Abstract
Type VI secretion system (T6SS) is a contractile nanoweapon employed by many Proteobacteria to deliver effectors to kill or inhibit their competitors. One T6SS gene, vgrG, encodes a spike protein for effector translocation and is often present as multiple copies in bacterial genomes. Our phylogenomic analyses sampled 48 genomes across diverse Proteobacteria lineages and found ∼70% of them encode multiple VgrGs, yet only four genomes have nearly identical paralogs. Among these four, Agrobacterium tumefaciens 1D1609 has the highest vgrG redundancy. Compared to A. tumefaciens model strain C58 which harbors two vgrG genes, 1D1609 encodes four vgrG genes (i.e., vgrGa-d) with each adjacent to different putative effector genes. Thus, 1D1609 was selected to investigate the functional redundancy and specificity of multiple vgrG genes and their associated effectors. Secretion assay of single and multiple vgrG deletion mutants demonstrated that these four vgrGs are functionally redundant in mediating T6SS secretion. By analyzing various vgrG mutants, we found that all except for the divergent vgrGb could contribute to 1D1609’s antibacterial activity. Further characterizations of putative effector-immunity gene pairs revealed that vgrGa-associated gene 2 (v2a) encodes an AHH family nuclease and serves as the major antibacterial toxin. Interestingly, C58’s VgrG2 shares 99% amino acid sequence identity with 1D1609’s VgrGa, VgrGc and VgrGd. This high sequence similarity allows 1D1609 to use an exogenous VgrG delivered from C58 to kill another competing bacterium. Taken together, Agrobacterium can use highly similar VgrGs, either produced endogenously or injected from its close relatives, for T6SS-mediated interbacterial competition.
Keywords: type VI secretion system, VgrG, effector, antibacterial, functional redundancy, Agrobacterium tumefaciens
Introduction
Bacteria have evolved many survival strategies, including pathogenesis, competition, and cooperation, to thrive in diverse and changing environments. One of the weapons that they use is type VI secretion system (T6SS), a machinery that delivers effectors to both eukaryotic and prokaryotic target cells. Its coding genes are present in approximately 25% of the Gram-negative bacterial genomes sequenced (Bingle et al., 2008).
T6SS has an important role in interbacterial competition and provides T6SS-possessing bacteria with competitive advantage in microbial community. It can deliver a variety of effectors such as nuclease, amidase, phospholipase, peptidoglycan hydrolase, muramidase, glycosidase, NADase, and ADP-ribosyltransferase into the target bacterial cell by a contractile machinery (Russell et al., 2014; Tang et al., 2018; Ting et al., 2018). Contraction of the sheath-like structure drives the inner tube composed of Hcp tipped by VgrG-PAAR and the associated effectors across bacterial membranes to extracellular milieu or into the target cell. After firing, the structure is immediately disassembled into its individual components, which can be recycled to assemble new machinery for continuous firing (Zoued et al., 2014).
The spike component VgrG is homologous to the gp27/gp5 complex or the tail spike of bacteriophage T4 and assembles into a trimeric complex (Leiman et al., 2009). Current knowledge based on studies from several bacterial systems suggests that VgrG is specifically required for delivery of cognate effector(s) encoded in the same vgrG genetic module (Hachani et al., 2014; Whitney et al., 2014). Furthermore, the C-terminal variable region of VgrG is the molecular determinant conferring specificity of effector delivery by binding to its cognate effector directly or via adaptor/chaperone that interacts with a specific effector (Bondage et al., 2016; Flaugnatti et al., 2016). T6SS adaptors/chaperones including DUF4123-, DUF1795-, and DUF2169-containing proteins are required for loading a specific effector onto the cognate VgrG for delivery (Lien and Lai, 2017).
Agrobacterium tumefaciens is a soil Alphaproteobacterium that infects a broad range of dicotyledonous plants and transfers T-DNA, an oncogenic DNA fragment, to plant’s nuclear chromosomes (Gelvin, 2000; Hwang et al., 2017). A. tumefaciens strain C58 encodes a single T6SS gene cluster and is equipped with three toxins namely type VI amidase effector (Tae), type VI DNase effector 1 and 2 (Tde1 and Tde2), in which its toxin activity can be neutralized by its cognate immunity. Tde is a major antibacterial weapon during in planta interbacterial competition and its associated VgrG is specifically required for Tde1/2-dependent bacterial killing (Ma et al., 2014; Bondage et al., 2016). A gene which encodes DUF2169 is always found between vgrG2 and tde2 orthologs across many Proteobacterial classes (Bondage et al., 2016). In A. tumefaciens strain C58, a DUF2169-containing protein encoded upstream of tde2 is required to stabilize Tde2 and for Tde2-mediated antibacterial activity. For vgrG1-associated Tde1, Tap1 (encoded upstream of tde1) is the specific chaperone/adaptor interacting with Tde1 prior to loading to VgrG1 for formation of Tde1-Tap1-VgrG-PAAR secretion complex (Bondage et al., 2016).
Aside from the representative A. tumefaciens model strain C58, there is little knowledge of T6SS available for other A. tumefaciens strains. Our recent comparative analysis of T6SS gene clusters from 11 A. tumefaciens strains with complete genome sequences revealed that T6SS is present in all sequenced strains belonging to different genomospecies (Wu et al., 2019b). The imp operon (impA-N) encoding core structural or regulatory components and the first five genes (clpV, tai, tae, hcp, vgrG) encoded in the hcp operon are highly conserved but the vgrG-associated downstream genes are variable. While all strains only harbor one vgrG gene encoded in the main T6SS gene cluster, additional orphan vgrG genes not genetically linked to the main T6SS gene cluster were often identified in some strains.
A T6SS-harboring bacterium can encode one to multiple VgrG proteins, in which several of them were demonstrated to be specifically required for delivery of cognate effector(s) encoded in the same vgrG genetic module (Hachani et al., 2014; Whitney et al., 2014). However, the prevalence and biological significance of vgrG redundancy has not been tackled. In this study, A. tumefaciens 1D1609 which has the highest number of nearly identical vgrG genes among all sampled Proteobacteria lineages was chosen to address this question. We investigated the functional redundancy and specificity of multiple VgrG in the effector delivery strategies of T6SS in the context of interbacterial competition. We generated single to multiple in-frame deletion of vgrG mutants to characterize the role of paralogous VgrG proteins and its associated effectors in 1D1609. We found that the four vgrGs are functionally redundant in mediating Hcp secretion but also exert both redundancy and specificity in mediating effector delivery for interbacterial competition. We also demonstrated that 1D1609 employs a nuclease effector as the major antibacterial toxin. Importantly, we provided experimental evidence that A. tumefaciens can use T6SS in the context of interbacterial competition to exchange VgrG as an effector carrier between its close siblings.
Results
Majority of T6SS-Possessing Proteobacterial Genomes Harbor Multiple vgrG Genes
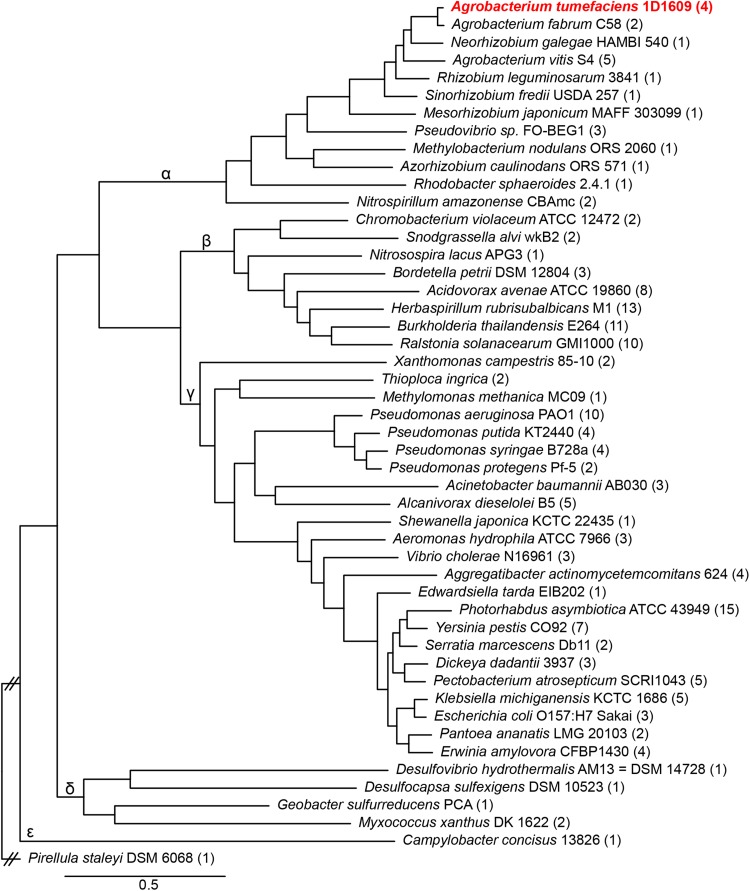
To survey the number of vgrG genes in T6SS-encoding bacterial genomes, 48 representative Proteobacteria harboring T6SS gene cluster(s) were selected for phylogenetic analysis. The result revealed that 33 of these genomes (∼70%) encode multiple vgrG genes. The vgrG copy numbers have no strong tie to the phylogenetic placement of individual genomes (Figure 1), suggesting that the copy number evolution is highly dynamic, even at the intra-genus level (e.g., Agrobacterium in Alphaproteobacteria or Pseudomonas in Gammaproteobacteria). The vgrG gene tree (Supplementary Figure S1) is not congruent with the species tree (Figure 1) and the patterns suggest that horizontal gene transfers across different classes are not rare events. Nonetheless, the species with multiple VgrG homologs mostly (∼64%) form a monophyletic clade at family levels. The genome of all the sampled Rhizobiaceae lineages harbors only one main T6SS gene cluster and forms a monophyletic clade except for Sinorhizobium fredii (Supplementary Figure S1) (Bladergroen et al., 2003; Wu et al., 2019b). This pattern suggests that gene duplication could be the major driving force for copy number increase within families.
FIGURE 1.
Bayesian phylogeny of representative Proteobacteria based on 120 shared single-copy genes. The labels on the branches indicate the five classes within this phylum: Alphaproteobacteria (α), Betaproteobacteria (β), Deltaproteobacteria (δ), Epsilonproteobacteria (ε) and Gammaproteobacteria (γ). The numbers in parentheses after strain names indicate the vgrG copy numbers.
We further asked, when there are multiple vgrG homologs in a genome, how often can we find highly similar genes that are likely to be functionally redundant. Using protein sequence identity ≥ 95% as the cut-off, only four genomes met the criteria (Supplementary Table S1). These include Alphaproteobacterium A. tumefaciens strain 1D1609 (three out of four with 99% identity, the remaining one is 93–94% identical), Betaproteobacterium Acidovorax avenae subsp. avenae ATCC 19860 (two out of eight with 99% identity), Gammaproteobacterium Aggregatibacter actinomycetemcomitans strain 624 (four copies belonging to two types: within-type = 99–100% identity, between-type = 90% identity), and Gammaproteobacterium Dickeya dadantii strain 3937 (two out of three with 97% identity, the remaining one is 78% identical). Among them 1D1609 has the highest redundancy.
1D1609 Has Four vgrG Genes With Each Genetically Linked to Specific Putative Effector Gene(s)
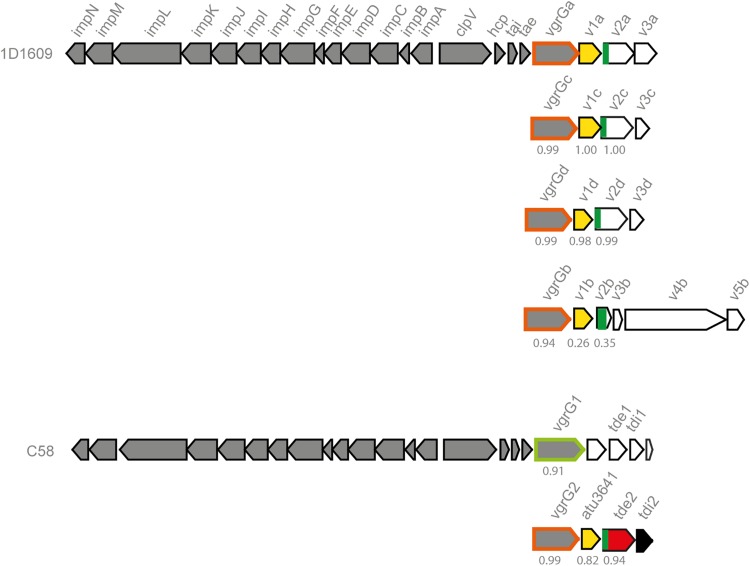
1D1609 genome encodes one main T6SS gene cluster including vgrGa (At1D1609_RS23245) that is encoded in hcp operon within the T6SS main gene cluster. The other three vgrGs – vgrGb (At1D1609_RS26290), vgrGc (At1D1609_RS22460) and vgrGd (At1D1609_RS18895) are encoded in different loci elsewhere (Figure 2). All four vgrG genes are genetically linked with distinct potential effector gene(s) and associated genes predicted to function as adaptor/chaperone or immunity (Wu et al., 2019b). The four vgrG genetic modules consist of the first three genes with conserved domain and same gene order encoding VgrG-DUF2169-DUF4150. The vgrGa-, vgrGc-, and vgrGd-associated genes (v2a, v2c, v2d) encode N-terminal PAAR-like DUF4150 domain followed by C-terminal putative effector domain. The vgrGb-linked v2b does not encode obvious effector domain and instead followed by downstream genes encoding putative effector domain, as predicted by Phyre or NCBI CDD search and BLASTP (Boratyn et al., 2013; Kelley et al., 2015; Marchler-Bauer et al., 2017) (Supplementary Table S2). The vgrGc-associated putative effector gene v2c does not encode any conserved domain, while the one (v2d) associated with vgrGd encodes a putative GH25 muramidase domain. Genes encoding DUF2169 domain are commonly found downstream of vgrG and upstream of DUF4150-containing effector genes. This gene order in vgrG genetic module is highly conserved in the vgrG2 locus of C58, in which DUF2169-containing protein (Atu3641) may function as adaptor/chaperone of Tde2 due to its requirement for Tde2 stability and Tde2-dependent bacterial killing (Bondage et al., 2016). Thus, DUF2169-containing V1 protein is likely to function as a chaperone/adaptor required for delivery of cognate effectors in conjunction with the associated VgrG.
FIGURE 2.
1D1609 encodes four vgrG genes, each is genetically linked with different potential effector gene(s). Genetic organization of A. tumefaciens 1D1609 and C58 T6SS gene cluster and vgrG genetic modules. C58 vgrG1 is outlined with green and C58 vgrG2 and 1D1609’s four vgrG genes are outlined in orange. The gene encoding DUF2169 domain is highlighted in yellow and DUF4150 (PAAR-like) domain is shown in dark green. The vgrG-associated genes (v) are numbered according to their position from vgrG. The percentage identity, which corresponds to Supplementary Table S3 is indicated below the gene cluster.
Unlike VgrG1 and VgrG2 which share 92% identity and are highly divergent at their C-terminus (Bondage et al., 2016), we found that the three VgrG proteins – VgrGa, VgrGc and VgrGd in 1D1609 share 99% overall amino acid sequence identity (and 100% identity in the C-terminal region). These are also highly similar with VgrG2 of C58 (Figure 2 and Supplementary Table S3). VgrGb shares 93–94% overall identity to the other three VgrGs and has a highly divergent C-terminal region (Supplementary Figure S2 and Supplementary Table S3). Consistent with the VgrG comparisons, the DUF2169 and DUF4150 proteins with vgrGa/c/d also share 98–100% and 99–100% amino acid sequence identity, respectively (Supplementary Table S3). In contrast, VgrGb-associated DUF2169 and DUF4150 share only 26 and 35% sequence identities to those encoded in the other three vgrG loci respectively. The four predicted effector domains fused to DUF4150 or Rhs do not share high sequence similarity to each other or to known effectors, which could equip 1D1609 with diverse effector activities to fight with a wide range of competing bacteria. Homologs of vgrGa-, vgrGb-, vgrGc-, and vgrGd-associated effector genes are widely found in species belonging to Rhizobiales, suggesting that these are common effectors in this group.
Four VgrG Proteins Are Functionally Redundant in Mediating Hcp Secretion
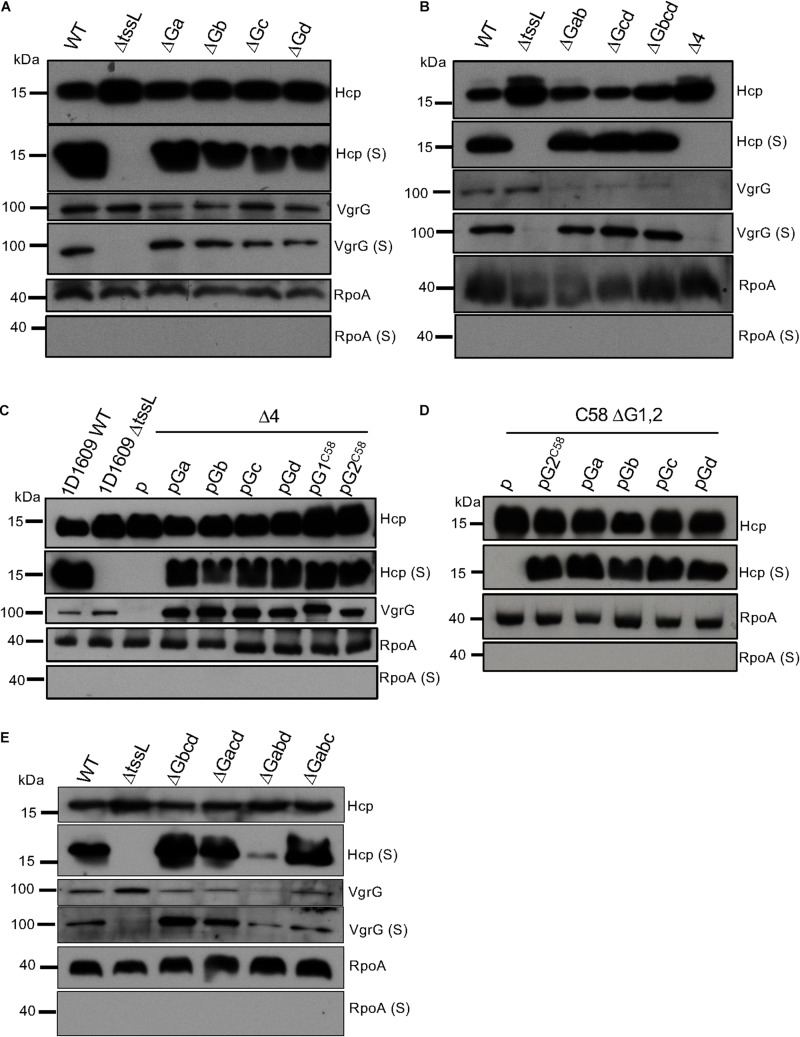
In C58, VgrG1 and VgrG2 are functionally redundant in mediating Hcp secretion but specifically required for delivery of the cognate effectors Tde1 and Tde2, respectively (Lin et al., 2013; Bondage et al., 2016). Each of in-frame deletion mutants of vgrGa, vgrGb, vgrGc and vgrGd were generated to determine their functions in mediating Hcp secretion, a hallmark of a functional T6SS assembly. The ΔtssL mutant, with deletion of gene encoding the essential T6SS membrane component TssL, is used as a negative control. The secretion assay showed that all single vgrG deletion mutants remain active in Hcp secretion, suggesting that they are functionally redundant for assembly of a functional T6SS (Figure 3A). We are able to detect secretion of VgrG proteins from 1D1609 using polyclonal antibody against VgrG1 protein of C58, which can recognize all four VgrG proteins produced in 1D1609 and VgrG2 of C58. The Hcp and VgrG proteins are secreted in a T6SS-dependent manner because no Hcp and VgrG signals are detected in the secretion fraction of ΔtssL and RpoA (RNA polymerase α-subunit), a non-secreted protein, is not detectable in any secretion fraction. Hcp secretion remains active in double and triple vgrG deletion mutants but is only completely abolished when all four vgrG genes was deleted (Figure 3B). The overexpression of each of all four vgrG on a plasmid in the quadruple vgrG mutant restores Hcp secretion (Figure 3C). Furthermore, trans complementation of each 1D1609 vgrG constitutively expressed on a plasmid can also restore Hcp secretion in C58 double vgrG deletion mutant (ΔvgrG1,2) (Figure 3D). This is consistent with a previous study that either VgrG1 or VgrG2 is sufficient in mediating the secretion of Hcp in C58 (Lin et al., 2013). Indeed, all triple vgrG mutants wherein only one VgrG is present remains capable of mediating Hcp and VgrG secretion (Figure 3E). In ΔvgrGabd mutant, wherein only VgrGc is produced, Hcp secretion level is at lesser amount as compared to WT. The transcript expression level of vgrGc is also low compared to the other vgrGs (Haryono et al., 2019). Altogether, our comprehensive secretion assays demonstrated that the four VgrGs are functionally redundant in mediating Hcp secretion. Single vgrG is sufficient for assembly of a functional T6SS although the degree of assembly efficiency may not be the same due to expression level or sequence divergence.
FIGURE 3.
The four VgrG proteins of 1D1609 are functionally redundant for Hcp secretion. Western blot analysis of cellular and secreted (S) proteins from single vgrG deletion mutants (A), multiple vgrG deletion mutants (B), overexpression of VgrG in 1D1609 quadruple vgrG mutant (Δ4) (C), and in C58 double vgrG deletion mutant (ΔG1,2) (D), and triple vgrG deletion mutants (E). Various A. tumefaciens strains were cultured in LB (A,B,E) or AK (C,E) broth at 25°C and cellular and secreted (S) fractions were subjected to western blotting using anti-Hcp, anti-VgrG, and RpoA as indicated. The T6SS inactive mutant ΔtssL is a negative control and RpoA serves as a non-secreted protein control. Protein markers (in kDa) are indicated on the left. Similar results were obtained from 2–4 independent experiments.
T6SS-Dependent Antibacterial Activity Against Escherichia coli Is Mainly Contributed by vgrGa and vgrGd
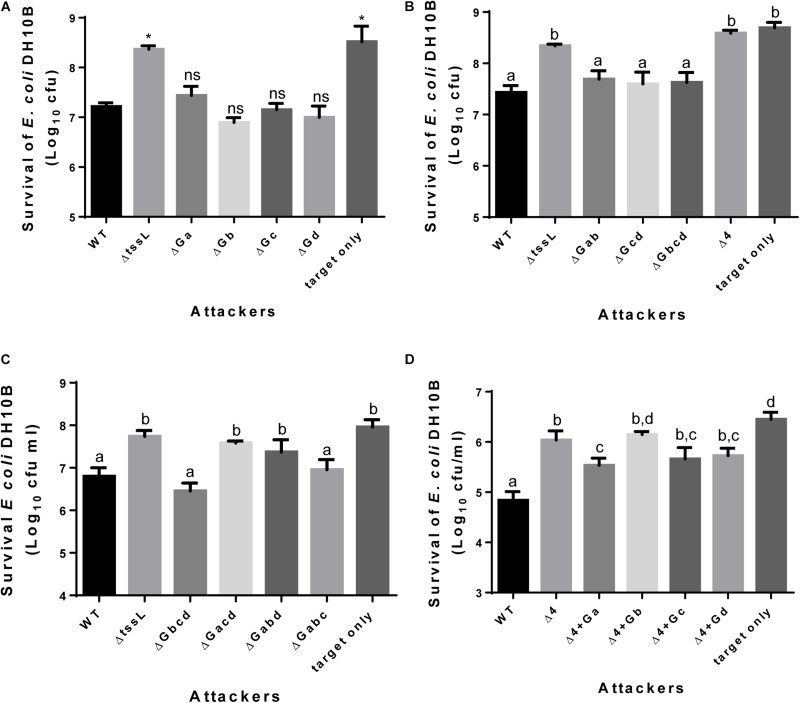
We previously showed that all tested A. tumefaciens strains including 1D1609 exhibit T6SS-mediated antibacterial activity against T6SS-negative E. coli (Wu et al., 2019b). Here, we further investigated the role of the four vgrG genes in interbacterial competition against E. coli. As a positive control, ∼10x reduction of surviving E. coli target cells was observed when co-cultured with 1D1609 WT as compared to that of ΔtssL. However, each of single vgrG deletion mutants remains to have similar antibacterial activity to WT (Figure 4A). Further antibacterial activity assay of double, triple and quadruple vgrG mutants showed that the antibacterial activity of 1D1609 is completely lost in ΔGacd, ΔGabd, or ΔGabcd, while ΔGbcd and ΔGabc triple vgrG mutants remain similar antibacterial activity to that of WT 1D1609 (Figures 4B,C). These results suggest that vgrGa and vgrGd alone is sufficient to contribute to the antibacterial activity of 1D1609. In contrast, vgrGb or vgrGc alone cannot exert any detectable antibacterial activity even though either vgrG alone (ΔGacd, ΔGabd) is sufficient in mediating Hcp and VgrG secretion (Figure 3E). However, when each single vgrG is constitutively overexpressed on a plasmid in quadruple vgrG mutant, vgrGa, vgrGc, and vgrGd but not vgrGb is able to partially restore antibacterial activity (Figure 4D). These results strongly suggest that endogenous vgrGa and vgrGd indeed play important roles for antibacterial activity while vgrGb plays no role in antibacterial activity to E. coli. The data that endogenous vgrGc plays no role but can exert its function in antibacterial activity when overexpressed are consistent with its low endogenous expression and high amino acid sequence identity to VgrGa and VgrGd.
FIGURE 4.
vgrGa and vgrGd contributes to antibacterial activity of 1D1609. Each of various A. tumefaciens 1D1609 strains (indicated in the x axis) was mixed with E. coli DH10B cells expressing pRL662 to confer gentamicin resistance at 30:1 ratio in LB (A,B) or AK (C,D) media for 18 h. The survival of E. coli cells was quantified as cfu as shown on the y axis. (A) Antibacterial activity of single vgrG deletion mutants. Data are mean ± SEM of three biological replicates, similar results were obtained from three independent experiments. Two-tailed Student’s t-test was used for statistical analyses. ∗p < 0.01. (B) Antibacterial activity in multiple vgrG deletion mutants. (C) Antibacterial activity using triple vgrG mutants. (D) Single vgrG expressed in trans using pRLBla in quadruple vgrG mutant (Δ4). Data are mean ± SEM of three biological replicates (B) or four biological replicates from two independent experiments (C,D). Different letters above the bar indicate statistically different groups of strains (P < 0.05) determined by Tukey’s HSD test, based on cfu of surviving E. coli cells.
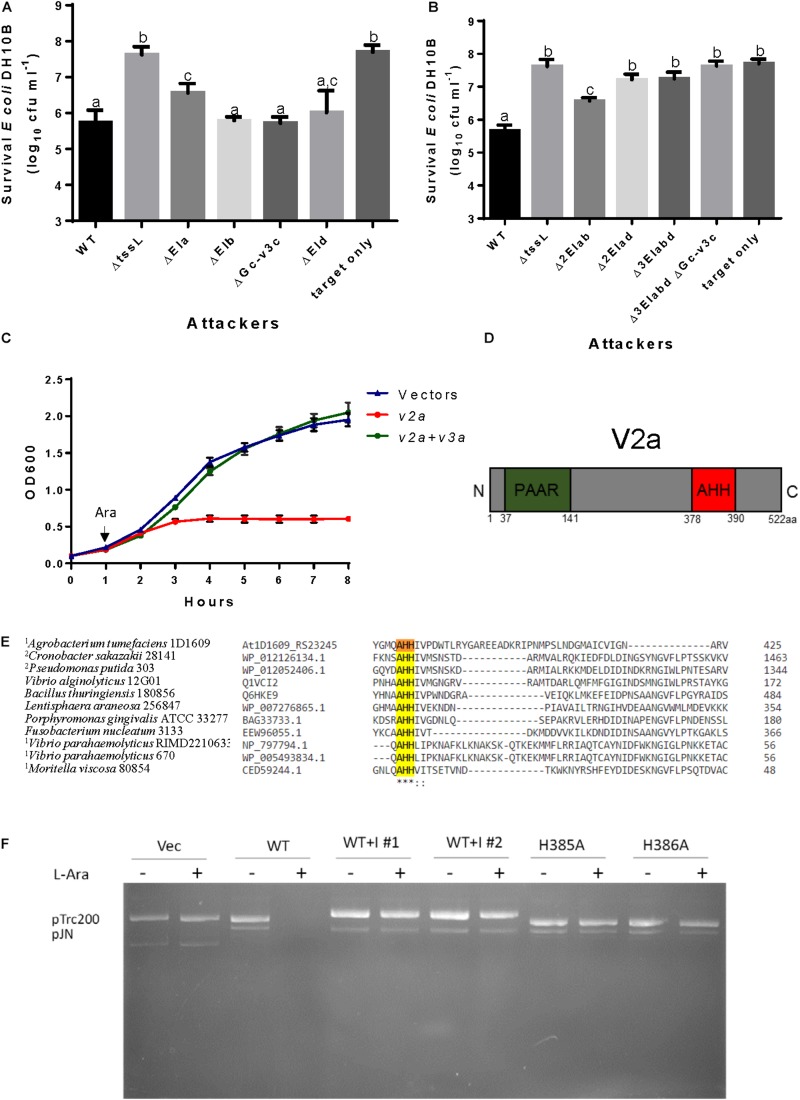
The vgrGa- and vgrGd-Associated EI Pairs Are Responsible for Antibacterial Activity of 1D1609 and V2a Is a Nuclease of HNH/ENDO VII Superfamily
We also generated deletion mutants of each putative effector-immunity (EI) gene pairs individually and mutants with deletions in multiple putative EI pairs. Due to failure in generating the vgrGc-associated v2c-v3c EI pair deletion mutant after several attempts, we instead generated the deletion of the whole vgrGc cluster to assay the antibacterial effect of vgrGc-associated effector. Deletion of vgrGa- and vgrGd-associated EI pair partially compromised antibacterial activity against E. coli while deletion of vgrGb-associated EI pair or vgrGc cluster/associated toxin has no effect in compromising 1D1609’s killing activity to E. coli, as compared to WT (Figure 5A). Importantly, any mutant minimally deleting both vgrGa- and vgrGd-associated EI pairs (Δ2EIad, Δ3EIabd, or Δ4EIabcd) completely abolished the antibacterial activity against E. coli (Figure 5B). These results suggest that vgrGa- and vgrGd-associated EI pairs contribute to antibacterial activity to E. coli.
FIGURE 5.
vgrGa- and vgrGd-associated EI pairs are responsible for antibacterial activity and V2a is a nuclease. A. tumefaciens 1D1609 strains (indicated in the x axis) was mixed with E. coli DH10B cells expressing pRL662 to confer Gentamicin resistance at 30:1 ratio in LB. The survival of E. coli cells was quantified as cfu as shown on the y axis when co-cultured with single (A) and multiple (B) EI pair deletion mutant. Data are mean ± SEM of four biological repeats from two independent experiments. Different letters above the bar indicate statistically different groups of strains (P < 0.05) determined by Tukey’s HSD test, based on cfu of surviving E. coli cells. (C) E. coli growth inhibition analysis in vgrGa-associated EI pair. E. coli cultures were induced with 1 mM IPTG at 0 h for the expression of putative immunity protein expressed on pTrc200 plasmid followed by L-arabinose (Ara) induction at 1 hr to induce putative toxin gene from pJN105 plasmid. Cell growth was recorded every hour at OD600. Empty vectors were used as controls. Data are mean ± SEM of three independent experiments. (D) Schematic diagram showing N-terminal PAAR region and C-terminal AHH nuclease domain of vgrGa-associated effector V2a. (E) Partial sequence alignment of representative nuclease family of HNH/ENDO VII superfamily with conserved AHH motif, modified from NCBI CDD sequence alignment of nuclease family of the HNH/ENDO VII superfamily with conserved AHH (pfam14412). The strain name and locus tag/accession number are on the left, and the conserved amino acid residues are indicated as ∗ (identical) and: (similar) respectively. AHH motif is highlighted in yellow with the two His targeted for mutagenesis. AHH domain linked to DUF4150 domain1 or Rhs2 is indicted. (F) Nuclease activity assay. E. coli cells with pTrc200 and pJN105 plasmid (Vec) or derivatives expressing v2a (WT), WT with immunity protein (WT + I; #1 and #2 are two independent constructs) and catalytic site mutants (H385A, H386A) were induced with (+) or without (–) L-arabinose (L-Ara) for 2 h. Plasmid DNA was extracted and the degradation pattern was observed in agarose gel.
Since single deletion of vgrGa-associated EI pair (ΔEIa) or in combination with additional EI-pair/s showed reduced or abolished antibacterial activity (Figures 5A,B), we considered vgrGa-associated effector V2a as the primary toxin and characterized its function. The putative effector gene v2a was expressed by an arabinose inducible promoter on plasmid pJN105 and its putative cognate immunity gene v3a was expressed constitutively on plasmid pTrc200 in E. coli. The results showed that the bacterial growth is inhibited when v2a is induced as compared to the vector control whereas co-expression with v3a restores the growth (Figure 5C), indicating their toxin-immunity relationship.
V2a is 522-aa in length and contains a N-terminal PAAR-like DUF4150 domain and a C-terminal nuclease domain belonging to HNH/ENDOVII superfamily (pfam14412) of the treble cleft fold (Marchler-Bauer et al., 2017) (Figure 5D). This nuclease family has a conserved motif AHH, which is a predicted toxin module found in bacterial polymorphic toxin systems. In the AHH motif, the first His forms one of the catalytic metal-chelating ligands and the second His contributes to the active site that directs the water for phosphoester hydrolysis (Zhang et al., 2011). Partial sequence alignment of representative nuclease family of HNH/ENDO VII superfamily with conserved AHH motif is shown in Figure 5E.
To determine its nuclease activity, we examined the degradation of the plasmid DNA extracted from E. coli cells after v2a is induced by arabinose for 2 h. We showed that both pTrc200 and pJN-derived plasmids were not detectable from v2a-containing cells induced by arabinose, in contrast to the presence of both plasmids in vector control or v2a-containing cells not treated with arabinose (Figure 5F). The plasmid degradation is indeed caused by nuclease activity of V2a because the plasmid degradation is no longer detectable when the cells expressing the catalytic site (H385A and H386A) mutant or co-expression of v3a (Figure 5F). Taken together, we demonstrated that V2a is a T6SS effector that requires its AHH motif for nuclease activity. Moreover, this toxicity can be specifically neutralized by its cognate immunity protein. While this study was under way, a type VI nuclease effector Tse7 with GHH catalytic site (Tox-GHH2, pfam14412) from the same HNH/ENDO VII superfamily was reported in P. aeruginosa (Pissaridou et al., 2018). In conclusion, HNH nuclease superfamily appeared to be a widespread T6SS nuclease toxin family for antibacterial activity. These results strongly suggested the v2a-v3a toxin-immunity relationship, which supported their contribution to 1D1609’s antibacterial activity to E. coli.
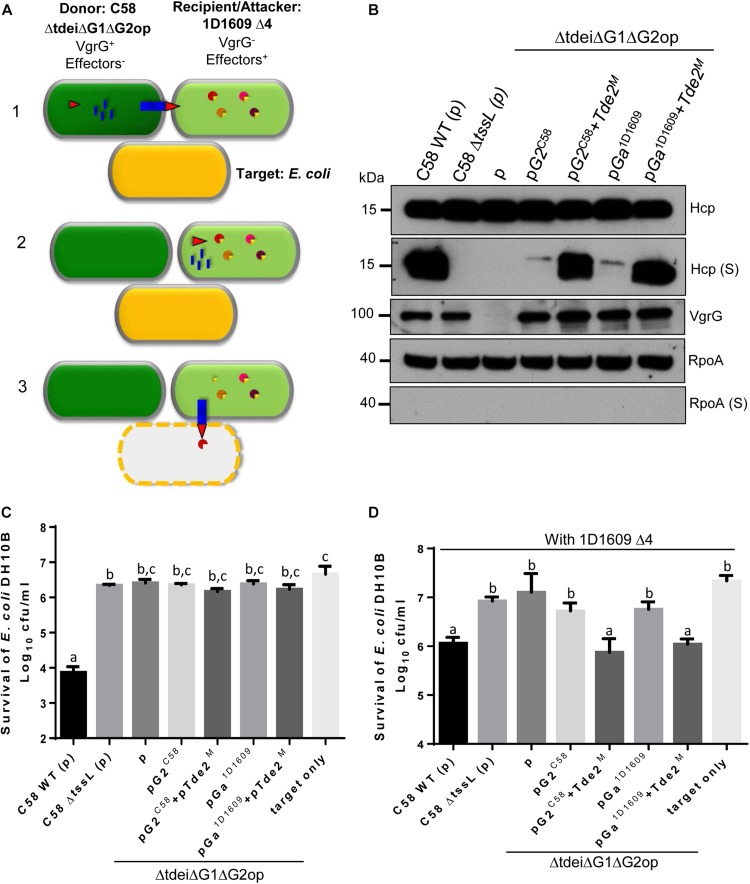
VgrG Can Be Exchanged Between C58 and 1D1609
Considering that VgrGa, VgrGc and VgrGd of 1D1609 and VgrG2 of C58 are highly similar with identical C-terminal region and their DUF2169 and DUF4150 share high sequence similarity (Supplementary Table S3), it is plausible that these VgrG proteins can substitute each other’s role in effector delivery. Previous work showed that Hcp and VgrG can be exchanged between Vibrio cholerae cells in a T6SS-dependent manner and can be reused to assemble a new T6SS (Vettiger and Basler, 2016). Thus, we hypothesized that Agrobacterium may take advantage of having multiple, highly similar VgrG to receive or donate this effector carrier from/to its siblings and the donated VgrG can be used by the recipient cell to kill another competing bacterial cell. To test this hypothesis, we designed a tripartite interbacterial transfer system that will allow us to see the killing of the target E. coli cell only when there is a functionally exchangeable VgrG protein translocated from C58 to 1D1609. The nearly identical VgrGs (G2C58 and Ga1D1609) expressed on a non-transferable pRL662 plasmid in C58 Δtdei1ΔG1ΔG2op, a mutant deleting vgrG1, tde1-tdi1, vgrG2 operon, serves as a donor cell. 1D1609 Δ4 with deletion of all four vgrG genes but is still armed with effectors is designated as a recipient cell as well as an attacker to kill E. coli (Figure 6A). Since no antibacterial activity can be detected in C58 Δtdei1ΔG1ΔG2op (Ma et al., 2014) and 1D1609 Δ4 (Figure 4B), 1D1609 Δ4 can only exhibit antibacterial activity by obtaining VgrG proteins from C58 donor and using it for effector delivery to kill E. coli.
FIGURE 6.
VgrG can be exchanged between C58 and 1D1609 for executing antibacterial activity. (A) Schematic diagram for tripartite interaction assay to determine the VgrG exchange/killing of E. coli by the recipient Agrobacterium. The donor C58 ΔtdeiΔG1ΔG2op was co-cultured with recipient 1D1609 Δ4 and E. coli target. The “exchangeable” VgrGs (G2C58 or Ga1D1609) was expressed alone or co-expressed with Atu3641 and Tde2 with catalytic site mutation (named as Tde2M) in donor C58 cells. The exchange/killing is divided in three steps: (1) Donor C58 expresses T6SS components without active toxin effectors and recipient 1D1609 expresses effectors complexed with cognate immunity proteins (EI pairs shown in circle with triangle) without T6SS assembly. T6SS is assembled in donor C58 for injecting Hcp (blue blocks) and VgrG (red triangle) into recipient 1D1609. (2) Recipient 1D1609 uses the translocated VgrG to assemble a new T6SS machine carrying 1D1609 effector. (3) Recipient 1D1609 injects the toxin effector and kills the target E. coli. (B) Hcp secretion in the donor C58. A. tumefaciens strain C58 or derivatives containing the plasmid only (p) or expressing the indicated genes were cultured in ABMES (pH 5.5) broth at 25°C and cellular and secreted (S) fractions were subjected to immunoblotting using anti-Hcp, anti-VgrG, and RpoA. The T6SS inactive mutant ΔtssL is a negative control and RpoA serves as a non-secreted protein control. (C,D) A. tumefaciens 1D1609 strains (indicated in the x axis) was mixed with E. coli DH10B cells expressing pRLBla to confer ampicillin resistance at 30:1 ratio in AK medium. The survival of E. coli cells was quantified as cfu as shown on the y axis when co-cultured with donor cells only (C) or with donor and recipient 1D1609 cells, at 1:10 ratio (D). Data are mean ± SEM of four biological repeats from three independent experiments. Different letters above the bar indicate statistically different groups of strains (P < 0.05) determined by Tukey’s HSD test, based on cfu of surviving E. coli cells.
During this course of study, our group also discovered that Tde effector loading onto VgrG is critical for T6SS assembly and Hcp secretion (Wu et al., 2019a). Thus, aside from expressing G2C58 or Ga1D1609 in C58 ΔtdeiΔG1ΔG2op donor strain, a plasmid expressing Tde2 with catalytic site mutation and its cognate DUF2169 chaperone/adaptor (pTde2M) was also included to ensure effective T6SS assembly and firing. Hcp secretion and antibacterial activity assays confirmed that the G2C58 or Ga1D1609 in C58 ΔtdeiΔG1ΔG2op donor strain remained active in Hcp secretion as WT level but completely lost the antibacterial activity against E. coli (Figures 6B,C). However, the donor-recipient-prey tripartite co-incubation showed that only the experimental set with C58 ΔtdeiΔG1ΔG2op donor strain harboring G2C58 or Ga1D1609 in the presence of pTde2M can allow 1D1609 Δ4 to exhibit antibacterial activity to E. coli (Figure 6D). 1D1609 Δ4 co-cultured with donor strains C58 ΔtssL, ΔtdeiΔG1ΔG2op harboring vector only, G2C58 or Ga1D1609 alone do not exhibit detectable killing activity to E. coli. These results suggest that 1D1609 can use the VgrG2C58 and VgrGa1D1609 delivered from C58 to kill E. coli and VgrG2C58 can function as an effector carrier in 1D1609.
Discussion
Functional redundancy is a useful strategy for the pathogen to build a robust system (Galán, 2009). In this study, our phylogenomic analyses revealed that the majority of T6SS-harboring Proteobacteria encode multiple vgrG genes. Among them, A. tumefaciens strain 1D1609 is unique for having multiple VgrG paralogs (VgrGa, VgrGc, and VgrGd) with high sequence identity (99%). The three vgrG genetic modules have the same gene orders encoding highly conserved VgrG-DUF2169-DUF4150. However the DUF4150-linked effectors do not share significant sequence similarities suggesting that these effectors may have distinct functions. This study provided genetic evidence that these nearly identical VgrG paralogs can carry not only their genetically linked cognate effector but also non-genetically linked effectors sharing the highly similar DUF4150. The functional replacement or exchange can occur for these proteins produced in the same bacterial cells or those taking from its isogenic sibling or close relatives during T6SS attacks. Thus, such functional redundancy may be a beneficial strategy for agrobacteria to compete in their ecological niche.
While several studies clearly demonstrated that VgrG is specifically required for delivery of cognate effector(s) encoded in the same vgrG genetic module (Hachani et al., 2014; Whitney et al., 2014) and C-terminal variable region of VgrG is the molecular determinant (Bondage et al., 2016; Flaugnatti et al., 2016), only few studies reveal the roles of highly homologous VgrG proteins in effector delivery. D. dadantii 3937 also encodes two highly similar VgrG homologs (VgrGA and VgrGB, Supplementary Table S1) which are both genetically linked to genes encoding homologous DUF1795-containing EagR chaperone and Rhs-linked effectors RhsA and RhsB (Koskiniemi et al., 2013). The homologous DUF1795 shares 97% amino acid sequence identity and RhsA and RhsB share 86% amino acid sequence identity with conserved N-terminal PAAR domain (99% identity). By sharing the highly similar chaperone and PAAR-Rhs regions, RhsA and RhsB effectors each harboring distinct C-terminal nuclease toxin domains could be delivered by not only its cognate but also non-cognate VgrG. Indeed, the competition assay revealed that either vgrGA or vgrGB is required for RhsB-mediated inhibition. The inhibitor cells without vgrGB in ΔrhsA/rhsB+ background can still have competitive advantage. The advantage is lost when both vgrGs are deleted and restored when vgrGB was expressed in double vgrG deletion mutant. Sharing the same VgrG carrier was also found in Serratia marcescens Db10, in which two different PAAR-domain containing Rhs effectors (RhS1 and RhS2) can functionally pair with the same VgrG protein (VgrG2) for delivery to target cells (Cianfanelli et al., 2016). The rhs2 effector gene is not genetically linked to vgrG2, but both Rhs1 and Rhs2 have the same DUF1795 (EagR)-PAAR-Rhs gene organization. The EagR1 shares only 26% amino acid sequence identity with EagR2 and each specifically interacts with its cognate Rhs and is required for delivery (Alcoforado Diniz and Coulthurst, 2015; Cianfanelli et al., 2016). VgrG2 can form VgrG2-Rhs1-EagR1 or VgrG2-Rhs2-EagR2 complexes even though the N-terminal PAAR and full-length amino acid sequence of Rhs1 and Rhs2 share only 34 and 30% identity, respectively. In this case, high amino acid sequence identity in chaperone and PAAR is not required for functional exchange. However, they may share structural similarity that is sufficient for interacting with the same VgrG.
A recent study demonstrated that T6SS components, Hcp and VgrG, can be transferred and reused by V. cholera cells (Vettiger and Basler, 2016). An isogenic LacZ+ reporter strain was used to measure the level of LacZ released for detection of cell lysis when the isogenic donor cells translocate Hcp and VgrG2 to isogenic recipient cells for T6SS assembly and firing. In our system, a tripartite interbacterial exchange/competition assay was designed to show evidence that highly similar VgrG proteins can be exchanged between two different A. tumefaciens genomospecies to kill the co-existing E. coli. Different A. tumefaciens genomospecies are known to co-exist in the same geographic area (Vogel et al., 2003). Several studies revealed that more than two genomospecies of A. tumefaciens can be found in the same crown gall or soil samples (Costechareyre et al., 2010; Bouri et al., 2016). Using tomato and maize seedlings, Gharsa et al. (2018) evaluated the competition of different A. tumefaciens genomospecies in soil and rhizosphere and concluded that related ecotypes can coexist. The initial competition alters the relative abundance, but does not eliminate the weaker strain. VgrG exchange may bring the advantages of sympatry and possessing different sets of T6SS gene clusters can maintain balance in sympatry. In V. cholerae, the bacterial strains that are incompatible (i.e., with different effector module sets), actively use their T6SS to kill susceptible strain during intra-species competition (Unterweger et al., 2014). However, in A. tumefaciens species, compatibility of T6SS EI pairs is not always predictive of competition outcomes (Wu et al., 2019b). C58 and 1D1609, which belongs to different genomospecies, can exhibit antibacterial activity against each other when the attacker is initiated with a higher relative abundance during an in planta interbacterial competition assay. However, C58 and 1D1609 can coexist with similar competitiveness when co-cultured at equal ratio despite their distinct EI pairs (Supplementary Figure S4). No significant difference of competitiveness (competitive index ∼1) could be observed when C58 WT is co-inoculated with equal number of 1D1609 WT or ΔtssL, or vice versa. This indicates that the two strains are proliferating equally in planta regardless of the presence or absence of a functional T6SS of Agrobacterium in different host plants. Interestingly, highly similar VgrGs are common in different A. tumefaciens genomospecies. In genomospecies 1 (G1), all (except 1D1108) share 100% identity while G8 (represented by C58 VgrG1 and two VgrGs in 1D132) share 95.7–99.8%. G7 (three VgrGs in 1D1609) and G8 (C58 VgrG2, 1D132 VgrG and three VgrGs in 1D1609) share 96.9–99.9% identity (Supplementary Figure S3). Thus, VgrG exchange might be common between different genomospecies, which may allow them to coexist and could in turn benefit A. tumefaciens sympatry in its ecological niche.
While not always evident by statistical analysis, deletion of single vgrGa (ΔvgrGa) or its associated effector-immunity (ΔEIa), but not other single vgrG or EI pair mutants, showed reduction of antibacterial activity (Figures 4A, 5A,B). The data suggest that vgrGa genetic module play the primary role in antibacterial activity of 1D1609. The vgrGa-associated effector V2a belongs to different type of nuclease toxin distinct from the Tde1 and Tde2 nuclease toxins (toxin_43 domain with HxxD catalytic motif) identified in C58 (Ma et al., 2014). The use of enzymatic toxins such as nuclease is the most prominent theme in bacterial warfare (Galán, 2009). Using bioinformatics analysis, Ma et al. (2017) found that among more than 2,500 T6SS-dependent Rhs-CTs with N-terminal PAAR in 143 bacterial species, nuclease represents the major group. Among these, the Rhs-AHH is the most dominant nuclease found in 66 bacterial species. Effectors with predicted AHH nuclease has been reported with antibacterial activity in V. parahaemolyticus VP1415 (Salomon et al., 2014) and Pectobacterium carotovorum subsp. brasiliense strain PBR 1692 (Bellieny-Rabelo et al., 2019) but no nuclease activity has been tested. In Acinetobacter baylyi, it was shown that the genomic DNA was completely degraded when Rhs2 with C-terminal AHH motif is expressed in E. coli (Fitzsimons et al., 2018). In this study, we characterize further to show that the AHH motif is indeed responsible for the DNase activity (Figure 5). The other conserved motif of HNH/ENDO VII superfamily nuclease has been reported in P. putida KT2440 Tke2 (Rhs-Tox-HNH) and Tke4 (Rhs-Tox-SHH) (Bernal et al., 2017) and functional characterization was done only in Tox-GHH2 in Tse7 in P. aeruginosa (Pissaridou et al., 2018). Among the 100 Soft-rot Enterobacteriaceae (SRE) genomes surveyed by Bellieny-Rabelo et al. (2019), about a quarter encodes AHH, suggesting that this nuclease is a common weapon deployed by both plant and animal pathogenic bacteria in interbacterial competition.
Among the four vgrG clusters encoded in 1D1609, vgrGb is distinct from the other three. Our data suggest that vgrGb and v4b encoding a putative toxin does not seem to play a role in antibacterial activity (Figures 4, 5A,B). This may be due to low expression level of this gene. Alternatively, V4b may target eukaryotic cells because it contains a nuclear localization signal and has an extensive structural similarity to the insecticidal TcdB2-TccC3 toxin of Photorhabdus luminescens and YenC2 toxin of Yersinia entomophaga (Supplementary Table S2). The activity of the remaining effectors remains unknown and is currently under investigation.
In conclusion, Agrobacterium has developed a flexible mode of T6SS effector delivery, by using highly similar VgrGs, either produced endogenously or injected from its close relatives, for T6SS assembly and firing. VgrG and PAAR are among the least abundant T6SS components (Lin et al., 2019) and limiting VgrG can result in reduced number of T6SS assemblies (Vettiger and Basler, 2016). Therefore, it is plausible that having multiple highly similar VgrGs may enhance robustness and abundance of T6SS assemblies. This flexibility could allow them to be an ally to its sister cells and an effective weapon against distantly related bacterial competitors. The knowledge gained in this study can help advance the understanding of mechanisms and physiological roles of multiple VgrG proteins and associated effectors and may provide new insights for sympatric speciation.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains, plasmids and primers used in this study are listed in Supplementary Tables S4, S5. A. tumefaciens strains were grown in 523 medium at 25°C while E. coli strains were grown in LB medium at 37°C. In-frame deletion mutants in Agrobacterium were generated using a suicide plasmid via double crossing over by electroporation or by conjugation (Ma et al., 2009). The mutants were confirmed by colony PCR and/or Southern blot analysis (Supplementary Figure S5). Site directed mutagenesis was done using overlapping PCR (Ho et al., 1989).
Phylogenetic and Sequence Analysis
All bioinformatics tools were used with the default parameters unless stated otherwise. The function of putative effectors were inferred from NCBI’s Conserved Domain Database (CDD) based on BLASTP searches (Boratyn et al., 2013; Marchler-Bauer et al., 2017) and Protein Homology/analogy Recognition Engine (Phyre2) (Kelley et al., 2015). SignalP was used to predict signal peptides (Petersen et al., 2011). PSORTb version 3.0.2 was used to predict sub-cellular localization of proteins (Yu et al., 2010).
For the phylogenomic analysis to investigate the VgrG diversity among Proteobacteria, the VgrG homologs from five representatives (i.e., A. tumefaciens 1D1609, Burkholderia thailandensis E264, Geobacter sulfurreducens PCA, Helicobacter cinaedi CCUG 18818, and P. aeruginosa PAO1) were used as queries to run BLASTP searches against the NCBI non-redundant protein database (e-value cutoff = 1e-15, max target sequences = 100,000). Spurious hits, defined as hits with the high scoring pairs (HSP) accounting for <74% of the query sequence or sequence similarity <48% within the HSP, were removed. The resulting lists from different query species were combined to remove redundant hits. The combined list was further manually curated to keep only the targets with complete genome sequences available in GenBank. Based on the taxonomy information of the targets, the list was iteratively trimmed to achieve a balance taxon sampling with ∼50 genomes to represent all major lineages within Proteobacteria. One Planctomycetes, Pirellula staleyi DSM 6068, was included as the outgroup to root the species phylogeny (Figure 1). In addition to the aforementioned data set for phylum-level (i.e., Proteobacteria) diversity, a second data set containing 11 Agrobacterium genomes (Wu et al., 2019b) was compiled to examine the genus-level VgrG diversity; Neorhizobium galegae HAMBI 540 was included as the outgroup for this second data set (Supplementary Figure S3).
After genome selection, the procedures for homologous gene identification and molecular phylogenetic analysis were based on those described in our previous studies (Lo et al., 2013; Lo and Kuo, 2017). Briefly, the homologous genes among all genomes were identified by using OrthoMCL (Li et al., 2003). The single-copy genes shared by all genomes were used for inferring the species phylogeny. Additionally, all vgrG homologs in these genomes, defined as the genes containing at least 80% of the VgrG domain (TIGR03361), were extracted for inferring the gene tree. The protein sequences were aligned using MUSCLE (Edgar, 2004) and the molecular phylogenies were inferred using MrBayes (Ronquist and Huelsenbeck, 2003). The amino acid substitution model was set to mix with gamma-distributed rate variation across sites and a proportion of invariable sites. The number of rate categories for the gamma distribution was set to four. The Markov chain Monte Carlo analysis was set to run for 1,000,000 generations and sampled every 100 generations. The first 25% of the samples were discarded as the burn-in. Furthermore, the protein sequence identity among all VgrG homologs were calculated using PROTDIST (Felsenstein, 1989). The amino acid sequence identity and similarity percentage of VgrG and VgrG-associated proteins were determined using Vector NTI Advance 11.5.4.
Secretion Assay
Secretion assay from liquid culture of A. tumefaciens grown in either LB rich medium or AB-MES minimal medium was done as previously described (Ma et al., 2009). Proteins from cellular and secreted fractions were resolved by SDS-PAGE and transferred onto a PVDF membrane by using a transfer apparatus (Bio-Rad). The membrane was probed with primary antibody against C58 VgrG1 (1:1,000), which recognizes the C58 VgrG2 and all four VgrGs of 1D1609, Hcp (1:2,500), and RpoA (1:7,500) (Lin et al., 2013), followed by incubation with horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:30,000) and visualized with the ECL system (PerkinElmer).
Interbacterial Competition Assay
Escherichia coli killing assay was performed as described previously (Bondage et al., 2016). In brief, overnight culture of A. tumefaciens and E. coli strain were adjusted to OD600 0.1 and incubated at 25°C for 4 h. A. tumefaciens and E. coli cells were mixed at a 30:1 ratio (OD 0.3:0.01) and spotted on LB agar (1.5%) plates. After 18 hr incubation at 25°C, the co-cultured cells were collected, serially diluted and plated on LB agar containing appropriate antibiotics to quantify surviving E. coli by counting colony forming unit (cfu). When enhanced killing activity is desired, an optimized growth medium, named as Agrobacterium Kill-triggering, AK medium (3 g K2HPO4, 1 g NaH2PO4, 1 g NH4Cl, 0.15 g KCl, 9.76 g MES in 900 mL ddH20, pH5.5) modified from AB-MES medium and solidified by 2% (w/v) agar, was used instead. Data are expressed as mean ± SEM (standard error of the mean) from three independent experiments. Statistics was done using one-way ANOVA and Tukey’s honestly significance difference (HSD) test1.
In planta bacterial competition assay was performed as described previously (Ma et al., 2014). A. tumefaciens strain C58 transformed with gentamicin resistance-conferring pRL662 plasmid and strain 1D1609 conferring spectinomycin resistance were mixed at 1:1 (OD 1) ratio and infiltrated into 6- to 7-week-old Nicotiana benthamiana and Medicago truncatula leaves. The competition outcome was quantified at 0 h and 24 h by counting cfu on LB plates with appropriate antibiotics. 1D1609/C58 ratio at t = 24 h was divided by 1D1609/C58 ratio at t = 0h to calculate the competitive index.
Growth Inhibition Assay
Growth inhibition assay was performed as described previously (Ma et al., 2014). In brief, overnight cultures of E. coli DH10B strain with vectors or their derivatives were adjusted to OD600 0.1. The expression of the tested immunity protein was induced by 1 mM IPTG for 1 h before L-arabinose (0.2% final concentration) was added to induce expression of the toxin.
Plasmid DNA Degradation Analysis in E. coli Cells
In vivo plasmid DNA degradation analysis was performed as described previously (Ma et al., 2014). In brief, overnight cultures of E. coli DH10B strain with empty vectors or derivatives expressing v2a and catalytic site mutant were harvested and adjusted to OD600 0.3. L-arabinose (0.2%, final concentration) was added to induce the expression and cultured for 2 h. Equal amounts of cells were used for plasmid DNA extraction and equal volume of extracted DNA was resolved in agarose gel followed by ethidium bromide staining.
Southern Blot Hybridization
Southern blot hybridization was performed as described (Sambrook, 2001). In brief, genomic DNA (gDNA) was extracted from overnight cultures of selected strains using Genomic DNA purification Kit (Promega) as per manufacturer’s instructions. About 35 μg of gDNA was digested using NEB restriction enzymes (Nsil and PvuII) and resolved in 0.8% agarose gel, 50 V, 3 h. The DNA fragments were transferred to a positively charged nylon membrane (Roche Diagnostics). Nylon membranes were cross-linked and used for hybridization with DIG-labeled probe (Roche). Hybridization was done overnight at 65°C using hybridization solution (FastHyb-Hybridization solution, BioChain). The washing, blocking and detection were done using Roche Wash and Block Buffer Set and DIG DNA Labeling and Detection Kit, according to manufacturer’s instructions (Roche). The membrane was exposed to X-ray for detection. The probe for the four vgrGs (vgrGa, vgrGb, vgrGc, and vgrGd) and putative toxin genes (v2a, v2c, v2d, and v4b) was prepared using the plasmid DNA vgrGa-pJN105, v2d-pJN105, and v4b-pJN105, respectively. The primer sets used are listed in Supplementary Table S5.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
E-ML and MS conceptualized the study. MS, S-TC, and C-JC worked on the data curation. E-ML, C-HK, C-FW, and MS worked on the methodology. C-HK, S-TC, MS, and C-JC carried out the investigation. E-ML was responsible for project administration and resources. E-ML and C-HK supervised the study. MS, C-HK, and E-ML wrote the manuscript. All authors reviewed and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Manda Yu for sharing the recipes of AK medium for enhanced T6SS killing assay and members of Lai lab for their help, encouragement, and stimulating discussion. We are grateful for Genomic Technology Core of Institute of Plant and Microbial Biology for DNA sequencing. This manuscript has been released as a Pre-Print at bioRxiv 740209; doi:10.1101/740209.
Funding. Funding for this project was provided by the Ministry of Science and Technology of Taiwan (MOST) Grant Nos. 104-2311-B-001-025-MY3 and 107-2311-B-001-019-MY3 to E-ML. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Open access publication fees will be paid by the Institute of Plant and Microbial Biology, Academia Sinica, Taiwan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03004/full#supplementary-material
Bayesian phylogeny of the VgrG homologs identified in the representative Proteobacteria genomes presented in Figure 1. The color coding is based on the class: Alphaproteobacteria (black), Betaproteobacteria (blue), Deltaproteobacteria (cyan), Epsilonproteobacteria (purple) and Gammaproteobacteria (green). Numbers on the branches indicate the support levels based on posterior probabilities, only values > 60% were shown.
Multiple sequence alignment of VgrG amino acid sequences of 1D1609 and C58. All VgrG homologs were aligned and conserved amino acid residues are shaded in yellow and variable residues are in blue or green, while the C-terminal extension of C58 VgrG1 not homologous to the other VgrGs is unshaded. The red and blue lines indicate the predicted gp27 and gp5 domains, respectively. The amino acid residue number is indicated.
VgrG tree of the 11 A. tumefaciens strains. All VgrG homologs in these 11 genomes were extracted for inferring the gene tree. The VgrG orthologs share ≥ 95% identity in amino acid sequence were highlighted. The genomospecies and strain name for each VgrG indicated by a locus tag are shown.
In planta competition experiments between C58 and 1D1609 at 1:1 ratio. A. tumefaciens strain C58 harboring gentamicin resistance-conferring pRL662 and strain 1D1609 which was selected in spectinomycin plate were mixed at 1:1 ratio and infiltrated into leaves of 6- to 7-week-old Nicotiana benthamiana (A) and Medicago truncatula (B). The competition outcome was quantified at 0 and 24 h post-infection by counting cfu on LB plates with appropriate antibiotics. Data are mean ± SEM, with each data point indicating the competitive index of 4–6 biological replicates from a total of 2–3 independent experiments. No statistical difference (P > 0.05) could be detected among different samples as determined by Tukey’s HSD test.
Southern blot analysis of mutants generated in this study. Schematic diagram showing probes used for Southern blotting, restriction enzyme cleavage sites and expected sizes. The genomic DNA is hybridized with (A) 821-bp vgrG probe with homology to all four vgrG genes (vgrGabcd) was used to confirm vgrG mutants; (B) 478-bp v2 probe with homology to v2acd was used to confirm v2a, v2c, and v2d mutant and (C) 778-bp v4b probe was used to confirm v4b mutant. Each lane contains 30 μg of genomic DNA digested with NsiI only or combined with PvuII. The expected size (in bp) is indicated on the side of the blot.
Percentage identity of VgrG within genomes with highly identical multiple VgrGs. The locus tag of each homolog is shown in rows and column and highlight shows ≥ 95% identity.
Predicted functions/domains of toxin/s associated with each vgrG based on Phyre or NCBI CDD search and BLASTP.
Amino acid sequence similarity/identity among the protein homologs encoded in the vgrG genetic modules in A. tumefaciens strains C58 and 1D1609.
Bacterial strains and plasmids.
Primers used in this study.
References
- Alcoforado Diniz J., Coulthurst S. J. (2015). Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J. Bacteriol. 197 2350–2360. 10.1128/JB.00199-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellieny-Rabelo D., Tanui C. K., Miguel N., Kwenda S., Shyntum D. Y., Moleleki L. N. (2019). Transcriptome and comparative genomics analyses reveal new functional insights on key determinants of pathogenesis and interbacterial competition in pectobacterium and Dickeya spp. Appl. Environ. Microbiol. 85: e02050–018. 10.1128/AEM.02050-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal P., Allsopp L. P., Filloux A., Llamas M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11 972–987. 10.1038/ismej.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle L. E., Bailey C. M., Pallen M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11 3–8. 10.1016/j.mib.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Bladergroen M. R., Badelt K., Spaink H. P. (2003). Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 16 53–64. 10.1094/mpmi.2003.16.1.53 [DOI] [PubMed] [Google Scholar]
- Bondage D. D., Lin J.-S., Ma L.-S., Kuo C.-H., Lai E.-M. (2016). VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor–effector complex. Proc. Natl. Acad. Sci. U.S.A. 113 E3931–E3940. 10.1073/pnas.1600428113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn G. M., Camacho C., Cooper P. S., Coulouris G., Fong A., Ma N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41 W29–W33. 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouri M., Chattaoui M., Gharsa H. B., McClean A., Kluepfel D., Nesme X., et al. (2016). Analysis of Agrobacterium populations isolated from tunisian soils: genetic structure, avirulent-virulent ratios and characterization of tumorigenic strains. J. Plant Pathol. 98 265–274. [Google Scholar]
- Cianfanelli F. R., Alcoforado Diniz J., Guo M., De Cesare V., Trost M., Coulthurst S. J. (2016). VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 12:e1005735. 10.1371/journal.ppat.1005735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costechareyre D., Rhouma A., Lavire C., Portier P., Chapulliot D., Bertolla F., et al. (2010). Rapid and efficient identification of agrobacterium species by recA Allele analysis. Microb. Ecol. 60 862–872. 10.1007/s00248-010-9685-7 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP - phylogeny inference package (Version 3.2). Cladistics 5 164–166. [Google Scholar]
- Fitzsimons T. C., Lewis J. M., Wright A., Kleifeld O., Schittenhelm R. B., Powell D., et al. (2018). Identification of novel Acinetobacter baumannii Type VI secretion system antibacterial effector and immunity pairs. Infect. Immun. 86:e00297-18. 10.1128/IAI.00297-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaugnatti N., Le T. T., Canaan S., Aschtgen M. S., Nguyen V. S., Blangy S., et al. (2016). A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol. Microbiol. 99 1099–1118. 10.1111/mmi.13292 [DOI] [PubMed] [Google Scholar]
- Galán J. E. (2009). Common themes in the design and function of bacterial effectors. Cell Host Microbe 5 571–579. 10.1016/j.chom.2009.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S. B. (2000). Agrobacterium and plant genes involved in t-dna transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 223–256. [DOI] [PubMed] [Google Scholar]
- Gharsa H. B., Bouri M., Glick B., Gannar A., Mougou Hamdane A., Rhouma A. (2018). Evaluation of the interspecific competition within Agrobacterium spp. in the soil and rhizosphere of tomato (Solanum lycopersicum) and maize (Zea mays). J. Plant Pathol. 100 505–511. 10.1007/s42161-018-0114-y [DOI] [Google Scholar]
- Hachani A., Allsopp L. P., Oduko Y., Filloux A. (2014). The VgrG proteins are “à la Carte” delivery systems for bacterial type VI effectors. J. Biol. Chem. 289 17872–17884. 10.1074/jbc.M114.563429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haryono M., Cho S.-T., Fang M.-J., Chen A.-P., Chou S.-J., Lai E.-M., et al. (2019). Differentiations in gene content and expression response to virulence induction between two Agrobacterium strains. Front. Microbiol. 10:1554 10.3389/fmicb.2019.01554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77 51–59. 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- Hwang H.-H., Yu M., Lai E.-M. (2017). Agrobacterium-mediated plant transformation: biology and applications. Arabidop. Book 15:e0186 10.1199/tab.0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi S., Lamoureux J. G., Nikolakakis K. C., t’Kint de Roodenbeke C., Kaplan M. D., Low D. A., et al. (2013). Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. U.S.A. 110 7032–7037. 10.1073/pnas.1300627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106 4154–4159. 10.1073/pnas.0813360106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Roos D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien Y. W., Lai E. M. (2017). Type VI secretion effectors: methodologies and biology. Front. Cell. Infect. Microbiol. 7:254. 10.3389/fcimb.2017.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-S., Ma L.-S., Lai E.-M. (2013). Systematic dissection of the agrobacterium type VI secretion system reveals machinery and secreted components for subcomplex formation. PLoS One 8:e67647. 10.1371/journal.pone.0067647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Lezan E., Schmidt A., Basler M. (2019). Abundance of bacterial Type VI secretion system components measured by targeted proteomics. Nat. Commun. 10:2584. 10.1038/s41467-019-10466-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.-S., Chen L.-L., Chung W.-C., Gasparich G. E., Kuo C.-H. (2013). Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genomics 14:22. 10.1186/1471-2164-14-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.-S., Kuo C.-H. (2017). Horizontal acquisition and transcriptional integration of novel genes in mosquito-associated spiroplasma. Genome Biol. Evol. 9 3246–3259. 10.1093/gbe/evx244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Sun M., Dong W., Pan Z., Lu C., Yao H. (2017). PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ. Microbiol. 19 345–360. 10.1111/1462-2920.13621 [DOI] [PubMed] [Google Scholar]
- Ma L.-S., Hachani A., Lin J.-S., Filloux A., Lai E.-M. (2014). Agrobacterium tumefaciens deploys a superfamily of Type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16 94–104. 10.1016/j.chom.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. S., Lin J. S., Lai E. M. (2009). An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191 4316–4329. 10.1128/JB.00029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J., Lu S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45 D200–D203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pissaridou P., Allsopp L. P., Wettstadt S., Howard S. A., Mavridou D. A. I., Filloux A. (2018). The Pseudomonas aeruginosaT6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proc. Natl. Acad. Sci. U.S.A. 115 12519–15524. 10.1073/pnas.1814181115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Russell A. B., Peterson S. B., Mougous J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Micro. 12 137–148. 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D., Kinch L. N., Trudgian D. C., Guo X., Klimko J. A., Grishin N. V., et al. (2014). Marker for type VI secretion system effectors. Proc. Natl. Acad. Sci. U.S.A. 111 9271–9276. 10.1073/pnas.1406110111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Tang J. Y., Bullen N. P., Ahmad S., Whitney J. C. (2018). Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system. J. Biol. Chem. 293 1504–1514. 10.1074/jbc.RA117.000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S.-Y., Bosch D. E., Mangiameli S. M., Radey M. C., Huang S., Park Y.-J., et al. (2018). Bifunctional immunity proteins protect bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell 175 1380.e–1392.e. 10.1016/j.cell.2018.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterweger D., Miyata S. T., Bachmann V., Brooks T. M., Mullins T., Kostiuk B., et al. (2014). The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat. Commun. 5:3549. 10.1038/ncomms4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettiger A., Basler M. (2016). Type VI secretion system substrates are transferred and reused among sister cells. Cell 167 99.e12–110.e12. 10.1016/j.cell.2016.08.023 [DOI] [PubMed] [Google Scholar]
- Vogel J., Normand P., Thioulouse J., Nesme X., Grundmann G. L. (2003). Relationship between spatial and genetic distance in Agrobacterium spp. in 1 cubic centimeter of soil. Appl. Environ. Microbiol. 69 1482–1487. 10.1128/aem.69.3.1482-1487.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney J. C., Beck C. M., Goo Y. A., Russell A. B., Harding B. N., De Leon J. A., et al. (2014). Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92 529–542. 10.1111/mmi.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-F., Lien Y.-W., Bondage D., Lin J.-S., Pilhofer M., Shih Y.-L., et al. (2019a). Effector loading onto the VgrG carrier activates type VI secretion system assembly. EMBO Rep. e47961 10.15252/embr.201947961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-F., Santos M. N. M., Cho S.-T., Chang H.-H., Tsai Y.-M., Smith D. A., et al. (2019b). Plant-pathogenic Agrobacterium tumefaciens strains have diverse type VI effector-immunity pairs and vary in in planta competitiveness. Mol. Plant Microbe Interact. 32 961–971. 10.1094/MPMI-01-19-0021-R [DOI] [PubMed] [Google Scholar]
- Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26 1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L. M., Aravind L. (2011). A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 39 4532–4552. 10.1093/nar/gkr036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoued A., Brunet Y. R., Durand E., Aschtgen M.-S., Logger L., Douzi B., et al. (2014). Architecture and assembly of the Type VI secretion system. Biochim. Biophys. Acta Mol. Cell Res. 1843 1664–1673. 10.1016/j.bbamcr.2014.03.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian phylogeny of the VgrG homologs identified in the representative Proteobacteria genomes presented in Figure 1. The color coding is based on the class: Alphaproteobacteria (black), Betaproteobacteria (blue), Deltaproteobacteria (cyan), Epsilonproteobacteria (purple) and Gammaproteobacteria (green). Numbers on the branches indicate the support levels based on posterior probabilities, only values > 60% were shown.
Multiple sequence alignment of VgrG amino acid sequences of 1D1609 and C58. All VgrG homologs were aligned and conserved amino acid residues are shaded in yellow and variable residues are in blue or green, while the C-terminal extension of C58 VgrG1 not homologous to the other VgrGs is unshaded. The red and blue lines indicate the predicted gp27 and gp5 domains, respectively. The amino acid residue number is indicated.
VgrG tree of the 11 A. tumefaciens strains. All VgrG homologs in these 11 genomes were extracted for inferring the gene tree. The VgrG orthologs share ≥ 95% identity in amino acid sequence were highlighted. The genomospecies and strain name for each VgrG indicated by a locus tag are shown.
In planta competition experiments between C58 and 1D1609 at 1:1 ratio. A. tumefaciens strain C58 harboring gentamicin resistance-conferring pRL662 and strain 1D1609 which was selected in spectinomycin plate were mixed at 1:1 ratio and infiltrated into leaves of 6- to 7-week-old Nicotiana benthamiana (A) and Medicago truncatula (B). The competition outcome was quantified at 0 and 24 h post-infection by counting cfu on LB plates with appropriate antibiotics. Data are mean ± SEM, with each data point indicating the competitive index of 4–6 biological replicates from a total of 2–3 independent experiments. No statistical difference (P > 0.05) could be detected among different samples as determined by Tukey’s HSD test.
Southern blot analysis of mutants generated in this study. Schematic diagram showing probes used for Southern blotting, restriction enzyme cleavage sites and expected sizes. The genomic DNA is hybridized with (A) 821-bp vgrG probe with homology to all four vgrG genes (vgrGabcd) was used to confirm vgrG mutants; (B) 478-bp v2 probe with homology to v2acd was used to confirm v2a, v2c, and v2d mutant and (C) 778-bp v4b probe was used to confirm v4b mutant. Each lane contains 30 μg of genomic DNA digested with NsiI only or combined with PvuII. The expected size (in bp) is indicated on the side of the blot.
Percentage identity of VgrG within genomes with highly identical multiple VgrGs. The locus tag of each homolog is shown in rows and column and highlight shows ≥ 95% identity.
Predicted functions/domains of toxin/s associated with each vgrG based on Phyre or NCBI CDD search and BLASTP.
Amino acid sequence similarity/identity among the protein homologs encoded in the vgrG genetic modules in A. tumefaciens strains C58 and 1D1609.
Bacterial strains and plasmids.
Primers used in this study.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.