Abstract
Cell-free DNA profiling using patient blood is emerging as a non-invasive complementary technique for cancer genomic characterization. Since these liquid biopsies will soon be integrated into clinical trial protocols for pediatric cancer treatment, clinicians should be informed about potential applications and advantages but also weaknesses and potential pitfalls. Small retrospective studies comparing genetic alterations detected in liquid biopsies with tumor biopsies for pediatric solid tumor types are encouraging. Molecular detection of tumor markers in cell-free DNA could be used for earlier therapy response monitoring and residual disease detection as well as enabling detection of pathognomonic and therapeutically relevant genomic alterations.
Conclusion: Existing analyses of liquid biopsies from children with solid tumors increasingly suggest a potential relevance for molecular diagnostics, prognostic assessment, and therapeutic decision-making. Gaps remain in the types of tumors studied and value of detection methods applied. Here we review the current stand of liquid biopsy studies for pediatric solid tumors with a dedicated focus on cell-free DNA analysis. There is legitimate hope that integrating fully validated liquid biopsy–based innovations into the standard of care will advance patient monitoring and personalized treatment of children battling solid cancers.
|
What is Known: • Liquid biopsies are finding their way into routine oncological screening, diagnosis, and disease monitoring in adult cancer types fast. • The most widely adopted source for liquid biopsies is blood although other easily accessible body fluids, such as saliva, pleural effusions, urine, or cerebrospinal fluid (CSF) can also serve as sources for liquid biopsies | |
|
What is New: • Retrospective proof-of-concept studies in small cohorts illustrate that liquid biopsies in pediatric solid tumors yield tremendous potential to be used in diagnostics, for therapy response monitoring and in residual disease detection. • Liquid biopsy diagnostics could tackle some long-standing issues in the pediatric oncology field; they can enable accurate genetic diagnostics in previously unbiopsied tumor types like renal tumors or brain stem tumors leading to better treatment strategies |
Keywords: Liquid biopsies, Pediatric solid tumors, Cell-free DNA profiling
Introduction
The analysis of circulating cell-free nucleic acids is being introduced in several medical fields. In obstetrics, non-invasive prenatal aneuploidy screening for trisomy 21 is well established and widely implemented with high sensitivity and specificity [1]. In transplantation medicine, the amount of circulating donor-derived cell-free DNA in the recipient is being explored as a surrogate marker for cellular damage in the donated organ [23, 26]. The analysis of tumor-derived cell-free DNA and RNA is emerging as an alternative to or complementary assay for molecular genetic analyses in tumor tissue biopsies. Commonly referred to as liquid biopsies, the most widely adopted source is blood although other easily accessible body fluids, such as saliva, pleural effusions, urine, or cerebrospinal fluid (CSF), can also serve as sources for liquid biopsies [27]. Moss et al. used cell type-specific methylation to track cell origin, identifying 55% of cell-free DNA in healthy individuals as originating from white blood cells, with contributions from erythrocyte progenitors (30%), vascular endothelial cells (10%), and hepatocytes (1%) [47]. Cell-free nucleic acids are thought to originate from apoptotic or necrotic tissue under physiological and pathological circumstances, but exact biological origins and roles are still under investigation (reviewed in [66]). The fraction of cell-free DNA originating from the tumor is sometimes referred to as circulating or cell-free tumor DNA both abbreviated as ctDNA, which we will use throughout this review. Circulating tumor cells and extracellular vesicles (30–100 nm diameter) originating from tumor cells known as exosomes are other biological sources for DNA, RNA, and proteins in liquid biopsies. This review is limited to the analysis of ctDNA, the most widely adopted fraction. Siravegna [61] and Wan [73] have comprehensively reviewed how other types of liquid biopsies can be exploited to guide patient care, while Merker et al. [61] reviewed the current information about clinical ctDNA assays. We refer the reader to these reviews for more information on those topics.
Opportunities for liquid biopsies in pediatric oncology
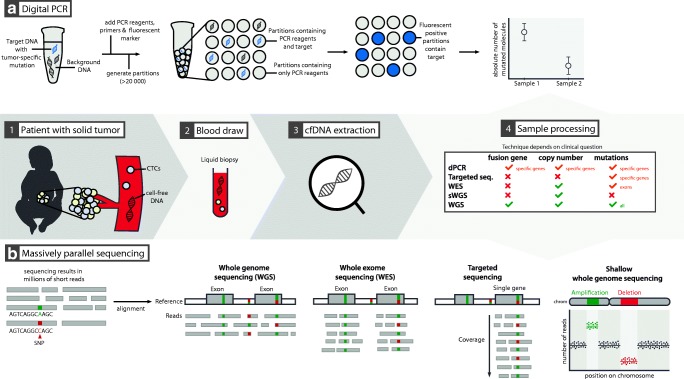
The discovery of ctDNA dates back to 1977 [40]. However, technological advances have only recently made routine and sensitive analysis feasible, with the advent of the digital polymerase chain reaction (PCR, Fig. 1a) and massively parallel sequencing (Fig. 1b) [27, 61]. The liquid biopsy approach has significant theoretical advantages over classical biopsies, which are often invasive, costly, and potentially harmful to patients. Liquid biopsies are relatively simple to obtain, making them less invasive and less expensive. The patient also benefits from the potential for improved care through finer time-resolved diagnostic monitoring. Liquid biopsies can facilitate increased diagnostic accuracy and enable therapy response monitoring and minimal residual disease (MRD) detection for solid tumors. Tissue biopsy also only reflects a subpopulation of the tumor cells, creating sampling bias. Liquid biopsy better detects spatial or subclonal tumor heterogeneity [15, 25]. Liquid biopsies also enable monitoring of tumor clonal evolution and detection of therapy-relevant novel mutations arising during treatment. Early cancer detection through liquid biopsy is also emerging as a way to perform population surveillance [2]. For many pediatric tumor types, it remains unknown if and to what degree liquid biopsy–based tests will contribute to diagnosis, treatment stratification, and follow-up monitoring. We will discuss the current state of liquid biopsy applications in pediatric oncology and indicate where opportunities can be found.
Fig. 1.
The optimal technique for cell-free DNA (cfDNA) analysis is chosen depending on the clinical question at hand. Commonly used techniques are digital PCR (dPCR) and massively parallel, or “next-generation,” sequencing. Digital PCR (panel a), with its unmatched sensitivity, is suited for monitoring known (hotspot) mutations, can be used to detect amplifications or losses of one or two pre-specified genomic regions and to detect pre-defined sites of genomic fusion. Massive parallel sequencing (panel b) is useful to detect all types of genomic alterations, depending on the sequencing strategy used. It can evaluate single nucleotide variants (SNVs), copy number aberrations (CNA), genomic fusions or a combination thereof. Whole-genome sequencing (WGS) results in uniform coverage across the entire genome. When performed at low coverage, the technique is termed shallow WGS, and is a cost-effective method to detect CNAs. Performed at higher coverage, the detailed analysis of mutations or translocations on a genome-wide scale is feasible. Whole-exome sequencing (WES) focuses the sequencing effort on the coding regions of the genome, but non-coding or structural variation is largely missed. Targeted sequencing will result in extremely high coverage over a small proportion of the genome, allowing the detection of variants in that specific region with high sensitivity. PCR, polymerase chain reaction. CTC, circulating tumor cells
Screening and early diagnosis
Little is known about the role of liquid biopsies in cancer screening for the pediatric population.
Screening for cancer requires an easy, low-cost, and low-impact test with a minimalized false detection rate. A test with very high sensitivity and specificity will still detect many false positives if a disease it screens for is very rare, as is the case for cancer in children. As an example, a hypothetical screening test with 99.9% specificity and sensitivity for neuroblastoma, which affects 9–12 children (< 15 years) per million, would generate 988 false positives per 1000 positively screened cases when screening all children under 15 years of age in the general population. Earlier attempts to screen populations for neuroblastoma by ultrasound [51, 57, 76] did not reduce mortality, but overdiagnosed benign adrenal masses as neuroblastomas. The utility of cancer screening in the general pediatric population remains dubious, since cancer screening only becomes clinically relevant if patient benefit can be achieved. Either preventive therapeutic options taken to avoid cancer development (e.g., colectomy in patients with familial adenomatous polyposis) or improved outcome due to treatment after early tumor detection can make screening useful. It is currently unclear what the gold standard is for early cancer detection programs in children with cancer predisposition syndromes stemming from gene mutations in the germline, such as Li-Fraumeni (TP53), Von Hippel Lindau (VHL), familial adenomatous polyposis (APC), DICER1 syndrome or subtypes of primary immunodeficiencies [34]. Liquid biopsies may play a role in this setting in the (near) future, but much controversy remains about correct follow-up since studies with high-quality follow-up remain scarce [39, 54, 65, 75]. Liquid biopsy–based screening could potentially be more accurate, simpler, and associated with less discomfort than current monitoring protocols. Children affected by constitutional mismatch repair deficiency, whose chance of developing cancer by 18 years of age is estimated to be as high as 80 percent [75], could benefit from early detection.
Diagnostics and therapeutic stratification
Data describing the diagnostic utility of liquid biopsies in pediatric oncology is accumulating. One multi-entity study by Kurihara et al. detected cell-free DNA in plasma samples from all 44 patients with a diverse range of pediatric tumors, but could not definitively show it was tumor-derived because of the paucity in genomic alterations [38]. No large-scale multi-entity studies assessing the diagnostic potential of ctDNA in patients with pediatric solid tumors have been completed to date. However, ultralow passage whole-genome sequencing conducted on 45 pediatric diagnostic pretreatment plasma samples [36] demonstrated the presence of ctDNA in more than half of samples from patients with osteosarcoma, neuroblastoma, Wilms tumor, and alveolar rhabdomyosarcoma. Changes in cell-free DNA plasma load also correlated with treatment response, with higher loads detected in patients with progressive disease. We discuss the studies conducted for single cancers in the context of each disease below. Further evidence of the feasibility of ctDNA detection across different pediatric cancers and different biological sources will come from the ongoing NGSkids (NCT02546453) and MICCHADO (NCT03496402) trials.
Neuroblastoma
Recent evidence demonstrates that copy number alterations, a mandatory analysis for risk stratification, can be determined from cell-free DNA in blood plasma from neuroblastoma patients [14, 68]. Chromosomal copy number profiles assessed from ctDNA by shallow whole-genome sequencing (at 0.4-fold genomic coverage) were highly concordant with profiles generated from the gold standard, array-based comparative genomic hybridization from the primary tumor biopsy. Work by Combaret et al. showed that the two activating ALK mutations commonly occurring in neuroblastomas can be detected in plasma by droplet digital PCR with high sensitivity and specificity (90–100%), producing results concordant to those achieved with deep sequencing [18]. Quantitative PCR-based detection of MYCN amplifications in peripheral blood from neuroblastoma patients was proven feasible in 2002, before the concept of cancer liquid biopsies was established [17]. Detection sensitivity and specificity is further improved by droplet digital PCR [41]. In all four of the above-mentioned studies, genomic alterations were detected in circulating cell-free DNA that were not detectable in the primary tumor biopsy, suggesting that liquid biopsy diagnostics may be better at capturing tumor heterogeneity or detecting alterations present in metastases.
Ewing sarcoma
The diagnostic hallmark for Ewing sarcoma is a rearrangement involving the EWSR1 gene, most commonly EWSR1-FLI1 and EWSR1-ERG rearrangements, while other rare translocation partners have been reported. EWSR1 fusion genes can be detected in circulating cell-free DNA with droplet digital PCR or targeted sequencing, providing a liquid biopsy–based diagnostic strategy [37, 60].
Lymphomas
Although no detailed genomic analysis was conducted, two studies detected significantly higher cell-free DNA loads in plasma from 201 pediatric patients with various lymphoma subtypes [49] and 155 patients with Hodgkin lymphoma [55] as compared with plasma from healthy controls. High circulating cell-free DNA levels correlated with poor prognosis in patients with Hodgkin lymphoma [49], and are present at diagnosis in plasma from patients with B cell non-Hodgkin lymphoma, but decrease during treatment [43]. Pathognomonic NPM-ALK fusion genes are readily detectable in plasma from patients with anaplastic large cell lymphoma [49].
Renal tumors
Pediatric renal tumors are most often not biopsied due to the risk of tumor rupture, which would spill tumor cells into the peritoneal cavity and require treatment intensification. This lack of histological confirmation at diagnosis can lead to misdiagnosis and suboptimal treatment of non-Wilms type tumors. Jimenez et al. [33] retrospectively examined plasma samples collected at diagnosis of different renal tumor types in 18 patients. Tumor-specific copy number and/or single-nucleotide alterations were detected in plasma from all but one patient. Molecular characterization of kidney tumors from plasma samples collected at diagnosis could, therefore, open the door to more appropriate and tumor-specific neoadjuvant chemotherapy. A small proof-of-concept study [67] developed and applied a PCR assay detecting internal tandem duplications in BCOR, a hallmark of clear cell sarcoma of the kidney, to plasma samples. This assay was used to pre-operatively differentiate clear cell sarcoma from nephroblastoma in four patients.
Brain tumors
Pathological examination is the gold standard for definitive brain tumor diagnosis and subtyping, but this is often challenging. The most recent WHO classification defines molecular parameters in addition to histopathology for diagnosis [42]. Classification of brain tumors by methylation analysis appears to outperform histopathological diagnostics at least for several tumor types [12, 52, 62]. This predicts a major role for ctDNA-based diagnosis of central nervous system tumors, facilitating early management and therapy, especially in cases where tumor localization prevents resection. Cell-free DNA load in blood plasma has only been tested in pediatric patients with medulloblastomas (among brain tumors) to date, where its presence was demonstrated in 40% of patients [7]. Tumor-derived histone H3 gene mutations were detected in blood plasma from pediatric patients with diffuse midline gliomas [31]. Martinez-Ricarte et al. were able to classify 17 of 20 patients (including 2 children) with diffuse gliomas by analyzing only 7 genes in cell-free DNA from CSF [44]. Paret et al. reported on one pediatric case of neuroepithelial high-grade tumor of the central nervous system showing a BCOR internal duplication, whose detection in plasma cell-free DNA correlated with relapse development [53]. The blood-brain barrier significantly restricts the amount of ctDNA entering the blood [7, 22]. An alternative source of ctDNA for brain tumors is CSF, which has been demonstrated to contain ctDNA to a certain extent in adult patients [58]. Many pediatric patients with brain tumors present with critically elevated intracranial pressure [50], in whom acute neurosurgical intervention is necessary. CSF can be safely obtained for ctDNA analysis during this procedure with no additional risk or burden to the patient. The diagnostic utility of this analysis across the range of both high- and low-grade pediatric brain tumors has not yet been explored. We expect this evidence to emerge within the next years, as techniques for cell-free DNA methylation detection are being further developed [21, 59]. CSF can also be obtained by lumbar puncture; while not minimally invasive, this technique is a relatively safe and often included in routine testing for neurological symptoms in pediatric patients and as a staging tool in brain tumors. When a CNS tumor is suspected, the benefit of a lumbar puncture to obtain CSF for ctDNA analysis might outweigh the risks associated with sampling.
Retinoblastoma
Although not minimally invasive or easily accessible, the vitreous fluid has been retrospectively examined in 26 patients with retinoblastomas. Tumor-specific copy number alterations and RB1 mutations detected in the vitreous fluid using shallow whole-genome sequencing strongly correlated with the need for eye enucleation. This testing may become a biomarker to guide the important decision whether to enucleate or salvage the eye in future trials [5, 6]. Blood-based liquid biopsies have not been explored for retinoblastoma.
Evaluating therapeutic response and clonal evolution
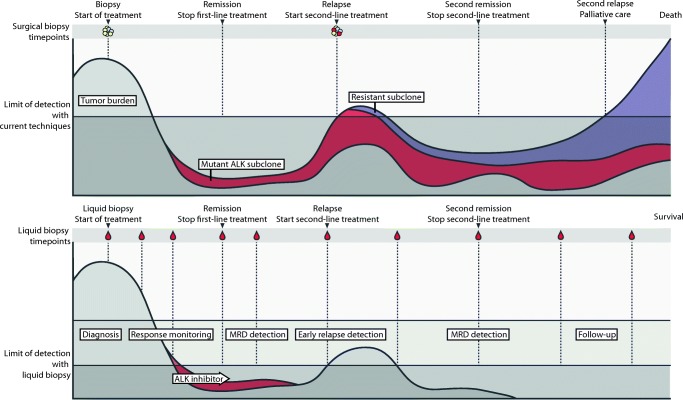
Liquid biopsy–based monitoring of therapy response in pediatric cancer patients has been evaluated in some studies in limited patient numbers, but clear evidence from prospectively validated large studies has not yet been published. Tumor heterogeneity and longitudinal follow-up of single-nucleotide variants and copy number aberrations in circulating cell-free DNA have best been explored to date in a study by Chicard et al. [15]. Using a combination of whole-exome and targeted sequencing of both the primary tumor and plasma samples collected at different time points, the authors demonstrated the subclonal makeup of neuroblastomas and the accumulation of additional genomic alterations during tumor evolution towards therapy-resistant disease. Some alterations were potentially targetable (MAPK pathway) suggesting an application for liquid biopsy diagnostics in therapy decisions and response evaluation [15]. We demonstrate the difference that could be made by liquid biopsy–based monitoring in the clinical course of a theoretical patient with neuroblastoma (Fig. 2).
Fig. 2.
Comparison for a theoretical patient with neuroblastoma: the course under current care protocols (top panel) of a patient with neuroblastoma and the projected course after the routine implementation of liquid biopsy diagnostics (bottom panel). Current treatment protocols evaluate the primary tumor only at diagnosis and relapse. A subclone harboring an ALK mutation is able to remain undetected until overt clinical relapse and is able to develop resistant to therapy. The tumor burden can be monitored more continuously using liquid biopsies. The subclone harboring the ALK mutation could be detected earlier, and treatment with an ALK inhibitor initiated without first observing a clinical overt relapse. Earlier treatment could possibly prevent a resistant subclone from expanding, thus, potentially improving outcome for the patient
Minimal residual disease and early relapse detection
Two major opportunities provided by liquid biopsies are to improve MRD monitoring under primary treatment and the early molecular-based diagnosis of relapse during follow-up. MRD detection is well established for children with acute lymphoblastic leukemia since the beginning of the 2010s, and is now part of routine follow-up in many frontline treatment protocols both to determine optimal treatment intensity and to diagnose relapse prior to the onset of clinical symptoms [11, 32]. Based on the analysis of plasma obtained from 44 pediatric patients with different solid tumors using next-generation sequencing and droplet digital PCR, Kurihara et al. suggest that total ctDNA amount can serve as a marker to evaluate how completely a pediatric tumor is resected following surgery [38]. Whole-genome profiling of primary neuroblastomas was used to generate tumor-specific DNA-based PCR assays for MRD monitoring in blood and bone marrow in eight patients, providing proof-of-concept for this paradigm in solid tumors. The MRD panel was capable of predicting disease relapse or bone marrow progression in four of five patients [69]. Further, different mRNA markers are being explored for MRD detection in blood (PHOX2B, TH, DDC, DBH, and CHRNA3) and bone marrow (PHOX2B, TH, DDC, CHRNA3, and GAP43) from neuroblastoma patients [63, 71]. This mRNA-based detection panel can be applied to assess treatment response as well as MRD detection, since high panel transcript levels at diagnosis, after induction therapy, and at the completion of treatment were associated with worse patient outcome [19, 64, 70, 72, 78]. Increasing evidence suggests that treatment follow-up using liquid biopsies is also feasible for patients with Ewing sarcoma. Measurement of the EWSR1 fusion gene copy numbers in 234 blood samples from 20 patients showed that recurring or increasing levels correlated with relapse [37]. In another study, patient-specific primers for use in droplet digital PCR were established to detect tumor-specific ESWR-ETS fusion gene breakpoint fragments in plasma samples from, to date, three patients with Ewing sarcoma. In two of these patients, fusion gene fragments were detected in plasma samples at a time when the disease was radiographically undetectable, altogether suggesting that measuring tumor-specific EWS-ETS fusion gene breakpoint fragments in the blood may serve as a reliable personalized biomarker for early relapse detection in patients with Ewing sarcoma [30].
Limitations and challenges
Quality issues and standards
The pre-analytical phase, including type and handling of the blood collection tube, storage temperature, time-to-processing and centrifugation speed, all influences the availability and composition of cell-free DNA in the sample [24]. For example, white blood cell lysis further increases their contribution to total cell-free DNA and dilutes the ctDNA fraction. Most studies conducted so far have utilized retrospectively collected plasma and blood samples, preventing investigation of pre-analytical variables that might interfere with the amount of ctDNA or total cell-free DNA. Standardizing pre-analytical variables will be necessary before liquid biopsies can enter routine clinical care. Preservation tubes for cell-free DNA analysis exist and have clearly proven capable of stabilizing cell-free DNA for longer periods [35, 45], but are more expensive and not often on hand in all hospitals. Further testing is necessary to arrive at standards to maintain quality for different liquid biopsy–based assays.
Physiology of cell-free DNA
Analogous to other pediatric biological variables, it is to be expected that processes regulating the shedding of cell-free DNA into the bloodstream and its metabolism may be different in children and adults, and may even vary between infants, young children, and adolescents. Cell-free DNA levels were shown to be higher in older individuals (19–30 and 67–97 age groups were compared) in the general population, and authors speculated that older people could have difficulty clearing cell-free DNA from the blood [47]. It is conceivable that health, similar to age, has a general impact on plasma cell-free DNA content. Comorbidities can also occur in pediatric cancer patients (e.g., acute and chronic kidney disease, sepsis) that have been shown to elevate total circulating cell-free DNA levels [16]. Data from adult patients show that the ctDNA compartment makes up a significantly higher fraction of circulating cell-free DNA in plasma from patients with high-stage and metastatic disease than low-stage disease [7], indicating liquid biopsies may be more relevant in high-stage and metastatic disease in children with cancer as well. The one study of retrospectively collected plasma samples from patients with multiple pediatric cancer types detected a significantly higher level of cell-free DNA in plasma from patients with neuroblastoma compared with patients with Ewing sarcoma, osteosarcoma, Wilms tumor, and alveolar rhabdomyosarcoma [36]. However, relative ctDNA plasma loads have not yet been thoroughly investigated across pediatric cancer types and stages. Circulating cell-free DNA has been most intensively studied in patients with neuroblastoma to date. Total cell-free DNA levels are approximately 100-fold higher in plasma from patients diagnosed with high-risk neuroblastoma than in healthy adults, indicating that tumor-derived DNA contributes largely to the circulating DNA in high-risk patients. Tumor-derived DNA was estimated to make up between 3 and 99% of circulating cell-free DNA in these patients [15, 36]. While these investigations indicate liquid biopsies may be more beneficial for patients with high-stage or metastatic disease, we should be careful of generalizing from the limited data available at this time. The extent to which patient age, tumor type, disease stage, or other variables impact the uses or usefulness of liquid biopsies will only become clear after their systematic integration in studies accompanying trials.
Low rates of recurrent genomic alterations common to pediatric cancers
In comparison with adult tumor types, hotspot mutations and recurrent genomic alterations are rare in pediatric cancer [28]. Recurrent genetic alterations or hotspot mutations that are tumor-specific are ideal targets for ctDNA diagnostics or MRD detection [13]. They offer the advantage that highly optimized assays, such as those applying droplet digital PCR, can be developed for their detection. Although altogether rare, some notable exceptions for hotspot mutations and recurrent genomic alterations that impact clinical care exist. This includes ALKF1174 and ALKR1275 mutations and MYCN amplifications in primary neuroblastomas [29, 56, 63], the BRAFV600E mutation in Langerhans cell histiocytosis [3], and EWSR1 translocations in Ewing sarcoma [20], to name a few.
Impact on therapy and outcome
Liquid biopsy–based diagnostics, when optimized, can support improved MRD monitoring and early relapse detection and provide information for therapy decisions. The value of monitoring recurrence will remain limited without the availability of adequate relapse treatments, but this limitation also exists for on-going large concerted biology-driven, early phase precision medicine trials for high-risk, relapsed or refractory pediatric cancers (eSMART: NCT02813135; INFORM [77]; Ped-MATCH: NCT03155620; PRISM: NCT03336931; iCat2 from the GAIN Consortium: NCT02520713). All these studies require extensive molecular profiling of relapsed tumor samples before trial entry. Some (early phase) clinical trials for relapsed patients now include ctDNA analysis to follow the evolution of tumor genetics during targeted treatment (eSMART; MAPPYACTS: NCT02613962).
Discussion and conclusions
Liquid biopsy applications for pediatric oncology are lagging behind their adult counterpart, and studies so far have mostly been retrospective proof-of-concept studies in small cohorts. Nevertheless, these proof-of-concept studies illustrate that the technology yields substantial potential (Table 1). Pediatric tumor types, consisting mainly of highly immature and fast-growing tumor cells, might even be better suited to liquid biopsy–based genomics than many cancers arising in adult patients. Larger and especially prospective clinical trials are needed to fully explore the potential of liquid biopsy–based diagnosis, therapy response monitoring, and residual disease detection. In comparison with adult oncology, where less than 5% of patients are enrolled in a randomized controlled trial [48], the majority of children with cancers are enrolled. The first randomized controlled trials in pediatric oncology that will collect liquid biopsies to explore its potential are currently being initiated (NCT02546453, NCT03496402, NCT03336931). The data generated will hopefully elucidate whether this novel technology is complementary to traditional diagnostic procedures and demonstrate what impact they can have on clinical decision making. We expect to see many novel frontline international treatment protocols make use of liquid biopsy diagnostics in the future. Blood-based liquid biopsies might be the first step, but in due time, could be complemented or replaced by urine [10] or other fluid sources, such as saliva or CSF [46], as has developed for specific applications in the adult population.
Table 1.
Overview of proof-of-concept studies of liquid biopsies involving pediatric oncology patients
| Author | Tumor entity | Number of pediatric patients | Technique | Biomaterial | Method | Clinical application |
|---|---|---|---|---|---|---|
| Chicard [14] | Neuroblastoma | 70 | OncoScan Array (Affymetrix) | Plasma | CNA profiling | Therapeutic and risk stratification |
| Van Roy [68] | Neuroblastoma | 37 | sWGS | Plasma | CNA profiling | Therapeutic and risk stratification |
| Combaret [17] | Neuroblastoma | 114 | ddPCR | Plasma | ALK hotspot mutation | Diagnostic and therapeutic stratification |
| Combaret [18] | Neuroblastoma | 102 | PCR | Plasma, serum | MYCN amplification | Diagnostic and therapeutic stratification |
| Lodrini [41] | Neuroblastoma | 10 | ddPCR | Plasma | ALK and MYCN copy number status | Diagnosis and therapeutic stratification, Monitoring disease progression |
| Chicard [15] | Neuroblastoma | 19 | WES, ands targeted resequencing | Plasma | SNV and CNA profiling at different timepoints |
Identifying treatment-resistant clones, Longitudinal follow-up |
| Jimenez [33] | Renal tumors | 18 | WES | Plasma | Somatic SNV and CNA profiling at diagnosis | Improved molecular diagnosis |
| Ueno-Yokohata [67] | Clear cell sarcoma of the kidney | 4 | PCR | Plasma | Detection of internal tandem duplication of BCOR | Improved molecular diagnosis |
| Krumbholz [37] | Ewing sarcoma | 20 | ddPCR | Plasma | EWSR1 fusion gene detection | Therapy monitoring |
| Shukla [60] |
Ewing sarcoma, Desmoplastic small round cell tumor |
7 |
ddPCR, Targeted resequencing |
Plasma | ESWR1 fusion gene detection | Disease monitoring |
| Hayashi [30] | Ewing sarcoma | 3 | ddPCR | Plasma | EWS-ETS fusion gene detection | Therapy monitoring |
| Barris [4] | Osteosarcoma | 4 | Targeted resequencing | Plasma | Patient-specific alterations in 7 genes | Disease monitoring |
| Mussolin [49] | Hodgkin and NHL | 201 | qPCR | Plasma | Presence of cell free DNA | Improved diagnostics |
| Machado [43] | B-NHL | 30 | qPCR | Plasma | Total cell-free and EBV virus DNA quantification | Disease detection and treatment, response monitoring |
| Bruscaggin [9] | Hodgkin lymphoma | 44 | CAPP-Seq | Plasma | Genotyping of newly diagnosed and refractory HL | Disease monitoring |
| Primerano [55] | Hodgkin lymphoma | 155 | qPCR | Plasma | Cell-free DNA quantification | Disease detection and treatment Response monitoring |
| Berry [5] | Retinoblastoma | 6 | sWGS, Sanger | Vitreous fluid | CNA profiling, RB1 mutation detection | Surrogate for tumor biopsy after salvage therapy |
| Berry [6] | Retinoblastoma | 26 | sWGS | Vitreous fluid | CNA profiling | Therapy response monitoring |
| Huang [31] | Diffuse midline gliomas | 11 | Nested PCR, Sanger | CSF | Histone H3 gene mutation in CSF | Alternative or complementary to tissue diagnosis |
| Martinez-Ricarte [44] | Gliomas | 2 | Targeted sequencing, ddPCR | CSF | Detection of IDH1/2, TP53, ATRX, TERT, H3F3A, HIST1H3B gene mutations in CSF | Facilitating diagnosis of diffuse gliomas |
| Paret [53] | HGNET-BCOR | 1 | Patient-specific PCR + Sanger | Plasma | Follow-up of a BCOR internal tandem duplication | Personalized treatment and therapy monitoring |
| Klega [36] | Osteosarcoma, neuroblastoma, alveolar rhabdomyosarcoma, Wilms tumor | 45 | sWGS | Plasma | CNA and translocation characterization | Disease detection, Risk stratification, Treatment response monitoring |
| Weaver [74] | Gliomas | 10 | Methylation-specific PCR | Plasma | Promoter methylation detection | Disease monitoring |
CSF, cerebrospinal fluid; HGNET-BCOR, high-grade neuroepithelial tumor of the central nervous system with BCOR alteration; PCR, polymerase chain reaction; SNV, single-nucleotide variant; WES, whole-exome sequencing; qPCR, quantitative PCR; ddPCR, droplet digital PCR: CNA, copy number alterations; NHL, non-Hodgkin lymphoma; EBV, Epstein-Barr virus; sWGS, shallow whole-genome sequencing; SV, structural variant
For specific disease types, liquid biopsy diagnostics may be able to resolve some long-standing issues. The discussion of whether to perform surgery first, or what type of chemotherapy should be applied for renal tumors might become resolved by liquid biopsy–based genomic profiling to improve diagnostics and risk stratification [8]. Identifying a kidney tumor as a Wilms or other tumor entity (e.g., differential diagnosis between Wilms tumor and clear cell sarcoma) from genomic profiling of circulating cell-free DNA at diagnosis would eliminate the need to resort to risky tissue biopsies with the potential to cause tumor rupture. Molecular classification of renal tumors through liquid biopsy approaches might directly impact the clinical course in up to 10% of children who currently succumb to a renal tumor. Additional valuable genomic insights might be available through liquid biopsy–based analysis of diffuse intrinsic pontine gliomas, for which no effective treatment strategies currently exist and biopsy at diagnosis is limited by the risks involved. Improved biological insights could drive development of novel, effective therapies. Liquid biopsies may even have an impact in general pediatrics, for example, in the diagnostic workup of an enlarged lymph node. A well-validated and highly sensitive test to detect lymphoma-associated genetic alterations might reduce the number of lymph node biopsies necessary to diagnose lymphadenitis as opposed to lymphoma. We expect the clinical impact of liquid biopsy diagnostics to become clear in the field of pediatric oncology in the coming decade. Well-organized prospective multicenter trials will be necessary to further delineate the potential applications and clinical utility of this novel technology.
Abbreviations
- CSF
Cerebrospinal fluid
- ctDNA
Circulating or cell-free tumor DNA
- MRD
Minimal residual disease
- PCR
Polymerase chain reaction)
Authors’ contributions
RVP and RV conceptualized and wrote the review, drafted the initial manuscript, and reviewed and revised the manuscript. KDP, NVR, FS, LW, TL, GL, GS, GT, KA, and HD reviewed the manuscript for important intellectual content. BDW conceptualized and coordinated the process and reviewed and revised the manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Funding information
R.V.P is funded by a predoctoral fellowship from the Research Foundation Flanders (FWO). B.D.W has an FWO fundamental clinical fellowship. T.L. is funded by vzw Kinderkankerfonds—a non-profit childhood cancer foundation under Belgian law. G.L. was funded by the Flemish League Against Cancer. The liquid biopsy studies by H.E.D. are supported by the Berlin Institute of Health (BIH) through TERMINATE-NB (CRG04, 1.1.4.4).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Ruben Van Paemel and Roos Vlug are shared first authors
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/23/2020
In the original version of this article, a reader pointed out that there was a mistake in the phrasing in a paragraph. This could potentially be harmful to children. The authors agree to change the wording. ���vitreous fluid��� will be changed to ���aqueous humor���.
Contributor Information
Ruben Van Paemel, Email: Ruben.VanPaemel@ugent.be.
Roos Vlug, Email: Roosvlug@gmail.com.
Katleen De Preter, Email: Katleen.DePreter@ugent.be.
Nadine Van Roy, Email: Nadine.VanRoy@ugent.be.
Frank Speleman, Email: franki.speleman@ugent.be.
Leen Willems, Email: Leen.Willems@uzgent.be.
Tim Lammens, Email: Tim.Lammens@ugent.be.
Geneviève Laureys, Email: Genevieve.Laureys@ugent.be.
Gudrun Schleiermacher, Email: gudrun.schleiermacher@curie.fr.
Godelieve A. M. Tytgat, Email: G.A.M.Tytgat@prinsesmaximacentrum.nl
Kathy Astrahantseff, Email: kathy.astrahantseff@uni-due.de.
Hedwig Deubzer, Email: hedwig.deubzer@charite.de.
Bram De Wilde, Email: Bram.DeWilde@uzgent.be.
References
- 1.Allyse M et al (2015) Non-invasive prenatal testing: a review of international implementation and challenges. Int J Women's Health 113. 10.2147/IJWH.S67124 [DOI] [PMC free article] [PubMed]
- 2.Aravanis AM, Lee M, Klausner RD. Next-generation sequencing of circulating tumor DNA for early cancer detection. Cell. 2017;168:571–574. doi: 10.1016/j.cell.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 3.Badalian-Very G, Vergilio JA, Degar BA, MacConaill L, Brandner B, Calicchio ML, Kuo FC, Ligon AH, Stevenson KE, Kehoe SM, Garraway LA, Hahn WC, Meyerson M, Fleming MD, Rollins BJ. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116:1919–1923. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barris DM et al (2018) Detection of circulating tumor DNA in patients with osteosarcoma. Oncotarget 9 [DOI] [PMC free article] [PubMed]
- 5.Berry J et al (2017) Aqueous humor as a surrogate liquid tumor biopsy in retinoblastoma. ISOO 2017, Abstract 0028
- 6.Berry Jesse L., Xu Liya, Kooi Irsan, Murphree A. Linn, Prabakar Rishvanth K., Reid Mark, Stachelek Kevin, Le Bao Han A., Welter Lisa, Reiser Bibiana J., Chévez-Barrios Patricia, Jubran Rima, Lee Thomas C., Kim Jonathan W., Kuhn Peter, Cobrinik David, Hicks James. Genomic cfDNA Analysis of Aqueous Humor in Retinoblastoma Predicts Eye Salvage: The Surrogate Tumor Biopsy for Retinoblastoma. Molecular Cancer Research. 2018;16(11):1701–1712. doi: 10.1158/1541-7786.MCR-18-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatnagar S. Management of Wilms’ tumor: NWTS vs SIOP. J Indian Assoc Pediatr Surg. 2009;14:6–14. doi: 10.4103/0971-9261.54811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruscaggin A, et al. Genotyping of classical Hodgkin lymphoma on the liquid biopsy. Hematol Oncol. 2017;35:64–65. doi: 10.1002/hon.2437_51. [DOI] [Google Scholar]
- 10.Burnham P, et al. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat Commun. 2018;9:2412. doi: 10.1038/s41467-018-04745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campana D. Minimal residual disease in acute lymphoblastic leukemia. Hematology. 2010;2010:7–12. doi: 10.1182/asheducation-2010.1.7. [DOI] [PubMed] [Google Scholar]
- 12.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, Kratz A, Wefers AK, Huang K, Pajtler KW, Schweizer L, Stichel D, Olar A, Engel NW, Lindenberg K, Harter PN, Braczynski AK, Plate KH, Dohmen H, Garvalov BK, Coras R, Hölsken A, Hewer E, Bewerunge-Hudler M, Schick M, Fischer R, Beschorner R, Schittenhelm J, Staszewski O, Wani K, Varlet P, Pages M, Temming P, Lohmann D, Selt F, Witt H, Milde T, Witt O, Aronica E, Giangaspero F, Rushing E, Scheurlen W, Geisenberger C, Rodriguez FJ, Becker A, Preusser M, Haberler C, Bjerkvig R, Cryan J, Farrell M, Deckert M, Hench J, Frank S, Serrano J, Kannan K, Tsirigos A, Brück W, Hofer S, Brehmer S, Seiz-Rosenhagen M, Hänggi D, Hans V, Rozsnoki S, Hansford JR, Kohlhof P, Kristensen BW, Lechner M, Lopes B, Mawrin C, Ketter R, Kulozik A, Khatib Z, Heppner F, Koch A, Jouvet A, Keohane C, Mühleisen H, Mueller W, Pohl U, Prinz M, Benner A, Zapatka M, Gottardo NG, Driever PH, Kramm CM, Müller HL, Rutkowski S, von Hoff K, Frühwald MC, Gnekow A, Fleischhack G, Tippelt S, Calaminus G, Monoranu CM, Perry A, Jones C, Jacques TS, Radlwimmer B, Gessi M, Pietsch T, Schramm J, Schackert G, Westphal M, Reifenberger G, Wesseling P, Weller M, Collins VP, Blümcke I, Bendszus M, Debus J, Huang A, Jabado N, Northcott PA, Paulus W, Gajjar A, Robinson GW, Taylor MD, Jaunmuktane Z, Ryzhova M, Platten M, Unterberg A, Wick W, Karajannis MA, Mittelbronn M, Acker T, Hartmann C, Aldape K, Schüller U, Buslei R, Lichter P, Kool M, Herold-Mende C, Ellison DW, Hasselblatt M, Snuderl M, Brandner S, Korshunov A, von Deimling A, Pfister SM. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, et al. Hotspot mutations delineating diverse mutational signatures and biological utilities across cancer types. BMC Genomics. 2016;17:394. doi: 10.1186/s12864-016-2727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chicard M, Boyault S, Colmet Daage L, Richer W, Gentien D, Pierron G, Lapouble E, Bellini A, Clement N, Iacono I, Bréjon S, Carrere M, Reyes C, Hocking T, Bernard V, Peuchmaur M, Corradini N, Faure-Conter C, Coze C, Plantaz D, Defachelles AS, Thebaud E, Gambart M, Millot F, Valteau-Couanet D, Michon J, Puisieux A, Delattre O, Combaret V, Schleiermacher G. Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin Cancer Res. 2016;22:5564–5573. doi: 10.1158/1078-0432.CCR-16-0500. [DOI] [PubMed] [Google Scholar]
- 15.Chicard Mathieu, Colmet-Daage Leo, Clement Nathalie, Danzon Adrien, Bohec Mylène, Bernard Virginie, Baulande Sylvain, Bellini Angela, Deveau Paul, Pierron Gaëlle, Lapouble Eve, Janoueix-Lerosey Isabelle, Peuchmaur Michel, Corradini Nadège, Defachelles Anne Sophie, Valteau-Couanet Dominique, Michon Jean, Combaret Valérie, Delattre Olivier, Schleiermacher Gudrun. Whole-Exome Sequencing of Cell-Free DNA Reveals Temporo-spatial Heterogeneity and Identifies Treatment-Resistant Clones in Neuroblastoma. Clinical Cancer Research. 2017;24(4):939–949. doi: 10.1158/1078-0432.CCR-17-1586. [DOI] [PubMed] [Google Scholar]
- 16.Clementi A, Virzì GM, Brocca A, Pastori S, de Cal M, Marcante S, Granata A, Ronco C. The role of cell-free plasma DNA in critically ill patients with sepsis. Blood Purif. 2016;41:34–40. doi: 10.1159/000440975. [DOI] [PubMed] [Google Scholar]
- 17.Combaret V, et al. Circulating MYCN DNA as a tumor-specific marker in neuroblastoma patients. Cancer Res. 2002;62:3646–3648. [PubMed] [Google Scholar]
- 18.Combaret V, et al. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med. 2015;4:540–550. doi: 10.1002/cam4.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corrias MV, et al. Event-free survival of infants and toddlers enrolled in the HR-NBL-1/SIOPEN trial is associated with the level of neuroblastoma mRNAs at diagnosis. Pediatr Blood Cancer. 2018;65:e27052. doi: 10.1002/pbc.27052. [DOI] [PubMed] [Google Scholar]
- 20.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, Kiezun A, Carter SL, Shukla SA, Mehta SS, Thorner AR, de Torres C, Lavarino C, Suñol M, McKenna A, Sivachenko A, Cibulskis K, Lawrence MS, Stojanov P, Rosenberg M, Ambrogio L, Auclair D, Seepo S, Blumenstiel B, DeFelice M, Imaz-Rosshandler I, Schwarz-Cruz Y Celis A, Rivera MN, Rodriguez-Galindo C, Fleming MD, Golub TR, Getz G, Mora J, Stegmaier K. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 21.De Koker A, Van Paemel R, De Wilde B, De Preter K, Callewaert N (2019) A versatile method for circulating cell-free DNA methylome profiling by reduced representation bisulfite sequencing. bioRxiv 663195. 10.1101/663195
- 22.De Mattos-Arruda L, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Vlaminck I, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77. doi: 10.1126/scitranslmed.3007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta. 2013;424:222–230. doi: 10.1016/j.cca.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald N, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gielis EM, Ledeganck KJ, de Winter BY, del Favero J, Bosmans JL, Claas FH, Abramowicz D, Eikmans M. Cell-free DNA: an upcoming biomarker in transplantation. Am J Transplant. 2015;15:2541–2551. doi: 10.1111/ajt.13387. [DOI] [PubMed] [Google Scholar]
- 27.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, Johann PD, Balasubramanian GP, Segura-Wang M, Brabetz S, Bender S, Hutter B, Sturm D, Pfaff E, Hübschmann D, Zipprich G, Heinold M, Eils J, Lawerenz C, Erkek S, Lambo S, Waszak S, Blattmann C, Borkhardt A, Kuhlen M, Eggert A, Fulda S, Gessler M, Wegert J, Kappler R, Baumhoer D, Burdach S, Kirschner-Schwabe R, Kontny U, Kulozik AE, Lohmann D, Hettmer S, Eckert C, Bielack S, Nathrath M, Niemeyer C, Richter GH, Schulte J, Siebert R, Westermann F, Molenaar JJ, Vassal G, Witt H, ICGC PedBrain-Seq Project. ICGC MMML-Seq Project. Burkhardt B, Kratz CP, Witt O, van Tilburg C, Kramm CM, Fleischhack G, Dirksen U, Rutkowski S, Frühwald M, von Hoff K, Wolf S, Klingebiel T, Koscielniak E, Landgraf P, Koster J, Resnick AC, Zhang J, Liu Y, Zhou X, Waanders AJ, Zwijnenburg DA, Raman P, Brors B, Weber UD, Northcott PA, Pajtler KW, Kool M, Piro RM, Korbel JO, Schlesner M, Eils R, Jones DTW, Lichter P, Chavez L, Zapatka M, Pfister SM. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 29.Hartomo TB, et al. Minimal residual disease monitoring in neuroblastoma patients based on the expression of a set of real-time RT-PCR markers in tumor-initiating cells. Oncol Rep. 2013;29:1629–1636. doi: 10.3892/or.2013.2286. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi M, Chu D, Meyer CF, Llosa NJ, McCarty G, Morris CD, Levin AS, Wolinsky JP, Albert CM, Steppan DA, Park BH, Loeb DM. Highly personalized detection of minimal Ewing sarcoma disease burden from plasma tumor DNA. Cancer. 2016;122:3015–3023. doi: 10.1002/cncr.30144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang TY, et al. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol Commun. 2017;5:28. doi: 10.1186/s40478-017-0436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 33.Jiménez Irene, Chicard Mathieu, Colmet-Daage Léo, Clément Nathalie, Danzon Adrien, Lapouble Eve, Pierron Gaelle, Bohec Mylène, Baulande Sylvain, Berrebi Dominique, Fréneaux Paul, Coulomb Aurore, Galmiche-Rolland Louise, Sarnacki Sabine, Audry Georges, Philippe-Chomette Pascale, Brisse Hervé J., Doz François, Michon Jean, Delattre Olivier, Schleiermacher Gudrun. Circulating tumor DNA analysis enables molecular characterization of pediatric renal tumors at diagnosis. International Journal of Cancer. 2018;144(1):68–79. doi: 10.1002/ijc.31620. [DOI] [PubMed] [Google Scholar]
- 34.Jonkman-Berk BM, van den Berg J, ten Berge I, Bredius RG, Driessen GJ, Dalm VA, van Dissel J, van Deuren M, Ellerbroek PM, van der Flier M, van Hagen P, van Montfrans J, Rutgers A, Schölvinck EH, de Vries E, van Beem R, Kuijpers TW. Primary immunodeficiencies in the Netherlands: National patient data demonstrate the increased risk of malignancy. Clin Immunol. 2015;156:154–162. doi: 10.1016/j.clim.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Kang Q, Henry NL, Paoletti C, Jiang H, Vats P, Chinnaiyan AM, Hayes DF, Merajver SD, Rae JM, Tewari M. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem. 2016;49:1354–1360. doi: 10.1016/j.clinbiochem.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Klega K et al (2018) Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis Oncol:1–13. 10.1200/PO.17.00285 [DOI] [PMC free article] [PubMed]
- 37.Krumbholz M, et al. Genomic EWSR1 fusion sequence as highly sensitive and dynamic plasma tumor marker in Ewing sarcoma. Clin Cancer Res. 2016;22:4356–4365. doi: 10.1158/1078-0432.CCR-15-3028. [DOI] [PubMed] [Google Scholar]
- 38.Kurihara S, Ueda Y, Onitake Y, Sueda T, Ohta E, Morihara N, Hirano S, Irisuna F, Hiyama E. Circulating free DNA as non-invasive diagnostic biomarker for childhood solid tumors. J Pediatr Surg. 2015;50:2094–2097. doi: 10.1016/j.jpedsurg.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 39.Lammens CRM, et al. Regular surveillance for Li-fraumeni syndrome: advice, adherence and perceived benefits. Familial Cancer. 2010;9:647–654. doi: 10.1007/s10689-010-9368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 41.Lodrini Marco, Sprüssel Annika, Astrahantseff Kathy, Tiburtius Daniela, Konschak Robert, Lode Holger N., Fischer Matthias, Keilholz Ulrich, Eggert Angelika, Deubzer Hedwig E. Using droplet digital PCR to analyze MYCN and ALK copy number in plasma from patients with neuroblastoma. Oncotarget. 2017;8(49):85234–85251. doi: 10.18632/oncotarget.19076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 43.Machado ASC, et al. Circulating cell-free and Epstein–Barr virus DNA in pediatric B-non-Hodgkin lymphomas. Leuk Lymphoma. 2010;51:1020–1027. doi: 10.3109/10428191003746331. [DOI] [PubMed] [Google Scholar]
- 44.Martínez-Ricarte Francisco, Mayor Regina, Martínez-Sáez Elena, Rubio-Pérez Carlota, Pineda Estela, Cordero Esteban, Cicuéndez Marta, Poca Maria A., López-Bigas Nuria, Ramon y Cajal Santiago, Vieito María, Carles Joan, Tabernero Josep, Vivancos Ana, Gallego Soledad, Graus Francesc, Sahuquillo Juan, Seoane Joan. Molecular Diagnosis of Diffuse Gliomas through Sequencing of Cell-Free Circulating Tumor DNA from Cerebrospinal Fluid. Clinical Cancer Research. 2018;24(12):2812–2819. doi: 10.1158/1078-0432.CCR-17-3800. [DOI] [PubMed] [Google Scholar]
- 45.Medina Diaz I, et al. Performance of Streck cfDNA blood collection tubes for liquid biopsy testing. PLoS One. 2016;11:e0166354. doi: 10.1371/journal.pone.0166354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, Zheng Y, Skakodub A, Mehta SA, Campos C, Hsieh WY, Selcuklu SD, Ling L, Meng F, Jing X, Samoila A, Bale TA, Tsui DWY, Grommes C, Viale A, Souweidane MM, Tabar V, Brennan CW, Reiner AS, Rosenblum M, Panageas KS, DeAngelis L, Young RJ, Berger MF, Mellinghoff IK. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565:654–658. doi: 10.1038/s41586-019-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moss J, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9:5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murthy VH, Krumholz HM, Gross CP. Participation in cancer clinical trials. JAMA. 2004;291:2720. doi: 10.1001/jama.291.22.2720. [DOI] [PubMed] [Google Scholar]
- 49.Mussolin L, Burnelli R, Pillon M, Carraro E, Farruggia P, Todesco A, Mascarin M, Rosolen A. Plasma cell-free DNA in paediatric lymphomas. J Cancer. 2013;4:323–329. doi: 10.7150/jca.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nejat F, El Khashab M, Rutka JT. Initial management of childhood brain tumors: neurosurgical considerations. J Child Neurol. 2008;23:1136–1148. doi: 10.1177/0883073808321768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishi M, Miyake H, Takeda T, Hanai J, Kikuchi Y, Takasugi N. Mass screening for neuroblastoma and mortality in birth cohorts. Int J Cancer. 1997;71:552–555. doi: 10.1002/(SICI)1097-0215(19970516)71:4<552::AID-IJC8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 52.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, Reimand J, Warnatz HJ, Ryzhova M, Mack S, Ramaswamy V, Capper D, Schweizer L, Sieber L, Wittmann A, Huang Z, van Sluis P, Volckmann R, Koster J, Versteeg R, Fults D, Toledano H, Avigad S, Hoffman LM, Donson AM, Foreman N, Hewer E, Zitterbart K, Gilbert M, Armstrong TS, Gupta N, Allen JC, Karajannis MA, Zagzag D, Hasselblatt M, Kulozik AE, Witt O, Collins VP, von Hoff K, Rutkowski S, Pietsch T, Bader G, Yaspo ML, von Deimling A, Lichter P, Taylor MD, Gilbertson R, Ellison DW, Aldape K, Korshunov A, Kool M, Pfister SM. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paret C et al (2017) Personalized therapy: CNS HGNET-BCOR responsiveness to arsenic trioxide combined with radiotherapy. Oncotarget 8 [DOI] [PMC free article] [PubMed]
- 54.Poulsen MLM, Budtz-Jørgensen E, Bisgaard ML. Surveillance in von Hippel-Lindau disease (vHL) Clin Genet. 2010;77:49–59. doi: 10.1111/j.1399-0004.2009.01281.x. [DOI] [PubMed] [Google Scholar]
- 55.Primerano S, Burnelli R, Carraro E, Pillon M, Elia C, Farruggia P, Sala A, Vinti L, Buffardi S, Basso G, Mascarin M, Mussolin L. Kinetics of circulating plasma cell-free dna in paediatric classical Hodgkin lymphoma. J Cancer. 2016;7:364–366. doi: 10.7150/jca.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, Kim J, Lawrence MS, Lichenstein L, McKenna A, Pedamallu CS, Ramos AH, Shefler E, Sivachenko A, Sougnez C, Stewart C, Ally A, Birol I, Chiu R, Corbett RD, Hirst M, Jackman SD, Kamoh B, Khodabakshi AH, Krzywinski M, Lo A, Moore RA, Mungall KL, Qian J, Tam A, Thiessen N, Zhao Y, Cole KA, Diamond M, Diskin SJ, Mosse YP, Wood AC, Ji L, Sposto R, Badgett T, London WB, Moyer Y, Gastier-Foster JM, Smith MA, Guidry Auvil JM, Gerhard DS, Hogarty MD, Jones SJ, Lander ES, Gabriel SB, Getz G, Seeger RC, Khan J, Marra MA, Meyerson M, Maris JM. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schilling FH, Spix C, Berthold F, Erttmann R, Fehse N, Hero B, Klein G, Sander J, Schwarz K, Treuner J, Zorn U, Michaelis J. Neuroblastoma screening at one year of age. N Engl J Med. 2002;346:1047–1053. doi: 10.1056/NEJMoa012277. [DOI] [PubMed] [Google Scholar]
- 58.Seoane J, De Mattos-Arruda L, Le Rhun E, Bardelli A, Weller M. Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann Oncol. 2019;30:211–218. doi: 10.1093/annonc/mdy544. [DOI] [PubMed] [Google Scholar]
- 59.Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, Kis O, Zhao Z, Spreafico A, Medina TDS, Wang Y, Roulois D, Ettayebi I, Chen Z, Chow S, Murphy T, Arruda A, O'Kane GM, Liu J, Mansour M, McPherson J, O'Brien C, Leighl N, Bedard PL, Fleshner N, Liu G, Minden MD, Gallinger S, Goldenberg A, Pugh TJ, Hoffman MM, Bratman SV, Hung RJ, de Carvalho DD. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579–583. doi: 10.1038/s41586-018-0703-0. [DOI] [PubMed] [Google Scholar]
- 60.Shukla NN et al (2017) Plasma DNA-based molecular diagnosis, prognostication, and monitoring of patients with EWSR1 fusion-positive Sarcomas. JCO Precis Oncol:1–11. 10.1200/PO.16.00028 [DOI] [PMC free article] [PubMed]
- 61.Siravegna Giulia, Marsoni Silvia, Siena Salvatore, Bardelli Alberto. Integrating liquid biopsies into the management of cancer. Nature Reviews Clinical Oncology. 2017;14(9):531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 62.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, Sill M, Buchhalter I, Northcott PA, Leis I, Ryzhova M, Koelsche C, Pfaff E, Allen SJ, Balasubramanian G, Worst BC, Pajtler KW, Brabetz S, Johann PD, Sahm F, Reimand J, Mackay A, Carvalho DM, Remke M, Phillips JJ, Perry A, Cowdrey C, Drissi R, Fouladi M, Giangaspero F, Łastowska M, Grajkowska W, Scheurlen W, Pietsch T, Hagel C, Gojo J, Lötsch D, Berger W, Slavc I, Haberler C, Jouvet A, Holm S, Hofer S, Prinz M, Keohane C, Fried I, Mawrin C, Scheie D, Mobley BC, Schniederjan MJ, Santi M, Buccoliero AM, Dahiya S, Kramm CM, von Bueren A, von Hoff K, Rutkowski S, Herold-Mende C, Frühwald MC, Milde T, Hasselblatt M, Wesseling P, Rößler J, Schüller U, Ebinger M, Schittenhelm J, Frank S, Grobholz R, Vajtai I, Hans V, Schneppenheim R, Zitterbart K, Collins VP, Aronica E, Varlet P, Puget S, Dufour C, Grill J, Figarella-Branger D, Wolter M, Schuhmann MU, Shalaby T, Grotzer M, van Meter T, Monoranu CM, Felsberg J, Reifenberger G, Snuderl M, Forrester LA, Koster J, Versteeg R, Volckmann R, van Sluis P, Wolf S, Mikkelsen T, Gajjar A, Aldape K, Moore AS, Taylor MD, Jones C, Jabado N, Karajannis MA, Eils R, Schlesner M, Lichter P, von Deimling A, Pfister SM, Ellison DW, Korshunov A, Kool M. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stutterheim J, Gerritsen A, Zappeij-Kannegieter L, Yalcin B, Dee R, van Noesel M, Berthold F, Versteeg R, Caron HN, van der Schoot C, Tytgat GA. Detecting minimal residual disease in neuroblastoma: the superiority of a panel of real-time quantitative PCR markers. Clin Chem. 2009;55:1316–1326. doi: 10.1373/clinchem.2008.117945. [DOI] [PubMed] [Google Scholar]
- 64.Stutterheim J, Zappeij-Kannegieter L, Versteeg R, Caron HN, van der Schoot C, Tytgat GA. The prognostic value of fast molecular response of marrow disease in patients aged over 1year with stage 4 neuroblastoma. Eur J Cancer. 2011;47:1193–1202. doi: 10.1016/j.ejca.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 65.Tabori U, Hansford JR, Achatz MI, Kratz CP, Plon SE, Frebourg T, Brugières L. Clinical management and tumor surveillance recommendations of inherited mismatch repair deficiency in childhood. Clin Cancer Res. 2017;23:e32–e37. doi: 10.1158/1078-0432.CCR-17-0574. [DOI] [PubMed] [Google Scholar]
- 66.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ueno-Yokohata Hitomi, Okita Hajime, Nakasato Keiko, Hishiki Tomoro, Shirai Ryota, Tsujimoto Shinichi, Osumi Tomoo, Yoshimura Satoshi, Yamada Yuji, Shioda Yoko, Kiyotani Chikako, Terashima Keita, Miyazaki Osamu, Matsumoto Kimikazu, Kiyokawa Nobutaka, Yoshioka Takako, Kato Motohiro. Preoperative diagnosis of clear cell sarcoma of the kidney by detection of BCOR internal tandem duplication in circulating tumor DNA. Genes, Chromosomes and Cancer. 2018;57(10):525–529. doi: 10.1002/gcc.22648. [DOI] [PubMed] [Google Scholar]
- 68.Van Roy Nadine, Van Der Linden Malaïka, Menten Björn, Dheedene Annelies, Vandeputte Charlotte, Van Dorpe Jo, Laureys Geneviève, Renard Marleen, Sante Tom, Lammens Tim, De Wilde Bram, Speleman Frank, De Preter Katleen. Shallow Whole Genome Sequencing on Circulating Cell-Free DNA Allows Reliable Noninvasive Copy-Number Profiling in Neuroblastoma Patients. Clinical Cancer Research. 2017;23(20):6305–6314. doi: 10.1158/1078-0432.CCR-17-0675. [DOI] [PubMed] [Google Scholar]
- 69.Van Wezel EM, et al. Whole-genome sequencing identifies patient-specific DNA minimal residual disease markers in neuroblastoma. J Mol Diagn. 2015;17:43–52. doi: 10.1016/j.jmoldx.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 70.van Wezel EM, et al. Neuroblastoma messenger RNA is frequently detected in bone marrow at diagnosis of localised neuroblastoma patients. Eur J Cancer. 2016;54:149–158. doi: 10.1016/j.ejca.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 71.Viprey VF, Corrias MV, Kagedal B, Oltra S, Swerts K, Vicha A, Ladenstein R, Burchill SA. Standardisation of operating procedures for the detection of minimal disease by QRT-PCR in children with neuroblastoma: Quality assurance on behalf of SIOPEN-R-NET. Eur J Cancer. 2007;43:341–350. doi: 10.1016/j.ejca.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 72.Viprey VF, Gregory WM, Corrias MV, Tchirkov A, Swerts K, Vicha A, Dallorso S, Brock P, Luksch R, Valteau-Couanet D, Papadakis V, Laureys G, Pearson AD, Ladenstein R, Burchill SA. Neuroblastoma mRNAs predict outcome in children with stage 4 neuroblastoma: a European HR-NBL1/SIOPEN study. J Clin Oncol. 2014;32:1074–1083. doi: 10.1200/JCO.2013.53.3604. [DOI] [PubMed] [Google Scholar]
- 73.Wan Jonathan C. M., Massie Charles, Garcia-Corbacho Javier, Mouliere Florent, Brenton James D., Caldas Carlos, Pacey Simon, Baird Richard, Rosenfeld Nitzan. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nature Reviews Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 74.Weaver KD, Grossman SA, Herman JG. Methylated tumor-specific DNA as a plasma biomarker in patients with Glioma. Cancer Investig. 2006;24:35–40. doi: 10.1080/07357900500449546. [DOI] [PubMed] [Google Scholar]
- 75.Wimmer K, Kratz CP, Vasen HF, Caron O, Colas C, Entz-Werle N, Gerdes AM, Goldberg Y, Ilencikova D, Muleris M, Duval A, Lavoine N, Ruiz-Ponte C, Slavc I, Burkhardt B, Brugieres L, EU-Consortium Care for CMMRD (C4CMMRD) Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium ‘Care for CMMRD’ (C4CMMRD) J Med Genet. 2014;51:355–365. doi: 10.1136/jmedgenet-2014-102284. [DOI] [PubMed] [Google Scholar]
- 76.Woods WG, et al. Screening of infants and mortality due to neuroblastoma. N Engl J Med. 2002;346:1041–1046. doi: 10.1056/NEJMoa012387. [DOI] [PubMed] [Google Scholar]
- 77.Worst BC, van Tilburg C, Balasubramanian GP, Fiesel P, Witt R, Freitag A, Boudalil M, Previti C, Wolf S, Schmidt S, Chotewutmontri S, Bewerunge-Hudler M, Schick M, Schlesner M, Hutter B, Taylor L, Borst T, Sutter C, Bartram CR, Milde T, Pfaff E, Kulozik AE, von Stackelberg A, Meisel R, Borkhardt A, Reinhardt D, Klusmann JH, Fleischhack G, Tippelt S, Dirksen U, Jürgens H, Kramm CM, von Bueren A, Westermann F, Fischer M, Burkhardt B, Wößmann W, Nathrath M, Bielack SS, Frühwald MC, Fulda S, Klingebiel T, Koscielniak E, Schwab M, Tremmel R, Driever PH, Schulte JH, Brors B, von Deimling A, Lichter P, Eggert A, Capper D, Pfister SM, Jones DT, Witt O. Next-generation personalised medicine for high-risk paediatric cancer patients - the INFORM pilot study. Eur J Cancer. 2016;65:91–101. doi: 10.1016/j.ejca.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 78.Yáñez Y, Hervás D, Grau E, Oltra S, Pérez G, Palanca S, Bermúdez M, Márquez C, Cañete A, Castel V. TH and DCX mRNAs in peripheral blood and bone marrow predict outcome in metastatic neuroblastoma patients. J Cancer Res Clin Oncol. 2016;142:573–580. doi: 10.1007/s00432-015-2054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]