Abstract
The fungi Fusarium oxysporum and Fusarium fujikuroi produce carotenoids, lipophilic terpenoid pigments of biotechnological interest, with xanthophyll neurosporaxanthin as the main end product. Their carotenoid biosynthesis is activated by light and negatively regulated by the RING-finger protein CarS. Global transcriptomic analysis identified in both species a putative 1-kb lncRNA that we call carP, referred to as Fo-carP and Ff-carP in each species, upstream to the gene carS and transcribed from the same DNA strand. Fo-carP and Ff-carP are poorly transcribed, but their RNA levels increase in carS mutants. The deletion of Fo-carP or Ff-carP in the respective species results in albino phenotypes, with strong reductions in mRNA levels of structural genes for carotenoid biosynthesis and higher mRNA content of the carS gene, which could explain the low accumulation of carotenoids. Upon alignment, Fo-carP and Ff-carP show 75–80% identity, with short insertions or deletions resulting in a lack of coincident ORFs. Moreover, none of the ORFs found in their sequences have indications of possible coding functions. We conclude that Fo-carP and Ff-carP are regulatory lncRNAs necessary for the active expression of the carotenoid genes in Fusarium through an unknown molecular mechanism, probably related to the control of carS function or expression.
Subject terms: Fungal genetics, Long non-coding RNAs
Introduction
The genus Fusarium comprises a large group of phytopathogenic fungi widely distributed in nature. Some species are well-known research models for basic biological processes, such as Fusarium oxysporum in phytopathogenesis1 and Fusarium fujikuroi in secondary metabolism2. F. oxysporum comprises many species-specific plant pathogens, called formae specialis, and some of them have been widely used for the study of pathogenesis mechanisms and fungus-plant interactions3,4. Fusarium fujikuroi was first known for the production of gibberellins, growth-promoting plant hormones, and has later been investigated for the synthesis of other metabolites, such as bikaverin and fusarins5,6.
Some Fusarium species are also models for the investigation of carotenoid biosynthesis and its regulation7. Early studies in Fusarium aquaeductuum revealed the accumulation of xanthophyll neurosporaxanthin (NX), a carboxylic apocarotenoid previously discovered in Neurospora crassa. Subsequent studies in Fusarium fujikuroi led to the identification of all the genes of the NX pathway7, carB, carRA, carT, and carD, which respectively encode a desaturase, a bifunctional phytoene synthase/carotene cyclase, a torulene cleaving dioxygenase, and an aldehyde dehydrogenase (Supplementary Fig. S1). The carB and carRA genes are organized in a coregulated cluster with another dioxygenase gene, carX8. The CarX enzyme is capable of cleaving β-carotene, a side product of NX biosynthesis9, into retinal10. A fourth gene in the cluster is carO, which encodes a photoactive rhodopsin that uses retinal as chromophore11.
The biosynthesis of carotenoids in Fusarium is upregulated by light12 through transcriptional activation of the structural genes of the pathway, in which the White Collar photoreceptor WcoA plays a central role13. The transcriptional response is severely impaired in the absence of a functional WcoA protein, but there is an apparent accumulation of carotenoids in wcoA mutants under illumination14 that involves the participation of at least a second photoreceptor. This could be DASH cryptochrome CryD13, as indicated by the slower accumulation of carotenoids in ΔcryD mutants despite activation by WcoA13. CryD photoactivity has been demonstrated biochemically15, but only minor effects were found in the mRNA levels of car genes in ΔcryD mutants after illumination, suggesting its participation in a post-transcriptional regulatory mechanism. Another photoreceptor protein that participates in the regulation of carotenogenesis by light in Fusarium is VvdA, an orthologue of the Vivid protein of N. crassa, which supposedly counteracts WcoA activity16.
A class of mutants of F. fujikuroi exhibits deep orange pigmentation in the dark due to the synthesis of large amounts of NX and carotenoid precursors17. These mutants, also found in F. oxysporum and generically called carS, are affected in a single gene that encodes a protein of the RING-finger family18,19. The deep orange phenotype is due to the presence of large amounts of transcripts for the structural genes of the pathway, especially those of the car cluster, which indicates that CarS is a negative regulator of the expression of the car genes. This cluster is not the only regulatory target of CarS; the carS mutation produces noticeable changes in the mRNA levels of hundreds of genes, a notable proportion of them also regulated by light20. The mechanism of action of CarS remains unknown, but RING-finger domains belong to a family of E3-like ligase involved in the labelling of target proteins by ubiquitylation, a regulatory signal that frequently leads to their degradation. The alleged ortholog of CarS in Mucor circinelloides, CrgA, has been the subject of detailed investigation and its ubiquitylation activity has been demonstrated21–23. CarS and CrgA also contain a LON protease domain, whose function has not been determined.
Due to its regulatory effects on a large collection of genes, we assume that carS expression may be subject to complex regulation. The carS gene is preceded by a 4-kb upstream sequence without predicted ORFs in genome annotations. A former study of two F. oxysporum T-DNA insertion mutants, which exhibited deregulation of carotenoid biosynthesis, revealed sequence alterations in this 4-kb DNA segment upstream of the carS gene18, and a recent RNA-seq study suggested the occurrence of an unidentified transcript in this genomic region with features consistent with a long noncoding RNA (lncRNA). LncRNAs are polyadenilated RNAs of more than 200 bp in length, transcribed by RNA polymerase II but without protein coding functions. It has been discovered that an increasing number of lncRNAs are involved in important biological processes in higher eukaryotes, such as development or cancer. They modulate the expression of target genes through a variety of mechanisms24–26, associated with their sequences or their structure, involving transcriptional, post-transcriptional, and epigenetic levels. Thus, they can interact with transcription factors, chromatin remodelling factors, microRNAs, or mRNAs, and in some cases they can act as enhancer-like RNAs27. However, there is very little information on the functions of lncRNAs in lower eukaryotes. We have found that the transcript found in the genomic region upstream of carS plays a key positive role in the regulation of carotenoid biosynthesis in F. oxysporum and F. fujikuroi, probably through the control of the carS gene. This is the first functional study of a lncRNA in a species of Fusarium.
Results
Identification of a transcript upstream of the carS gene
In a search for putative regulatory elements in the 4-kb upstream sequence of carS in F. oxysporum, a bioinformatic microRNA prediction tool identified a putative precursor microRNA sequence, which we will call here carP*. This alleged microRNA was not confirmed by a specific RNA-seq study for small RNAs (J. Pardo-Medina, unpublished), but a recent RNA-seq analysis on the effect of light and carS mutations in F. fujikuroi and F. oxysporum20 revealed a transcript in both species, which overlaps with the previous sequence of carP* in F. oxysporum (Fig. 1) and which we have called carP. The transcripts of carP have a length of 1195 bp in F. oxysporum and 1357 pb in F. fujikuroi, considering as initial and final nucleotides those present in at least 2 readings of the RNA-seq data in different strains and culture conditions (Supplementary Fig. S2A). To distinguish the versions of carP in both species, we call them Fo-carP and Ff-carP.
Figure 1.
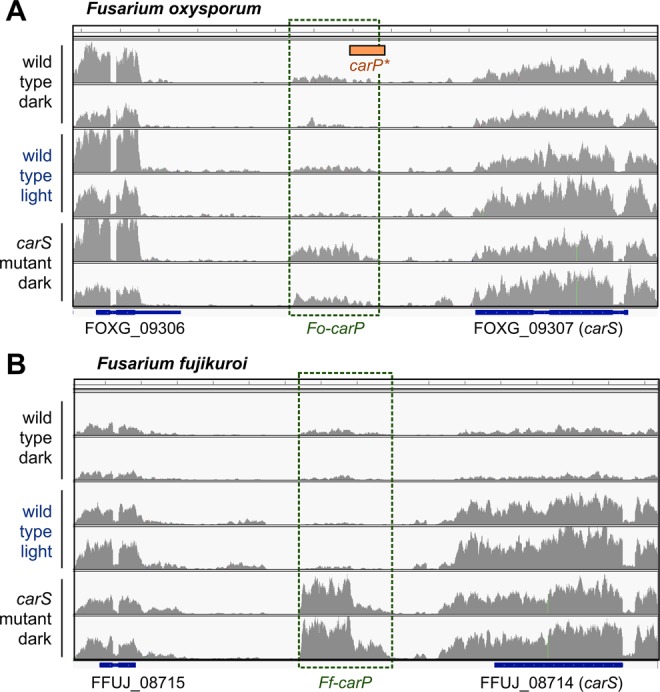
Effect of light (one-hour illumination) and carS mutation on transcript readings according to RNA-Seq data in the genomic region between FOXG_09306 and carS (FOXG_09307) genes in F. oxysporum (panel A) (carS mutant: SX1), and between Ffuj_08715 and carS (FFUJ_08714) genes in F. fujikuroi (panel B) (carS mutant: SG39). The location of a putative microRNA precursor sequence found in F. oxysporum, called carP*, is indicated with an orange box. In each condition, the profiles obtained with two independent biological samples are shown. The readings were represented with IGV program.
To determine the orientation of carP transcription, a PCR assay was carried out to identify strand-specific PCR products. For this purpose, we obtained cDNA samples using mixtures of primers for each DNA strand of the carP sequence and we used them as substrates for amplification with specific primers of Fo-carP and Ff-carP. As a control, the same protocol was used to determine the orientation of the β-tubulin gene. The results (Fig. 2) indicated that both genes encoding carP are transcribed from the same strand as the neighbouring carS gene.
Figure 2.
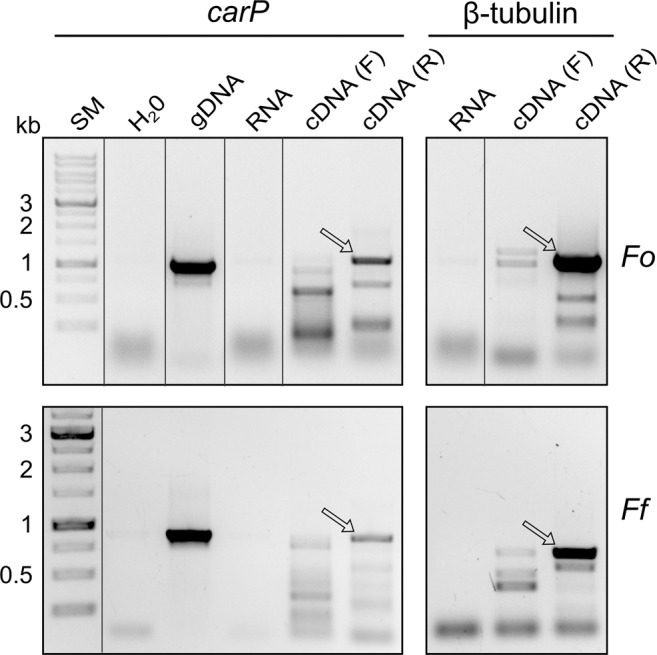
Determination of the orientation of the transcription of carP in F. oxysporum (Fo) and F. fujikuroi (Ff) through the amplification from specific single stranded DNA (ssDNA). SM: size markers. gDNA: amplification of the carP transcript using genomic DNA of the wild strain grown in the dark (positive control) as template; RNA: lack of amplification of the carP transcript using as template total RNA (negative PCR control and DNA contamination control in the ssDNA samples). cDNA (F): lack of amplification of carP from ssDNA obtained from a retrotranscription of total RNA with a mixture of forward carP primers. cDNA (R): amplification of carP from ssDNA obtained from a retrotranscription of total RNA with a mixture of reverse carP primers. Results are also shown for amplification of the β-tubulin gene as a control with a known transcriptional orientation. Lanes cropped from different positions of the original gels are separated by a black line. Full-length blots/gels are presented in Supplementary Fig. S13. The sizes of the expected PCR products are 1099 bp for carP and 1169 bp for β-tubulin gene in F. oxysporum, which correspond to 910 and 693 bp in F. fujikuroi. The primers used are described in Supplementary Table S5A. Genomic DNA and total RNA samples were obtained from wild strains grown for 3 days in darkness.
Sequence features of carP
A Clustal alignment between the carP sequences of F. oxysporum and F. fujikuroi revealed high conservation, with 929 matches along an overlapping stretch that covers 1165 bp in Fo-carP and 1238 bp in Ff-carP (Supplementary Fig. S2B), with a non-homogenous distribution of coincidences, and the occurrence of numerous gaps (Fig. 3A). To consider their possible coding functions, open reading frames (ORFs) in both chains with a threshold of 25 consecutive amino acids were determined. The results (Supplementary Fig. S3) showed 11 ORFs for the Fo-carP sequence, 7 of them in the forward direction (Fig. 3B), and 12 ORFs in Ff-carP, of which 5 in the forward direction (Fig. 3C). The longest ORFs encoded 131 residues in Fo-carP and 107 in Ff-carP, located in different regions of carP. ORF F4 of Fo-carP was previously annotated as hypothetical proteins FOTG_04033 in F. oxysporum f.sp. vasinfectum, and FOVG_10518 in F. oxysporum f.sp. pisi, but they were not annotated in other Fusarium genomes. Comparisons of the ORF sequences with those of the protein databases did not identify known conserved sequences or domains. E.g, the best match of the Fo-carP sequence gave an E value of 0.005. Most significantly, with the exception of some short segments (Supplementary Fig. S3), there are no coincident ORFs in the carP sequences between both species.
Figure 3.
Sequence features of Fo-carP and Ff-carP. (A) Schematic representation of the clustal alignment between Fo-carP (red) and Ff-carP (blue). Gaps resulting from the alignment are indicated. The matching bases are shown below as long grey lines. (B,C) Open reading frames (ORFs) in Fo-carP (B, in red) and Ff-carP (C, in blue). The positions of each of the ORFs are represented in the boxed panels (F: forward, in dark color; R: reverse, in pale color). Residues corresponding to forward ORFs are shown below each boxed panel. The analysis of the ORFs was done through the ORFfinder tool.
Taking into account the possible occurrence of intron sequences, we achieved BlastX ORF-independent searches with whole carP sequences against the NCBI nr database. Without considering the two annotated carP segments of F. oxysporum, the maximum E value was 0.3, found against a cyclase of Streptomyces sp. As alternative approaches, the coding capacity of carP was verified through other sequence characteristics. Analyses based on 3-base periodicity (Supplementary Fig. S4) or codon usage (Supplementary Fig. S5) did not provide evidence of coding functions. Other methods used to assess the protein-coding potential of carP were two versions of Coding Potential Calculator: CPC28 and CPC229, based on intrinsic sequence features, and RNAcode, based on comparative sequence data30. None of these methods provided any support to the occurrence of putative coding sequences in Fo-carP or Ff-carP (Supplementary Tables S1 and S2).
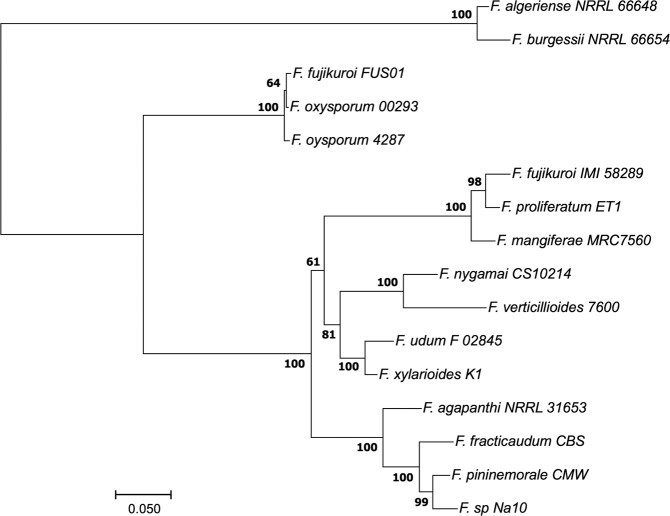
Taken together, all the data suggest that the carP transcript does not have a protein-coding function and, therefore, could play a role as regulatory RNA. The carP sequence is too long for reliable 2D predictions, but if its function depends on a conserved secondary conformation, the structure could be inferred from comparison of the orthologs of carP in different Fusarium species. We have used the RNAcode procedure, capable of detecting possible coding features based on multiple alignment, to obtain a consensus structure. The alignment was performed with the LocARNA-P method31, using 15 carP sequences (Supplementary Fig. S6 and Table S3), including Fo-carP and Ff-carP. The resulting maximum-likelihood tree (Fig. 4) showed that most of the carP sequences investigated are closer to Ff-carP than Fo-carP. The rapid evolution of carP makes it possible to build a reliable phylogenetic tree for such closely related species, which contrasts with the lower resolution of the trees based on gene sequences usually used for this purpose, as those of the Tef1 gene (supplementary Fig. S7).
Figure 4.
Maximum-likelihood tree of the carP sequences listed in Fig. S6. The tree derives from multiple alignment based on structures made with the locARNA program used for the RNAcode test. Branch support analysis was evaluated by 1000 ultrafast bootstrap replicates. The tree indicates that FUS01 is actually a F. oxysporum strain, wrongly classified as F. fujikuroi.
The finding of a consensus secondary structure (Supplementary Fig. S8) is consistent with a hypothetical structural role but does not prove it. Some lncRNAs act directly on mRNAs of the target genes through pairing, affecting positively or negatively their stability or their translation. A search with the Clustal Omega program on possible target mRNAs, such as those of the carS or wcoA/wco1 regulatory genes, or those of the structural genes of carotenogenesis, revealed the absence of sequences of more than 20 bp with more than 50% identity, a threshold below which coincident segments are found in any random gene. This result makes unlikely a direct function of carP by pairing on mRNAs of the genes of interest.
Occurrence of carP in other species
NCBI megablast with Fo-carP versus the nr database of all species, except fungi, did not give significant matches (E < 10). Similarly, no significant match was obtained with an NCBI megablast with Fo-carP or Ff-carP versus the nr database restricted to all fungi, except those of the genus Fusarium, and the same result was obtained with blastn of Fo-carP in the JGI Mycocosm database. Therefore, carP is specific to Fusarium among the fungi and is not found outside the fungal kingdom. Moreover, the carP sequence is widespread but not universal in Fusarium species: A blastn search with Fo-carP against the 249 Fusarium whole genome sequences (WGS) available at NCBI gave only positive hits in 166, using as selection criteria E < 0.001 and query cover >25% (Supplementary Table S4). Therefore, carP was not identified in 1/3 of the available Fusarium WGS. In contrast, orthologs for the carRA and carB carotenoid pathway genes, as well as for the carS regulatory gene, are found in all of them. However, when carP was present, it exhibited a high sequence conservation. Thus, when compared to at least 97% of the Fo-carP sequence, the percentages of carP identity in other Fusarium species varied from 68,6% in Fusarium verticillioides to 97,1% in Fusarium solani.
Genomic organization of carP in relation to carS in Fusarium
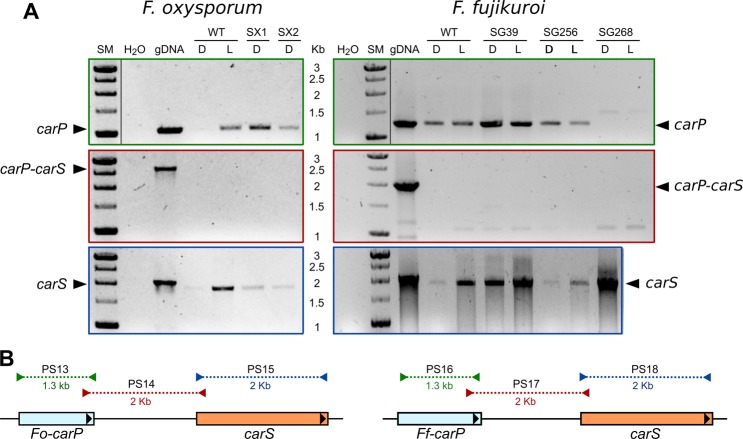
The proximity of carP to carS and its transcription on the same strand suggest a functional connection. We revised the information available for Fusarium species on prediction of transcripts in the carS region through the Browser FungiDB, using Fusarium oxysporum f. sp. lycopersici 4287 as a reference. The result showed considerable variations in 5′ and 3′-UTR carS extensions between different Fusarium species, as well as carS antisense transcripts from the 3′ extensions of the neighbouring gene in some species (Supplementary Fig. S9). In some cases, the 5′ end of the carS transcript extended to the 4-kb upstream region, and at least in the case of F. verticillioides, it overlapped with the carP sequence. This raises the question of whether Fo-carP and Ff-carP may be the result of eventual transcription of carS from a 5′ distant site, resulting in a long carP-containing 5′-UTR. To test this hypothesis, PCR experiments were carried out with combinations of primers from internal regions of carP and carS to verify their occurrence in single transcripts using cDNA obtained from mycelia of wild type and mutant strains of F. oxysporum and F. fujikuroi, grown in darkness or under light. PCR amplification between the carS and carP sequences was obtained from the genomic DNA, but not from the cDNA of any of the strains and species tested, while the amplifications were detected for the internal carP or carS sequences (Fig. 5). We conclude that carP and carS derive from separate transcription events, and that carP is an independent lncRNA.
Figure 5.
(A) PCR verification of carP and carS as independent transcripts in F. oxysporum (left panels) and F. fujikuroi (right panels). Amplifications were performed by PCR on cDNA samples in the indicated strains and conditions using primers from internal sequences of carP (upper panels), internal sequences of carS (lower panels) or connecting internal sequences of carP and carS (central panels). SM: size markers. H2O: test for lack of amplification without template DNA. gDNA: amplification from genomic DNA from the wild strain (positive control), WT: Wild strain. SX1, SX2, and SG39: carS mutants. SG256: SG39 complemented with wild carS. SG268: mutant lacking the carP sequence (ΔcarP) as an additional negative control in F. fujikuroi. Lanes cropped from different positions of the original gels are separated by a black line. Full-length blots/gels are presented in Supplementary Fig. S13. (B) Schematic representation of sizes and locations of PCR products obtained with the indicated primers sets (PS) in F. oxysporum (left) and F. fujikuroi (right).
Expression of carP
The RNA-seq data displayed in Fig. 1 show that the carS mutation results in enhanced transcript levels for carP in both Fusarium species. However, due to the low content of carP RNA in wild strains, the effect of light was not clear. To learn about the regulation of carP transcription, the effects of light and carS mutation on carP RNA levels were investigated by RT-qPCR in different strains and conditions in F. oxysporum and F. fujikuroi, and the analysis was extended to other relevant genes.
In wild F. oxysporum, transcript levels of Fo-carP increased appreciably in light in relation to darkness (Fig. 6A). The result was similar with primers for carP* or for another carP sequence, as expected for two regions of the same transcript. Photoinduction was less apparent in the carS mutants SX1 and SX2, which exhibited higher levels of carP RNA in the dark and after illumination. In contrast, exposure to light produced a similar five-fold increase in carS transcript, regardless of the presence of the carS mutation. SX1 and SX2 have point mutations in the coding sequence of carS18, and therefore are expected to contain the complete transcripts of the gene. In the case of SX2, a mutation resulted in a premature stop codon in the carS coding sequence that is expected to generate a truncated polypeptide that lacks approximately 50% of the protein. The results with Fo-carP were similar to those obtained with the structural genes carRA and carB, but in this case the effect of light in the wild strain or the effect of the carS mutation were much greater. As expected, the carotenoid content correlated with the mRNA levels of the carRA and carB genes, but with higher carotenoid content in carS mutants than in the wild strain under illumination.
Figure 6.
(A) Effect of light and carS mutation (carS mutants SX1 and SX2) on transcript levels for genes carP, carS, carRA, and carB (light: 1-hour illumination) and carotenoid content (light: continuous illumination) in F. oxysporum. In the case of carP, the RT-qPCR primers were selected from two different carP regions, one of them contained in the carP* sequence. (B) Effect of light (1-hour illumination) or carS mutation (carS mutant SG39 and carS complemented strain SG256) on transcript levels for the carP and carB genes in F. fujikuroi. In both species, for RT-qPCR analyses the strains were grown in DGasn broth for 3 days in the dark (dark bars) and exposed for 1 hour to light (light bars). RT-qPCR data show the mean and standard error of RT-qPCR data from three independent experiments. The relative mRNA levels are referred to the mRNA content of the wild strain in darkness. Differences found to be significant according to the t tests are indicated (P-values, *p < 0.033; **p < 0.002; ***p < 0.001).
The expression pattern of carP in F. fujikuroi differed from that of F. oxysporum (Fig. 6B), a result already suggested by the comparison of their RNA-seq data (Fig. 1). Ff-carP RNA levels were hardly affected by light in the wild strain investigated. In this case, to test the effect of the carS mutation, the study focused on a carS mutant, SG39, and its carS-complemented strain SG256, in which a functional carS gene has been reintroduced. carP transcript levels were higher in the carS mutant than in the strains with a functional carS gene, especially without illumination.
Phenotype of a ΔcarP and ΔcarP* mutants in F. oxysporum
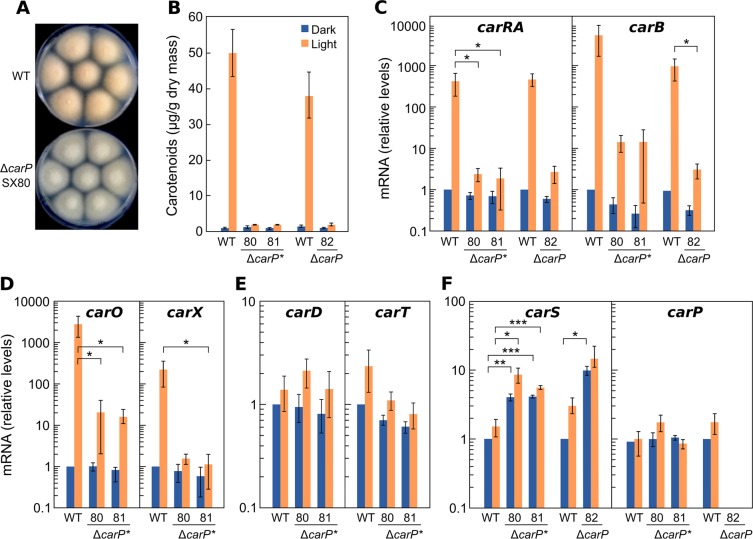
To determine the function of carP in F. oxysporum, a ΔcarP mutant (SX82) and two ΔcarP* mutants (SX80 and SX81) were obtained. All three mutants had the same morphology and growth rate as the wild strain. However, they exhibited albino mycelia under illumination (SX80 shown as an example in Fig. 7A), consistent with a defect in carotenoid production, while presenting a wild-type appearance in the dark. As expected, carotenoid analyses showed a strong photoinduction of the carotenoid content in the wild strain, but only traces of carotenoids in the three mutants in the light, with levels only slightly higher than those in the dark (Fig. 7B).
Figure 7.
Effect of carP deletion in F. oxysporum. (A) Aspect of surface colonies of the wild strain and a representative ΔcarP mutant grown on minimal medium for 1 week under light. (B) Carotenoid content in the wild strain and mutants ΔcarP* and ΔcarP grown for 1 week in the dark or under light. (C–F) Transcript levels for the carRA, carB, carO, carX, carD, carT, carS, and carP genes in the same strains grown in the dark or exposed for 1 hour to light. RT-qPCR data show the mean and standard error of three independent experiments. Relative mRNA levels refer to the mRNA content of the wild strain in the dark. Differences found to be significant according to the t tests are indicated (P-values, *p < 0.033; **p < 0.002; ***p < 0.001).
To determine whether the phenotype in the light was due to reduced expression of the structural car genes, their mRNA levels were quantified by RT-qPCR in the dark or after one hour of illumination and compared with those of the wild strain. The results showed a sharp decrease in the transcript levels of carRA and carB genes in the ΔcarP mutants compared to the wild strain (Fig. 7C). Such descent was similar in the deletion strains ΔcarP and ΔcarP*, indicating that the absence of carP* was sufficient for the total loss of Fo-carP function. However, although the mutants exhibited a 100-fold decrease in the mRNA levels for these genes, a minor photoinduction was still apparent compared to the levels in the dark, which was barely reflected in the actual carotenoid content in the light. On the other hand, the mRNA content of carRA and carB did not suffer significant variations in the dark compared to those of the wild strain, indicating the participation of Fo-carP in the transcriptional photoinduction of the structural car genes in F. oxysporum.
The deletion of carP also produced clear reductions in the photoinduction of the carX and carO genes (Fig. 7D), organized in a coregulated cluster with carRA and carB (Supplementary Fig. S1). The effect on carO was similar to that of carB, adjacent in the car cluster, and the effect on carX was similar to that of carRA, transcribed divergently from a common regulatory region. In contrast, the effect of carP deletion was hardly apparent for the carT and carD genes (Fig. 7E), involved in the last steps of NX production. This is consistent with the lower photoinduction exhibited by these genes compared to carRA and carB, which may explain the poor influence of carP deletion. Interestingly, the transcript levels of the carS gene increased appreciably in the ΔcarP and ΔcarP* mutants, in the dark or after illumination (Fig. 7F). However, the ΔcarP* mutation did not affect the levels of the transcription of carP itself.
Phenotype of ΔcarP mutants in F. fujikuroi
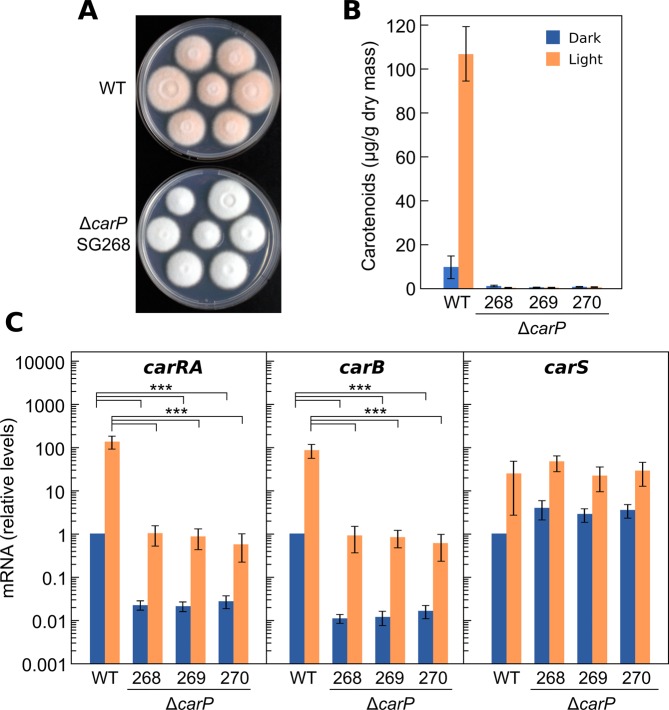
To determine the function of the carP gene in F. fujikuroi, three ΔcarP strains (SG268, SG269, and SG270) were obtained. As already observed in F. oxysporum, the colonies of the three ΔFf-carP strains exhibited morphologies and growth capacities similar to those of the wild strain but lacked the characteristic orange pigmentation in the light (SG268 shown in Fig. 8A). However, analysis of their carotenoid contents revealed a more drastic effect on carotenogenesis than that observed in F. oxysporum. The wild strain of F. fujikuroi had detectable amounts of carotenoids in the dark, which increased approximately tenfold under illumination, but the ΔFf-carP mutants contained only traces of carotenoids either in the light or in darkness (Fig. 8B). However, mRNA levels for the carRA and carB genes decreased about 100-fold but exhibited the same pattern of photoinduction. Interestingly, the ΔFf-carP mutants contained in the light the same amounts of carRA and carB transcripts as the wild strain in the dark, but despite that, their carotenoid content was much lower. In contrast to the effect on carRA and carB, the ΔFf-carP mutants exhibited a modest increase on carS mRNA levels in the dark, which was not apparent after illumination.
Figure 8.
Effect of carP deletion in F. fujikuroi. (A) Aspect of surface colonies of the wild strain and a representative ΔcarP mutant grown on minimal medium for 1 week under light. (B) Carotenoid content in the wild strain and three ΔcarP mutants grown for 1 week in the dark or under light. (C) Transcript levels for the carRA, carB, and carS genes in the same strains grown in the dark or exposed for 1 hour to light. RT-qPCR data show the mean and standard error of three independent experiments. Relative mRNA levels refer to the mRNA content of the wild strain in darkness. Differences found to be significant according to the t tests are indicated (P-values, *p < 0.033; **p < 0.002; ***p < 0.001).
Effect of carP deletion on the expression of photoreceptor genes
As already mentioned, the regulation of carotenogenesis by light is mediated in F. fujikuroi by the WcoA photoreceptor, called Wc1 in F. oxysporum32, with the accessory participation of the CryD and VvdA photoreceptors16. For that reason, we investigated whether the reduced photoinduction of car structural genes as a result of the deletion of carP is due to an alteration in expression of the genes for these photoreceptors. The study of mRNA levels of the wcoA/wc1, cryD, and vvdA genes by RT-qPCR in the wild strain and ΔcarP mutants showed the same expression patterns for the three genes in F. fujikuroi and F. oxysporum (Supplementary Fig. S10). The transcript levels of the cryD and vvdA genes were strongly photoinduced, but this photoinduction was maintained in the ΔcarP mutants, indicating that carP is not necessary for this photoresponse. As additional evidence, the effect of carP deletion was also checked in the F. oxysporum FOXG_01269 gene, ortholog of the photoregulated con-10 gene of N. crassa33. FOXG_01269 maintained a robust photoinduction in the ΔcarP mutant, indicating that carP is not a general regulator of photoinducible genes in Fusarium.
Discussion
Most studies on lncRNAs have been dedicated to higher eukaryotes, especially mammals, but they are present in all taxonomic groups, including plants34 and microorganisms, such as the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe35. The occurrence of lncRNAs has also been reported in some filamentous fungi. In N. crassa, a study of massive RNA sequencing revealed 939 lncRNAs, more than half of them antisense of annotated genes36. In Fusarium graminearum, a search identified 2574 lncRNAs, of which 1040 were antisense transcripts, and 547 exhibited differential expression associated to the formation of fruiting bodies37. F. oxysporum induces changes in the formation of lncRNAs by plants during the infection process as a defence mechanism38,39, but the presumable changes in lncRNA production in the fungus associated to pathogenesis have not been investigated.
To date, only one lncRNA has been characterized in filamentous fungi, HAX1 of T. reesei40, which plays a regulatory role in the expression of cellulases, and possibly other lytic enzymes. The HAX1 gene was identified from phenotypic alterations resulting from insertional mutants, and its function was confirmed by targeted mutation and overexpression, but its mechanism of action remains unknown. In this work, we have demonstrated for the first time the participation of a Fusarium lncRNA, that we called carP, in a specific metabolic function. The lack of translation of the ORFs in the lncRNAs is difficult to prove. In our case, the lack of correlation between ORFs in two carP versions investigated: Fo-carP in F. oxysporum and Ff-carP in F. fujikuroi, whose deletions produce similar phenotypes, is a strong indication of the non-participation of a protein in the regulatory function of carP. This lack of correlation is basically due to the occurrence of gains or losses of few bases, as revealed by the alignment of Fo-carP and Ff-carP, which indicates that their sequences have evolved maintaining their function despite the alteration of their ORFs. Accordingly, none of the various informatic tools used to investigate its sequence found any indication of protein coding functions either in Fo-carP or Ff-carP. Considered globally, the available information strongly supports the non-coding nature of carP in Fusarium.
The absence of relevant carP matching sequences in predictable target genes suggests that carP does not exert its function by modulating the translation or stability of mRNAs involved in carotenogenesis. Other types of mechanisms, such as those based on a specific three-dimensional conformation of the transcript, are not ruled out. carP RNA could interfere with the function of a regulatory protein, which could be CarS. E.g., carP RNA could bind CarS and block its repressor function, so that the absence of carP would leave CarS free to suppress carotenoid synthesis. In this case, the light could perform its activating function, at least partially, facilitating the binding of carP to CarS. As an example of protein inactivation by binding of a lncRNA, the 600-nt gas5 ncRNA binds to the DNA-binding domain of the glucocorticoid receptor under nutrient starvation and inhibits glucocorticoid-regulated transcription in human cells41. Other mechanisms of action based on carP interaction with regulatory proteins cannot be ruled out. E.g., carP might act as a scaffold to facilitate the assembly of histone modification enzymes and alter the expression of target genes. This is the case of the human HOTAIR lncRNA, which binds to two independent protein complexes and results in histone H3 lysine 27 methylation and lysine 4 demethylation42.
A possible regulatory mechanism of lncRNAs is the interference of its transcription on the start of transcription of a neighbor gene from a downstream promoter, resulting in a down-regulation. A well-known example is the regulation of the SER3 gene by the upstream SRG1 ncRNA in S. cerevisiae43. This could be the case of the carP gene, whose transcription could interfere negatively with the transcription of carS. The increase in carS mRNA levels found in the ΔFo-carP mutants is consistent with this hypothesis. However, this effect is not so evident in the ΔFf-carP mutants, which again suggests a different mechanism of action between carP RNAs in both species. Moreover, the ΔFo-carP* deletion affects only the 3′ segment of carP, but the phenotype resulting of this mutation is very similar to the elimination of the entire carP segment. Approximately 56% of the carP sequence (659 of 1178 bp) is conserved in the ΔFo-carP* mutant. This should be interpreted as that the 3′ end region of carP plays an essential role in its function, but it does not mean that the rest of the transcript is not essential, as would happen if the function depended on the overall structure of the RNA.
The orientation of the carP transcript in the upstream region of carS in Fusarium species suggests an evolutionary origin from a long 5′-UTR region of the carS gene. This hypothesis is supported by the finding of the carP sequence in a single transcript with carS in F. verticillioides. 5′-UTRs are usually shorter than 3′-UTRs in eukaryotic transcripts, even in higher eukaryotes44. The length of the 5′-UTR of the carS gene in F. verticillioides would be very exceptional compared to usual 5′-UTR lengths. It should also be noted that, according to RNA-seq data, the levels of Fo-carP and Ff-carP are very low, and different from those of carS. This indicates a low transcription of carP or a very short life for this lncRNA. However, the levels of Fo-carP and Ff-carP are higher in carS mutants than in wild strains, suggesting a regulatory loop between both genes. This could be consistent with a direct role of carP on the CarS protein, that could result in the accumulation of carP in the absence of a functional CarS polypeptide.
Despite the functional similarity between Fo-carP and Ff-carP, there are some discrepancies between them that suggest differences in their mechanisms of action. RT-qPCR analyses show that in wild strains, Fo-carP is apparently induced by light, but Ff-carP is not. On the other hand, the ΔFo-carP mutants have lost the photoinduction of carotenoid synthesis, although they retain a slight photoinduction of the structural car genes, whereas the ΔFf-carP mutants show a clear decay in carotenoid synthesis in both light and in darkness and a drastic drop in mRNA levels of structural car genes, but they maintain a more clear photoinduction. ΔcarP phenotypes indicate that carP is necessary for high expression of car genes in both species, but its influence on photoinduction is only partial in F. oxysporum, and almost non-existent in F. fujikuroi. This is consistent with the lack of effect of the deletion of carP on the expression of the genes involved in the photoreceptor machinery, either in F. oxysporum or in F. fujikuroi. In summary, the characteristics described for carP conform to the requirements to be considered as a lncRNA that acts as an up-regulatory element in Fusarium carotenogenesis. Its mechanism of action is unknown, and it will be the focus of future work.
Methods
Strains and culture conditions
Wild strain of Fusarium oxysporum f. sp. lycopersici 4287 (race 2) was kindly provided by A. Di Pietro (Universidad de Córdoba, Spain), and carS mutants SX1 and SX2 were obtained from strain 4287 by chemical mutagenesis18. Wild strain of Fusarium fujikuroi IMI58289 was obtained from the Imperial Mycological Institute (Kew, Surrey, England), and the carS mutant SG39 was obtained from IMI58289 by chemical mutagenesis17. The carS-complemented strain SG256 was obtained through introduction of a wild carS allele in SG3919 and subsequent loss of the mutant allele20.
For routine maintenance and to obtain conidia, F. oxysporum strains were grown on PDB medium, either prepared in the laboratory (200 g potatoes were peeled, sliced, boiled in 1 l of distilled water for 60 minutes, and filtered. 20 g of glucose were added, and the volume was completed with distilled water to 1 l) or commercially obtained (24 g/l of PDB, FORMEDIUM LTD, 16 g/l agar). To obtain conidia, the strains were grown for 3–4 days with shaking at 200 rpm at 30 °C in 250 ml Erlenmeyer flasks with 100 ml of culture medium, inoculated with 108 conidia. The conidia were collected through a filter (Monodur 10–15 μm) and used immediately or kept frozen in 30% glycerol at −80 °C. For expression studies the strains were grown on DGasn minimal medium, containing 30 g/l glucose, 3 g/l asparagine, 1 g/l KH2PO4, 0.5 g/l KCl, 0.5 g/l MgSO4·7H2O, and microelements (adapted from DG medium45).
Because of their lower conidiation levels, conidia of F. fujikuroi strains were obtained after growth on sporulation agar, consisting of 1 g/l glucose, 1 g/l yeast extract, 1 g/l NO3NH4, 1 g/l KH2PO4, 0.5 g/l MgSO4·7H2O, 16 g agar. To collect the conidia, the culture surface was washed with water, the mycelial debris was removed by filtration, and the resulting conidia were counted in a haemocytometer (Bürker chamber, Blau Brand, Germany). For phenotypic characterizations and expression studies, F. fujikuroi strains were grown on DG medium, identical to DGasn medium but with 3 g/l NaNO3 instead of asparagine45.
For expression studies, in the case of F. oxysporum, Petri dishes of 15 cm diameter with 80 ml DGasn liquid medium were inoculated with 106 fresh conidia of the corresponding strains and incubated in the dark for three days. At this time, they were eventually exposed to light for one hour. In the case of F. fujikuroi, 500-ml Erlenmeyer flasks with 100 ml of DG medium were inoculated with 106 conidia of the corresponding strains and cultured in the dark for three days on an orbital shaker at 150 rpm. Subsequently, 25-ml samples were transferred to standard Petri dishes under red safelight and incubated for one hour in the dark or under white light. In both species, light exposures were performed under a platform with 4 fluorescent tubes (Philips TL-D 18 W/840) at a distance of ca. 60 cm, providing a light intensity of 7 w/m2 (420 Lm/w). Upon incubation, the mycelium samples were removed by filtration, washed with distilled water, frozen in liquid nitrogen, and stored at −80 °C.
For analysis of carotenoid production, standard 9-cm diameter Petri dishes with 25 ml solid medium were incubated at 30 °C in the dark or under light as described above. Each Petri dish was inoculated with 7 symmetrically distributed punctures with sterile sticks previously punctured on isolated fresh colonies.
PCR assays and determination of carP transcript orientation
Depending on the purpose of the experiment, different thermostable polymerases were used for PCR reactions: BIOTAQTM DNA polymerase (Bioline, Memphis, TN, U.S.A.) for PCR checks, Expand High Fidelity PCR System (Roche, Mannheim, Germany) for vector construction, and Velocity DNA polymerase (Bioline) for expected products larger than 5 kb. Reaction conditions were those indicated in manufacturer’s instructions. The reactions were carried out in total volumes of 25 µl with an amount of template DNA ranging from 10 to 50 ng for genomic DNA and from 1 to 10 ng for plasmid DNA. The sets of primers used are described in Supplementary Table S5.
To analyse the transcription orientation of the carP gene, a variant of the cDNA synthesis protocol was used. DNase-treated RNA was reverse-transcribed with the Transcriptor First Strand cDNA synthesis kit (Roche), following the manufacturer’s instructions. The polyT primer, normally used in the synthesis of cDNA, was replaced with a mixture of specific primers for each of the chains, so that only the cDNA substrate corresponding to the orientation of the transcript was obtained. Each primer mix consists of 3–4 different primers, described for each species of Fusarium in Supplementary Table S5, which cover different parts of the transcript in a single orientation. Subsequently, a PCR was performed for the carP gene with both substrates (primer sets 3 and 9) and it was only expected that the PCR product would be formed from the substrate generated by the mixture of primers complementary to the orientation of the transcript. As a control, the same analysis was performed in parallel with the β-tubulin gene in each species (primers 4, 5, 10, and 11, and primer sets 6 and 12).
Plasmid constructions and transformation
Plasmids for deletion of target sequences in F. oxysporum and F. fujikuroi were constructed by homologous recombination of four DNA segments, with overlapping end-sequences of at least 20 bases. One DNA segment was the 5.7-kb yeast vector pRS426 (Fungal Genetics Stock Center46), which contains the URA3 gene of S. cerevisiae, digested with XhoI and EcoRI at its multicloning site. Another DNA segment was the hygR cassette, which contains the hph gene of E. coli under control of regulatory sequences of Aspergillus nidulans, obtained by PCR from plasmid pCSN44 (Fungal Genetics Stock Center) with primer set 19. Finally, two DNA segments were obtained on both sides of the target sequence of carP by PCR using primers with tails overlapping with the ends of XhoI/EcoRI digested pRS426 on one side, and with the ends of the HygR resistance cassette on the other side. The four fragments were introduced by transformation into the strain FY834 of S. cerevisiae (MATα, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2Δ202), where they recombined through the coinciding end-sequences to generate circular plasmids.
Using this method, the 8.9-kb plasmid pDul8 was obtained through the joining of the linear pRS426 and HygR cassette segments with a DNA fragment upstream to carP*, obtained with primer set 23, and with a DNA fragment downstream of carP*, obtained with primer set 24. Similarly, the 9.8-kb plasmid pDul14 was obtained by joining the linear pRS426 and HygR cassette segments with a DNA fragment upstream to the entire Fo-carP sequence, obtained with primer set 20; and with a DNA fragment downstream of Fo-carP, obtained with primer set 21. pDul8 and pDul14 were used to generate the carP* and carP deletions in F. oxysporum, respectively. The same procedure was followed to obtain the 9.3-kB plasmid pcarPhyg, used to obtain the carP deletion in F. fujikuroi. In this case, the DNA segments corresponding to the pRS426 and the HygR cassette were joined with DNA fragments upstream (obtained with primer set 26) and downstream (primer set 27) to Ff-carP. Transformations were achieved with linear replacement cassettes obtained from plasmids pDul14, pDul8, and pcarHyg with primers sets 22, 25, and 28, respectively. Sizes of the 5′ and 3′ carP border regions in the replacement cassettes were (bp): 1429 (5′) and 1508 (3′) for carP, and 1193 (5′) and 790 (3′) for carP* in F. oxysporum, and 1175 (5′) and 1257 (3′) for carP in F. fujikuroi.
Protoplasts of the wild strain of F. fujikuroi were obtained and transformed as described47. The same protocol was used for the wild strain of F. oxysporum, except that for the formation of protoplast 5 × 108 spores were inoculated into 100 ml YPED-2G (3 g/l yeast extract, 10 g/l peptone, 20 g/l glucose) and incubated for 10–12 hours at 22 °C. Germlings were collected by filtration, washed with 0.7 M NaCl, and transferred to 15 ml of an enzyme solution in 0.7 M NaCl containing 13 g/l of lysing enzymes of Trichoderma harzianum (Sigma-Aldrich, St. Louis, MO, USA), 33 g/l of driselase from basidiomycetes (Sigma-Aldrich) and 60 mg/l of chitinase from Streptomyces griseus (Sigma-Aldrich). The enzymatic treatment lasted between 30 minutes and 1 hour until protoplast formation.
Generation of ΔcarP and ΔcarP* mutants in F. oxysporum
To obtain ΔFo-carP mutant strains, wild protoplasts were transformed with the linear replacement cassette derived from plasmid pDul14. Five transformants were obtained by selection on hygromycin-supplemented medium and, after three selection steps through uninucleate spores to ensure homokaryosis, they were analysed by PCR using primers external to the deleted region. Only one of the 5 transformants (#3) gave the 4.4 kb band corresponding to the replacement of carP with the HygR cassette, the same size of the DNA product obtained with the plasmid pDul14 (Supplementary Fig. S11A). In the other 4 transformants, a 4.2 kb band similar to that found in the wild strain and corresponding to the intact Fo-carP sequence was obtained, indicating heterologous integrations of the plasmid. As an additional verification, a Southern blot hybridization of genomic DNA from the wild strain and transformant #3 was performed (Supplementary Fig. S11B) using as a probe a DNA segment next to the deleted region (Supplementary Fig. S11C). Genomic DNA samples were digested with AvaI restriction enzyme, for which there is a target within the carP gene, but not in the HygR cassette. The result showed a 3.3-kb band in the transformant, indicating the presence of the HygR cassette instead of Fo-carP, and a 1.8 kb band in the wild strain, as expected from the presence of the Fo-carP sequence. This ΔcarP transformant was named SX82.
In addition, ΔFo-carP* mutants were obtained by transformation of wild protoplasts with the linear replacement cassette obtained from plasmid pDul8. As a result, eight transformants were obtained, passed three times through uninucleate spores, and checked by PCR with primers surrounding the replaced region. Five transformants (#2, #3, #4, #5, #8) showed the expected 2.7 kb band instead of 2 kb of the wild strain, indicating the replacement of carP* by the HygR cassette (Supplementary Fig. S11D). Two positive transformants were chosen for verification by Southern blot using as a probe a DNA segment adjacent to the deleted carP* region (Supplementary Fig. S11C). This hybridization gave a 4-kb band in the case of the two transformants analysed (#3 and #4), consistent with the replacement of carP* by the HygR cassette, compared to the 1.8 kb band obtained with the wild strain (Supplementary Fig. S11E). These ΔcarP* transformants were called SX80 and SX81.
Generation of ΔcarP mutants in F. fujikuroi
Wild protoplasts were transformed with the linear replacement cassette obtained from plasmid pcarPhyg, and twenty transformants were obtained upon selection on hygromycin-supplemented medium. After three passes through uninucleate spores, the replacement of the carP sequence by the HygR cassette in the candidate transformants was analyzed by PCR using different primer combinations (Supplementary Fig. S12A). In eleven of the transformants, a 3.7 kb band was obtained with primer set 33, corresponding to an amplification from a sequence external to carP and absent in pcarPhyg, and a terminal HygR sequence, indicating the desired loss of the carP sequence (positive transformants #1, #2, #4, and #10 shown in Supplementary Fig. S12B). Southern blot hybridization of genomic DNA from the wild strain, three of the positive transformants (#1, #2, and #4) and one negative (#6), was carried out using as a probe a DNA segment near the deleted region (Supplementary Fig. S12A). Genomic DNA samples were digested with restriction enzymes XhoI and EcoRI, chosen for their opposite presence in the carP gene and in the HygR cassette. The results in the EcoRI-digested samples showed a 4-kb band in the wild strain and in the negative transformant, and the expected 1.9-kb band in the positive transformants (Supplementary Fig. S12C). In samples digested with XhoI, the 4.9-kb band corresponding to the presence of the HygR cassette was found in the positive transformants, while the expected 3.1-kb band for the carP sequence was found in the wild strain and in the negative transformant, confirming the results of the PCR tests and the hybridization of the samples digested with EcoRI. Therefore, the transformants #1, #2, and #4 had lost the Ff-carP sequence and were chosen for detailed phenotypic characterization. These strains were called SG268, SG269, and SG270, respectively.
Southern blot of transformants
Southern blot analyses were achieved essentially as described48. Samples of at least 10 μg of genomic DNA were treated with restriction enzymes and the resulting products were electrophoresed on a 0.7% agarose gel and subjected to the Southern protocol using a positively charged nylon membrane (Hybond-N from Amersham). The probes used in F. oxysporum were obtained from genomic DNA sequences by PCR with primer set 30 in the case of the southern for ΔcarP mutants, and primer set 32 in the case of the southern for ΔcarP* mutants. The probe used in the southern of the F. fujikuroi transformants was obtained with primer set 37. The probes were labelled with 32P dCTP and the membranes were exposed for radioactivity detection in a radioisotope imaging system (FujiFilm FLA 5100, Life Science, Cambridge, MA, USA).
Expression analysis
Transcript levels for the genes of interest were determined by reverse transcription qPCR (RT-qPCR) with a LightCycler 480 real-time equipment (Roche). Total RNA samples were extracted with the RNeasy RNA isolation kit (Qiagen, Chatsworth, CA, USA). RNA concentrations were determined with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). Samples of 2.5 μg of RNA were retrotranscribed to cDNA with Transcriptor first-strand cDNA synthesis kit (Roche), and final cDNA concentrations were adjusted to 25 ng/μl. LightCycler 480 SYBR Green I Master (Roche) was used for amplification and detection following the manufacturer’s protocols. Genes and primer sets (forward vs reverse in 5′- >3′ orientation) for F. oxysporum genes carP, carS, carRA, carB, carO, carX, carD, carT, cryD, vvdA, wc1, and the con-10 ortholog FOXG_01269, and for F. fujikuroi genes carP, carS, carRA, carB, cryD, vvdA, and wcoA, are listed in Supplementary Table S5E (primers sets 38–50 and 52–59). Transcript levels were normalized against the β-tubulin gene FOXG_06228 of F. oxysporum (primer set 51) and FFUJ_04397 of F. fujikuroi (primer set 61). In some cases, the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene gpdh1, FFUJ_13490, was used as a second reference gene in F. fujikuroi (primer set 60).
Statistical significance of differences between mRNA values was checked with the Welch’s t test using the GraphPad QuickCalcs program (www.graphpad.com/quickcalcs/contMenu). Since data were relativized to the value of the wild strain grown in the dark, tests against these values were carried out with the one sample t test.
Bioinformatic analyses
The identification of ORFs in carP was carried out with ORFfinder49 and the detection of potential protein domains was performed with InterproScan50. Clustal alignments were done through the EMBL/EBI server (https://www.ebi.ac.uk/Tools/msa/clustalo). We used different approaches to check the coding capability of carP. Three-base periodicity was checked through TestCode statistics51 with the Tcode program of EMBOSS52, using a window of 200 and a step of 3. Codon usage statistics was plotted with the program Syco of EMBOSS52 using a codon usage table generated by the Cusp program of EMBOSS52 from the F. oxysporum CDSs. The protein-coding potential of carP was also analysed using the CPC28 and CPC229 algorithms. Coding capability can be also detected by evolutionary conservation. For that, we first made a structure-based alignment of carP homologous sequences with LocARNA31. Then we studied coding capability by evolutionary conservation with RNAcode30.
For the prediction of secondary structure for carP, we used LocARNA31 plotted with VARNA53. Maximum-likelihood trees were generated with the IQ-Tree server54. ModelFinder, implemented at the IQ-Tree server, was used to find the free rate heterogeneity substitution model that best fit the alignments55. Maximum-likelihood tree of carP was generated with the carP sequences shown in Fig. S6. The tree derives from the multiple alignment based on the structures carried out with the locARNA program used for the RNAcode test. The maximum-likelihood tree for species phylogeny was based on the translation elongation factor 1-alpha (TEF1). TEF1 was selected as species barcoding gene because rRNAs of the studied species are very similar and ITSs showed intrastrain sequence heterogeneity. TEF1 has proven to be one of the best barcoding genes in fungi56. Multiple alignment was performed with ProbCons57. In both trees, branch support analysis was evaluated by 1000 ultrafast bootstrap replicates.
Supplementary information
Acknowledgements
This work was funded by the Spanish Government (Ministerio de Economía y Competitividad, projects BIO2015-69613-R and RTI2018-101902-B-I00). G.G. was funded by project BIO2015-67148-R. Spanish grants included support from the Regional Development Fund (ERDF) of the European Union. Authors belong to the Spanish Carotenoid Network (CaRed) funded by the Spanish MINECO (Ministry of Economy, Industry and Competitiveness) grant BIO2015-71703-REDT, and the European Carotenoid Network (EuroCaroten) funded by the European Commission COST Action CA15136. We thank L. Roberto Rodríguez-Ortiz for previous work in the detection of the putative microRNA in F. oxysporum.
Author contributions
O.P.-R. and J.P.-M. performed research; G.G. carried out the bioinformatic analyses of the carP sequence, J.A. and M.C.L. supervised the work and wrote the manuscript. All authors reviewed and revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Obdulia Parra-Rivero and Javier Pardo-Medina.
Supplementary information
is available for this paper at 10.1038/s41598-020-57529-2.
References
- 1.Gordon TR. Fusarium oxysporum and the Fusarium wilt syndrome. Annu. Rev. Phytopathol. 2017;55:23–39. doi: 10.1146/annurev-phyto-080615-095919. [DOI] [PubMed] [Google Scholar]
- 2.Wiemann P, et al. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9:e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Pietro A, Madrid MP, Caracuel Z, Delgado-Jarana J, Roncero MIG. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 2003;4:315–325. doi: 10.1046/j.1364-3703.2003.00180.x. [DOI] [PubMed] [Google Scholar]
- 4.Turrà D, Di Pietro A. Chemotropic sensing in fungus-plant interactions. Curr. Opin. Plant Biol. 2015;26:135–140. doi: 10.1016/j.pbi.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Niehaus, E.-M. et al. Fusarins and Fusaric Acid in Fusaria. In Biosynthesis and Molecular Genetics of Fungal SecondaryMetabolites (eds. Martín, J.-F., García-Estrada, C. & Zeilinger, S.) 239–262 (Springer New York, 2014).
- 6.Studt, L. & Tudzynski, B. Gibberellins and the red pigments bikaverin and fusarubin. In Biosynthesis and Molecular Genetics of Fungal SecondaryMetabolites (eds. Martín, J.-F., García-Estrada, C. & Zeilinger, S.) 209–238 (Springer New York, 2014).
- 7.Avalos J, et al. Carotenoid biosynthesis in Fusarium. J. Fungi. 2017;3(39):1–16. doi: 10.3390/jof3030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thewes S, Prado-Cabrero A, Prado MM, Tudzynski B, Avalos J. Characterization of a gene in the car cluster of Fusarium fujikuroi that codes for a protein of the carotenoid oxygenase family. Mol. Genet. Genomics. 2005;274:217–228. doi: 10.1007/s00438-005-0015-6. [DOI] [PubMed] [Google Scholar]
- 9.Prado-Cabrero A, et al. Deviation of the neurosporaxanthin pathway towards β-carotene biosynthesis in Fusarium fujikuroi by a point mutation in the phytoene desaturase gene. FEBS J. 2009;276:4582–4597. doi: 10.1111/j.1742-4658.2009.07164.x. [DOI] [PubMed] [Google Scholar]
- 10.Prado-Cabrero A, Scherzinger D, Avalos J, Al-Babili S. Retinal biosynthesis in fungi: Characterization of the carotenoid oxygenase CarX from Fusarium fujikuroi. Eukaryot. Cell. 2007;6:650–657. doi: 10.1128/EC.00392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Martínez J, Brunk M, Avalos J, Terpitz U. The CarO rhodopsin of the fungus Fusarium fujikuroi is a light-driven proton pump that retards spore germination. Sci. Rep. 2015;5:7798. doi: 10.1038/srep07798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avalos J, Estrada AF. Regulation by light in Fusarium. Fungal Genet. Biol. 2010;47:930–938. doi: 10.1016/j.fgb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Castrillo M, Avalos J. The flavoproteins CryD and VvdA cooperate with the white collar protein WcoA in the control of photocarotenogenesis in Fusarium fujikuroi. PloS One. 2015;10:e0119785. doi: 10.1371/journal.pone.0119785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estrada AF, Avalos J. The White Collar protein WcoA of Fusarium fujikuroi is not essential for photocarotenogenesis, but is involved in the regulation of secondary metabolism and conidiation. Fungal Genet. Biol. 2008;45:705–718. doi: 10.1016/j.fgb.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Castrillo M, Bernhardt A, Avalos J, Batschauer A, Pokorny R. Biochemical characterization of the DASH-type cryptochrome CryD from Fusarium fujikuroi. Photochem. Photobiol. 2015;91:1356–1367. doi: 10.1111/php.12501. [DOI] [PubMed] [Google Scholar]
- 16.Castrillo M, Avalos J. Light-mediated participation of the VIVID-like protein of Fusarium fujikuroi VvdA in pigmentation and development. Fungal Genet. Biol. 2014;71:9–20. doi: 10.1016/j.fgb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Avalos J, Cerdá-Olmedo E. Carotenoid mutants of Gibberella fujikuroi. Curr. Genet. 1987;11:505–511. doi: 10.1007/BF00384613. [DOI] [Google Scholar]
- 18.Rodríguez-Ortiz R, Michielse C, Rep M, Limón MC, Avalos J. Genetic basis of carotenoid overproduction in Fusarium oxysporum. Fungal Genet. Biol. 2012;49:684–696. doi: 10.1016/j.fgb.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez-Ortiz R, Limón MC, Avalos J. Functional analysis of the carS gene of Fusarium fujikuroi. Mol. Genet. Genomics. 2013;288:157–173. doi: 10.1007/s00438-013-0739-7. [DOI] [PubMed] [Google Scholar]
- 20.Ruger-Herreros M, et al. Comparative transcriptomic analysis unveils interactions between the regulatory CarS protein and light response in Fusarium. BMC Genomics. 2019;20:67. doi: 10.1186/s12864-019-5430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro E, et al. A negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol. Genet. Genomics. 2001;266:463–470. doi: 10.1007/s004380100558. [DOI] [PubMed] [Google Scholar]
- 22.Lorca-Pascual JM, Murcia-Flores L, Garre V, Torres-Martínez S, Ruiz-Vázquez RM. The RING-finger domain of the fungal repressor crgA is essential for accurate light regulation of carotenogenesis. Mol. Microbiol. 2004;52:1463–1474. doi: 10.1111/j.1365-2958.2004.04070.x. [DOI] [PubMed] [Google Scholar]
- 23.Silva F, et al. A RING-finger protein regulates carotenogenesis via proteolysis-independent ubiquitylation of a White Collar-1-like activator. Mol. Microbiol. 2008;70:1026–1036. doi: 10.1111/j.1365-2958.2008.06470.x. [DOI] [PubMed] [Google Scholar]
- 24.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. TIG. 2018;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Tao Y, Liao Q. Long noncoding RNA: a crosslink in biological regulatory network. Brief. Bioinform. 2018;19:930–945. doi: 10.1093/bib/bbx042. [DOI] [PubMed] [Google Scholar]
- 28.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang Y-J, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Washietl S, et al. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA N. Y. N. 2011;17:578–594. doi: 10.1261/rna.2536111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA N. Y. N. 2012;18:900–914. doi: 10.1261/rna.029041.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Roldán MC, Garre V, Guarro J, Mariné M, Roncero MI. Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot. Cell. 2008;7:1227–1230. doi: 10.1128/EC.00072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corrochano LM, Lauter FR, Ebbole DJ, Yanofsky C. Light and developmental regulation of the gene con-10 of Neurospora crassa. Dev. Biol. 1995;167:190–200. doi: 10.1006/dbio.1995.1016. [DOI] [PubMed] [Google Scholar]
- 34.Quan M, Chen J, Zhang D. Exploring the secrets of long noncoding RNAs. Int. J. Mol. Sci. 2015;16:5467–5496. doi: 10.3390/ijms16035467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita A, Shichino Y, Yamamoto M. The long non-coding RNA world in yeasts. Biochim. Biophys. Acta. 2016;1859:147–154. doi: 10.1016/j.bbagrm.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Arthanari Y, Heintzen C, Griffiths-Jones S, Crosthwaite SK. Natural antisense transcripts and long non-coding RNA in Neurospora crassa. PloS One. 2014;9:e91353. doi: 10.1371/journal.pone.0091353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, W., Miguel-Rojas, C., Wang, J., Townsend, J. P. & Trail, F. Developmental dynamics of long noncoding RNA expression during sexual fruiting body formation in Fusarium graminearum. mBio9 (2018). [DOI] [PMC free article] [PubMed]
- 38.Zhu Q-H, Stephen S, Taylor J, Helliwell CA, Wang M-B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014;201:574–584. doi: 10.1111/nph.12537. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Li C, Li S, Peng M. Long noncoding RNAs that respond to Fusarium oxysporum infection in ‘Cavendish’ banana (Musa acuminata) Sci. Rep. 2017;7:16939. doi: 10.1038/s41598-017-17179-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Till P, Pucher ME, Mach RL, Mach-Aigner AR. A long noncoding RNA promotes cellulase expression in Trichoderma reesei. Biotechnol. Biofuels. 2018;11:78. doi: 10.1186/s13068-018-1081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai M-C, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 44.Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Avalos J, Casadesús J, Cerdá-Olmedo E. Gibberella fujikuroi mutants obtained with UV radiation and N-methyl-N’-nitro-N-nitrosoguanidine. Appl. Environ. Microbiol. 1985;49:187–191. doi: 10.1128/AEM.49.1.187-191.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCluskey K, Wiest A, Plamann M. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 2010;35:119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- 47.Marente, J., Ortega, P., Pardo-Medina, J., Avalos, J. & Limón, M. C. Modulation of activity of a carotenoid pathway through the use of the TET-on regulatory system: application in the fungus Fusarium fujikuroi. Methods Mol. Biol. In press (2019). [DOI] [PubMed]
- 48.Sambrook, J. & Russell, D. W. Molecular cloning: a laboratory manual. (Cold Spring Harbor Laboratory Press, 2001).
- 49.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics Oxf. Engl. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fickett JW. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. TIG. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 53.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics Oxf. Engl. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu J. Fungal DNA barcoding. Genome. 2016;59:913–932. doi: 10.1139/gen-2016-0046. [DOI] [PubMed] [Google Scholar]
- 57.Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.