Abstract
Schizophrenia is a complex neurodevelopmental disorder with high rate of morbidity and mortality. While the heritability rate is high, the precise etiology is still unknown. Although schizophrenia is a central nervous system disorder, studies using peripheral tissues have also been established to search for patient specific biomarkers and to increase understanding of schizophrenia etiology. Among all peripheral tissues, fibroblasts stand out as they are easy to obtain and culture. Furthermore, they keep genetic stability for long period and exhibit molecular similarities to cells from nervous system. Using a unique set of fibroblast samples from a genetically isolated population in northern Sweden, we performed whole transcriptome sequencing to compare differentially expressed genes in seven controls and nine patients. We found differential fibroblast expression between cases and controls for 48 genes, including eight genes previously implicated in schizophrenia or schizophrenia related pathways; HGF, PRRT2, EGR1, EGR3, C11orf87, TLR3, PLEKHH2 and PIK3CD. Weighted gene correlation network analysis identified three differentially co-expressed networks of genes significantly-associated with schizophrenia. All three modules were significantly suppressed in patients compared to control, with one module highly enriched in genes involved in synaptic plasticity, behavior and synaptic transmission. In conclusion, our results support the use of fibroblasts for identification of differentially expressed genes in schizophrenia and highlight dysregulation of synaptic networks as an important mechanism in schizophrenia.
Subject terms: Gene expression, Neurodevelopmental disorders
Introduction
Schizophrenia is a neurodevelopmental disorder with lifetime risk of about 1%1,2. Due to the high rate of morbidity and mortality, schizophrenia is classified as a severe psychiatric disorder2,3. It is thought to be initiated by brain development disruption triggered by genetics, or environment, or both4. The heritability of schizophrenia has been estimated to up to 80%5,6, and over the past decade genetic studies have started to elucidate the complex genetic etiology of schizophrenia. Genome-wide association studies (GWAS) and structural variation studies have given rise to numerous findings of genetic contribution7. The largest GWAS study to date resulted in more than 100 distinct associated loci, with the majority not previously identified7–9. Genomic studies have also identified rare recurrent copy number variants (CNVs) which contribute to schizophrenia at high risk10–13. Similarly, whole genome sequencing (WGS) and exome sequencing have identified enrichment of de novo mutations and rare disruptive variants in genes belonging to specific neurodevelopmental pathways7,14. One large transcriptome study together with genetic data also provided further insight for potential underlying mechanisms of three major psychiatric disorders including schizophrenia15. The genetic data thus indicate both common variants conferring low risk and rare variants of strong effect.
Although schizophrenia is considered a central nervous system disorder, studies using peripheral tissues in search for disease-associated biomarkers have also been established. Identification of biomarkers that are specific to patients and can differentiate patients from controls with high sensitivity can lead to better understanding of central- and peripheral-related features. This is of importance also at the clinical level since it can be very beneficial for diagnostics and prognostics, and also providing better treatments for these patients16–18. Among all peripheral tissues, fibroblasts stand out as they are readily available, easy to culture under controlled conditions, and also showed similar expression profiles to post mortem brain in several studies of psychiatric disorders19–21. Using non-neuronal cells as a proxy to investigate the pathophysiology of neurodevelopmental disorders has long been established. Relevant biochemical processes can be investigated easier in peripheral tissues. Many neurodevelopmental disorders are caused by dysregulation of genes and pathways expressed also in fibroblasts or even ubiquitously. As an example, defects in basic molecular mechanisms like DNA damage and repair is common across all cell types22.
Other options for studies of brain related disorders include animal models, which may shed light on brain structure and function alterations and also behavioral disturbance23–25. However, since the disorder is unique to humans not all symptoms, such as hallucinations, are reproducible in animals and not all symptoms observed in animals are fully reflected in humans26. Using brain biopsies in neuropsychiatric disorders is another option but it also comes with its own challenges. Factors such as individual lifestyle, history of drug abuse and medications, cause of death, and postmortem interval can affect the findings and make their interpretation more complicated27. Furthermore, post-mortem biopsies cannot be used for diagnostic and therapeutic approaches28.
Taking all these limitations together, the interest in using patient-derived peripheral tissues, especially human dermal fibroblasts, in psychiatric disorders has increased over the last decades. Fibroblasts are easy to obtain, culture, and store. They keep genetic stability for up to 15–20 passages29. They have molecular similarities with cells from the central nervous system28, and also express transcription factors, receptors and signaling pathways similar to neurons to a high extent30. Fibroblasts can be used to establish induced pluripotent stem cells (iPSCs), which can then be differentiated into neurons. This has successfully been used as a model system to study schizophrenia31. On the other hand, as the cells grow on an artificial surface, they differ from neurons in some aspects including lack of in vivo signals, synapse formation, neurotransmitter release, and ion channel expression28,32. All these features establish fibroblasts as a complementary cell model to post-mortem and animal studies of neuropsychiatric disorders.
A classical approach to study genetic diseases is the analysis of large pedigrees with many affected members. Collection and analysis of such pedigrees was attempted in early days of psychiatric genetic research, focusing on linkage studies33,34. However, these samples have often not been analyzed using modern genomic approaches. Increased prevalence of schizophrenia in large pedigrees or isolated regions may be due to an increased burden of common risk factors, but could also be explained by one or a few strong risk factors. Analysis of large pedigrees may provide an alternative for identification of rare risk factors or disrupted signaling pathways.
Several epidemiological studies in northern Sweden have reported high prevalence of neuropsychiatric and neurodevelopmental disorders, including schizophrenia35. Previous studies have also described isolated regions with prevalence of psychiatric disorders several times higher than in the general Swedish population36–38. One of these genetically isolated regions with an increased prevalence of schizophrenia has been studied using various approaches ranging from epidemiology to genetics over the last 60 years37. The first founders of the population were three closely related Finnish families who settled in the region in early 17th century37.
In this study, we aimed to identify genes or gene networks associated with schizophrenia in a unique set of fibroblast samples from the genetically isolated region in northern Sweden, where the population has been shown to exhibit an increased prevalence of schizophrenia37. We sequenced RNA extracted from cultured fibroblasts from 16 individuals, seven controls and nine patients to compare differentially expressed genes. Data was analyzed for differential gene expression and co-expression networks were then constructed using weighted gene correlation network analysis (WGCNA)39.
Results
Identification of differentially expressed genes
In total, 48 DEGs with adjusted p-value < 0.1 were identified in patients compared to controls (Supplementary Table S2). Among these 48 DEGs, eight genes were identified in previous studies of schizophrenia or schizophrenia related pathways (Table 1): HGF, PRRT2, EGR1, EGR3, C11orf87, TLR3, PLEKHH2 and PIK3CD (Fisher exact test, P = 0.00021).
Table 1.
Differentially-expressed genes that have overlap with other schizophrenia-related studies (Fisher exact test, p-value = 0.00021).
| Ensemble Gene ID | Gene Symbol | Log2Fold Change | P-value | adj. P-value | Main finding | Study type |
|---|---|---|---|---|---|---|
| ENSG00000019991 | HGF | 3.81 | 9.27E-07 | 0.004 | Reported upregulation in schizophrenia by CMC40 | Expression |
| ENSG00000167371 | PRRT2 | 1.38 | 3.75E-05 | 0.031 | CNVs at this gene genomic position have been associated with schizophrenia48,49 | Genetic |
| ENSG00000120738 | EGR1 | 1.36 | 6.24E-05 | 0.045 | First reported to be associated with schizophrenia by PGC8 Reported upregulation in schizophrenia51 | Genetic Expression |
| ENSG00000171608 | PIK3CD | −0.79 | 6.23E-05 | 0.045 | Evidence for link between the NRG1-ErbB4 signaling pathway and PIK3CD in schizophrenia70 | Expression |
| ENSG00000185742 | C11orf87 | 1.00 | 6.77E-05 | 0.046 | First reported to be associated with schizophrenia by PGC8 | Genetic |
| ENSG00000164342 | TLR3 | 2.34 | 0.00022 | 0.09 | Reported de novo missense mutations associated with schizophrenia62 | Genetic |
| ENSG00000152527 | PLEKHH2 | 1.79 | 0.00026 | 0.097 | Reported de novo missense mutations in schizophrenia patients67 | Genetic |
| ENSG00000179388 | EGR3 | 2.72 | 0.00025 | 0.097 | Reported SNPs associated with schizophrenia56–59 | Genetic |
CMC: CommonMind Consortium; PGC: Psychiatric Genomics Consortium.
Co-expression network analysis
To study networks of genes that were expressed together, we further analyzed our data with WGCNA. Using the R library WGCNA to construct the co-expression networks, 32 modules were detected in total. Using a p-value cut-off of <0.05 for condition (affected vs unaffected) three modules were identified as significant: module skyblue with 176 genes, module floralwhite with 79 genes, and module lightcyan with 293 genes in total (Supplementary Fig. S1). Of these three modules, MEskyblue also showed significant results for age.
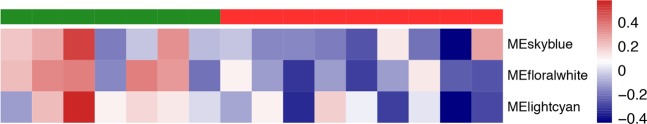
To investigate the overall expression profile of the genes in each module, the eigengene was calculated. All these three modules, MEskyblue, MEfloralwhite, and MElightcyan were generally downregulated in patient fibroblasts (red) compared to controls (green) (Fig. 1).
Figure 1.
The eigengene co-expression modules heatmap. Top legend: cases are colored in red and controls in green. Side legend: eigengene values showing the direction of co-expression in modules.
The genes identified in each module are listed in Supplementary Table S3.
Gene ontology and functional enrichment
We further performed functional enrichment analysis to investigate if co-expressed genes in these three modules were significantly enriched in certain biological pathways relevant to schizophrenia pathophysiology. Module skyblue had positive regulation of axon extension as the most significant GO category, including the genes FN1, MACF1, NDEL1 and SEMA7A. Module floralwhite was highly enriched in biological pathways including regulation of synaptic plasticity, behavior and synaptic transmission. The top four enriched biological pathways of floralwhite module and their associated genes have been illustrated in Fig. 2. Supplementary Table S4 shows the top enriched biological pathways in each module.
Figure 2.
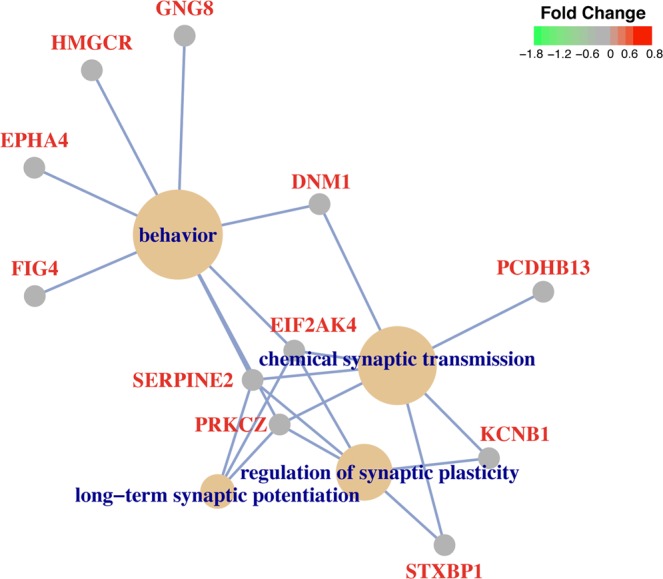
Top 4 enriched biological pathways in the floralwhite module and the genes associated with each GO category.
Discussion
In this study we used fibroblasts collected from a unique population in northern Sweden. The genetically isolated region has been the focus of studies on neuropsychiatric and neurodevelopmental disorders since more than 50 years37, due to an increased prevalence of neurodevelopmental disorders in the region. The increased genetic risk is presumed to be explained by a small founder population carrying multiple risk variants. Genetic analyses have shown that the inheritance is complex and not limited to one strong risk factor37. Over the last decades, increased mobility in and out of the previously isolated region has led to a reduction in risk for schizophrenia and developmental disorders. The fibroblasts used in the current study were initially collected in 1978 making them a unique resource for analysis of schizophrenia. A drawback of using this resource is that the number of available samples is highly limited and we recognize that the limited sample size lowers the power to detect subtle changes in expression.
In this transcriptome study, we found differential fibroblast expression between cases and controls for 48 genes, including eight genes previously implicated in schizophrenia. The genes HGF, PRRT2, EGR1, EGR3, C11orf87, TLR3 and PLEKHH2 have been reported in either genetic analysis or gene expression analysis of schizophrenia, and were all upregulated in fibroblasts from patients in our study. In addition, we found downregulation of PIK3CD, also previously associated with schizophrenia.
Hepatocyte growth factor (HGF), located within the genomic position 7q21.11, showed the largest fold change and was upregulated in cases versus controls. The upregulation of HGF in schizophrenia affected individuals was also reported in the CommonMind Consortium (CMC) study40. Hepatocyte growth factor/scatter factor (HGF/SF) is a key factor in proper migration of interneurons from ganglionic eminence (GE) to cerebral cortex41. HGF/SF displays differentiation and mitogenic activities by signaling through its receptor, MET, in neuronal42,43 and non-neuronal tissues44,45.
The expression of PRRT2, located within the genomic position 16p11.2 microduplication region, was also elevated significantly in cases versus controls. Several studies implicated the association of recurrent structural mutations in this genomic hotspot in individuals with schizophrenia. Mutations at 16p11.2 cause high risk of schizophrenia and microduplication of this region is associated with an 8–24 folds increased risk of the disorder10,12,46–50.
Early growth response 1 (EGR1) is located within the genomic position 5q31.2 and C11orf87 within 11q22.3. These loci were first reported to be associated with schizophrenia by the Schizophrenia Working Group of the Psychiatric Genomics Consortium in 20148. EGR1 was also reported as a potential biomarker to differentiate major psychoses, as its upregulation in fibroblasts and peripheral blood cells was specific to schizophrenia compared to Major Depressive Disorder (MDD) and Bipolar Disorder (BD)51. EGR1, a part of the immediate early genes (IEGs) family, is an early response gene to different growth stimuli52. It is involved in neuronal plasticity pathway though the precise underlying mechanism is still unknown53,54. Early growth response 3 (EGR3), located within genomic position 8p21.3, is another member of IEGs family and its expression is regulated downstream of neuregulin 1 (NRG1) signaling cascade in some cell types55. Single nucleotide polymorphisms (SNPs) in EGR3 in different populations of Japanese, Korean, Han Chinese, and European origin have been previously reported in association with schizophrenia56–59. In our study, we observed upregulation of EGR3 in schizophrenia patients compared to controls.
The C11orf87, also known as neuronal integral membrane protein 1 (NEURIM1) is primarily expressed in brain tissue. It is the only gene present within a locus (chr11:109285471-109610071) identified to harbor variation associated with schizophrenia in several genome-wide association studies8,60 in different populations, as well as association with self-reported educational attainment61.
One study in 2014, using exome sequencing of 623 schizophrenia trios, implicated Toll-like receptor 3 (TLR3) de novo missense mutation associated with schizophrenia62. TLR3 is located within the genomic position 4q35.1. Viral infection during pregnancy is associated with increased risk of neuropsychiatric disorders including schizophrenia in offspring63,64. Exposing pregnant mice to a synthetic virus led to inhibited cortical neurogenesis and behavioral abnormalities in offspring in a TLR3 dependent manner and through activation of innate immunity65. Another study using cultured neurons and mouse brain showed that TLR3 suppresses disrupted in schizophrenia 1 (Disc1) expression and consequently neuronal development66.
In an exome sequencing study of 42 sporadic and 15 familial schizophrenia trios, de novo missense SNV of PLEKHH2, located at genomic position 2p21, in an individual diagnosed with schizophrenia was ranked in the top 15% of probable haploinsufficient genes among candidate mutations based on functional impact67.
We also identified downregulation of phosphatidylinositol 3-kinase catalytic delta (PIK3CD), located at genomic position 1p36.22, among DEGs in patients versus controls. The signaling pathway of NRG1 and its receptor ErbB4 plays an important role in neural development and synaptic plasticity68 and has been associated with schizophrenia before69. One study in 2012, provides evidence for link between the NRG1-ErbB4 signaling pathway and PIK3CD in schizophrenia. They also describe PIK3CD as a potential therapeutic target for psychiatric disorders. Using a mouse model in their study to inhibit PIK3CD, they could block the effect of amphetamine in the animal and reverse the schizophrenia-related behaviors70.
We also applied WGCNA to identify differentially co-expressed networks of genes. As a result, differential expression in three modules were detected as significantly associated with schizophrenia. All three modules were significantly suppressed in patients compared to control, and one module (floralwhite) was highly enriched of genes involved in synaptic plasticity, behavior, synaptic transmission, GABAergic pathways. All these pathways and their underlying mechanisms have been implicated in schizophrenia before62. The critical role of synaptic plasticity in learning, memory, and also formation of mature neural circuits has made some scientists to think of synaptic plasticity as a key pathogenic feature in schizophrenia71. Post-mortem72,73 and large-scale genetic studies13,74 have constantly shown synaptic plasticity impairment in schizophrenia patients compared to controls. Additionally, cognitive functions including working memory are mediated by dorsolateral prefrontal cortex and, as a core feature of schizophrenia, have been impaired in patients75,76. This impairment is partly due to abnormalities in GABA-mediated circuitry77,78. A recent genetic analysis of another large pedigree from Northern Sweden showed that the members had an increased risk for schizophrenia based on polygenic risk scores, but did not carry rare variants contributing significant increase in risk38. Our results support a similar etiology in the present pedigree, where we find general support for modules with genes previously implicated in schizophrenia by both genetic and expression studies, but no single genes that stand out as rare and strong contributors to risk.
Our results support the use of fibroblast cell lines as an important resource for studies of schizophrenia. Although fibroblasts can never represent the diversity and complexity of brain tissue, many pathways of importance in neuropsychiatric disorders are expressed. We show that relevant pathways are detected also in this limited sample size. Previous studies of fibroblasts from schizophrenia patients and controls have shown mixed results, with one study found several differentially expressed genes (including EGR1 mentioned above) and two other studies finding no differentially expressed genes79,80. It is possible that the genetically isolated population from where these samples were collected represents a less heterogeneous sample compared to a general European population sample, and this homogeneity may have increased our ability to detect relevant gene expression differences between patient and control samples.
The cell lines used in this study were collected in 1978, and this has to be taken into consideration as a potential limitation to the study. According to published literatures the actual time of storage in liquid nitrogen has very limited impact, while number of passages and freeze/thaw cycles play a larger role. In one study where blood samples were stored at −80 °C and in liquid nitrogen separately and each for the maximum of 19 years, there was no systematic effect of the storage time on RNA quality81. This finding was supported by another study where RNA extracted from endocrine tissue stored at −80 °C for 27 years has been assessed based on RNA integrity number (RIN)82. In 2019, the Ontario Tumour Bank also published a report of tissue quality control over 11 years of storage in liquid nitrogen. Their results show no time-dependent decline in sample quality, and they concluded that extended periods of storage at cryogenic temperature is suitable for sample preservation in biobanks83. The fibroblasts used here have all gone through a very limited number of passages and have been stored under optimal conditions. All samples were collected in the same year and there has been no difference in sample handling between patient and control cell lines. We note that fibroblasts are also a valuable resource as they may differentiate into neuronal cells, either directly or via de-differentiation into stem cells. This would represent an interesting avenue forward for these cell lines in order to understand the role of the differentially expressed genes in an even more relevant cell model.
In conclusion, we performed transcriptome study of fibroblasts from patients and controls collected in a region with increased prevalence of schizophrenia. Our results support the findings of several genes previously implicated in schizophrenia and highlight dysregulation of synaptic networks as an important mechanism in schizophrenia. The results further lend support to the collection and use of fibroblasts as an important resource for studies of neuropsychiatric disorders.
Materials and Methods
Clinical materials
The donors of the fibroblast samples included in this study belong to three very large, interrelated pedigrees originated from Finnish ancestors37. All patients fulfilled the criteria for schizophrenia according to DSM-III-R, as well as according to84 and85. Healthy individuals included in this study have been followed up to age 65 and beyond for evaluation of psychiatric health problems. Skin biopsies were collected for establishment of primary fibroblast cultures in 1978. Doctor Jan Arvid Böök, performed the skin biopsies in the patient’s home. The patients and their healthy control parents and siblings were generally examined on the same day. A limited portion of the skin on the person’s forearm was first anesthetized. A punch blade, which was attached to a pencil-like handle, was rotated through the skin to the subcutaneous fat layer, giving a cylindrical tissue core. Minimal bleeding was noted and the area was left to heal without any complications. The skin samples were immediately sent from North Sweden to Berta Santesson at Uppsala University for culturing of fibroblasts. Divided portions of the fibroblast cultures were frozen on liquid nitrogen over the years until a small portion of the specific samples were thawed for the present study.
In this study, fibroblast cultures from 16 individuals including seven healthy and nine schizophrenics were analyzed. Supplementary Table S1 shows the demographic data of all 16 individuals. All samples were collected with informed consent and in accordance with the Declaration of Helsinki. The study has been approved by the Regional Ethical Review Board in Uppsala with approval dnr 2014/263. All research was performed in accordance with relevant regulations.
Sample preparation
Fibroblast cells were maintained in high glucose DMEM medium (Sigma) supplemented with 10% fetal bovine serum (FBS), 1% MEM non-essential amino acids, 2 mM of L-glutamine and 1× penicillin/streptomycin (Sigma) under optimal conditions, 37 °C and 5% CO2. When they became confluent, the cells were split to larger culture flasks. All fibroblasts were cultured just one more passage after thaw and were homogenized in trizol (TRI) for RNA extraction afterward. Total RNA was extracted from homogenized cells using Ribopure kit (Ambion) and treated with TURBO DNase (Ambion) according to the manufacturer’s instructions.
RNA sequencing
Sequencing libraries for 16 individual samples were prepared from 180–280 ng total RNA using the TruSeq stranded mRNA library preparation kit (Cat# RS-122- 2101/2102, Illumina Inc.) including polyA selection according to the manufacturer’s protocol. Sequencing was then performed on HiSeq. 2500 machine by the SNP&SEQ Technology Platform in Uppsala and data from three lanes of sequencing was uploaded on Uppmax.
Reads mapping and differential expression analysis
RNAseq data containing adapter sequences and low-quality reads were filtered out with Trimmomatic 0.3286. Filtered reads were mapped to human reference genome hg19 with STAR 2.5.1b software87, with an average of 29 million uniquely mapped reads per sample. Genes annotated in ENSEMBL annotation v75 were quantified with python framework HTSeq. 0.6.188. Reads mapping to multiple positions in the genome were not counted. Next, reads mapping to technical replicates were summed gene wise. After normalization, genes with <5 reads in 40% of the samples were removed. DESeq. 2 v1.16.1 was used for differential expression analysis89. After differential expression analysis, the resulting p-values were adjusted for multiple testing using the Benjamini–Hochberg procedure.
Co-expression network analysis
Co-expression networks were constructed using the R library WGCNA39. Briefly, the analysis was performed using same samples and number of genes that were used for differential expression analysis. Following this the DESeq. 2 implementation of variance stabilizing transform was used on the read counts for each gene. To find the correct power to use when constructing the network, soft thresholding powers for one to 20 were calculated using a signed network. As RNA sequencing data are likely to contain outliers the bi-weight mid-correlation was used90. After reviewing the resulting scale free topology index for the powers one to 20, a power of four was selected as this power gave the best tradeoff between network free topology and connectivity. A signed hybrid co-expression network was then calculated using standard settings. Modules were identified from the network and the eigenvectors of each module were correlated with the variables age, passage, sex, and condition (patient or control).
Gene ontology enrichment analysis
GO enrichment was performed on the genes with significant differential expression using an R package GOseq, controlling for gene length biases91. As background for the GO analysis all genes where DESeq2 performed a test for differential expression were used. Functional annotation enrichment was also performed for the genes resulting from WGCNA analysis. Overlaps with different gene sets for each significant module was assessed using Fisher’s exact test. The genes in each module were used to define a gene set, and each such gene set was tested for overlap with the gene set of DEGs in our fibroblast study, 693 DEGs in schizophrenia from the CommonMind Consortium (CMC)40, and curated genetic associations with schizophrenia8,48,49,62,67,92,93. Briefly, the genetic associations with schizophrenia were derived from: 108 discovered in a common variant GWAS8, 12 CNVs49, 756 nonsynonymous de novo mutations and 114 rare variants with loss of function, collated from several studies48,62,67,92,93, and are available at CMC knowledge portal (http://commonmind.org). A literature review of the differentially expressed genes was also performed to investigate their link to schizophrenia in previous studies.
Supplementary information
Acknowledgements
Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. Computational analyses were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX). The research was funded by the European Research Council ERC Starting Grant Agreement n. 282330 (to LF) and the Swedish Medical Research Council Grant 2017-01861 (to LF). Open access funding provided by Uppsala University.
Author contributions
Authors M.E., A.N. and L.F. had full access to all study data and take responsibility for the integrity of the data. Author L.W. performed the samples collection. Author M.E. performed experimental work. Author A.N. performed the data analysis and prepared figures. Authors M.E., A.N. and L.F. drafted the manuscript. All authors approved the final version of the paper for submission.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57467-z.
References
- 1.Rund BR. The research evidence for schizophrenia as a neurodevelopmental disorder. Scand. J. Psychol. 2018;59:49–58. doi: 10.1111/sjop.12414. [DOI] [PubMed] [Google Scholar]
- 2.Kahn RS, et al. Schizophrenia. Nat. Rev. Dis. Prim. 2015;1:15067. doi: 10.1038/nrdp.2015.67. [DOI] [PubMed] [Google Scholar]
- 3.Brennand KJ, Simone A, Tran N, Gage FH. Modeling psychiatric disorders at the cellular and network levels. Mol. Psychiatry. 2012;17:1239–1253. doi: 10.1038/mp.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388:86–97. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girard SL, Dion PA, Rouleau GA. Schizophrenia genetics: putting all the pieces together. Curr. Neurol. Neurosci. Rep. 2012;12:261–266. doi: 10.1007/s11910-012-0266-7. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schizophrenia Working Group of the Psychiatric Genomics, C Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardinas AF, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rees E, et al. CNV analysis in a large schizophrenia sample implicates deletions at 16p12.1 and SLC1A1 and duplications at 1p36.33 and CGNL1. Hum. Mol. Genet. 2014;23:1669–1676. doi: 10.1093/hmg/ddt540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levinson DF, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am. J. Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall CR, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandal Michael J., Zhang Pan, Hadjimichael Evi, Walker Rebecca L., Chen Chao, Liu Shuang, Won Hyejung, van Bakel Harm, Varghese Merina, Wang Yongjun, Shieh Annie W., Haney Jillian, Parhami Sepideh, Belmont Judson, Kim Minsoo, Moran Losada Patricia, Khan Zenab, Mleczko Justyna, Xia Yan, Dai Rujia, Wang Daifeng, Yang Yucheng T., Xu Min, Fish Kenneth, Hof Patrick R., Warrell Jonathan, Fitzgerald Dominic, White Kevin, Jaffe Andrew E., Peters Mette A., Gerstein Mark, Liu Chunyu, Iakoucheva Lilia M., Pinto Dalila, Geschwind Daniel H. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(6420):eaat8127. doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz E, et al. Validation of a blood-based laboratory test to aid in the confirmation of a diagnosis of schizophrenia. Biomark Insights. 2010;5:39–47. doi: 10.4137/BMI.S4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz E, et al. Identification of a biological signature for schizophrenia in serum. Mol. Psychiatry. 2012;17:494–502. doi: 10.1038/mp.2011.42. [DOI] [PubMed] [Google Scholar]
- 18.Harris LW, et al. Comparison of peripheral and central schizophrenia biomarker profiles. PLoS One. 2012;7:e46368. doi: 10.1371/journal.pone.0046368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manier DH, et al. Human fibroblasts as a relevant model to study signal transduction in affective disorders. J. Affect. Disord. 2000;61:51–58. doi: 10.1016/S0165-0327(99)00190-1. [DOI] [PubMed] [Google Scholar]
- 20.Akin D, Manier DH, Sanders-Bush E, Shelton RC. Signal transduction abnormalities in melancholic depression. Int. J. Neuropsychopharmacol. 2005;8:5–16. doi: 10.1017/S146114570400478X. [DOI] [PubMed] [Google Scholar]
- 21.Kalman S, et al. Metabolic stress-induced microRNA and mRNA expression profiles of human fibroblasts. Exp. Cell Res. 2014;320:343–353. doi: 10.1016/j.yexcr.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahadik SP, Mukherjee S. Cultured skin fibroblasts as a cell model for investigating schizophrenia. J. Psychiatr. Res. 1996;30:421–439. doi: 10.1016/S0022-3956(96)00025-8. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt MJ, Mirnics K. Modeling interneuron dysfunction in schizophrenia. Dev. Neurosci. 2012;34:152–158. doi: 10.1159/000336731. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt MJ, et al. Modulation of behavioral networks by selective interneuronal inactivation. Mol. Psychiatry. 2014;19:580–587. doi: 10.1038/mp.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown JA, et al. Inhibition of parvalbumin-expressing interneurons results in complex behavioral changes. Mol. Psychiatry. 2015;20:1499–1507. doi: 10.1038/mp.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat. Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt A, Parlapani E, Bauer M, Heinsen H, Falkai P. Is brain banking of psychiatric cases valuable for neurobiological research? Clin. (Sao Paulo) 2008;63:255–266. doi: 10.1590/S1807-59322008000200015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalman S, Garbett KA, Janka Z, Mirnics K. Human dermal fibroblasts in psychiatry research. Neuroscience. 2016;320:105–121. doi: 10.1016/j.neuroscience.2016.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanzelmann S, et al. Replicative senescence is associated with nuclear reorganization and with DNA methylation at specific transcription factor binding sites. Clin. Epigenetics. 2015;7:19. doi: 10.1186/s13148-015-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rieske P, Krynska B, Azizi SA. Human fibroblast-derived cell lines have characteristics of embryonic stem cells and cells of neuro-ectodermal origin. Differentiation. 2005;73:474–483. doi: 10.1111/j.1432-0436.2005.00050.x. [DOI] [PubMed] [Google Scholar]
- 31.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaros T, Larsen M, Li L. Macrophages and fibroblasts during inflammation, tissue damage and organ injury. Front. Biosci. (Landmark Ed.) 2009;14:3988–3993. doi: 10.2741/3506. [DOI] [PubMed] [Google Scholar]
- 33.Sherrington R, et al. Localization of a susceptibility locus for schizophrenia on chromosome 5. Nature. 1988;336:164–167. doi: 10.1038/336164a0. [DOI] [PubMed] [Google Scholar]
- 34.Blouin JL, et al. Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat. Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- 35.Torrey EF. Prevalence studies in schizophrenia. Br. J. Psychiatry. 1987;150:598–608. doi: 10.1192/bjp.150.5.598. [DOI] [PubMed] [Google Scholar]
- 36.van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 37.Book JA, Wetterberg L, Modrzewska K. Schizophrenia in a North Swedish geographical isolate, 1900–1977. Epidemiology, genetics and biochemistry. Clin. Genet. 1978;14:373–394. doi: 10.1111/j.1399-0004.1978.tb02105.x. [DOI] [PubMed] [Google Scholar]
- 38.Szatkiewicz J, et al. The genomics of major psychiatric disorders in a large pedigree from Northern Sweden. Transl. Psychiatry. 2019;9:60. doi: 10.1038/s41398-019-0414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell EM, Mars WM, Levitt P. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron. 2001;30:79–89. doi: 10.1016/S0896-6273(01)00264-1. [DOI] [PubMed] [Google Scholar]
- 42.Ebens A, et al. Hepatocyte growth factor/scatter factor is an axonal chemoattractant and a neurotrophic factor for spinal motor neurons. Neuron. 1996;17:1157–1172. doi: 10.1016/S0896-6273(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 43.Maina F, et al. Multiple roles for hepatocyte growth factor in sympathetic neuron development. Neuron. 1998;20:835–846. doi: 10.1016/S0896-6273(00)80466-3. [DOI] [PubMed] [Google Scholar]
- 44.Stoker M, Gherardi E, Perryman M, Gray J. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature. 1987;327:239–242. doi: 10.1038/327239a0. [DOI] [PubMed] [Google Scholar]
- 45.Matsumoto, K. & Nakamura, T. HGF: its organotrophic role and therapeutic potential. Ciba Found Symp212, 198-211; discussion 211–194 (1997). [DOI] [PubMed]
- 46.McCarthy SE, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 48.Gulsuner S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirov G, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol. Psychiatry. 2014;75:378–385. doi: 10.1016/j.biopsych.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang H, Li L, Li M, Xiao X. Rare and common variants at 16p11.2 are associated with schizophrenia. Schizophr. Res. 2017;184:105–108. doi: 10.1016/j.schres.2016.11.031. [DOI] [PubMed] [Google Scholar]
- 51.Cattane N, et al. Altered gene expression in schizophrenia: findings from transcriptional signatures in fibroblasts and blood. PLoS One. 2015;10:e0116686. doi: 10.1371/journal.pone.0116686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sukhatme VP, et al. A novel early growth response gene rapidly induced by fibroblast, epithelial cell and lymphocyte mitogens. Oncogene Res. 1987;1:343–355. [PubMed] [Google Scholar]
- 53.Cole AJ, Saffen DW, Baraban JM, Worley PF. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- 54.Duclot F, Kabbaj M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front. Behav. Neurosci. 2017;11:35. doi: 10.3389/fnbeh.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hippenmeyer S, et al. A role for neuregulin1 signaling in muscle spindle differentiation. Neuron. 2002;36:1035–1049. doi: 10.1016/S0896-6273(02)01101-7. [DOI] [PubMed] [Google Scholar]
- 56.Yamada K, et al. Genetic analysis of the calcineurin pathway identifies members of the EGR gene family, specifically EGR3, as potential susceptibility candidates in schizophrenia. Proc. Natl Acad. Sci. USA. 2007;104:2815–2820. doi: 10.1073/pnas.0610765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim SH, et al. EGR3 as a potential susceptibility gene for schizophrenia in Korea. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:1355–1360. doi: 10.1002/ajmg.b.31115. [DOI] [PubMed] [Google Scholar]
- 58.Zhang R, et al. Genetic evidence for the association between the early growth response 3 (EGR3) gene and schizophrenia. PLoS One. 2012;7:e30237. doi: 10.1371/journal.pone.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huentelman MJ, et al. Association of SNPs in EGR3 and ARC with Schizophrenia Supports a Biological Pathway for Schizophrenia Risk. PLoS One. 2015;10:e0135076. doi: 10.1371/journal.pone.0135076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 2017;49:1576–1583. doi: 10.1038/ng.3973. [DOI] [PubMed] [Google Scholar]
- 61.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Babulas V, Factor-Litvak P, Goetz R, Schaefer CA, Brown AS. Prenatal exposure to maternal genital and reproductive infections and adult schizophrenia. Am. J. Psychiatry. 2006;163:927–929. doi: 10.1176/ajp.2006.163.5.927. [DOI] [PubMed] [Google Scholar]
- 64.Brown AS, et al. Maternal exposure to respiratory infections and adult schizophrenia spectrum disorders: a prospective birth cohort study. Schizophr. Bull. 2000;26:287–295. doi: 10.1093/oxfordjournals.schbul.a033453. [DOI] [PubMed] [Google Scholar]
- 65.De Miranda, J. et al. Induction of Toll-like receptor 3-mediated immunity during gestation inhibits cortical neurogenesis and causes behavioral disturbances. MBio1, 10.1128/mBio.00176-10 (2010). [DOI] [PMC free article] [PubMed]
- 66.Chen CY, Liu HY, Hsueh YP. TLR3 downregulates expression of schizophrenia gene Disc1 via MYD88 to control neuronal morphology. EMBO Rep. 2017;18:169–183. doi: 10.15252/embr.201642586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCarthy SE, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li B, Woo RS, Mei L, Malinow R. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54:583–597. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nicodemus KK, et al. Biological validation of increased schizophrenia risk with NRG1, ERBB4, and AKT1 epistasis via functional neuroimaging in healthy controls. Arch. Gen. Psychiatry. 2010;67:991–1001. doi: 10.1001/archgenpsychiatry.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Law AJ, et al. Neuregulin 1-ErbB4-PI3K signaling in schizophrenia and phosphoinositide 3-kinase-p110delta inhibition as a potential therapeutic strategy. Proc. Natl Acad. Sci. USA. 2012;109:12165–12170. doi: 10.1073/pnas.1206118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Forsyth JK, Lewis DA. Mapping the Consequences of Impaired Synaptic Plasticity in Schizophrenia through Development: An Integrative Model for Diverse Clinical Features. Trends Cogn. Sci. 2017;21:760–778. doi: 10.1016/j.tics.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacDonald ML, et al. Altered glutamate protein co-expression network topology linked to spine loss in the auditory cortex of schizophrenia. Biol. Psychiatry. 2015;77:959–968. doi: 10.1016/j.biopsych.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinberger DR, Berman KF, Zec RF. Physiologic dysfunction of dorsolateral prefrontal cortex in schizophrenia. I. Regional cerebral blood flow evidence. Arch. Gen. Psychiatry. 1986;43:114–124. doi: 10.1001/archpsyc.1986.01800020020004. [DOI] [PubMed] [Google Scholar]
- 76.Goldman-Rakic PS. Working memory dysfunction in schizophrenia. J. Neuropsychiatry Clin. Neurosci. 1994;6:348–357. doi: 10.1176/jnp.6.4.348. [DOI] [PubMed] [Google Scholar]
- 77.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat. Rev. Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 78.Hashimoto T, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol. Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matigian NA, et al. Fibroblast and lymphoblast gene expression profiles in schizophrenia: are non-neural cells informative? PLoS One. 2008;3:e2412. doi: 10.1371/journal.pone.0002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang L, et al. Expression profiling of fibroblasts identifies cell cycle abnormalities in schizophrenia. J. Proteome Res. 2010;9:521–527. doi: 10.1021/pr900867x. [DOI] [PubMed] [Google Scholar]
- 81.Hebels DG, et al. Performance in omics analyses of blood samples in long-term storage: opportunities for the exploitation of existing biobanks in environmental health research. Env. Health Perspect. 2013;121:480–487. doi: 10.1289/ehp.1205657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andreasson A, Kiss NB, Juhlin CC, Hoog A. Long-term storage of endocrine tissues at − 80 degrees C does not adversely affect RNA quality or overall histomorphology. Biopreserv Biobank. 2013;11:366–370. doi: 10.1089/bio.2013.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kelly Rachel, Albert Monique, de Ladurantaye Manon, Moore Melissa, Dokun Olusola, Bartlett John M.S. RNA and DNA Integrity Remain Stable in Frozen Tissue After Long-Term Storage at Cryogenic Temperatures: A Report from the Ontario Tumour Bank. Biopreservation and Biobanking. 2019;17(4):282–287. doi: 10.1089/bio.2018.0095. [DOI] [PubMed] [Google Scholar]
- 84.Taylor MA, Abrams R, Gaztanaga P. Manic-depressive illness and schizophrenia: a partial validation of research diagnostic criteria utilizing neuropsychological testing. Compr. Psychiatry. 1975;16:91–96. doi: 10.1016/0010-440X(75)90025-5. [DOI] [PubMed] [Google Scholar]
- 85.Feighner JP, et al. Diagnostic criteria for use in psychiatric research. Arch. Gen. Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- 86.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Langfelder, P. & Horvath, S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J. Stat Softw46 (2012). [PMC free article] [PubMed]
- 91.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 93.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.