Abstract
Sarcomatoid clear-cell renal cell carcinomas (sRCC) are associated with dismal prognosis. Genomic alterations associated with sarcomatoid dedifferentiation are poorly characterized. We sought to define the genomic landscape of sRCC and uncover potentially actionable therapeutic targets. We assessed the genomic landscape of sRCC using targeted panel sequencing including patients with microdissected sarcomatoid and epithelial components. Along with common genomic alterations associated with clear-cell histology, we found that Hippo was one of the most frequently altered pathways in these tumours. Hippo alterations were differentially enriched in sRCC compared to non-sRCC. Functional analysis showed that Hippo members mutations were associated with higher nuclear accumulation of YAP/TAZ, core effectors of the Hippo pathway. In a NF2-mutant sRCC model, YAP1 knockdown and NF2 reconstitution suppressed cell proliferation, tumour growth and invasion, both in vitro and in vivo. Overall, we show that Hippo pathway alterations are a feature of sRCC, and enable the exploration of the Hippo pathway as a novel potential therapeutic target.
Subject terms: Cancer genetics, Cancer
Introduction
Sarcomatoid dedifferentiation has been reported in ~5% of clear-cell renal cell carcinomas (ccRCC) and is considered one of the most aggressive features of the disease1. Sarcomatoid ccRCCs (sRCC) have been associated with high tumour burden and frequent metastatic dissemination, with a synchronous metastasis rate at diagnosis of 70%, compared to 30% in non-sRCCs2. Patient prognosis is dismal, with median overall survival duration less than 12 months in patients treated with targeted molecular therapies3.
The mechanism underlying sarcomatoid dedifferentiation in ccRCC remains poorly defined. Several recent studies tried to define the genomic landscape of these tumours, with discordant results. Molecular profiling of 26 sarcomatoid renal cell carcinomas with mixed clear cell and non–clear cell histological subtypes identified recurrent alterations of TP53, Hippo pathway member NF2 and cell cycle gene CDKN2A4. An analysis of another 65 sarcomatoid kidney tumours of varying histological subtypes5 reported increased mutations of PTEN and chromatin remodelling genes BAP1 and SETD2 in clear cell subtypes, increased NF2 mutations in papillary subtypes and increased TP53 alterations in both. In one study that performed systematic microdissection of epithelial and sarcomatoid components of renal cell carcinomas6, 21 tumours with mixed histology had sarcomatoid components harbouring a greater mutational load than paired epithelial components, as well as greater numbers of mutations of TP53, chromatin modifiers BAP1 and ARID1 and Hippo pathway gene FAT2. Overall, these studies displayed different pictures of sarcomatoid kidney cancers, and key events leading to sarcomatoid dedifferentiation remain unsettled.
Thus, we sought to understand the molecular events associated with sarcomatoid dedifferentiation in ccRCC, looking for alterations that might affect the therapeutic management of these tumours. We show that alterations in the Hippo pathway are frequent in sRCCs, and that inhibition of downstream Hippo effectors inhibit cell proliferation, tumour growth and invasion. Ours is the first report of Hippo pathway alterations as a candidate actionable driver of sarcomatoid dedifferentiation in ccRCC.
Results
Targeted sequencing of sRCCs uncovers recurrent genomic alterations
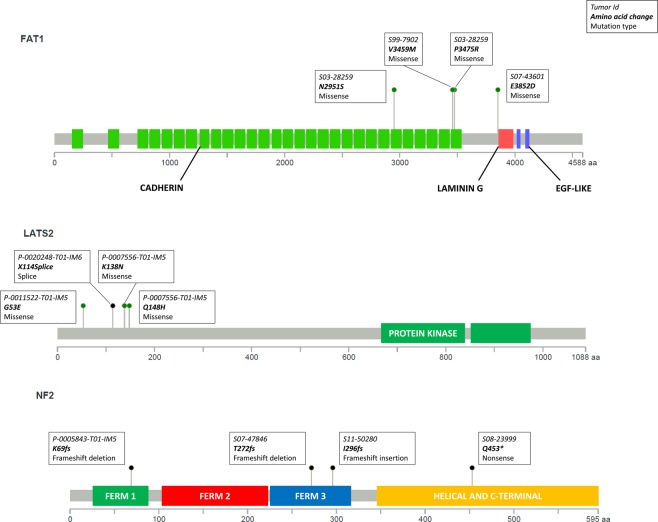
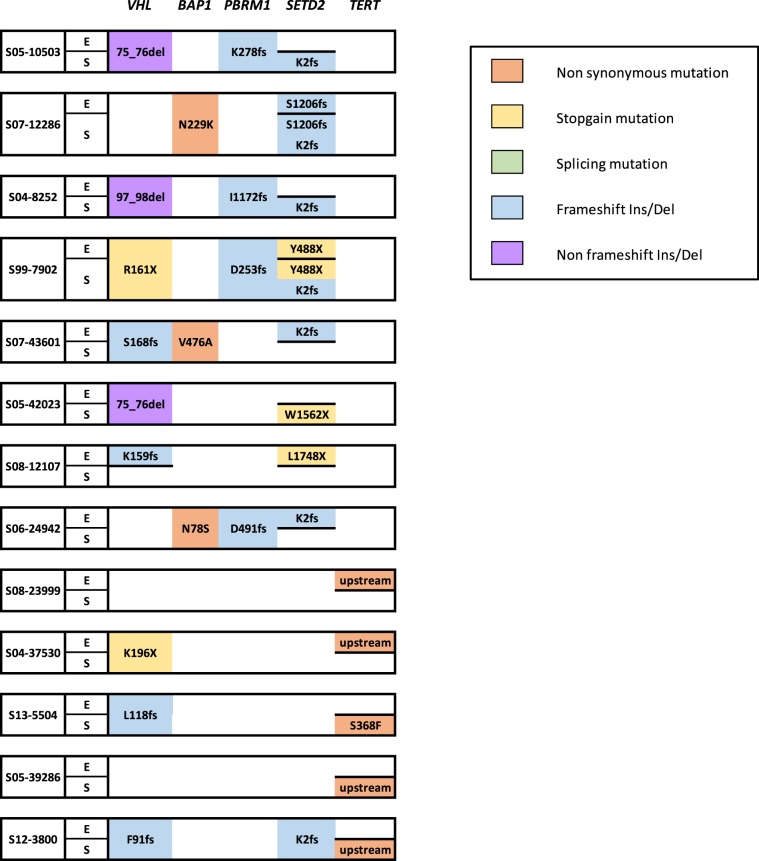
We first performed targeted sequencing in 50 samples from 27 sRCCs that had been microdissected (Fig. 1A and Supplementary Table S1). Paired sequencing of mesenchymal and epithelial components was available for 23 of these tumours, while 2 had only a mesenchymal component and 2 only an epithelial component available. Across all samples, recurrent oncogenic alterations involved key tumour suppressor genes in ccRCC, including VHL (36/50, 72%) and chromatin remodelling genes SETD2 (20/50, 40%), PBRM1 (17/50, 34%) and BAP1 (13/50, 26%). Interestingly, 9 of the 20 mutations of SETD2 involved a hotspot frameshift mutation (K2fs). Recurrent mutations of TERT were identified in 9 samples (18%), including 6 C228T hotspot mutations and 2 C250T hotspot mutations, both located in the TERT promoter7,8. Additional mutations found in >10% of samples involved MTOR pathway members PTEN (7/50, 14%) and TSC2 (6/50, 12%), as well as Hippo pathway members NF2 (5/50, 10%) and FAT1 (5/50, 10%). As recurrent mutations of the Hippo pathway were not previously described with this high rate in ccRCC, we assessed the functional impact of these mutations and found that all mutations of NF2 and FAT1 were deleterious and affected functional domains of the proteins (Fig. 2 and Supplementary Table S2).
Figure 1.
Genomic alterations in sRCCs by targeted panel sequencing (A) Genomic alterations identified by targeted sequencing in microdissected sRCCs (N = 27). Epithelial component is labeled E, mesenchymal (sarcomatoid) component is labeled S. (B) Genomic alterations identified by targeted sequencing in non-microdissected sRCCs (N = 22).
Figure 2.
Mapping of Hippo protein alterations in sRCCs.
We then analyzed mutated genes in consideration with putative oncogenic mechanisms in these samples (Fig. 1A). Along with VHL mutations, the most frequent alterations affected chromatin remodelling genes (36/50, 72%). In addition to recurrent mutations of BAP1, PBRM1 and SETD2 previously described, we found mutations of SWI/SNF members ARID1B in 3 samples and ARID1A in one, as well as mutations of epigenetic regulators KDM6A (4%), MLL (4%), ASXL1 (4%), NSD1 (4%), DNMT3A (2%), MLL2 (2%) and MLL3 (2%). Notably, 2 paired epithelial and mesenchymal samples from one tumour presented with convergent mutations of SETD2 and NSD1, which have been described to be associated with an aggressive phenotype in ccRCC9. Other frequent alterations involved the MTOR pathway (25/50, 50%), DNA repair (15/50, 30%) and the Hippo pathway (10/50, 20%).
We then compared the genomic profiles of the 23-paired epithelial and mesenchymal components (Fig. 1A and Supplementary Table S1). Alterations of SETD2 and TERT repeatedly differed between mesenchymal and epithelial components, with 8/20 mutations of SETD2 and 5/9 mutations of TERT not shared (Fig. 3). These observations are in line with recent studies reporting that SETD2 alterations are associated with high rates of subclonality10,11. Overall, 16 of 23 sRCCs had at least one putative oncogenic mutation specifically found in the mesenchymal component (Fig. 1A). Apart from known ccRCC oncogenic alterations, one tumour harboured NF2 and CDKN2A mutations exclusively in its mesenchymal component, and 2 tumours had TP53 mutations that were present exclusively in the mesenchymal component of these tumours.
Figure 3.
Differential alterations of SETD2 and TERT in epithelial and mesenchymal components of sRCCs.
In 22 additional sRCCs, targeted sequencing was performed without prior microdissection (Fig. 1B and Supplementary Table S1). The genomic profiles of these additional samples were concordant with previous findings, with key oncogenic alterations of VHL in 68%, of chromatin remodelling genes in 73% and of the MTOR pathway in 50%. In addition, TP53 alterations were reported in 27%. TERT mutations and DNA repair pathway alterations were reported in 18% and 14%, respectively, of these tumours. Interestingly, we again found frequent Hippo pathway alterations (18%). Notably, 3 tumours harboured deleterious mutations of the core Hippo pathway member LATS2, one tumour with a splicing mutation and 2 with missense mutations. One additional tumour had a frameshift mutation of NF2 (Fig. 2).
Overall, in both microdissected and non-microdissected tumours, 10 of the 49 sRCCs displayed deleterious Hippo pathway alterations (20%) in at least one tumour section. We then investigated whether Hippo pathway alterations were more frequent in sRCCs than in the 268 non-sRCCs. The non-sRCCs were predominantly higher risk tumours; only 14 of the 268 (5%) had Hippo pathway alterations, involving NF2 (6/268), FAT1 (3/268), LATS1 (2/268), LATS2 (3/268) and YAP1 (1/268). Thus, the frequency of Hippo pathway mutations was significantly higher in sRCCs than in non-sRCCs (p = 0.001).
YAP/TAZ is upregulated in Hippo-mutant sRCCs
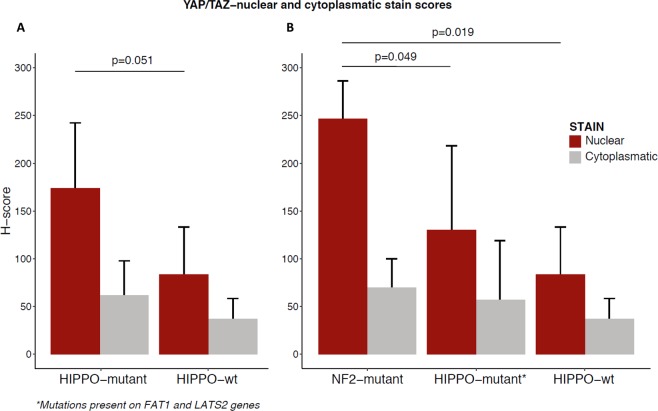
Several reports showed that NF2 is a potent suppressor of hippo signalling through phosphorylation of YAP/TAZ leading to their sequestration in the cytoplasm and their degradation by the proteasome, thereby blocking oncogenic transcription associated with YAP/TAZ nuclear translocation12. To explore the relevance of mutations affecting Hippo genes in sRCC, we determined YAP/TAZ protein expression and intracellular localization by immunohistochemistry in 8 Hippo-mutated sRCC and 8 Wild-type sRCC. As expected, there was a trend toward increased nuclear YAP/TAZ signal in Hippo-mutant cases as compared to wild-type cases (p = 0.051) (Fig. 4A). Strikingly, when we focused on NF2-mutant cases (n = 3), we observed a stronger nuclear YAP/TAZ protein as compared to wild-type sRCC (p = 0.019) and other Hippo-mutants (LATS1 and FAT1) (p = 0.049) (Fig. 4B), suggesting distinct functional importance of mutations affecting Hippo pathway components. Of note, no difference was observed between cytoplasmic YAP/TAZ signal between Hippo-mutant cases and Wild-type cases (p = 0.35).
Figure 4.
Immunohistochemistry staining for YAP/TAZ in sarcomatoid components of Hippo-mutant (N = 8) and wild-type (N = 8) sRCCs A, expression of YAP/TAZ in Hippo-mutant and Hippo wild-type sRCCs B, expression of YAP/TAZ according to Hippo-mutation type.
YAP1 knockout and NF2 reconstitution inhibit proliferation and invasion of sRCC
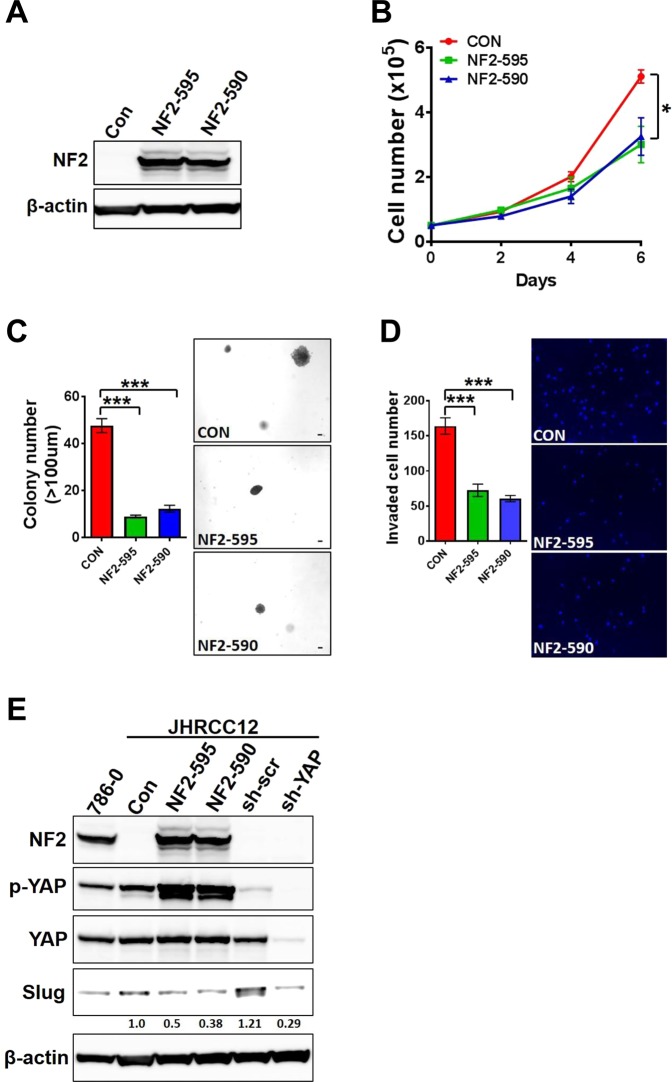
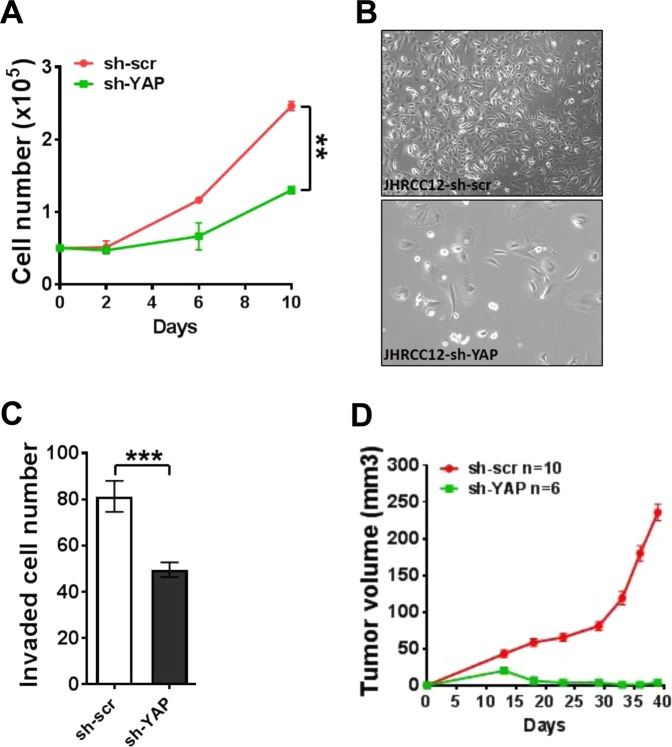
To analyse the functional role of hippo core genes mutations, we used JHRCC12, a NF2-mutant sRCC cell line, as a model. Notably, NF2 reconstitution (Fig. 5A) suppressed cell proliferation, transformation and invasion (Fig. 5B–D). As YAP1 expression has been shown to strongly induce expression of mesenchymal genes such as SLUG (SNAI2)13, we then investigated the effect of NF2 reconstitution and YAP1 knockdown on SLUG expression; as expected, we found decrease of SLUG which was more profound with YAP1 knockdown than with NF2 reconstitution (Fig. 5E). We then asked whether interference with hippo pathway could disrupt tumour growth in vitro and in vivo. YAP1 knockdown in JHRCC12 cells suppressed cell proliferation, invasion, and tumour growth and induced change in their morphology (Fig. 6A–C). Finally, to explore the effect of YAP1 interference in vivo, we performed loss-of-function experiment in the male immunocompromised NOD-SCID IL2Rg−/− (NSG) mice using JHRCC12 xenografts. Notably, we observed that YAP1 suppression blocked tumour growth (Fig. 6D), suggesting that proliferation in vivo occurs in cells with higher YAP1 activity. Together, the in vitro and in vivo observations suggest that the interference with Hippo pathway might have therapeutic relevance.
Figure 5.
NF2 reconstitution suppressed cell proliferation, transformation and invasion in NF2-mutant sRCC. (A) Immunoblot of NF2 reconstitution expression in JHRCC12 cells. (B) Cell proliferation after NF2 reconstitution was performed by cell counting on day 0, 2, 4, and 6. Data are presented as mean ± SD. *P < 0.05. (C) Soft agar assay for control and NF2 reconstituted JHRCC12 cells. Scale bar is 100um. Data presented are mean ± SD. ***P < 0.001. (D) Invasion assay for control and NF2 reconstituted JHRCC12 cells. Data presented are mean ± SD. ***P < 0.001. (E) Immunoblot of NF2, p-YAP, YAP, Slug and β-actin in 786-0, control, NF2 reconstitution and YAP1 knockdown JHRCC12 cells. Slug expression level was normalized to control expression sample.
Figure 6.
YAP1 knockdown suppressed cell proliferation, invasion and tumour growth in NF2-mutant sRCC. (A) Cell proliferation of YAP1 knockdown in JHRCC12 cells. Data are presented as mean ± SD. **P < 0.005. (B) Cell number and morphology change after YAP1 knockdown. (C) Invasion assay for scramble-control (sh-scr) and YAP1 knockdown (sh-YAP) in JHRCC12 cells. Data presented are mean ± SD. ***P < 0.001. (D) Tumour growth curve of scramble-control (sh-scr) and YAP1 knockdown (sh-YAP) JHRCC12 cells.
Discussion
Our study highlights for the first time an association between alterations of the Hippo pathway, a key regulator of organ growth, and sarcomatoid dedifferentiation in ccRCC. Mutations of NF2, FAT1 and LATS2 were the most frequent Hippo pathway alterations14. Most notably, sRCCs exhibited more frequent mutations in Hippo genes than non-sRCC controls. We showed that downstream effectors of the Hippo pathway were upregulated in patients with alteration in Hippo pathway genes. Finally, mechanistic studies in vitro and in vivo demonstrated that YAP1 inhibition or NF2 reconstitution impaired tumour growth and proliferation. Thus, Hippo pathway mutations appear as one of the most frequent potential oncogenic mechanisms in sRCC. The finding of one tumour with deleterious NF2 mutation in its sarcomatoid component only, suggest that Hippo pathway mutations might be late events in ccRCCs with sarcomatoid dedifferentiation.
The prevalence of Hippo pathway gene alterations in this study is supported by previous observations from Bi et al. of increased FAT family member mutations in the sarcomatoid component of kidney tumours6. However, various observations in our study contrast with previous molecular studies. Notably, the role of tumour suppressor TP53 in sRCCs is still unclear, as we found a lower frequency of TP53 alterations than expected, reported in 19% of sRCCs. Conversely, TP53 mutations were found in 31% and 42%, respectively, of sarcomatoid kidney tumours with mixed histology in the studies by Bi et al.6 and Malouf et al.4. Such differences might be explained by the small numbers of patients, and heterogeneity of histologies in the previously published papers. Along with TP53, TERT was also recurrently altered in our sRCC cohorts, while such alterations were found in less than 3% of lower grade tumours datasets15,16. It is yet unknown whether this high rate of TERT and TP53 alterations is related to subclonal evolution in aggressive and heterogeneous tumors, or could be directly associated with epithelial mesenchymal transition mechanisms. Thus, the importance of TP53 and TERT in sRCC should be further explored in follow-up studies.
Our study also highlighted recurrent mutations of NF2 in sRCCs, which contrasts with previous reports of NF2 alterations that were specific to sarcomatoid tumours with papillary histology5. In addition, recent work suggested that the Hippo pathway is recurrently disrupted in aggressive, unclassified kidney tumours. In that study, loss of NF2 and concurrent accumulation of YAP/TAZ in the nucleus occurred in 26 of 62 (26%) unclassified kidney cancers17. These data pinpoint that Hippo alterations might be a marker of tumour aggressiveness regardless of histology.
The putative role of the Hippo pathway in sarcomatoid dedifferentiation is supported by the promotion of epithelial-mesenchymal transition (EMT) by the core members of the Hippo pathway, YAP113,18,19 and TAZ20,21. YAP1 and TAZ are transcriptional regulators reported to be expressed upon TGF-β pathway activation, and are both associated with EMT signatures in solid tumours13 through upregulation of EMT genes vimentin, fibronectin, SLUG and ZEB1. An interplay between MAPK signalling and YAP1 is also reported in various studies13,19. Our study confirm the importance of Hippo core pathway member YAP1, but also of upstream effector NF2 for the regulation of tumor morphology and invasive properties.
A comprehensive molecular characterization of 19 Hippo core genes was performed on 9,125 tumour samples across 33 cancer types from The Cancer Genome Atlas22. Among the hyper-altered cancer types, mesotheliomas and papillary renal cell carcinomas were the most significant, and this finding was likely related to a high frequency of NF2, LATS2, and SAV1 mutations. Of note, YAP1/TAZ activation has been previously shown to confer malignant phenotypes to mesothelial cells and conditional (mesothelium-specific) NF2 knockout mouse models exhibited an increased incidence of mesothelioma development23. Although function of the Hippo pathway in the kidney has been unknown for long time, recent studies have identified a key role of YAP1 signalling in glomerular embryonic development, maintenance of podocyte homeostasis, and renal fibrogenesis24. Whether conditional (kidney-specific) Hippo knockout mouse models would be associated with kidney cancer development and/or sarcomatoid dedifferentiation remain unknown to last of our knowledge.
Targeting the Hippo pathway can be a valid approach in Hippo-driven malignancies. Currently, different approaches to block YAP1/TAZ interactions with their target transcription factors TEADs are explored. Verteporfin was the first small molecule to act as a YAP1-TEAD binding inhibitor; Other YAP1-TEAD inhibitors include the bioengineer-designer cyclic YAP-1 like peptides as well as the natural YAP1 antagonist VGLL412. In a preclinical model of papillary renal cell carcinomas harbouring NF2 loss25, inhibition of the YAP1 partner YES1 by dasatinib or saracatinib led to repression of Hippo transcriptional targets and provided potent antitumour activity. In addition, several distinct small molecules including dasatinib, pazopanib, statins, and ivermectin, have also been shown to inhibit YAP1/TAZ by drug screening in human cancer cell lines26. These data suggest that our findings can have immediate perspectives for clinical care.
Overall, our study identifies the Hippo pathway as a key alteration in sRCCs. These findings advocate for new therapeutic developments targeting the Hippo pathway in sarcomatoid renal cell carcinomas, and further investigation of this pathway’s oncogenic potential across other cancer subtypes.
Methods
Sample collection
Frozen or formalin-fixed, paraffin-embedded (FFPE) sRCC, tissue samples were collected from 49 patients who underwent nephrectomy in Memorial Sloan Kettering Cancer Center, NY, USA. All were de-identified. Dedicated genitourinary pathologists reviewed all samples to confirm the diagnosis of sRCC, defined as clear-cell RCC associated with any percentage of sarcomatoid component according to the International Society of Urological Pathology guidelines27. All patients had previously provided written informed consent for tumour collection and analysis.
We performed laser-assisted microdissection of samples of 27 tumours allowing targeted sequencing of the epithelial and mesenchymal components. We expanded that cohort with 22 primary sRCC samples that did not undergo microdissection. Local controls comprised non-sRCC samples from 268 patients.
The Memorial Sloan Kettering Cancer Center’s Institutional Review Board approved the study, and the collection and use of tissues followed ethical procedures formulated in the Helsinki Declaration
Targeted sequencing
We performed targeted sequencing using the MSK-IMPACT assay28. Samples from the 27 patients were analysed on a panel of 341 key cancer-associated genes, and samples from the other 22 patients were analysed by an updated assay that includes 410 genes. Full lists of the genes in both assays are included as Supplementary Table S3. Barcoded sequences were prepared and captured by hybridization with custom biotinylated DNA probes for all exons and selected introns of the selected genes using 100–500 ng of input DNA. Captured libraries were sequenced on an Illumina HiSeq (2 × 100 bp paired-end reads). The raw reads were aligned to the human genome (hg19) using the Burrows-Wheeler Alignment Tool, followed by duplicate read removal, base recalibration and indel realignment using GATK (v 2.6–5)57. Somatic variants were called using MuTect (v 1.1.4) for single-nucleotide variants and Somatic Indel Detector (GATK 2.3–9) for indels; they were then annotated by Annovar for cDNA and amino-acid changes as well as their presence in the dbSNP database (v137) and COSMIC database (v68) and in the 1000 Genomes database for minor allele frequencies. IMPACT was designed to focus on somatic mutation detection by filtering out alterations also present in normal samples. We assessed the functional impact of single nucleotide variants using 2 independent web-based tools, Mutation Assessor29 and Polyphen230.
Statistical analysis
Mapping of the Hippo protein alterations was performed using MutationMapper v1.0.131,32. We associated mutated genes with oncogenic pathways according to comprehensive analysis of the published literature14,33–36. Comparisons of clinical, pathological and molecular data were performed with the Fisher exact test or Wilcoxon test, depending on the study variable. All statistical tests were 2-sided and conducted at the significance level of 0.05.
Animal studies
All animal work was performed in accordance with institutional guidelines and Institutional Animal Care and Use Committee (IACUC) approval. Five-to-six-week-old male immunocompromised NOD-SCID IL2Rg−/− (NSG) mice (Jackson Laboratory) were used for the study. To generate xenograft tumour growth curve, one million cells resuspended in PBS:matrigel (1:1) were injected into the flank of the mice subcutaneously. Two flank tumours were placed per mouse. Tumour volume was measured using calipers (calculated as 0.5 × L xW2), and growth curves were generated using GraphPad Prism software.
Plasmid constructs and virus production
Full-length NF2 isoforms 1 (amino acid 1–595) and 2 (amino acid 1–590) were digested and cloned into pBabe-puro plasmid at the BamHI and EcoRI sites. pLKO1-shYAP plasmid (addgene 27369) was used for YAP1 knockdown. Both retrovirus for NF2 reconstitution and lentivirus for YAP1 knockdown were generated by transfecting HEK293T cells with Lipofectamine 2000 reagent (Invitrogen). Cell culture medium with virus was collected and centrifuged at 3000 g for 5 minutes at 4 °C. The supernatant was used for target cell infection with 8ug/ml polybrene. The infected cells were selected in 2 ug/ml puromycin in culture medium and used for assays.
Cell culture, proliferation and western blot
Cell culture was performed as describe before37. For cell proliferation assay, 5 × 104 cells were seeded in a 6-cm dishes in triplicate on day0 and counted on different time points indicated in the figures by cell counter (Nexcelom, USA). Cell proliferation curves were generated with GraphPad Prism software v.5.02 (La Jolla, CA, USA). For western blot, cells were lysed in standard RIPA buffer. Antibodies for NF2 (abcam ab88957), p-YAP (Cell Signaling 13008), YAP (Cell Signaling 12395), Slug (Cell Signaling 9585) and β-actin (Sigma A1978) were used for western blot.
Soft agar assay and invasion assay
For soft agar assay, 1 × 105 cells were seeded onto a 6-cm dish containing a top layer of 0.3% noble agar and a bottom layer of 0.6% noble agar base. Cells were fed with media every 3 days. After 3 weeks, colonies with diameter larger than 100 μm were scored. Three independent triplicate experiments were performed. Scale bar is 100um. Data presented are mean ± SD of three independent experiments.
For invasion assay, 2 × 104 cells seeded in each transwell of 24-well (8um) Corning invasion chamber in culture medium with 0.1% FBS. Full culture medium with 10% FBS was added in each well of the plate as the attractant. After 24-hour incubation at 37 °C, cells in each transwell were removed and the membrane was fixed in 10% formalin and stained with DAPI solution. Invaded cell number was counted in 9 random views from triplicate under fluorescence microscope.
Immunohistochemistry
Immunohistochemistry was conducted in 5 µm formalin-fixed paraffin-embedded (FFPE) whole tissue sections. Staining for YAP/TAZ (D24E4, Cell Signalling Technology, 1:50) was performed using an automated Ventana Discovery system (Ventana). Immunostaining scores (H-scores) for YAP/TAZ nuclear and cytoplasmic staining in the sarcomatoid areas were assessed separately and calculated as [H = intensity (0-3) × percentage of positive cells (1–100)].
Supplementary information
Acknowledgements
Patients were supported in part by MSKCC Support Grant/Core Grant (P30 CA008748).
Author contributions
G.G.M., J-P.S., D.K., J.A.K., N.M.T. and A.A.H. designed the study. E.C., M.R., R. Mano, J.A.K. and P.R. collected samples. Y.D., Y.B.C., K.A.B. and A.A.H. performed the experiments. G.G.M., R.F., R.G.D., X.S., E.C., M.R., H.Y., R. Mouawad, T.H.H., S.K.T., J.J.H. and A.A.H. participated in data analysis and interpretation. All authors participated in manuscript writing. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gabriel G. Malouf, Ronan Flippot, Yiyu Dong, Nizar M. Tannir and Abraham A. Hakimi.
Contributor Information
Gabriel G. Malouf, Email: maloufg@igbmc.fr
Nizar M. Tannir, Email: ntannir@mdanderson.org
Abraham A. Hakimi, Email: hakimia@mskcc.org
Supplementary information
is available for this paper at 10.1038/s41598-020-57534-5.
References
- 1.Kunene V, et al. Sarcomatoid renal cell carcinoma: clinical outcome and survival after treatment with sunitinib. Clin. Genitourin. Cancer. 2014;12:251–255. doi: 10.1016/j.clgc.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Shuch B, et al. Impact of pathological tumour characteristics in patients with sarcomatoid renal cell carcinoma. BJU Int. 2012;109:1600–1606. doi: 10.1111/j.1464-410X.2011.10785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golshayan AR, et al. Metastatic sarcomatoid renal cell carcinoma treated with vascular endothelial growth factor-targeted therapy. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009;27:235–241. doi: 10.1200/JCO.2008.18.0000. [DOI] [PubMed] [Google Scholar]
- 4.Malouf GG, et al. Genomic Characterization of Renal Cell Carcinoma with Sarcomatoid Dedifferentiation Pinpoints Recurrent Genomic Alterations. Eur. Urol. 2016;70:348–357. doi: 10.1016/j.eururo.2016.01.051. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, et al. Sarcomatoid renal cell carcinoma has a distinct molecular pathogenesis, driver mutation profile and transcriptional landscape. Clin. Cancer Res. clincanres. 2017;1057:2017. doi: 10.1158/1078-0432.CCR-17-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi M, et al. Genomic characterization of sarcomatoid transformation in clear cell renal cell carcinoma. Proc. Natl. Acad. Sci. USA. 2016;113:2170–2175. doi: 10.1073/pnas.1525735113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, et al. TERT promoter mutations in renal cell carcinomas and upper tract urothelial carcinomas. Oncotarget. 2014;5:1829–1836. doi: 10.18632/oncotarget.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casuscelli Jozefina, Becerra Maria F., Manley Brandon J., Zabor Emily C., Reznik Ed, Redzematovic Almedina, Arcila Maria E., Tennenbaum Daniel M., Ghanaat Mazyar, Kashan Mahyar, Stief Christian G., Carlo Maria, Voss Martin H., Feldman Darren R., Motzer Robert J., Chen Yingbei, Reuter Victor E., Coleman Jonathan A., Russo Paul, Hsieh James J., Hakimi Abraham Ari. Characterization and Impact of TERT Promoter Region Mutations on Clinical Outcome in Renal Cell Carcinoma. European Urology Focus. 2019;5(4):642–649. doi: 10.1016/j.euf.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X, et al. NSD1 Inactivation and SETD2 Mutation Drive a Convergence toward Loss of Function of H3K36 Writers in Clear Cell Renal Cell Carcinomas. Cancer Res. 2017;77:4835–4845. doi: 10.1158/0008-5472.CAN-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turajlic S, et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell. 2018;173:581–594.e12. doi: 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanu N, et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene. 2015;34:5699–5708. doi: 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekido Yoshitaka. Targeting the Hippo Pathway Is a New Potential Therapeutic Modality for Malignant Mesothelioma. Cancers. 2018;10(4):90. doi: 10.3390/cancers10040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao DD, et al. KRAS and YAP1 Converge to Regulate EMT and Tumor Survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 17.Chen, Y.-B. et al. Molecular analysis of aggressive renal cell carcinoma with unclassified histology reveals distinct subsets. Nat. Commun. 7 (2016). [DOI] [PMC free article] [PubMed]
- 18.Lamar JM, et al. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl. Acad. Sci. 2012;109:E2441–E2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, et al. YAP modulates TGF-β1-induced simultaneous apoptosis and EMT through upregulation of the EGF receptor. Sci. Rep. 2017;7:45523. doi: 10.1038/srep45523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, et al. The Hippo transducer TAZ promotes epithelial to mesenchymal transition and cancer stem cell maintenance in oral cancer. Mol. Oncol. 2015;9:1091–1105. doi: 10.1016/j.molonc.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Liu J, Ying X, Lin PC, Zhou BP. Twist-mediated Epithelial-mesenchymal Transition Promotes Breast Tumor Cell Invasion via Inhibition of Hippo Pathway. Sci. Rep. 2016;6:24606. doi: 10.1038/srep24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 2018;25:1304–1317.e5. doi: 10.1016/j.celrep.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jongsma J, et al. A conditional mouse model for malignant mesothelioma. Cancer Cell. 2008;13:261–271. doi: 10.1016/j.ccr.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 24.Wong JS, Meliambro K, Ray J, Campbell KN. Hippo signaling in the kidney: the good and the bad. Am. J. Physiol. - Ren. Physiol. 2016;311:F241–F248. doi: 10.1152/ajprenal.00500.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sourbier C, et al. Targeting loss of the Hippo signaling pathway in NF2-deficient papillary kidney cancers. Oncotarget. 2018;9:10723–10733. doi: 10.18632/oncotarget.24112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oku Y, et al. Small molecules inhibiting the nuclear localization of YAP/TAZ for chemotherapeutics and chemosensitizers against breast cancers. FEBS Open Bio. 2015;5:542–549. doi: 10.1016/j.fob.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delahunt B, et al. The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am. J. Surg. Pathol. 2013;37:1490–1504. doi: 10.1097/PAS.0b013e318299f0fb. [DOI] [PubMed] [Google Scholar]
- 28.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118–e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerami E, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banumathy G, Cairns P. Signaling pathways in renal cell carcinoma. Cancer Biol. Ther. 2010;10:658–664. doi: 10.4161/cbt.10.7.13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang L, Nogales E, Ciferri C. Structure and Function of SWI/SNF Chromatin Remodeling Complexes and Mechanistic Implications for Transcription. Prog. Biophys. Mol. Biol. 2010;102:122–128. doi: 10.1016/j.pbiomolbio.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair SS, Kumar R. Chromatin remodeling in Cancer: A Gateway to regulate gene Transcription. Mol. Oncol. 2012;6:611–619. doi: 10.1016/j.molonc.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong Y, et al. Tumor Xenografts of Human Clear Cell Renal Cell Carcinoma But Not Corresponding Cell Lines Recapitulate Clinical Response to Sunitinib: Feasibility of Using Biopsy Samples. Eur. Urol. Focus. 2017;3:590–598. doi: 10.1016/j.euf.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.