Abstract Abstract
Chinese chestnut (Castanea mollissima) is an important crop tree species in China. In the present study, Cytospora specimens were collected from Chinese chestnut trees and identified using molecular data of combined ITS, LSU, ACT and RPB2 loci, as well as morphological features. As a result, two new Cytospora species and four new host records were confirmed, viz. C. kuanchengensissp. nov., C. xinglongensissp. nov., C. ceratospermopsis, C. leucostoma, C. myrtagena and C. schulzeri.
Keywords: Castanea mollissima, Cytosporaceae , Diaporthales , systematics, taxonomy
Introduction
Chinese chestnut (Castanea mollissima) is a widely cultivated crop tree species in China, producing nutritious and delicious nuts for humans (Lu and Guo 2016). However, Cryphonectria parasitica and several fungi are causing severe chestnut diseases worldwide, which reduce the nut production, even killing the hosts. (Aghayeva et al. 2017, Shuttleworth and Guest 2017, Jiang et al. 2018a, Rigling and Prospero 2018). Recently, several diaporthalean species were described from Chinese chestnut trees for the clear taxonomic concepts of families, genera and species in Diaporthales (Rossman et al. 2007, Senanayake et al. 2017, 2018), including species of Aurantiosacculus, Coryneum, Cryphonectria, Dendrostoma, Endothia, Gnomoniopsis, Neopseudomelanconis and Ophiognomonia (Gong et al. 2017, Jiang et al. 2018b, 2018c, 2019a, 2019b, Jiang and Tian 2019).
Cytospora (Cytosporaceae, Diaporthales) is a widely distributed genus worldwide, occurring on a broad range of hosts (Sarma and Hyde 2001, Yang et al. 2015, Lawrence et al. 2017, Norphanphoun et al. 2017, 2018, Wijayawardene et al. 2018, Jayawardena et al. 2019, Phookamsak et al. 2019, Fan et al. 2020). Some species can cause severe canker diseases on woody trees, such as Cytospora chrysosperma, which is a commom pathogen on the commercial tree genera, Populus and Salix (Fan et al. 2014b, Zhang et al. 2014, Kepley et al. 2015, Wang et al. 2015). Host affiliation was considerd as the main evidence for separating species in Cytospora before DNA sequences were used; however, morphology combined with phylogeny has revealed many cryptic species. For example, 28 Cytospora species were discovered from Eucalyptus from South Africa (Adams et al. 2005) and six from apple trees in Iran (Mehrabi et al. 2011), three from Chinese scholar tree (Fan et al. 2014a), four from walnut tree (Fan et al. 2015a), six from anti-desertification plants in China (Fan et al. 2015b) and two from grapevine in North America (Lawrence et al. 2017). Several recent studies discovered new species of Cytospora using multiphasic analyses (Lawrence et al. 2018, Norphanphoun et al. 2017, 2018, Senanayake et al. 2017, 2018, Pan et al. 2018, Zhu et al. 2018, Zhang et al. 2019).
During our investigations of chestnut disease in China from 2016 to 2019, diseased branches with typical Cytospora fruiting bodies were discovered and collected (Fig. 1). In the present study, Cytospora species from Castanea mollissima were identified using a combined method of morphology and phylogeny.
Figure 1.
Canker symptoms on Castanea mollissima caused by Cytospora spp.
Materials and methods
Sample collections and isolations
Chinese chestnut has a wide distribution in China. In the present study, we surveyed Hebei, Shaanxi and Shandong Provinces from 2016 to 2019. Dead and dying branches with typical Cytospora fruiting bodies were collected and packed in paper bags. Isolates were obtained by removing the ascospores or conidial masses from the fruiting bodies on to clean PDA plates and incubating at 25 °C until spores germinated. Single germinated spores were transferred on to the new PDA plates and incubated at 25 °C in the dark. Specimens were deposited in the Museum of the Beijing Forestry University (BJFC) and axenic cultures are maintained in the China Forestry Culture Collection Centre (CFCC).
Morphological analysis
Observation and description of Cytospora species from Castanea mollissima was based on fruiting bodies formed on tree barks. Ascomata and conidiomata from tree barks were sectioned by hand using a double-edged blade and strctures were observed under a dissecting microscope. At least 10 conidiostromata/ascostromata, 10 asci and 50 conidia/ascospores were measured to calculate the mean size and standard deviation. Measurements are reported as maximum and minimum in parentheses and the range representing the mean plus and minus the standard deviation of the number of measurements is given in parentheses (Voglmayr et al. 2017). Microscopy photographs were captured with a Nikon Eclipse 80i compound microscope equipped with a Nikon digital sight DS-Ri2 high definition colour camera, using differential interference contrast illumination. Introduction of the new species, based on molecular data, follow the recommendations of Jeewon and Hyde (2016).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from young mycelium growing on PDA plates following Doyle and Doyle (1990). PCR amplifications were performed in a DNA Engine Peltier Thermal Cycler (PTC-200; Bio-Rad Laboratories, Hercules, CA, USA). The primer pair ITS1/ITS4 (White et al. 1990) was used to amplify the ITS region. The primer pair LR0R/LR5 (Vilgalys and Hester 1990) was used to amplify the LSU region. The primer pair ACT512F/ACT783R (Carbone and Kohn 1999) was used to amplify ACT gene. The primer pair dRPB2-5f/dRPB2-7r (Voglmayr et al. 2016) was used to amplify the RPB2 gene. The polymerase chain reaction (PCR) assay was conducted as described in Fan et al. (2020). PCR amplification products were assayed via electrophoresis in 2% agarose gels. DNA sequencing was performed using an ABI PRISM 3730XL DNA Analyzer with a BigDye Terminater Kit v.3.1 (Invitrogen, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
Phylogenetic analyses
The preliminary identities of the isolates sequenced were obtained by conducting a standard nucleotide BLAST search using ITS, LSU, ACT and RPB2. Then all Cytospora isolates were selected to conduct phylogenetic analyses, based on sequence datasets from Fan et al. (2020). Diaporthe vaccinia (CBS 160.32) in Diaporthaceae was selected as the outgroup taxon. All sequences were aligned using MAFFT v. 6 (Katoh and Toh 2010) and edited manually using MEGA v. 6 (Tamura et al. 2013). Phylogenetic analyses were performed using PAUP v. 4.0b10 for Maximum Parsimony (MP) analysis (Swofford 2003) and PhyML v. 3.0 for Maximum Likelihood (ML) analysis (Guindon et al. 2010).
MP analysis was run using a heuristic search option of 1000 search replicates with random-additions of sequences with a tree bisection and reconnection algorithm. Maxtrees were set to 5000, branches of zero length were collapsed and all equally parsimonious trees were saved. Other calculated parsimony scores were tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency (RC). ML analysis was performed using a GTR site substitution model including a gamma-distributed rate heterogeneity and a proportion of invariant sites (Guindon et al. 2010). The branch support was evaluated using a bootstrapping method of 1000 replicates (Hillis and Bull 1993). Phylograms were shown using FigTree v. 1.4.3 (Rambaut 2016). Novel sequences, generated in the current study, were deposited in GenBank (Table 1) and the aligned matrices used for phylogenetic analyses in TreeBASE (accession number: S25160).
Table 1.
Strains used in the phylogenetic tree and their culture accession and GenBank numbers. Strains from this study are in bold and ex-strains are marked with *.
| Species | Strain | Host | Origin | GenBank accession numbers | |||
|---|---|---|---|---|---|---|---|
| ITS | LSU | ACT | RPB2 | ||||
| Cytospora ailanthicola | CFCC 89970* | Ailanthus altissima | China | MH933618 | MH933653 | MH933526 | MH933592 |
| Cytospora abyssinica | CMW 10181* | Eucalyptus globulus | Ethiopia | AY347353 | NA | NA | NA |
| Cytospora acaciae | CBS 468.69 | Ceratonia siliqua | Spain | DQ243804 | NA | NA | NA |
| Cytospora ampulliformis | MFLUCC 16-0583* | Sorbus intermedia | Russia | KY417726 | KY417760 | KY417692 | KY417794 |
| MFLUCC 16-0629 | Acer platanoides | Russia | KY417727 | KY417761 | KY417693 | KY417795 | |
| Cytospora amygdali | CBS 144233* | Prunus dulcis | USA | MG971853 | NA | MG972002 | NA |
| CFCC 89615 | Juglans regia | China | KR045618 | KR045700 | KF498673 | KU710946 | |
| CFCC 89616 | Juglans regia | China | KR045619 | KR045701 | KF498674 | KU710947 | |
| Cytospora atrocirrhata | CFCC 89615 | Juglans regia | China | KR045618 | KR045700 | KF498673 | KU710946 |
| Cytospora austromontana | CMW 6735* | Eucalyptus pauciflora | Australia | AY347361 | NA | NA | NA |
| Cytospora beilinensis | CFCC 50493* | Pinus armandii | China | MH933619 | MH933654 | MH933527 | NA |
| CFCC 50494 | Pinus armandii | China | MH933620 | MH933655 | MH933528 | NA | |
| Cytospora berberidis | CFCC 89927* | Berberis dasystachya | China | KR045620 | KR045702 | KU710990 | KU710948 |
| CFCC 89933 | Berberis dasystachya | China | KR045621 | KR045703 | KU710991 | KU710949 | |
| Cytospora berkeleyi | StanfordT3* | Eucalyptus globulus | USA | AY347350 | NA | NA | NA |
| UCBTwig3 | Eucalyptus globulus | USA | AY347349 | NA | NA | NA | |
| Cytospora brevispora | CBS 116811* | Eucalyptus grandis × tereticornis | Congo | AF192315 | NA | NA | NA |
| CBS 116829 | Eucalyptus grandis | Venezuela | AF192321 | NA | NA | NA | |
| Cytospora bungeanae | CFCC 50495* | Pinus bungeana | China | MH933621 | MH933656 | MH933529 | MH933593 |
| CFCC 50496 | Pinus bungeana | China | MH933622 | MH933657 | MH933530 | MH933594 | |
| Cytospora californica | CBS 144234* | Juglans regia | USA | MG971935 | NA | MG972083 | NA |
| Cytospora carbonacea | CFCC 89947 | Ulmus pumila | China | KR045622 | KP310812 | KP310842 | KU710950 |
| Cytospora carpobroti | CMW 48981* | Carpobrotus edulis | South Africa | MH382812 | MH411216 | NA | NA |
| Cytospora castaneae | AUCCT/DBT 183* | Castanea sativa | India | KC963921 | NA | NA | NA |
| Cytospora cedri | CBS 196.50 | NA | Italy | AF192311 | NA | NA | NA |
| Cytospora celtidicola | CFCC 50497* | Celtis sinensis | China | MH933623 | MH933658 | MH933531 | MH933595 |
| CFCC 50498 | Celtis sinensis | China | MH933624 | MH933659 | MH933532 | MH933596 | |
| Cytospora centrivillosa | MFLUCC 16-1206* | Sorbus domestica | Italy | MF190122 | MF190068 | NA | MF377601 |
| MFLU 17-0887 | Sorbus domestica | Italy | MF190123 | MF190069 | NA | NA | |
| MFLUCC 17-1660 | Sorbus domestica | Italy | MF190124 | MF190070 | NA | MF377600 | |
| Cytospora ceratosperma | CFCC 89624 | Juglans regia | China | KR045645 | KR045724 | NA | KU710976 |
| CFCC 89625 | Juglans regia | China | KR045646 | KR045725 | NA | KU710977 | |
| Cytospora ceratospermopsis | CFCC 89626* | Juglans regia | China | KR045647 | KR045726 | KU711011 | KU710978 |
| CFCC 89627 | Juglans regia | China | KR045648 | KR045727 | KU711012 | KU710979 | |
| CFCC 52471 | Castanea mollissima | China | MK432629 | MK429899 | MK442953 | MK578087 | |
| CFCC 52472 | Castanea mollissima | China | MK432630 | MK429900 | MK442954 | MK578088 | |
| Cytospora chrysosperma | CFCC 89629 | Salix psammophila | China | KF765673 | KF765689 | NA | KF765705 |
| CFCC 89981 | Populus alba subsp. pyramidalis | China | MH933625 | MH933660 | MH933533 | MH933597 | |
| CFCC 89982 | Ulmus pumila | China | KP281261 | KP310805 | KP310835 | NA | |
| Cytospora cinerostroma | CMW 5700* | Eucalyptus globulus | Chile | AY347377 | NA | NA | NA |
| Cytospora cotini | MFLUCC 14-1050* | Cotinus coggygria | Russia | KX430142 | KX430143 | NA | KX430144 |
| Cytospora curvata | MFLUCC 15-0865* | Salix alba | Russia | KY417728 | KY417762 | KY417694 | KY417796 |
| Cytospora davidiana | CXY 1350* | Populus davidiana | China | KM034870 | NA | NA | NA |
| CXY 1374 | Populus davidiana | China | KM034869 | NA | NA | NA | |
| Cytospora diatrypelloidea | CMW 8549* | Eucalyptus globulus | Australia | AY347368 | NA | NA | NA |
| Cytospora disciformis | CMW 6509* | Eucalyptus grandis | Uruguay | AY347374 | NA | NA | NA |
| CMW 6750 | Eucalyptus globulus | Australia | AY347359 | NA | NA | NA | |
| Cytospora donetzica | MFLUCC 15-0864 | NA | NA | KY417729 | KY417763 | KY417695 | KY417797 |
| MFLUCC 16-0574* | Rosa sp. | Russia | KY417731 | KY417764 | KY417696 | KY417798 | |
| Cytospora elaeagni | CFCC 89632 | Elaeagnus angustifolia | China | KR045626 | KR045706 | KU710995 | KU710955 |
| CFCC 89633 | Elaeagnus angustifolia | China | KF765677 | KF765693 | KU710996 | KU710956 | |
| Cytospora eriobotryae | IMI 136523* | Eriobotrya japonica | India | AY347327 | NA | NA | NA |
| Cytospora erumpens | CFCC 50022 | Prunus padus | China | MH933627 | MH933661 | MH933534 | NA |
| MFLUCC 16-0580* | Salix × fragilis | Russia | KY417733 | KY417767 | KY417699 | KY417801 | |
| Cytospora eucalypti | CBS 144241 | Eucalyptus globulus | USA | MG971907 | NA | MG972056 | NA |
| LSEQ | Sequoia sempervirens | USA | AY347340 | NA | NA | NA | |
| Cytospora eucalypticola | ATCC 96150* | Eucalyptus nitens | Australia | AY347358 | NA | NA | NA |
| CMW 5309 | Eucalyptus grandis | Uganda | AF260266 | NA | NA | NA | |
| Cytospora eucalyptina | CMW 5882 | Eucalyptus grandis | Columbia | AY347375 | NA | NA | NA |
| Cytospora eugeniae | CMW 7029 | Tibouchina sp. | Australia | AY347364 | NA | NA | NA |
| CMW 8648 | Eugenia sp. | Indonesia | AY347344 | NA | NA | NA | |
| Cytospora euonymicola | CFCC 50499* | Euonymus kiautschovicus | China | MH933628 | MH933662 | MH933535 | MH933598 |
| CFCC 50500 | Euonymus kiautschovicus | China | MH933629 | MH933663 | MH933536 | MH933599 | |
| Cytospora euonymina | CFCC 89993* | Euonymus kiautschovicus | China | MH933630 | MH933664 | MH933537 | MH933600 |
| CFCC 89999 | Euonymus kiautschovicus | China | MH933631 | MH933665 | MH933538 | MH933601 | |
| Cytospora fraxinigena | MFLUCC 14-0868* | Euonymus kiautschovicus | China | MH933631 | MH933665 | MH933538 | MH933601 |
| Cytospora friesii | CBS 194.42 | Abies alba | Switzerland | AY347328 | NA | NA | NA |
| Cytospora fugax | CXY 1381 | NA | NA | KM034853 | NA | NA | NA |
| Cytospora germanica | CXY 1322 | Elaeagnus oxycarpa | China | JQ086563 | JX524617 | NA | NA |
| Cytospora gigalocus | CFCC 89620* | Juglans regia | China | KR045628 | KR045708 | KU710997 | KU710957 |
| CFCC 89621 | Juglans regia | China | KR045629 | KR045709 | KU710998 | KU710958 | |
| Cytospora gigaspora | CFCC 50014 | Juniperus procumbens | China | KR045630 | KR045710 | KU710999 | KU710959 |
| CFCC 89634* | Salix psammophila | China | KF765671 | KF765687 | KU711000 | KU710960 | |
| Cytospora granati | CBS 144237* | Punica granatum | USA | MG971799 | NA | MG971949 | NA |
| Cytospora hippophaës | CFCC 89639 | Hippophaë rhamnoides | China | KR045632 | KR045712 | KU711001 | KU710961 |
| CFCC 89640 | Hippophaë rhamnoides | China | KF765682 | KF765698 | KF765730 | KU710962 | |
| Cytospora japonica | CFCC 89956 | Prunus cerasifera | China | KR045624 | KR045704 | KU710993 | KU710953 |
| CFCC 89960 | Prunus cerasifera | China | KR045625 | KR045705 | KU710994 | KU710954 | |
| Cytospora joaquinensis | CBS 144235* | Populus deltoides | USA | MG971895 | NA | MG972044 | NA |
| Cytospora junipericola | MFLU 17-0882* | Juniperus communis | Italy | MF190125 | MF190072 | NA | NA |
| Cytospora juniperina | CFCC 50501* | Juniperus przewalskii | China | MH933632 | MH933666 | MH933539 | MH933602 |
| CFCC 50503 | Juniperus przewalskii | China | MH933634 | MH933668 | MH933541 | MH933604 | |
| Cytospora kantschavelii | CXY 1383 | Populus maximowiczii | China | KM034867 | NA | NA | NA |
| Cytospora kuanchengensis | CFCC 52464* | Castanea mollissima | China | MK432616 | MK429886 | MK442940 | MK578076 |
| CFCC 52465 | Castanea mollissima | China | MK432617 | MK429887 | MK442941 | MK578077 | |
| Cytospora kunzei | CBS 118556 | Pinus radiata | South Africa | DQ243791 | NA | NA | NA |
| Cytospora leucosperma | CFCC 89622 | Pyrus bretschneideri | China | KR045616 | KR045698 | KU710988 | KU710944 |
| CFCC 89894 | Pyrus bretschneideri | China | KR045617 | KR045699 | KU710989 | KU710945 | |
| Cytospora leucostoma | CFCC 50018 | Prunus serrulata | China | MH933636 | MH933670 | MH933543 | NA |
| CFCC 50019 | Rosa helenae | China | MH933637 | MH933671 | MH933544 | NA | |
| CFCC 50021 | Prunus salicina | China | MH933639 | MH933673 | MH933546 | NA | |
| CFCC 50023 | Cornus alba | China | KR045635 | KR045715 | KU711003 | KU710964 | |
| CFCC 52461 | Castanea mollissima | China | MK432624 | MK429894 | MK442948 | NA | |
| CFCC 52462 | Castanea mollissima | China | MK432625 | MK429895 | MK442949 | NA | |
| Cytospora longiostiolata | MFLUCC 16-0628* | Salix × fragilis | Russia | KY417734 | KY417768 | KY417700 | KY417802 |
| Cytospora longispora | CBS 144236* | Prunus domestica | USA | MG971905 | NA | MG972054 | NA |
| Cytospora lumnitzericola | MFLUCC 17-0508* | Lumnitzera racernosa | Tailand | MG975778 | MH253461 | MH253457 | MH253453 |
| Cytospora mali | CFCC 50028 | Malus pumila | China | MH933641 | MH933675 | MH933548 | MH933606 |
| CFCC 50029 | Malus pumila | China | MH933642 | MH933676 | MH933549 | MH933607 | |
| Cytospora melnikii | MFLUCC 15-0851* | Malus domestica | Russia | KY417735 | KY417769 | KY417701 | KY417803 |
| MFLUCC 16-0635 | Populus nigra var. italica | Russia | KY417736 | KY417770 | KY417702 | KY417804 | |
| Cytospora mougeotii | ATCC 44994 | Picea abies | Norway | AY347329 | NA | NA | NA |
| Cytospora multicollis | CBS 105.89T | Quercus ilex subsp. rotundifolia | Spain | DQ243803 | NA | NA | NA |
| Cytospora myrtagena | CBS 116843* | Tibouchiina urvilleana | USA | AY347363 | NA | NA | NA |
| CFCC 52454 | Castanea mollissima | China | MK432614 | MK429884 | MK442938 | MK578074 | |
| CFCC 52455 | Castanea mollissima | China | MK432615 | MK429885 | MK442939 | MK578075 | |
| Cytospora nitschkii | CMW 10180* | Eucalyptus globulus | Ethiopia | AY347356 | NA | NA | NA |
| CMW 10184 | Eucalyptus globulus | Ethiopia | AY347355 | NA | NA | NA | |
| Cytospora nivea | CFCC 89641 | Elaeagnus angustifolia | China | KF765683 | KF765699 | KU711006 | KU710967 |
| CFCC 89643 | Salix psammophila | China | KF765685 | KF765701 | NA | KU710968 | |
| Cytospora oleicola | CBS 144248* | Olea europaea | USA | MG971944 | NA | MG972098 | NA |
| Cytospora palm | CXY 1280* | Cotinus coggygria | China | JN411939 | NA | NA | NA |
| Cytospora parakantschavelii | MFLUCC 15-0857* | Populus × sibirica | Russia | KY417738 | KY417772 | KY417704 | KY417806 |
| MFLUCC 16-0575 | Pyrus pyraster | Russia | KY417739 | KY417773 | KY417705 | KY417807 | |
| Cytospora parapersoonii | T28.1* | Prunus persica | USA | AF191181 | NA | NA | NA |
| Cytospora parapistaciae | CBS 144506* | Pistacia vera | USA | MG971804 | NA | MG971954 | NA |
| Cytospora parasitica | MFLUCC 15-0507* | Malus domestica | Russia | KY417740 | KY417774 | KY417706 | KY417808 |
| Cytospora paratranslucens | MFLUCC 15-0506* | Populus alba var. bolleana | Russia | KY417741 | KY417775 | KY417707 | KY417809 |
| MFLUCC 16-0627 | Populus alba | Russia | KY417742 | KY417776 | KY417708 | KY417810 | |
| Cytospora pini | CBS 197.42 | Pinus sylvestris | Switzerland | AY347332 | NA | NA | NA |
| CBS 224.52* | Pinus strobus | USA | AY347316 | NA | NA | NA | |
| Cytospora pistaciae | CBS 144238* | Pistacia vera | USA | MG971802 | NA | MG971952 | NA |
| Cytospora platanicola | MFLU 17-0327* | Platanus hybrida | Italy | MH253451 | MH253452 | MH253449 | MH253450 |
| Cytospora platycladi | CFCC 50504* | Platycladus orientalis | China | MH933645 | MH933679 | MH933552 | MH933610 |
| CFCC 50505 | Platycladus orientalis | China | MH933646 | MH933680 | MH933553 | MH933611 | |
| CFCC 50506 | Platycladus orientalis | China | MH933647 | MH933681 | MH933554 | MH933612 | |
| Cytospora platycladicola | CFCC 50038* | Platycladus orientalis | China | KT222840 | MH933682 | MH933555 | MH933613 |
| CFCC 50039 | Platycladus orientalis | China | KR045642 | KR045721 | KU711008 | KU710973 | |
| Cytospora plurivora | CBS 144239* | Olea europaea | USA | MG971861 | NA | MG972010 | NA |
| Cytospora populicola | CBS 144240* | Populus deltoides | USA | MG971891 | NA | MG972040 | NA |
| Cytospora populina | CFCC 89644* | Salix psammophila | China | KF765686 | KF765702 | KU711007 | KU710969 |
| Cytospora populinopsis | CFCC 50032* | Sorbus aucuparia | China | MH933648 | MH933683 | MH933556 | MH933614 |
| CFCC 50033 | Sorbus aucuparia | China | MH933649 | MH933684 | MH933557 | MH933615 | |
| Cytospora predappioensis | MFLUCC 17-2458* | Platanus hybrida | Italy | MG873484 | MG873480 | NA | NA |
| Cytospora prunicola | MFLU 17-0995* | Prunus sp. | Italy | MG742350 | MG742351 | MG742353 | MG742352 |
| Cytospora pruinopsis | CFCC 50034* | Ulmus pumila | China | KP281259 | KP310806 | KP310836 | KU710970 |
| CFCC 50035 | Ulmus pumila | China | KP281260 | KP310807 | KP310837 | KU710971 | |
| Cytospora pruinosa | CFCC 50036 | Syringa oblata | China | KP310800 | KP310802 | KP310832 | NA |
| CFCC 50037 | Syringa oblata | China | MH933650 | MH933685 | MH933558 | NA | |
| Cytospora prunicola | MFLU 17-0995* | Prunus sp. | Italy | MG742350 | MG742351 | MG742353 | MG742352 |
| Cytospora punicae | CBS 144244 | Punica granatum | USA | MG971943 | NA | MG972091 | NA |
| Cytospora quercicola | MFLU 17-0881 | Quercus sp. | Italy | MF190129 | MF190074 | NA | NA |
| MFLUCC 14-0867* | Quercus sp. | Italy | MF190128 | MF190073 | NA | NA | |
| Cytospora rhizophorae | MUCC302 | Eucalyptus grandis | Australia | EU301057 | NA | NA | NA |
| Cytospora ribis | CFCC 50026 | Ulmus pumila | China | KP281267 | KP310813 | KP310843 | KU710972 |
| CFCC 50027 | Ulmus pumila | China | KP281268 | KP310814 | KP310844 | NA | |
| Cytospora rosae | MFLU 17-0885 | Rosa canina | Italy | MF190131 | MF190076 | NA | NA |
| Cytospora rostrata | CFCC 89909* | Salix cupularis | China | KR045643 | KR045722 | KU711009 | KU710974 |
| CFCC 89910 | Salix cupularis | China | KR045644 | KR045723 | KU711010 | KU710975 | |
| Cytospora rusanovii | MFLUCC 15-0853 | Populus × sibirica | Russia | KY417743 | KY417777 | KY417709 | KY417811 |
| MFLUCC 15-0854* | Salix babylonica | Russia | KY417744 | KY417778 | KY417710 | KY417812 | |
| Cytospora salicacearum | MFLUCC 16-0576 | dead aerial branch | Russia | KY417747 | KY417781 | KY417713 | KY417815 |
| MFLUCC 15-0509* | Salix alba | Russia | KY417746 | KY417780 | KY417712 | KY417814 | |
| MFLUCC 15-0861 | Salix × fragilis | Russia | KY417745 | KY417779 | KY417711 | KY417813 | |
| MFLUCC 16-0587 | NA | NA | KY417748 | KY417782 | KY417714 | KY417816 | |
| Cytospora salicicola | MFLUCC 14-1052* | Salix alba | Russia | KU982636 | KU982635 | KU982637 | NA |
| MFLUCC 15-0866 | Salix alba | Russia | KY417749 | KY417783 | KY417715 | KY417817 | |
| Cytospora salicina | MFLUCC 15-0862* | Salix alba | Russia | KY417750 | KY417784 | KY417716 | KY417818 |
| MFLUCC 16-0637 | Salix × fragilis | Russia | KY417751 | KY417785 | KY417717 | KY417819 | |
| Cytospora schulzeri | CFCC 50040 | Malus domestica | China | KR045649 | KR045728 | KU711013 | KU710980 |
| CFCC 50042 | Malus asiatica | China | KR045650 | KR045729 | KU711014 | KU710981 | |
| CFCC 52468 | Castanea mollissima | China | MK432626 | MK429896 | MK442950 | MK578084 | |
| CFCC 52469 | Castanea mollissima | China | MK432627 | MK429897 | MK442951 | MK578085 | |
| CFCC 52470 | Castanea mollissima | China | MK432628 | MK429898 | MK442952 | MK578086 | |
| Cytospora sibiraeae | CFCC 50045* | Sibiraea angustata | China | KR045651 | KR045730 | KU711015 | KU710982 |
| CFCC 50046 | Sibiraea angustata | China | KR045652 | KR045731 | KU711015 | KU710983 | |
| Cytospora sophorae | CFCC 50048 | Magnolia grandiflora | China | MH820401 | MH820394 | MH820409 | MH820397 |
| CFCC 89598 | Styphnolobium japonicum | China | KR045654 | KR045733 | KU711018 | KU710985 | |
| Cytospora sophoricola | CFCC 89595* | Styphnolobium japonicum var. pendula | China | KR045655 | KR045734 | KU711019 | KU710986 |
| CFCC 89596 | Styphnolobium japonicum var. pendula | China | KR045656 | KR045735 | KU711020 | KU710987 | |
| Cytospora sophoriopsis | CFCC 89600* | Styphnolobium japonicum | China | KR045623 | KP310804 | KU710992 | KU710951 |
| Cytospora sorbi | MFLUCC 16-0631* | Sorbus aucuparia | Russia | KY417752 | KY417786 | KY417718 | KY417820 |
| Cytospora sorbicola | MFLUCC 16-0584* | Acer pseudoplatanus | Russia | KY417755 | KY417789 | KY417721 | KY417823 |
| MFLUCC 16-0633 | Cotoneaster melanocarpus | Russia | KY417758 | KY417792 | KY417724 | KY417826 | |
| Cytospora spiraeae | CFCC 50049* | Spiraea salicifolia | China | MG707859 | MG707643 | MG708196 | MG708199 |
| CFCC 50050 | Spiraea salicifolia | China | MG707860 | MG707644 | MG708197 | MG708200 | |
| Cytospora tamaricicola | CFCC 50507 | Rosa multifolora | China | MH933651 | MH933686 | MH933559 | MH933616 |
| CFCC 50508* | Tamarix chinensis | China | MH933652 | MH933687 | MH933560 | MH933617 | |
| Cytospora tanaitica | MFLUCC 14-1057* | Betula pubescens | Russia | KT459411 | KT459412 | KT459413 | NA |
| Cytospora thailandica | MFLUCC 17-0262* | Xylocarpus moluccensis | Thailand | MG975776 | MH253463 | MH253459 | MH253455 |
| MFLUCC 17-0263 | Xylocarpus moluccensis | Thailand | MG975777 | MH253464 | MH253460 | MH253456 | |
| Cytospora tibouchinae | CPC 26333* | Tibouchina semidecandra | France | KX228284 | KX228335 | NA | NA |
| Cytospora translucens | CXY 1351 | Populus davidiana | China | KM034874 | NA | NA | NA |
| Cytospora ulmi | MFLUCC 15-0863* | Ulmus minor | Russia | KY417759 | NA | NA | NA |
| Cytospora ulmicola | MFLUCC 18-1227* | Ulmus pumila | Russia | MH940220 | MH940218 | MH940216 | NA |
| Cytospora valsoidea | CMW 4309* | Eucalyptus grandis | Indonesia | AF192312 | NA | NA | NA |
| CMW 4310 | Eucalyptus grandis | Indonesia | AF192312 | NA | NA | NA | |
| Cytospora variostromatica | CBS 118086 | Eucalyptus grandis | South Africa | AF260264 | NA | NA | NA |
| CMW 1240 | Eucalyptus grandis | South Africa | AF260263 | NA | NA | NA | |
| CMW 6766* | Eucalyptus globulus | Australia | AY347366 | NA | NA | NA | |
| Cytospora vinacea | CBS 141585* | Vitis interspecific hybrid ‘Vidal’ | USA | KX256256 | NA | NA | NA |
| Cytospora viticola | CBS 141586* | Vitis vinifera ’Cabernet Franc’ | USA | KX256239 | NA | NA | NA |
| Cytospora xinglongensis | CFCC 52458* | Castanea mollissima | China | MK432622 | MK429892 | MK442946 | MK578082 |
| CFCC 52459 | Castanea mollissima | China | MK432623 | MK429893 | MK442947 | MK578083 | |
| Cytospora xylocarpi | MFLUCC 17-0251* | Xylocarpus granatum | Thailand | MG975775 | MH253462 | MH253458 | MH253454 |
| Diaporthe vaccinii | CBS 160.32 | Vaccinium macrocarpon | USA | KC343228 | NA | JQ807297 | NA |
Results
Phylogenetic analyses
The alignment based on the combined sequence dataset (ITS, LSU, ACT and RPB2) included 124 ingroup taxa and one outgroup taxon, comprising 2097 characters in the aligned matrix. Of these, 1375 characters were constant, 89 variable characters were parsimony-uninformative and 663 characters were parsimony informative. The MP analysis resulted in 14 equally most parsimonious trees and the first tree (TL = 3270, CI = 0.344, RI = 0.815, RC = 0.281) was present as in Fig. 2. The ML analysis yielded a tree with a likelihood value of ln: -18627.915604 and the following model parameters: alpha: 0.181328; Π(A): 0.246855, Π(C): 0.260898, Π(G): 0.272379 and Π(T): 0.219868. Isolates from Castanea mollissima formed six clades in Fig. 2, representing two undescribed species and four known species.
Figure 2.
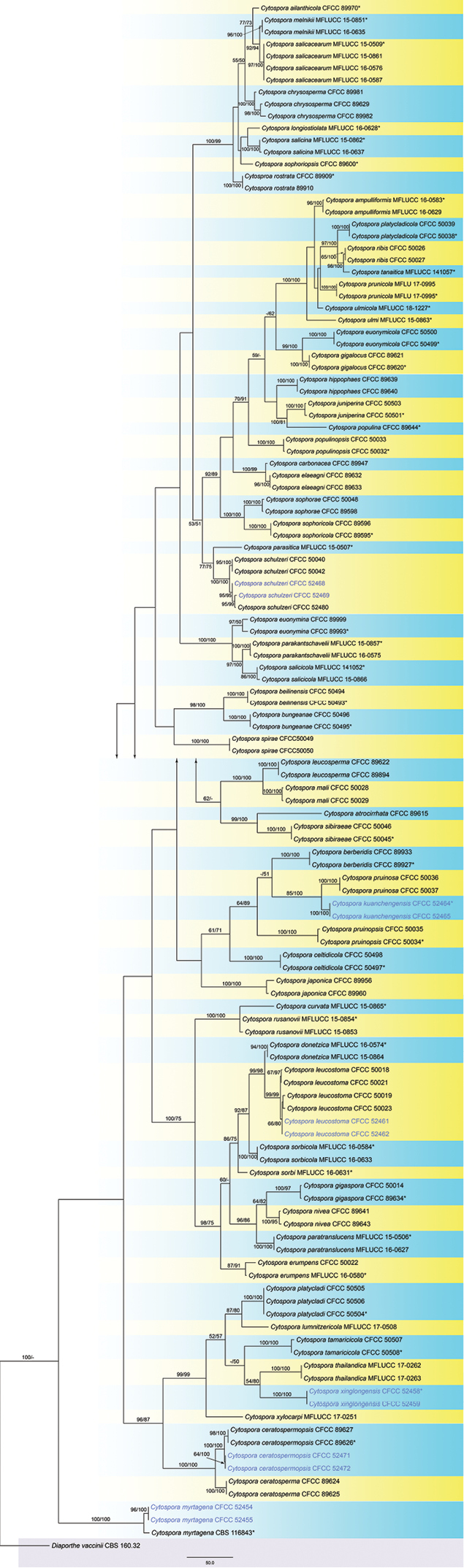
Maximum parsimony phylogram of Cytospora obtained from the combined matrix of ITS, LSU, ACT and RPB2 genes. Bootstrap value ≥ 50% for MP and ML analyses are presented at the first and second position. Scale bar = 200 nucleotide substitutions. The strains in the current study are in blue and ex-strains are marked with *.
Taxonomy
Cytospora ceratosp ermopsis
C.M. Tian & X.L. Fan, Persoonia 45: 19. 2020
990B2D8E-95BC-5250-884C-64AF73BCEF11
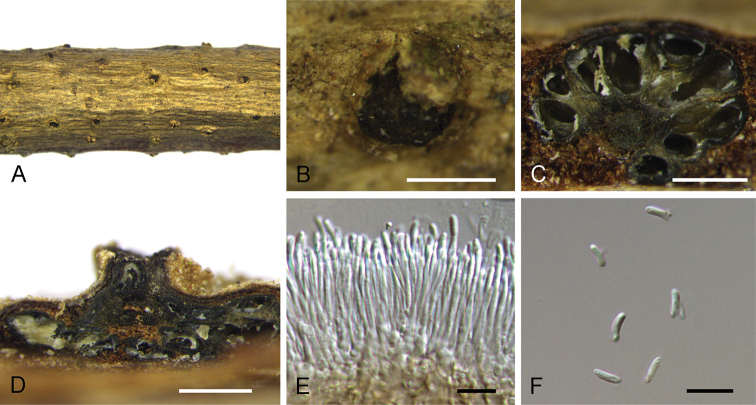
Figure 3.
Cytospora ceratospermopsis on Castanea mollissima (BJFC-S1699, BJFC-S1700). A, C Habit of conidiomata on branches B habit of ascomata on branches D transverse section of conidiomata E transverse section of ascomata F longitudinal section through conidiomata G longitudinal section through ascomata H, I asci J ascospores K conidiogenous cells with attached conidia L conidia. Scale bars: 500 μm (C–G), 10 μm (H–L).
Description.
Sexual morph: Ascostromata immersed in the bark, erumpent through the surface of bark, scattered, (350–)550–900(–1300) µm diam., with 15–40 perithecia arranged circularly or irregularly. Conceptacle absent. Ectostromatic disc black, usually surrounded by tightly ostiolar necks, circular to ovoid, (180–)240–410(–450) µm diam. Ostioles black, at the same level as the disc or slightly above, concentrated, dark brown to black, arranged circularly in a disc, (55–)60–85(–110) µm diam. Perithecia dark brown, flask-shaped to spherical, arranged circularly or irregularly, (255–)280–350(–420) µm diam. Asci clavate to elongate obovoid, 8-spored, (20.5–)27–35.5(–43) × (4–)4.5–6.5(–8) μm (x̄= 31.2 × 5.6 μm). Ascospores biseriate, elongate-allantoid, thin-walled, hyaline, aseptate, (5.8–)7.5–9.2(–11.5) × (3–)3.2–4.8(–5.5) μm (x̄ = 8.6 × 4.1 μm). Asexual morph: Pycnidial stromata ostiolated, immersed in bark, scattered, erumpent through the surface of bark, discoid to conical, with multiple locules. Conceptacle absent. Ectostromatic disc light brown to grey, circular to ovoid, (230–)280–360(–480) µm diam., with one ostiole per disc. Ostiole in the centre of the disc, dark grey to black, conspicuous, at the same level as the disc, (60–)75–110(–135) μm diam. Locule numerous, arranged circularly or elliptically with independent walls, (300–)350–600(–950) µm diam. Peridium comprising few layers of cells of textura angularis, with innermost layer brown, outer layer brown to dark brown. Conidiophores hyaline, branched or not, thin walled, filamentous. Conidiogenous cells enteroblastic polyphialidic, (6.5–)8.5–15.5(–18) × 1.5–2.5 μm (x̄ = 12.2 × 1.9 μm). Conidia hyaline, allantoid, smooth, aseptate, thin-wall, (4.5–)5–6.5(–7) × 1–1.5 μm (x̄ = 5.9 × 1.3 μm).
Culture characters.
On PDA at 25 °C in darkness. Cultures are initially white, becoming olivaceous buff in centre after 7 d and finally olivaceous at 30 d. The colony is flat, thin with a felt and tight texture in centre. Pycnidia distributed irregularly on medium surface.
Specimens examined.
China, Hebei Province, Chengde City, Xinglong County, chestnut plantation, 40°24'32"N, 117°27'56"E, on branches of Castanea mollissima, 11 October 2017, N. Jiang (BJFC-S1699, living culture CFCC 52471 from the ascospore; BJFC-S1700, living culture CFCC 52472 from the conidium).
Notes.
Fresh specimens with both sexual and asexual morphs were collected from cankered branches of Castanea mollissima and two isolates were obtained from the ascospore and conidium, respectively. Phylogenically, the two isolates were close to Cytospora ceratospermopsis represented by CFCC 89626 and CFCC 89627 (Fig. 2). We compared their sequences and found no differences in LSU and RPB2, but 2 bp differences in ITS and 3 bp differences in ACT. Fan et al. (2020) reported the asexual morph of Cytospora ceratospermopsis from Juglans regia in China with conidial size in 4.5–6 × 1–1.5 μm, which is exactly matched with the asexual characters observed in the present study. Hence, we described the asexual morph of Cytospora ceratospermopsis in its sexual morph for the first time and reported a new host, Castanea mollissima.
Cytospora kuanchengensis
C.M. Tian & N. Jiang sp. nov.
2BB85219-15AA-55BA-8D48-E0C738D2DA36
829514
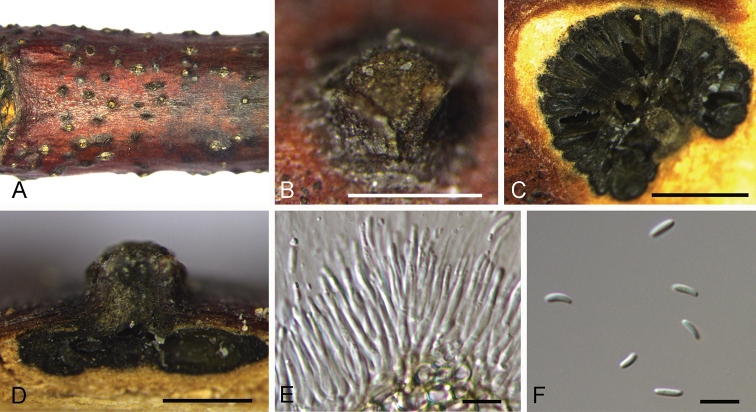
Figure 4.
Cytospora kuanchengensis on Castanea mollissima (BJFC-S1695). A, B Habit of conidiomata on branches C longitudinal section through conidiomata D transverse section of conidiomata E peridium F, G conidiogenous cells attached with conidia H conidia. Scale bars: 500 μm (B–D), 10 μm (E–G), 5 μm (H).
Diagnosis.
Cytospora kuanchengensis can be distinguished from C. oleicola and C. pruinose by longer conidia.
Etymology.
Named after the county where it was collected, Kuancheng County.
Description.
Sexual morph: not observed. Asexual morph: Pycnidial stromata ostiolated, immersed in bark, scattered, erumpent through the surface of bark, discoid, with multiple locules. Conceptacle black, circular surrounded stromata. Ectostromatic disc black, circular to ovoid, (350–)455–540(–575) µm diam., with 1–7 ostiole per disc. Ostioles black, at the same level as the disc, (40–)60–85(–115) μm diam. Locule numerous, arranged circularly or elliptically with independent walls, (285–)355–520(–605) µm diam. Peridium comprising few layers of cells of textura angularis, with innermost layer brown, outer layer brown to dark brown. Conidiophores hyaline, unbranched, thin walled, filamentous. Conidiogenous cells enteroblastic polyphialidic, (6.5–)8.5–11(–15) × 1–1.5 μm (x̄ = 9.8 × 1.3 μm). Conidia hyaline, allantoid, smooth, aseptate, thin-walled, (5.5–)6–7.5(–8) × 1–2 μm (x̄ = 6.9 × 1.6 μm).
Culture characters.
On PDA at 25 °C in darkness. Cultures are initially white, producing pale brown pigment after 10 d. The colony is flat, felt-like, with concentric circular texture. Pycnidia distributed irregularly on medium surface.
Specimens examined.
China, Hebei Province, Chengde City, Kuancheng County, chestnut plantation, 40°38'37"N, 118°27'54"E, on branches of Castanea mollissima, 13 October 2017, N. Jiang (holotypeBJFC-S1695, ex-type living culture CFCC 52464; paratypeBJFC-S1696, living culture CFCC 52465).
Notes.
Cytospora kuanchengensis is associated with canker disease of Castanea mollissima in China. Cytospora kuanchengensis differs from its phylogenetically closely species, C. pruinosa, by ITS and ACT loci (7/470 in ITS and 21/245 in ACT). Morphologically, C. kuanchengensis has slightly larger conidia than C. pruinose (5.5–8 × 1–2 μm in Cytospora kuanchengensis vs. 5–7.5 × 1–1.5 μm in C. pruinosa) (Fan et al. 2020).
Cytospora leucostoma
(Pers.) Sacc., Michelia 2(7): 264. 1881.
3D4086B0-49AA-50B7-A2BE-60091113DAAA
Figure 5.
Cytospora leucostoma on Castanea mollissima (BJFC-S1697). A, B Habit of conidiomata on branches C transverse section of conidiomata D longitudinal section through conidiomata E conidiogenous cells attached with conidia F conidia. Scale bars: 500 μm (B–D), 10 μm (E, F).
Description.
Sexual morph: not observed. Asexual morph: Pycnidial stromata ostiolated, immersed in bark, scattered, erumpent through the surface of bark, with multiple locules. Conceptacle black. Ectostromatic disc black, circular to ovoid, (150–)250–300(–375) µm diam., with one ostiole per disc. Ostioles black, at the same level as the disc, (40–)50–85(–115) μm diam. Locule numerous, arranged circularly or elliptically with independent walls, (550–)700–1200(–1350) µm diam. Peridium comprising few layers of cells of textura angularis, with innermost layer brown, outer layer brown to dark brown. Conidiophores hyaline, unbranched, thin walled, filamentous. Conidiogenous cells enteroblastic polyphialidic, (7.5–)9.5–21(–22.5) × 1–1.5 μm (x̄ = 15.2 × 1.3 μm). Conidia hyaline, allantoid, smooth, aseptate, thin-walled, (3.5–)4.5–5.5(–7) × 1–1.5 μm (x̄ = 4.9 × 1.3 μm).
Specimens examined.
China, Hebei Province, Chengde City, Kuancheng County, chestnut plantation, 40°38'37"N, 118°27'5"E, on branches of Castanea mollissima, 13 October 2017, N. Jiang (BJFC-S1697, living culture CFCC 52461; BJFC-S1698, living culture CFCC 52462).
Notes.
Cytospora leucostoma is a common species causing canker disease on Rosaceae in China (Teng 1963, Tai 1979, Wei 1979, Fan et al. 2020). In this study, fresh specimens were collected from diseased branches of the Chinese chestnut for the first time and identified as Cytospora leucostoma, based on strictly matched asexual morph (4–5.5 × 1–2 μm from Castanea mollissima in this study vs. 4.5–5.5 × 1–1.5 μm from multiple specimens in Fan et al. 2020) and phylogenic analysis (Fig. 2).
Cytospora myrtagena
(G.C. Adams & M.J. Wingf.) G.C. Adams & Rossman, IMA Fungus 6 (1): 147. 2015.
8A67297D-DD92-575B-B7B2-A24925C785AD
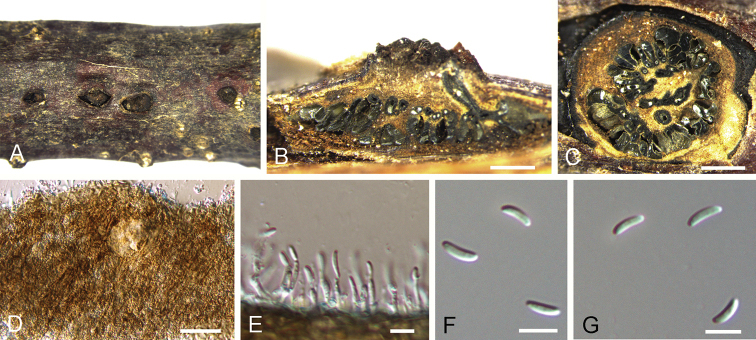
Figure 6.
Cytospora myrtagena on Castanea mollissima (BJFC-S1704). A, B Habit of conidiomata on branches C, E transverse section of conidiomata D longitudinal section through conidiomata F, G conidiogenous cells attached with conidia H conidia. Scale bars: 500 μm (B–D), 5 μm (E, G), 10 μm (H).
Description.
Sexual morph: not observed. Asexual morph: Pycnidial stromata pulvinate, immersed in bark, scattered, erumpent through the surface of bark. Conceptacle absent. Ostiole dark grey to black, conspicuous, at the same level as the disc, (50–)65–75(–82) μm diam. Locules undivided, circular to ovoid, (430–)550–720(–810) µm diam. Peridium comprising few layers of cells of textura angularis, with innermost layer brown, outer layer brown to dark brown. Conidiophores hyaline, unbranched, thin-walled, filamentous. Conidiogenous cells enteroblastic polyphialidic, (6.5–)8.4–12.5(–15.3) × 0.9–1.4 μm (x̄ = 10.2 × 1.2 μm). Conidia hyaline, allantoid, smooth, aseptate, thin-walled, (3.2–)3.4–5.4(–6.2) × 1–1.5 μm (x̄ = 4.7 × 1.3 μm).
Culture characters.
On PDA at 25 °C in darkness. Cultures are initially white, becoming olivaceous buff in centre after 7 d and finally olivaceous at 30 d. The colony is flat, thin with a felt and tight texture in centre. Pycnidia distributed irregularly on medium surface.
Specimens examined.
China, Shaanxi Province, Ankang City, Xiangxidong forest park, 32°40'33"N, 109°18'57"E, on stem barks of Castanea mollissima, 1 July 2017, N. Jiang (BJFC-S1704, living culture CFCC 52454; BJFC-S1705, living culture CFCC 52455).
Notes.
Cytospora myrtagena was introduced from Eucalyptus and Tibouchina in America and Indonesia (Adams et al. 2005). Two ITS sequences of Cytospora myrtagena were available, AY347363 from the type strain CBS 116843 and AY347380 from CBS 117013. However, there are 14 bp differences between AY347363 and AY347380. Cytospora tibouchinae was introduced as a phylogenically close species to Cytospora myrtagena (Suppl. material 1: Fig. S1), with 21 bp differences to CBS 116843 and 14 bp bp differences to CBS 117013 (Crous et al. 2016). Two isolates from Castanea mollissima in the present study were close to Cytospora myrtagena and Cytospora tibouchinae (Suppl. material 1: Fig. S1), with 22 bp differences to CBS 116843, 15 bp differences to CBS 117013 and 6 bp differences to Cytospora tibouchinae. Morphologically, they have similar conidial sizes (3.4–5.4 × 1–1.5 μm in BJFC-S1704 vs. 3–4 × 1 μm in C. myrtagena vs. 3–4 × 1.5–2 μm in C. tibouchinae) (Adams et al. 2005, Crous et al. 2016). Hence, it is hard to identify our isolates to C. myrtagena or C. tibouchinae, for the large differences between two ITS sequences in C. myrtagena provided by Adams et al. (2005) and absence of ACT and RPB2 loci in C. myrtagena and C. tibouchinae. We give the name Cytospora myrtagena to our isolates provisionally, and hope for more studies on this species.
Cytospora schulzeri
Sacc. & P. Syd., Syll. fung. (Abellini) 14(2): 918. 1899.
17EB9794-D05F-5D4A-A3D3-7DDD2A3E30A1
Figure 7.
Cytospora schulzeri on Castanea mollissima (BJFC-S1702). A, B Habit of conidiomata on branches C transverse section of conidiomata D longitudinal section through conidiomata E conidiogenous cells attached with conidia F conidia. Scale bars: 500 μm (B–D), 10 μm (E, F).
Description.
Sexual morph: not observed. Asexual morph: Pycnidial stromata ostiolated, immersed in bark, scattered, erumpent through the surface of bark, flat, discoid, with multiple locules. Conceptacle absent. Ectostromatic disc brown, circular to ovoid, (250–)300–400(–475) µm diam., with 1–5 ostiole per disc. Ostioles black, at the same level as the disc, (40–)50–85(–115) μm diam. Locule numerous, arranged circularly with common walls, (600–)700–1500(–1750) µm diam. Peridium comprising a few layers of cells of textura angularis, with innermost layer brown, outer layer brown to dark brown. Conidiophores hyaline, unbranched, thin walled, filamentous. Conidiogenous cells enteroblastic polyphialidic, (6.5–)8.5–18.5(–21) × 1–2 μm (x̄ = 13.1 × 1.6 μm). Conidia hyaline, allantoid, smooth, aseptate, thin-walled, (3.5–)4.5–6.5(–7) × 1–1.5 μm (x̄ = 5.2 × 1.3 μm).
Specimens examined.
China, Hebei Province, Chengde City, Kuancheng County, chestnut plantation, 40°38'37"N, 118°27'54"E, on branches of Castanea mollissima, 13 October 2017, N. Jiang (living culture CFCC 52468; BJFC-S1702, living culture CFCC 52469; BJFC-S1703, living culture CFCC 52470).
Notes.
Cytospora schulzeri is a common species causing apple canker disease in China (Teng 1963, Tai 1979, Wei 1979, Zhuang 2005, Fan et al. 2020). In this study, fresh specimens were collected from diseased branches of chestnut trees and identified as Cytospora schulzeri, based on the strictly matched asexual morph (4.5–6.5 × 1–2 μm from Castanea mollissima in this study vs. 4–7 × 1–1.5 μm from multiple specimens in Fan et al. (2020)) and phylogenic analysis (Fig. 2).
Cytospora xinglongensis
C.M. Tian & N. Jiang sp. nov.
D2D4ECD5-0D36-53AC-B6D1-29F9A3AA6879
829517
Figure 8.
Cytospora xinglongensis on Castanea mollissima (BJFC-S1706). A Habit of conidiomata on branches B longitudinal section through conidiomata C transverse section of conidiomata D peridium E conidiogenous cells attached with conidia F, G conidia. Scale bars: 500 μm (B, C), 10 μm (E–G).
Diagnosis.
Cytospora xinglongensis can be distinguished from C. californica and C. eucalypti by longer conidia.
Etymology.
Named after the county where it was collected, Xinglong County.
Description.
Sexual morph: not observed. Asexual morph: Pycnidial stromata immersed in bark, erumpent through the surface of bark, discoid, with a solitary undivided locule. Conceptacle black, circular surrounded stromata. Ostiole inconspicuous. Locules undivided, circular to ovoid, (480–)540–685(–755) µm diam. Conidiophores hyaline, unbranched. Peridium comprising a few layers of cells of textura angularis, with innermost layer brown, outer layer brown to dark brown. Conidiogenous cells enteroblastic polyphialidic, (4.5–)6.5–8.5(–12) × 1–1.5 μm (x̄ = 7.4 × 1.3 μm). Conidia hyaline, allantoid, eguttulate, smooth, aseptate, thin-walled, (7.5–)8.5–9.5(–10.5) × 1–1.5 μm (x̄ = 8.9 × 1.3 μm).
Culture characters.
On PDA at 25 °C in darkness. Cultures are white. The colony is flat, thin with a uniform texture, lacking aerial mycelium. Pycnidia distributed uniformly on medium surface.
Specimens examined.
China, Hebei Province, Chengde City, Xinglong County, chestnut plantation, 40°24'32"N, 117°28'56"E, on branches of Castanea mollissima, 11 October 2017, N. Jiang (holotypeBJFC-S1706, ex-type living culture CFCC 52458; paratypeBJFC-S1707, living culture CFCC 52459).
Notes.
Cytospora xinglongensis is associated with canker disease of Castanea mollissima in China. Cytospora xinglongensis can be distinguished from its phylogenetically closely species C. thailandica by having much longer conidia (8.5–9.5 μm in C. xinglongensis vs. 3.3–4 μm in C. thailandica) (Norphanphoun et al. 2018). In addition, Cytospora xinglongensis differs from C. thailandica by ITS, ACT and RPB2 loci (16/470 in ITS, 22/245 in ACT and 52/726 in RPB2).
Discussion
In the present study, an important fruit tree species, Castanea mollissima was investigated and Cytospora canker was found as a commom disease in plantations in Hebei Province. Identification was conducted based on 13 isolates from fruiting bodies using both morphological and molecular methods. As a result, six Cytospora species were confirmed. Cytospora kuanchengensis and C. xinglongensis are introduced as new species, C. ceratospermopsis, C. leucostoma, C. myrtagena and C. schulzeri are firstly reported on Castanea mollissima.
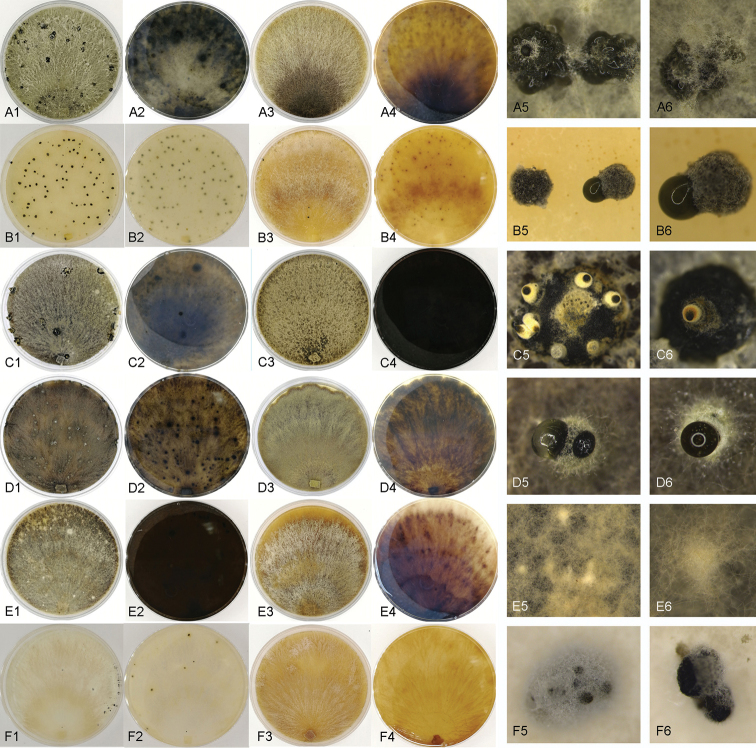
These six chestnut Cytospora species can be easily distinguished using DNA sequences of single ITS sequence or combined sequences of ITS, LSU, ACT and RPB2 (Fig. 2; Suppl. material 1: Fig. S1). In addition, colonies on PDA and MEA of these six species are also different (Fig. 9). Cytospora xinglongensis never produce fruiting bodies on PDA or MEA, while the other five species form conidiomata in one month (Fig. 9). Morphologically, Cytospora xinglongensis has obviously longer conidia than others. However, the conidial dimension can hardly distinguish C. ceratospermopsis, C. kuanchengensis, C. leucostoma, C. myrtagena and C. schulzeri.
Figure 9.
Cultures of Cytospora species from Castanea mollissima after 1 month at 25 °C. AC. myrtagenaBC. kuanchengensisCC. ceratospermopsisDC. leucostomaEC. xinglongensisFC. schulzeriA1–G2 cultures on PDA A3–G4 cultures on MEA A5–G6 fruiting bodies or hyphal masses produced on cultures
Dar and Rai reported Cytospora diseases on Castanea sativa in India, causing perennial cankers on stems and branches (Dar and Rai 2014). The Cytospora isolates were identified mainly based on ITS sequence data, which were introduced as a new species named Cytospora castaneae (wrongly wrriten as Cytospora castanae in the original paper) (Dar and Rai 2014). However, further study is required to confirm the species position within the genus, including detailed morphogical features and sequences of high quality.
Cytospora canker is a common disease on chestnut trees, but there are few formal reports. In China, this disease is known amongst phytopathologists, but no-one conducted accurate identifications. Hence, this paper is the first formal report of Cytospora chestnut canker in China. From our investigations of chestnut diseases in China, Cytospora species are closely associated with canker diseases in chestnut plantations. In most cases, they infect twigs or small branches, causing necrotic lesions (Fig. 1A), finially forming fruiting bodies on dead tissues (Fig. 1D). However, Cytospora myrtagena was discovered on stems of a 15-year-old chestnut tree, causing typical Cytospora canker symptoms. More works should be conducted on the newly emerging pathogens from several aspects.
As the species concept of Cytospora has been improved a lot by using molecular data (Yang et al. 2015, Lawrence et al. 2017, Norphanphoun et al. 2017, 2018, Jayawardena et al. 2019, Fan et al. 2020), many Cytospora canker diseases and new species have been discovered and reported in recent years. Further studies are, however, now required to confirm their pathogenicity.
Supplementary Material
Acknowledgements
This study is financed by National Natural Science Foundation of China (Project No.: 31670647). We are grateful to Chungen Piao, Minwei Guo (China Forestry Culture Collection Center (CFCC), Chinese Academy of Forestry, Beijing.
Citation
Jiang N, Yang Q, Fan X-L, Tian C-M (2020) Identification of six Cytospora species on Chinese chestnut in China. MycoKeys 62: 1–25. https://doi.org/10.3897/mycokeys.62.47425
Funding Statement
National Natural Science Foundation of China
Supplementary materials
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ning Jiang, Qin Yang, Xin-Lei Fan, Cheng-Ming Tian
Figure S1
Data type: (phylogram of Cytospora)
Explanation note: Phylogram of Cytospora obtained from the ITS gene.
References
- Adams GC, Roux J, Wingfield MJ, Roux J. (2005) Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Studies in Mycology 52: 1–144. [Google Scholar]
- Adams GC, Roux J, Wingfield MJ. (2006) Cytospora species (Ascomycota, Diaporthales, Valsaceae), introduced and native pathogens of trees in South Africa. Australasian Plant Pathology 35: 521–548. 10.1071/AP06058 [DOI] [Google Scholar]
- Aghayeva DN, Rigling D, Meyer JB, Mustafabeyli E. (2017) Diversity of fungi occurring in the bark of Castanea sativa in Azerbaijan. In VI International Chestnut Symposium 1220, 79–86. 10.17660/ActaHortic.2018.1220.12 [DOI]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 3: 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM. et al. (2016) Fungal Planet description sheets: 400–468. Persoonia 36: 316–458. 10.3767/003158516X692185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar MA, Rai MK. (2014) Occurrence of Cytospora castanae sp nov., associated with perennial cankers of Castanea sativa. Mycosphere 5: 747–757. 10.5943/mycosphere/5/6/5 [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. 10.2307/2419362 [DOI] [Google Scholar]
- Fan XL, Bezerra JDP, Tian CM, Crous PW. (2020) Cytospora (Diaporthales) in China. Persoonia 45: 1–45. 10.3767/persoonia.2020.45.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XL, Hyde KD, Liu M, Liang YM, Tian CM. (2015a) Cytospora species associated with walnut canker disease in China, with description of a new species C. gigalocus. Fungal Biology 119: 310–319. 10.1016/j.funbio.2014.12.011 [DOI] [PubMed] [Google Scholar]
- Fan XL, Hyde KD, Yang Q, Liang YM, Ma R, Tian CM. (2015b) Cytospora species associated with canker disease of three anti-desertification plants in northwestern China. Phytotaxa 197: 227–244. 10.11646/phytotaxa.197.4.1 [DOI] [Google Scholar]
- Fan XL, Liang YM, Ma R, Ma R, Tian CM. (2014a) Morphological and phylogenetic studies of Cytospora (Valsaceae, Diaporthales) isolates from Chinese scholar tree, with description of a new species. Mycoscience 55: 252–259. 10.1016/j.myc.2013.10.001 [DOI] [Google Scholar]
- Fan XL, Tian CM, Yang Q, Liang YM, You CJ, Zhang YB. (2014b) Cytospora from Salix in northern China. Mycotaxon 129: 303–315. 10.5248/129.303 [DOI] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Gong S, Zhang X, Jiang S, Chen C, Ma H, Nie Y. (2017) A new species of Ophiognomonia from Northern China inhabiting the lesions of chestnut leaves infected with Diaporthe eres. Mycological Progress 16: 83–91. 10.1007/s11557-016-1255-z [DOI] [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Jayawardena RS, Hyde KD, McKenzie EHC. et al. (2019) One stop shop III: taxonomic update with molecular phylogeny for important phytopathogenic genera: 51–75. Fungal Diversity 98: 1–84. 10.1007/s13225-019-00433-6 [DOI] [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Jiang N, Fan XL, Tian CM. (2019a) Identification and pathogenicity of Cryphonectriaceae species associated with chestnut canker in China. Plant Pathology 68: 1132–1145. 10.1111/ppa.13033 [DOI] [Google Scholar]
- Jiang N, Fan XL, Crous PW, Tian CM. (2019b) Species of Dendrostoma (Erythrogloeaceae, Diaporthales) associated with chestnut and oak canker diseases in China. Mycokeys 48: 67–96. 10.3897/mycokeys.48.31715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Fan XL, Yang Q, Du Z, Tian CM. (2018a) Two novel species of Cryphonectria from Quercus in China. Phytotaxa 347: 243–250. 10.11646/phytotaxa.347.3.5 [DOI] [Google Scholar]
- Jiang N, Li J, Piao CG, Guo MW, Tian CM. (2018b) Identification and characterization of chestnut branch-inhabiting melanocratic fungi in China. Mycosphere 9: 1268–1289. 10.5943/mycosphere/9/6/14 [DOI] [Google Scholar]
- Jiang N, Tian CM. (2019) An Emerging Pathogen from Rotted Chestnut in China: Gnomoniopsis daii sp. nov. Forests 10: 1016. 10.3390/f10111016 [DOI]
- Jiang N, Voglmayr H, Tian CM. (2018c) New species and records of Coryneum from China. Mycologia 110: 1172–1188. 10.1080/00275514.2018.1516969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepley JB, Reeves FB, Jacobi WR, Adams GC. (2015) Species associated with Cytospora canker on Populus tremuloides. Mycotaxon 130: 783–805. 10.5248/130.783 [DOI] [Google Scholar]
- Lawrence DP, Holland LA, Nouri MT, Travadon R, Abramians A, Michailides TJ, Trouillas FP. (2018) Molecular phylogeny of Cytospora species associated with canker diseases of fruit and nut crops in California, with the descriptions of ten new species and one new combination. IMA Fungus 9: 333–370. 10.5598/imafungus.2018.09.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence DP, Travadon R, Pouzoulet J, Rolshausen PE, Wilcox WF, Baumgartner K. (2017) Characterization of Cytospora isolates from wood cankers of declining grapevine in North America, with the descriptions of two new Cytospora species. Plant Pathology 66: 713–725. 10.1111/ppa.12621 [DOI] [Google Scholar]
- Lu C, Guo SJ. (2016) Analysis on the nutritional characters and comprehensive evaluation of 16 chestnut germplasm resources. Science and Technology of Food Industry 37: 357–376. [Google Scholar]
- Mehrabi ME, Mohammadi GE, Fotouhifar KB. (2011) Studies on Cytospora canker disease of apple trees in Semirom region of Iran. Journal of Agricultural Technology 7: 967–982. [Google Scholar]
- Norphanphoun C, Doilom M, Daranagama DA, Phookamsak R, Wen TC, Bulgakov TS, Hyde KD. (2017) Revisiting the genus Cytospora and allied species. Mycosphere 8: 51–97. 10.5943/mycosphere/8/1/7 [DOI] [Google Scholar]
- Norphanphoun C, Raspé O, Jeewon R, Wen TC, Hyde KD. (2018) Morphological and phylogenetic characterisation of novel Cytospora species associated with mangroves. Mycokeys 38: 93–120. 10.3897/mycokeys.38.28011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Zhu HY, Tian CM, Alvarez LV, Fan XL. (2018) Cytospora piceae sp. nov. associated with canker disease of Picea crassifolia in China. Phytotaxa 383: 181–196. 10.11646/phytotaxa.383.2.4 [DOI] [Google Scholar]
- Phookamsak R, Hyde KD, Jeewon R. et al. Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Diversity 95: 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Rambaut A. (2016) FigTree, version 1.4.3. University of Edinburgh, Edinburgh.
- Rigling D, Prospero S. (2018) Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Molecular Plant Pathology 19: 7–20. 10.1111/mpp.12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY, Farr DF, Castlebury LA. (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48: 135–144. 10.1007/S10267-007-0347-7 [DOI] [Google Scholar]
- Sarma VV, Hyde KD. (2001) A review on frequently occurring fungi in mangroves. Fungal Diversity 8: 1–34. [Google Scholar]
- Senanayake IC, Crous PW, Groenewald JZ. et al. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86: 217–296. 10.1016/j.simyco.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Jeewon R, Chomnunti P. et al. (2018) Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity 93: 241–443. 10.1007/s13225-018-0410-z [DOI] [Google Scholar]
- Shuttleworth LA, Guest DI. (2017) The infection process of chestnut rot, an important disease caused by Gnomoniopsis smithogilvyi (Gnomoniaceae, Diaporthales) in Oceania and Europe. Australasian Plant Pathology 46: 397–405. 10.1007/s13313-017-0502-3 [DOI] [Google Scholar]
- Swofford DL. (2003) PAUP*: Phylogenetic Analyses Using Parsimony, * and Other Methods, Version 4.0b10. Sinauer Associates, Sunderland.
- Tai FL. (1979) Sylloge fungorum sinicorum. Science Press, 1–1527.
- Teng SC. (1963) Fungi of China. Science Press, 1–808. 10.1136/bmj.1.5333.808 [DOI]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Akulov OY, Jaklitsch WM. (2016) Reassessment of Allantonectria, phylogenetic position of Thyronectroidea, and Thyronectria caraganae sp. nov. Mycological Progress 15: 921–937. 10.1007/s11557-016-1218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Castlebury LA, Jaklitsch WM. (2017) Juglanconis gen. nov. on Juglandaceae, and the new family Juglanconidaceae (Diaporthales). Persoonia 38: 136–155. 10.3767/003158517X694768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Lu Q, Decock C, Li YX, Zhang XY. (2015) Cytospora species from Populus and Salix in China with C. davidiana sp. nov. Fungal Biology 119: 420–432. 10.1016/j.funbio.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Wei JC. (1979) Identification of Fungus Handbook. Science Press, 1–780.
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R. (2018) Outline of Ascomycota: 2017. Fungal Diversity 88: 167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- Yang Q, Fan XL, Crous PW, Liang YM, Tian CM. (2015) Cytospora from Ulmus pumila in northern China. Mycological Progress 14: 74. 10.1007/s11557-015-1096-1 [DOI]
- Zhang LX, Alvarez LV, Bonthond G, Tian CM, Fan XL. (2019) Cytospora elaeagnicola sp. nov. associated with narrow-leaved oleaster canker disease in China. Mycobiology 47: 319–328. 10.1080/12298093.2019.1633902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YB, You CJ, Fan XL, Tian CM. (2014) Taxonomy and phylogeny of Cytospora in Northeast China. Mycosystema 33: 806–818. [Google Scholar]
- Zhu HY, Fan XL, Tian CM. (2018) Multigene phylogeny and morphology reveal Cytospora spiraeae sp. nov. (Diaporthales, Ascomycota) in China. Phytotaxa 338: 49–62. 10.11646/phytotaxa.338.1.4 [DOI] [Google Scholar]
- Zhuang WY. (2005) Fungi of Northwestern China, New York, USA. Ithaca, Mycotaxon, Ltd, 1–430.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ning Jiang, Qin Yang, Xin-Lei Fan, Cheng-Ming Tian
Figure S1
Data type: (phylogram of Cytospora)
Explanation note: Phylogram of Cytospora obtained from the ITS gene.