, and Mexican hairless mini-pigs24,28,32. Allele 07:02 was common for Duroc, Berkshire, and Landrace, and 12:01 for KNP, Yorkshire, and Landrace (Table S6). In the remaining alleles, 16 were shared by only 2 breeds (Table S6) and 19 were breed specific (Table S7). Yorkshire harbors the largest number of breed-specific alleles (n = 6). Duroc showed only a single breed-specific allele. Taken together, our results showed the presence of significant genetic difference in SLA-1 among the different breeds of pigs.
Increased level of SLA-1 mRNA expression by gene duplication
The relationship between SLA-1 copy numbers and corresponding expression levels has not been clearly illuminated. Therefore, we identified pig cells with different SLA-1 copy numbers and evaluated their expression level using real-time PCR (Fig. 4). The average level of SLA-1 transcripts from cells with three copies of SLA-1 (CCL-166, CRL-2528) was 2.8 times higher than those with two copies (CL-184 and CRL-1746, p = 0.0025), suggesting a possible importance of SLA-1 duplication for immune responses in pigs. We also observed variations in the level of SLA-1 expression in cells with identical copy numbers. This could be resulted from the difference in the characteristics of cells or alleles (Table S3). However, we were unable to conclude the difference in the expression level of SLA-1 on the protein level among the cells with different SLA-1 copy numbers because the signals were not only from SLA-1 but also from SLA-2 and -3 due to the unavailability of SLA-1-specific antibodies.
Figure 4.
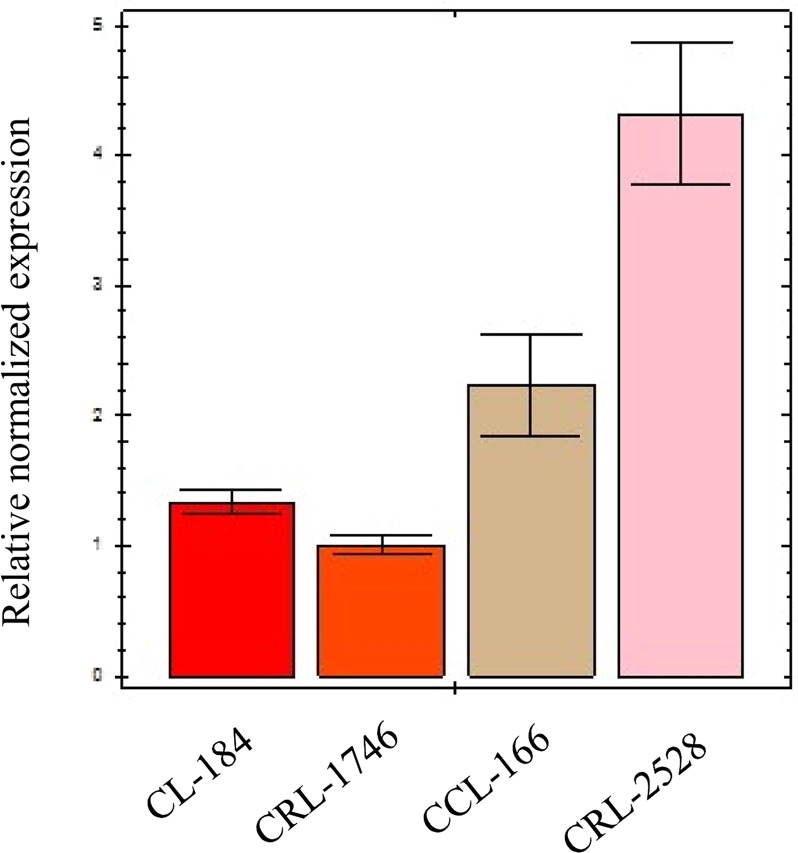
Comparison of the expression levels of SLA-1 between cells with different SLA-1 copy numbers. Four fibroblast cell lines of pigs were evaluated for each of different SLA-1 copy numbers, CL-184 and CRL-1746 for 2 copies, and CCL-166 and CRL-2528 for 3 copies. Cell names are indicated in the bottom.
Discussion
The complexity of the SLA class I region caused by extreme intra-locus sequence variations and inter-locus sequence similarity, presence of pseudogenes, and locus duplication makes the region difficult to study. To overcome the difficulty, we systemically developed a genomic DNA-based high resolution typing method for SLA-1 and presented the analysis results from a large-scale typing on diverse pig breeds including the population level analysis of SLA-1 CNVs.
Considering the existence of over 700 pig breeds or lines worldwide2, the number of uncovered alleles still could be significant in numbers. Considering the extreme polymorphisms of SLA-1, unraveling the information will not only reveal the genetic diversity of the gene but also contribute to the illumination of the immunogenetic aspects of pig evolution and relationships among them during breed formation33.
SLA-1*04:01 was shared by multiple breeds and suggests the early origin of this allele in pigs and the possible importance of the allele in antigen recognition. However, a large number of breed specific alleles (19 alleles, 36.54%) observed in this study may also indicate the influence of recent artificial selections through animal breeding or barriers in gene transfer among the breeds.
SLA-2 also showed a lower allele sharing among breeds with a large number (n = 28) of breed-specific alleles21. The allele SLA-2*04:01 was the most shared allele for SLA-2 among the major pig breeds and belongs to the same haplotype as SLA-1*04:01 (Hp-4)28. This indicated the maintenance of this major class I haplotype during breed diversification. The pattern, however, differs in SLA class II genes such as SLA-DRB1 and DQB1. The much lower rate of breed-specific alleles was identified from both SLA-DRB1 and DQB1, than in SLA class I genes18,20. This could be associated with less polymorphic nature of SLA class II genes in comparison with class I genes, suggesting that the analysis of SLA class I genes may harbor more information to present the history of the species than SLA class II genes.
The MHC class I region appears to have undergone repeated duplication and loss34, resulting in three functional classical class I genes in most mammals35. However, SLA-1 was further duplicated in certain haplotypes and more than three functional MHC class I genes are present in individuals with the duplicated SLA-1. Duplication of SLA-1 was previously reported from Duroc, Sinclair, Hanford, Westran, Belgian, Danish, Yucatan, Kenyan, and Bama pigs, indicating the broad prevalence of SLA-1 duplication from the early period of pig speciation28,32. The distant phylogenetic relationships between the two alleles belonging to the same haplotype is also consistent with the history of SLA-1 duplication in pigs (Table 3, Fig. S3).
SLA-5 and SLA-12 were also duplicated among the SLA genes but they remain nonfunctional due to the presence of the premature stop codons in their coding region36. In contrast, multiple copies of SLA-1 which are functional could serve as a unique system to increase the potential for presenting diverse peptides to cytotoxic T cells.
The higher frequency of SLA-1 duplication in the miniature pigs used in biomedical studies, such as SNU (100%) and NIH (76%), than in other pig breeds, was observed. It is interesting to note that similar results were reported for the Sinclair and Hanford breeds37.
Several inbred lines were selected as homozygous for SLA in NIH miniature pigs for biomedical studies38. Previous and current studies showed the establishment of homozygosity in class II (observed heterozygosity, DRB1-0%; DQA1-0%; DQB1-34.72%) but not in class I genes (observed heterozygosity, SLA-1, 83% and SLA-2, 46.6%)18,20–22 for the breed. Our analysis on SLA-1 diversity in NIH miniature pigs, however, showed that the high SLA-1 (83%) heterozygosity was not due to the actual genetic diversity at the chromosomal level but to the presence of a haplotype with duplicated SLA-1 (Table 3).
In general, the increase of genetic diversity could benefit organismal fitness and disease resistance39. However, studies also suggested that there might be trade-offs between MHC variations and immune capacity by T-cell, deletion and reduced immunocompetence, dominant MHC susceptibility alleles to infectious and autoimmune diseases, MHC cell-surface concentration, and T-cell activation33,40.
The high prevalence of SLA-1 duplication makes it difficult to determine the allelic constitutions of SLA-1 in pigs. Here we observed SLA-1 duplication from a large number of animals (n = 123) and were able to analyze their haplotypes according to our haplotype phasing strategy (Fig. S5). Our results from the analysis of pig cell lines showed that both CNVs and allelic variations may contribute to the level of SLA-1 expression (Fig. 4). However, further studies are necessary to address the underlying mechanism for the allelic difference in SLA-1 expression.
Classical class I MHC molecules need to be associated with beta 2 microglobulin (B2M) to become functional. Interestingly, B2M was also duplicated in pigs41. Thus, dosage increase in one of the heavy chains to form MHC class I complexes balances with the duplication of the light chain, B2M.
The reported SLA-1 typing method in this study requires a single PCR and two sequencing reactions. The procedure for allele discrimination based on local BLAST is simple if the allele database is well prepared. In our method, the most laborious step is the characterization of new alleles that requires cloning and additional sequencing. With further improvements in the list of characterized SLA-1 alleles and duplicated haplotypes, our method can be applicable to high throughput typing.
To infer the complete SLA haplotypes including both class I and II genes from diverse pig breeds could deepen our understanding on highly polymorphic MHC genes in pigs and may reveal the importance of their variations to adaptive immune response. Although genomic DNA-based high-resolution typing is currently available for major SLA genes except SLA-3, simultaneous typing of major SLA genes with high accuracy against a large number of animals is still challenging.
As the cost of next generation sequencing decreases, several recent reports addressed the simultaneous capturing of multiple MHC loci in humans42–44 and other species including pigs32. However, this approach requires both the precise computational procedures to assemble short read sequences and sufficient preceded information on the allelic diversity of MHC genes to avoid errors in haplotype phasing and allelic designation45,46. Therefore, our results could serve as the basis for the high throughput analysis of the SLA system in the future.
Conclusions
We developed a comprehensive high-resolution typing of SLA-1 using single-genomic PCR and subsequent direct sequencing, and reported the identification of new alleles and duplication haplotypes from the typing of over 300 animals from diverse breeds and cell lines. There was a significant difference in the distribution of SLA-1 alleles among the pig breeds including breed-specific alleles. We presented the results of the population level analysis of SLA-1 duplication, for the first time. The high frequency of SLA-1 duplication was observed in the miniature pig breeds use in biomedical studies. We also showed that the duplication of SLA-1 results in an increase of SLA-1 expression in pig cells. In our opinion, this is the most comprehensive and large scale study of SLA-1.
Materials and Methods
Animals and cells
Experiments were conducted using DNA from a total of 295 pigs from 14 different breeds or genetic backgrounds, including 114 Korean native pigs (KNP), 52 Seoul National University (SNU) miniature pig originated from Minnesota miniature pigs47, 29 NIH miniature pigs, 21 Duroc, 19 Berkshire, 19 Yorkshire, 19 Landrace, 6 Landrace x KNP cross, 5 Lanyu, 5 Ossabaw Island hog, 4 Meishan, and 2 American Guinea Hog (AGH). Among the animals used in this study, 48 had pedigree information (Supplementary Table S2). In addition, 8 pig cell lines (CCL-166, CRL-1746, CL-184, CRL-2528, CRL-2842, CRL-6489, CL-101, and CCL-33) from the American Type Culture Collection (ATCC, Manassas, VA, USA) and 4 primary porcine alveolar macrophages (PAM) cells were also analyzed for validation of the typing results. ATCC fibroblast cell lines were cultured in DMEM/high glucose media (Hyclone, UT, USA) supplemented with 10% FBS (Hyclone), 1% penicillin-streptomycin (Gibco, NY, USA), and 2 mM L-glutamine (Gibco) with 5% CO2 and 37 °C. PAMs were isolated from ten 10-week-old euthanized healthy Yorkshire x Landrace cross pigs raised at a local farm using bronchoalveolar lavage through a conventional method48. All experiments were approved and performed accordance with the guidelines and regulations set by Institute of Animal Care and Use Committee and the Center for Research Ethics of Konkuk University
Specific amplification of SLA-1 from genomic DNA
The preparation of genomic DNA and mRNA is described as Supplementary Information. To determine SLA-1 specific primer binding sites, 2086 bp amplicons corresponding to the region from 5′ UTR to the middle of exon 4 of SLA class I genes were produced by PCR with 0.5 μM SLA class I specific primers (SLA-CATF and SLA-R, Table 1) as described previously21. For genomic DNA-based typing, SLA-1 specific amplicons were generated in a 20-μL reaction containing 50–100 ng of genomic DNA, 0.5 μM specific primers (SLA1-e1F1 and SLA e4R4) (Table 1), 200 μM dNTPs, 1 × PCR buffer, and 0.5 U of Supertherm™ DNA polymerase (JMR Holdings, Kent, UK) using a ABI 9700 Thermocycler (Applied Biosystems). The cycling profile consisted of an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 65 °C for 1 min, and extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min. The PCR products were checked by electrophoresis on a 1.5% agarose gel.
Specific amplification of SLA-1 from cDNA
Reverse transcription was carried out according to the manufacturer’s instructions in a 20-μL reaction with oligo(dT)15 and SuperScript™ III reverse transcriptase (Invitrogen, Carlsbad, CA) as described previously21. Two microliters of cDNA were used to amplify the SLA-1 transcripts using SLA1-e1F1 and SLA-e4R4 primers (Table 1) with a cycling profile of an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 65 °C for 1 min, and extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min.
DNA sequencing for SLA-1 typing
The preparation of templates for direct sequencing of PCR products was as described previously18. Sequencing reactions were performed using the ABI PRISM BigDye™ Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) with the sequencing primers SLA1-Seq. 2-F, SLA1-Seq. 2-R, SLA1-Seq. 3-F, and SLA1-Seq. 3-R (Table 1). The RT-PCR products cloned in the PCR-Script Amp SK( + ) cloning vector were bidirectionally sequenced using the T3 and T7 universal sequencing primers. Bidirectional sequencing of the cloned inserts within the pGEM-T Easy Vector for new allele confirmation was carried out using the T7 and SP6 universal primers. The procedures for plasmid sequencing was the same as those of direct sequencing except that the step for removing the unincorporated primers was excluded. A total of 8~10 clones were sequenced bidirectionally for each ligation to eliminate possible sequencing artifacts. For the samples containing new alleles, independent PCR products were sequenced at least twice.
Allelic discrimination of SLA-1
The SLA-1 alleles were discriminated using nucleotide sequence alignment as described previously21. Briefly, chromatograms with sequence quality values >20 were used for the allelic designation of the homozygotes. To separate and identify each allele in the heterozygotes, the PCR products were cloned when the pattern was observed for the first time, and the individual clones of each allele were sequenced. Since a single sequencing template contains information of both exons 2 and 3, the exonal combination was not an issue in our typing method. The sequencing results from 5ʹ and 3ʹ directions were imported into the CLC main workbench version 7.8.1 (CLC Bio, Aarhus, Denmark) for assembly. The resulting 546-bp sequence containing both SLA-1 exon 2 and exon 3 was used in a BLAST search against existing alleles in the local SLA database, which contains information of all reported alleles in public databases and new alleles from our SLA-1 typing. The best five matches were selected and aligned to the query sequence. The allele with any discrepancy compared to the query sequence was removed reiteratively and the final filtered alignment will contain the allele set of the typed sample. The sequence information of the new alleles was submitted to Genbank and the official allele names were designated by the International Society of Animal Genetics (ISAG) SLA nomenclature committee under established guidelines49.
Determination of SLA-1 duplication and haplotypes
The typing results of more than 2 alleles indicates the presence of two copies of SLA-1 alleles on the same chromosome caused by SLA-1 duplication. The determination of genetic linkage between alleles of duplicated SLA-1 are necessary for genetic analyses. The putative haplotypes for duplicated SLA-1 loci was established when the specific allele combination of previously reported24,28 was observed or a specific allele combination was always observed together in the typing results, and the strategy was described in Fig. S5. Subsequently, the identified haplotypes of SLA-1 duplication were applied to deduce allelic combinations of the typing results.
Population genetics analysis
Because of the SLA-1 duplication, the determination of genetic parameters including allele frequency and observed heterozygosity differs from that of a physically single locus. In our analysis, we calculated the values from typing results consisting of all detected SLA-1 in a given individual regardless of the copy numbers of SLA-1. Therefore, the heterozygosity indicates the presence of more than one allele regardless of the duplication. The expected heterozygosity was calculated according to Hartl et al.50. The pairwise Nei’s genetic identity51, Shannon’s diversity index, and allelic evenness52, were calculated as previously reported on the basis of the determined allele frequencies, which was calculated by dividing the total number of the observed allele with the total number of alleles while considering the number of involved SLA-1 loci from the duplication (Table S1). The breed specificity of SLA-1 alleles was visualized using the Jvenn software (http://bioinfo.genotoul.fr/jvenn/index.html). Phylogenetic analysis was performed using the Neighbor Joining method under the general time reversible model as the best fit model in CLC main workbench (CLC Bio).
Quantification of SLA-1 expression using real-time PCR
cDNA was prepared as described above. Primers, SLA1-e1F1 and SLA-e12R, were designed for the specific amplification (137 bp) of SLA-1 transcripts (Table 1). SLA-e12R was located at the junction of SLA-1 exons 1 and 2 to avoid amplification from the genomic DNA (Table 1, Fig. 2). Four ATCC pig cell lines with confirmed SLA-1 types, CL-184 and CRL-17462 with two copies, and CCL-166 and CRL-2528 with 3 copies were used as controls29 (Table S3). Real-time PCR was carried out in a 25-μL reaction containing 1 μL of synthesized cDNA and 0.25 μM of each primer in a 1 × solution of SsoAdvanced™ Universal SYBR® Green Supermix using CFX ConnectTM RealTime System (Bio-Rad, CA, US). The cycling condition of the two-step amplification consisted of an initial denaturation at 94 °C for 3 min; 40 cycles of denaturation at 94 °C for 15 s; annealing for 30 s at 65 °C for SLA-1 and 53 °C for GAPDH; followed by melting curve analysis from 65 °C to 95 °C with an increment of 0.5 °C in step of 0.05 s. The reaction was repeated three times for each sample. The PCR results were analyzed by Bio-Rad CFX Manager, version 3.1 (Bio-Rad). The 2−ΔΔCt method was used for the relative quantification to determine the relative expression level. GAPDH was used as the control gene and was amplified as a 123-bp amplicon using the primers GAPDH-F2 and GAPDH-R2 (Table 1). Statistical analysis was conducted using the Student’s t-test.
Supplementary information
Acknowledgements
The authors thank Dr. Lawrence Schook at University of Illinois Urbana-Champaign for the use of DNA from rare breed pigs. This work was supported by grants from the Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (No. 116134-3) of the Ministry of Agriculture, Food, and Rural Affairs, Republic of Korea and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (No. 2015R1A5A1009701), Republic of Korea.
Author contributions
Conceptualization and sample collection: Minh Thong Le, Hojun Choi, Jin-Hoi Kim, Chankyu Park. Data curation: Minh Thong Le, Hyejeong Lee, Chak-Sum Ho, Chankyu Park. Bioinformatic analysis: Minh Thong Le, Byeongyong Ahn. Methodology: Minh Thong Le, Van Chanh Quy Le, Hyejeong Lee. Manuscript writing: Minh Thong Le, Hojun Choi, Chankyu Park. Comments and discussion: Kwonho Hong, Hyuk Song.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57712-5.
References
- 1.Schook L, et al. Swine in biomedical research: creating the building blocks of animal models. Anim. Biotechnol. 2005;16:183–190. doi: 10.1080/10495390500265034. [DOI] [PubMed] [Google Scholar]
- 2.Chen K, Baxter T, Muir WM, Groenen MA, Schook LB. Genetic resources, genome mapping and evolutionary genomics of the pig (Sus scrofa) Int. J. Biol. Sci. 2007;3:153–165. doi: 10.7150/ijbs.3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vodicka P, et al. The miniature pig as an animal model in biomedical research. Ann. N. Y. Acad. Sci. 2005;1049:161–171. doi: 10.1196/annals.1334.015. [DOI] [PubMed] [Google Scholar]
- 4.VanderWaal K, Deen J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA. 2018 doi: 10.1073/pnas.1806068115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lunney JK, Murrell KD. Immunogenetic analysis of Trichinella spiralis infections in swine. Vet. Parasitol. 1988;29:179–193. doi: 10.1016/0304-4017(88)90125-2. [DOI] [PubMed] [Google Scholar]
- 6.Lumsden JS, Kennedy BW, Mallard BA, Wilkie BN. The influence of the swine major histocompatibility genes on antibody and cell-mediated immune responses to immunization with an aromatic-dependent mutant of Salmonella typhimurium. Can. J. Vet. Res. 1993;57:14–18. [PMC free article] [PubMed] [Google Scholar]
- 7.Lunney JK, Ho C-S, Wysocki M, Smith DM. Molecular genetics of the swine major histocompatibility complex, the SLA complex. Dev. Comp. Immunol. 2009;33:362–374. doi: 10.1016/j.dci.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Serão NVL, et al. Genetic and genomic basis of antibody response to porcine reproductive and respiratory syndrome (PRRS) in gilts and sows. Genetics Selection Evolution. 2016;48:51. doi: 10.1186/s12711-016-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janeway, C. A. Jr., Travers, P., Walport, M. & Shlomchik, M. J. The major histocompatibility complex and its functions. (2001).
- 10.Trowsdale J, Knight JC. Major Histocompatibility Complex Genomics and Human Disease. Annual Review of Genomics and Human Genetics. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wieczorek, M. et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol8, 10.3389/fimmu.2017.00292 (2017). [DOI] [PMC free article] [PubMed]
- 12.Robinson J, et al. The IPD and IMGT/HLA database: allele variant databases. Nucl. Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ando A, et al. Rapid assignment of the swine major histocompatibility complex (SLA) class I and II genotypes in Clawn miniature swine using PCR-SSP and PCR-RFLP methods. Xenotransplantation. 2005;12:121–126. doi: 10.1111/j.1399-3089.2005.00204.x. [DOI] [PubMed] [Google Scholar]
- 14.Ho C-S, Rochelle ES, Martens GW, Schook LB, Smith DM. Characterization of swine leukocyte antigen polymorphism by sequence-based and PCR-SSP methods in Meishan pigs. Immunogenetics. 2006;58:873–882. doi: 10.1007/s00251-006-0145-y. [DOI] [PubMed] [Google Scholar]
- 15.Martens GW, Lunney JK, Baker JE, Smith DM. Rapid assignment of swine leukocyte antigen haplotypes in pedigreed herds using a polymerase chain reaction-based assay. Immunogenetics. 2003;55:395–401. doi: 10.1007/s00251-003-0596-3. [DOI] [PubMed] [Google Scholar]
- 16.Smith DM, Martens GW, Ho CS, Asbury JM. DNA sequence based typing of swine leukocyte antigens in Yucatan Miniature Pigs. Xenotransplantation. 2005;12:481–488. doi: 10.1111/j.1399-3089.2005.00252.x. [DOI] [PubMed] [Google Scholar]
- 17.Ando A, et al. Genetic polymorphism of the swine major histocompatibility complex (SLA) class I genes, SLA-1, -2 and -3. Immunogenetics. 2003;55:583–593. doi: 10.1007/s00251-003-0619-0. [DOI] [PubMed] [Google Scholar]
- 18.Thong LM, et al. Systematic analysis of swine leukocyte antigen-DRB1 nucleotide polymorphisms using genomic DNA-based high-resolution genotyping and identification of new alleles. Tissue antigens. 2011;77:572–583. doi: 10.1111/j.1399-0039.2011.01662.x. [DOI] [PubMed] [Google Scholar]
- 19.Le M, et al. Development of a simultaneous high resolution typing method for three SLA class II genes, SLA-DQA, SLA-DQB1, and SLA-DRB1 and the analysis of SLA class II haplotypes. Gene. 2015;564:228–232. doi: 10.1016/j.gene.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 20.Park K, et al. Simple and comprehensive SLA-DQB1 genotyping using genomic PCR and direct sequencing. Tissue Antigens. 2010;76:301–310. doi: 10.1111/j.1399-0039.2010.01522.x. [DOI] [PubMed] [Google Scholar]
- 21.Choi H, et al. Sequence variations of the locus-specific 5′ untranslated regions of SLA class I genes and the development of a comprehensive genomic DNA-based high-resolution typing method for SLA-2. Tissue Antigens. 2015;86:255–266. doi: 10.1111/tan.12648. [DOI] [PubMed] [Google Scholar]
- 22.Le MT, et al. Comprehensive and high-resolution typing of swine leukocyte antigen DQA from genomic DNA and determination of 25 new SLA class II haplotypes. Tissue Antigens. 2012;80:528–535. doi: 10.1111/tan.12017. [DOI] [PubMed] [Google Scholar]
- 23.Maccari Giuseppe, Robinson James, Ballingall Keith, Guethlein Lisbeth A., Grimholt Unni, Kaufman Jim, Ho Chak-Sum, de Groot Natasja G., Flicek Paul, Bontrop Ronald E., Hammond John A., Marsh Steven G. E. IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Research. 2016;45(D1):D860–D864. doi: 10.1093/nar/gkw1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DM, et al. Nomenclature for factors of the SLA class-I system, 2004. Tissue Antigens. 2005;65:136–149. doi: 10.1111/j.1399-0039.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee J-H, et al. Characterization of the swine major histocompatibility complex alleles at eight loci in Westran pigs. Xenotransplantation. 2005;12:303–307. doi: 10.1111/j.1399-3089.2005.00231.x. [DOI] [PubMed] [Google Scholar]
- 26.Soe OK, et al. Assignment of the SLA alleles and reproductive potential of selective breeding Duroc pig lines. Xenotransplantation. 2008;15:390–397. doi: 10.1111/j.1399-3089.2008.00499.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, et al. Development of a simple SLA-1 copy-number-variation typing and the comparison of typing accuracy between real-time quantitative and droplet digital PCR. Anim. Genet. 2019;50:315–316. doi: 10.1111/age.12785. [DOI] [PubMed] [Google Scholar]
- 28.Ho CS, et al. Nomenclature for factors of the SLA system, update 2008. Tissue Antigens. 2009;73:307–315. doi: 10.1111/j.1399-0039.2009.01213.x. [DOI] [PubMed] [Google Scholar]
- 29.Ho CS, Franzo-Romain MH, Lee YJ, Lee JH, Smith DM. Sequence-based characterization of swine leucocyte antigen alleles in commercially available porcine cell lines. Int J Immunogenet. 2009;36:231–234. doi: 10.1111/j.1744-313X.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- 30.Gao, C. et al. Swine Leukocyte Antigen Diversity in Canadian Specific Pathogen-Free Yorkshire and Landrace Pigs. Front Immunol8, 10.3389/fimmu.2017.00282 (2017). [DOI] [PMC free article] [PubMed]
- 31.Yeom S-C, Park C-G, Lee B-C, Lee W-J. SLA typing using the PCR-SSP method and establishment of the SLA homozygote line in pedigreed SNU miniature pigs. Anim. Sci. J. 2010;81:158–164. doi: 10.1111/j.1740-0929.2009.00727.x. [DOI] [PubMed] [Google Scholar]
- 32.Sørensen, M. R. et al. Sequence-Based Genotyping of Expressed Swine Leukocyte Antigen Class I Alleles by Next-Generation Sequencing Reveal Novel Swine Leukocyte Antigen Class I Haplotypes and Alleles in Belgian, Danish, and Kenyan Fattening Pigs and Göttingen Minipigs. Front Immunol.8, 10.3389/fimmu.2017.00701 (2017). [DOI] [PMC free article] [PubMed]
- 33.Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Frontiers in Zoology. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flajnik MF, Kasahara M. Comparative Genomics of the MHC: Glimpses into the Evolution of the Adaptive Immune System. Immunity. 2001;15:351–362. doi: 10.1016/S1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- 35.Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H. Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol. Rev. 2002;190:95–122. doi: 10.1034/j.1600-065X.2002.19008.x. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka-Matsuda M, Ando A, Rogel-Gaillard C, Chardon P, Uenishi H. Difference in number of loci of swine leukocyte antigen classical class I genes among haplotypes. Genomics. 2009;93:261–273. doi: 10.1016/j.ygeno.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Ho C-S, et al. Swine leukocyte antigen (SLA) diversity in Sinclair and Hanford swine. Developmental & Comparative Immunology. 2010;34:250–257. doi: 10.1016/j.dci.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez, K., Dicks, N., Glanzner, W. G., Agellon, L. B. & Bordignon, V. Efficacy of the porcine species in biomedical research. Front Genet6, 10.3389/fgene.2015.00293 (2015). [DOI] [PMC free article] [PubMed]
- 39.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum. Mol. Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 40.Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. Biol. Sci. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le TM, et al. β2-microglobulin gene duplication in cetartiodactyla remains intact only in pigs and possibly confers selective advantage to the species. PLOS ONE. 2017;12:e0182322. doi: 10.1371/journal.pone.0182322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittig M, et al. Development of a high-resolution NGS-based HLA-typing and analysis pipeline. Nucl. Acids Res. 2015;43:e70–e70. doi: 10.1093/nar/gkv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erlich RL, et al. Next-generation sequencing for HLA typing of class I loci. BMC Genomics. 2011;12:42. doi: 10.1186/1471-2164-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danzer M, et al. Rapid, scalable and highly automated HLA genotyping using next-generation sequencing: a transition from research to diagnostics. BMC Genomics. 2013;14:221. doi: 10.1186/1471-2164-14-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ka S, et al. HLAscan: genotyping of the HLA region using next-generation sequencing data. BMC Bioinformatics. 2017;18:258. doi: 10.1186/s12859-017-1671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosomichi K, Shiina T, Tajima A, Inoue I. The impact of next-generation sequencing technologies on HLA research. J Hum Genet. 2015;60:665–673. doi: 10.1038/jhg.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Setcavage TM, Kim YB. Variability of the immunological state of germfree colostrum-deprived Minnesota miniature piglets. Infect Immun. 1976;13:600–607. doi: 10.1128/IAI.13.2.600-607.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Leengoed LA, Kamp EM. A method for bronchoalveolar lavage in live pigs. Vet Q. 1989;11:65–72. doi: 10.1080/01652176.1989.9694201. [DOI] [PubMed] [Google Scholar]
- 49.Smith DM, et al. Nomenclature for factors of the swine leukocyte antigen class II system, 2005. Tissue Antigens. 2005;66:623–639. doi: 10.1111/j.1399-0039.2005.00492.x. [DOI] [PubMed] [Google Scholar]
- 50.Hartl, D. L. & Clark, A. G. Principles of Population Genetics. 4th edition edn. (Sinauer Associates is an imprint of Oxford University Press, 2006).
- 51.Nei M. Genetic Distance between Populations. The American Naturalist. 1972;106:283–292. doi: 10.1086/282771. [DOI] [Google Scholar]
- 52.Shannon CE. A Mathematical Theory of Communication. Bell System Technical Journal. 1948;27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.