Abstract
Background
Super‐resolution microscopy has enabled high‐resolution imaging of the actin cytoskeleton in megakaryocytes and platelets. These technologies have extended our knowledge of thrombopoiesis and platelet spreading using megakaryocytes and platelets cultured in vitro on matrix proteins. However, for better understanding of megakaryocytopoiesis and platelet production, high‐resolution imaging of cells in an in vivo bone marrow microenvironment is required. Development of Kawamoto’s film method greatly advanced the techniques of thin cryosectioning of hard tissues such as undecalcified bones. One obstacle that remains is the spherical aberration that occurs due to the difference in the refractive index for the light path, limiting the usage of Kawamoto’s film method to lower magnification observation.
Objectives
To overcome the weakness of the conventional Kawamoto’s film method for higher magnification observation of undecalcified bone marrow.
Methods
We have modified the original method with a very simple method: flipping the film at the step of mounting the sections on the glass.
Results and Conclusions
This new method successfully led to the adjustment of the refractive index and enabled super‐resolution imaging of megakaryocytes in undecalcified mouse femurs. Our modified method will expand the application of Kawamoto’s film method and enable precise analysis of megakaryocytopoiesis and platelet production in the bone marrow microenvironment under pathophysiological conditions.
Keywords: bone marrow, frozen sections, histological techniques, megakaryocytes, single‐molecule imaging
Essentials.
Kawamoto’s film method is widely used for thin sectioning of frozen tissues.
A modified method was developed to overcome difficulties in imaging at higher magnifications.
Flipping the film and adjusting the refractive index improved the optical property and resolution.
Imaging beyond the diffraction limit in undecalcified bone is possible for megakaryocyte studies.
1. INTRODUCTION
Seeing is believing; optical imaging is one of the most powerful tools in biology. These data bring convincing evidence, novel insights, or in‐depth understanding in biology. Especially in histological analysis, visualization of antigens or colocalization provides a snapshot of physiological phenomena in vivo. Recent advances in optical technologies have brought revolutionary improvements in resolution. Super‐resolution microscopy, which won the Nobel Prize in Chemistry in 2014, is the representative technology.1, 2, 3 Super‐resolution imaging has been applied to in vitro cell culture, including megakaryocytes and platelets.4, 5 Some researchers have reported super‐resolution imaging in soft tissue such as brain,6 cardiac tissue,7 or rectal cancer.8 However, there have been no reports of super‐resolution imaging that visualized megakaryocytes in the intact structure of bone marrow.
One major barrier is that the parenchyma of bone marrow is surrounded by hard bone tissue, making it difficult to make sections compared to other tissue samples. Without special treatment or techniques, it is impossible to make a thin‐slice section while maintaining the integrity of the structure for histological analysis of bone marrow. One alternative method is to flush the bone marrow from the femur and then make thin sections.9 Another example is decalcification of the bone marrow.10 However, there is a trade‐off in these techniques. Flushing the bone marrow requires expertise to obtain a reliable sample and may still have the potential to destroy the micro‐architecture of hematopoietic cells. In the study of megakaryocyte migration and platelet generation, as well as in the stem cell research, analysis of the localization of cells in the bone marrow microenvironment is crucial.11, 12 On the other hand, the decalcification procedure requires harsh processing, which may influence the protein epitopes in the samples and recognition by the antibody.13, 14 Furthermore, decalcification is a time‐consuming process, and it takes several days to complete the EDTA decalcification process.
As is well known, Kawamoto and Kawamoto15 have developed a unique method using a special adhesive film to maintain the structure of the specimen. This method enabled us to make frozen sections easily from soft tissue, hard tissue including bone, or plant tissue as thin as several microns without needing decalcification. With the advantage that the proteins of specimens can be kept intact, this method has been applied in a wide range of research such as laser microdissection,16 enzymatic histochemistry,17 or mass spectrometry.18 The biggest advantage of this method is that thin‐sliced cryosections can be prepared in the same way as soft tissue, maintaining the antigen epitopes and expanding the options of antibodies for staining. The only concern is that looking at the specimen through the film is not compatible for high magnification or high numerical aperture (NA) objective lens because of the properties of the film. Therefore, most studies using Kawamoto’s film method have shown imaging captured at lower magnification.19, 20
From this context, we have developed a simple method to overcome the weakness of the conventional method while keeping the great advantage of the films. We discovered a way to modify the process of mounting the films to adjust the refractive index to minimize light scattering. Here, we report a modified application of Kawamoto’s film method, which greatly improved the resolution of megakaryocyte over the diffraction limit in undecalcified bone tissue.
2. MATERIAL AND METHODS
2.1. Sample preparation
All animal studies were performed according to protocol approved by the Scripps Research Institute and Institutional Animal Care and Usage Committee. According to the previous report of Kawamoto’s film method,17 bone marrow sections were prepared. Briefly, the limb was taken from 8‐ to 10‐week‐old male mice of C57BL/6J wild type, then directly snap‐frozen, soaking in hexane with dry ice and embedded in embedding medium. Samples were sectioned at 5 μm using a cryostat. Sections were fixed with 4% paraformaldehyde for 5 minutes and washed, then followed by immunostaining.
2.2. Immunostaining
Sections were fixed with 4% paraformaldehyde, then blocked and permeabilized with 0.1% Triton X‐100 PHEM buffer containing 5% normal goat serum (Invitrogen, Waltham, MA, USA). CD41 was stained with rat anti‐mouse Integrin αIIb (MWReg30; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and visualized with goat anti‐rat IgG secondary antibody Alexa Fluor 488 (Invitrogen). ß‐tubulin was stained with mouse anti‐mouse ß‐tubulin (TUB 2.1; Santa Cruz Biotechnology) and visualized with goat anti‐mouse IgG secondary antibody Alexa Fluor 555 (Invitrogen). F‐actin was stained with Alexa Fluor 647 phalloidin (Invitrogen). The nuclei were counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI) (Dojindo Molecular Technologies, Kumamoto, Japan).
2.3. Mounting procedure
For the conventional Kawamoto’s film method, after placing an adequate amount of mounting media, the film with the specimen was directly attached onto a glass slide. For the modified Kawamoto’s film method, the film with the specimen was first attached to a precision coverslip #1.5H (THORLABS, Newton, NJ, USA) with mounting media. The coverslip and film were subsequently placed onto a glass slide. To avoid air bubbles, additional mounting media was supplemented between the coverslip and film. Nail polish (Electron Microscopy Sciences, Hatfield, PA, USA) was used for sealing the edges of the coverslip. Mounting media was Prolong Glass (Invitrogen) for both methods. After 24 hours for curing of the mounting media, the following microscopic observation was conducted.
2.4. Wide‐field epi‐fluorescence microscopy
For wide‐field imaging, images were acquired with BZ‐X700 all‐in‐one fluorescence micro×scope (Keyence Corp., Osaka, Japan). Plan‐apochromat 10×/0.45 dry and plan‐apochromat 60×/1.40 oil immersion objective lens were used for the observation. The images were analyzed with BZ‐X Analyzer software (Keyence Corp.).
2.5. Confocal laser scanning microscopy
Confocal laser scanning microscopy (CLSM) was performed with an LSM 880 with Airyscan module (Carl Zeiss AG, Oberkochen, Germany). The objective lens was an alpha Plan‐Apochromat 100×/1.46 DIC M27 Elyra oil immersion objective lens. The image was analyzed with Fiji image analysis software.21 For the deconvolution of F‐action and ß‐tubulin signal, super‐resolution radial fluctuation (SRRF) was applied using NonoJ‐SRRF (ImageJ plugin, NIH, Bethesda, MD, USA).22
2.6. Stochastic optical reconstruction microscopy
For stochastic optical reconstruction microscopy (STORM) imaging, the mounting medium was changed to Vectashield (Vector Laboratories, Burlingame, CA, USA) according to the previous report.23 CFI Apochromat TIRF 100×/1.49 oil immersion objective lens on a Nikon Eclipse TE2000 inverted microscope equipped with a Nikon N‐STORM system was used. The image was analyzed with NIS‐Elements AR (Nikon Inc., Melville, NY, USA).
2.7. Quantification of images
For quantification of the fluorescence intensity, ImageJ was used. For absolute measurement of image focus quality, the images were analyzed by Microscope Image Focus Quality Classifier (ImageJ plugin).24 Data were analyzed by Prism7 (GraphPad Software, La Jolla, CA, USA). Significant differences were determined by the Student’s t‐test.
3. RESULTS AND DISCUSSION
The overview of our method is depicted in Figure 1. In the conventional Kawamoto’s film method, the film was directly attached onto the glass slide, and the section was preserved between the film and the glass slide. The film was in the middle of the microscope’s light path to the specimen. Because of the film’s property, the film can cause spherical aberrations or light scattering, which leads to a decrease of signal intensity and blurriness. To avoid this issue, the film was inversely placed by attaching the film to the coverslip, allowing the light path to travel without scattering. Also, to match the refractive index close to the coverslip, we used Prolong Glass, which has a 1.52 refractive index, similar to the mounting media. To compare the 2 methods, we used the femurs of wild‐type C57BL/6J mice, and stained CD41 and nuclei to visualize the megakaryocytes in the bone marrow. In the wide‐field microscopic observation performed by 10× NA = 0.45 dry objective lens, there was no obvious difference between the 2 methods (Figure 2A‐C). However, at higher magnification captured with 60× NA = 1.40 oil immersion objective lens, fluorescence intensity and focus quality were significantly higher with the specimens prepared with our modified method as compared to those obtained via the conventional Kawamoto’s film method (Figure 2A‐C).
Figure 1.
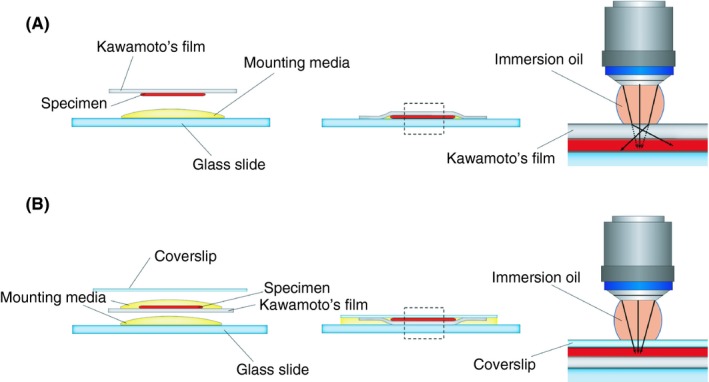
Schematic illustration of the conventional Kawamoto’s film method and the modified Kawamoto’s film method. (A) conventional Kawamoto’s film method; (B) modified Kawamoto’s film method. Dotted line area is shown in the right panel as the magnified figure. In the conventional Kawamoto’s film method, the film with the specimen was directly attached on the glass slide. In the modified Kawamoto’s film method, the film with the specimen was inverted and sandwiched between the coverslip and glass slide
Figure 2.
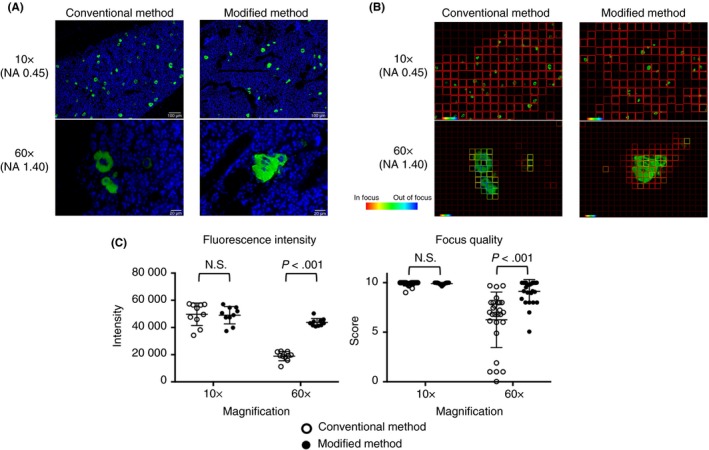
Image comparison of megakaryocytes in the bone marrow between the conventional Kawamoto’s film method and the modified Kawamoto’s film method in the wide‐field microscopy. (A) Merged images of DAPI (blue) and CD41 (green). Conventional Kawamoto’s film method (left column), modified Kawamoto’s film method (right column). Images were taken with 10× NA = 0.45 dry objective lens (upper row), with 60× NA = 1.40 oil immersion objective lens (lower row). All samples were prepared in the same condition and all images were captured by Epi‐fluorescence microscope (BZ‐X700) in the same setting. (B) Representative images of the absolute measurement of focus quality. The focus quality for CD41 signal was analyzed. (C) Quantitative analysis of the fluorescence intensity (left, n = 10 fields) and the image focus quality (right, n = 27 fields). DAPI, 4′,6‐diamidino‐2‐phenylindole
Next, we investigated whether the modified Kawamoto’s film method is compatible with super‐resolution microscopy. To make the signals of F‐actin and ß‐tubulin more precise, we processed the deconvolution by SRRF. With this method, F‐actin filament and ß‐tubulin in a megakaryocyte could be visualized with super‐resolution quality (Figure 3A). We further investigated the availability of STORM with Kawamoto’s film method. We were not able to perform STORM with the conventional Kawamoto’s method due to the blurriness of the images. With the modified Kawamoto’s method, after detecting megakaryocyte in the bone marrow by CD41, STORM imaging was conducted for the signals of AlexaFluor 647 phalloidin (Figure 3B). In the enlarged figure, the arrows indicate the accumulation of the phalloidin signals.
Figure 3.

Super‐resolution image of megakaryocytes in the bone marrow. (A) Images captured by LSM 880 with Airyscan module. Images are merged DAPI with CD41 (left), ß‐tubulin (middle) and F‐actin (right). Scale bar represents 5 μm. (B) STORM imaging of megakaryocytes in the bone marrow. Dotted area is enlarged in the right panel. The signal of phalloidin is shown in gray scale. DAPI, 4′,6‐diamidino‐2‐phenylindole; STORM, stochastic optical reconstruction microscopy
Kawamoto’s film method has the strong advantage that enables us to stain a hard tissue sample without a decalcifying process in a simple way. By virtue of that, it has provided enormous histologic evidence and has played a pivotal role in wide‐ranging research areas. However, on account of its film property, it has not been compatible with higher magnification observation as shown in Figure 2. To obtain clear, optimal images, elaborate sample preparation is required as subtle optical obstruction will influence the quality of the images, especially at higher magnification. The refractive index mismatch between the immersion oil and specimen can cause spherical aberrations, which impedes the resolution of microscopy, leading to the deterioration of the resolution and fluorescence intensity to become dim.25 Conventional Kawamoto’s film method uses the film not only as a supportive material of the section but also as a coverslip. Therefore, the difference in the refractive index between the immersion oil and the film, or the film and the mounting medium, can cause spherical aberrations (Figure 1A), resulting in signal reduction and blurriness in high‐magnification observation (Figure 2A‐C). For minimizing spherical aberrations, we have modified the method by (1) flipping the film to keep it away from the light path; (2) adjusting the refractive index using a mounting medium of which refractive index is close to 1.52; and (3) using the high‐precision coverslip, which has uniform thickness.
The majority of studies on megakaryocytes that use super‐resolution microscopy have been performed in vitro,26 which may have “artificial” conditions. With the proposed method, the molecular behavior of megakaryocytes will be able to be visualized in super‐resolution imaging in an intact bone marrow structure. Especially, the technique might have a potential to contribute to thrombosis and hemostasis research from the viewpoint of cytoskeletal proteins and megakaryocytopoiesis as shown in Figure 3.
In this paper, we showed staining of megakaryocytes only as an example of immunofluorescence of the undecalcified bone marrow; however, this modified method is expected to expand the usage of Kawamoto’s method for super‐resolution microscopy of various tissue samples. Kawamoto’s film method has been used for hematologic research as well as orthopedic research27 or cancer research such as bone metastasis.28 Additionally, Kawamoto’s film method is very useful for fragile tissue, such as lipid‐rich tissue or lymph nodes, because this method can confer the stability of tissue sectioning. Our modified method will be able to provide an opportunity to conduct super‐resolution microscopic analysis of these challenging tissues as well.
Kawamoto’s film method can be a compatible and feasible technique even in this super‐resolution era. To our best knowledge, this is the first report that achieved super‐resolution in undecalcified bone marrow tissue samples. Kawamoto’s film method has the great advantage of making sections of hard tissue without decalcification; however, we speculate that the film itself had suboptimal optics. We were able to overcome the optical issue of conventional Kawamoto’s film method by flipping the direction and adjusting the refractive index. Super‐resolution microscopy enables us to see subcellular imaging not only in in vitro cell culture but also in tissue specimens. This proposed method might be useful for further experimental or clinical research among bone‐ or bone marrow–related research fields, especially in hematologic research.
4. RELATIONSHIP DISCLOSURES
T. Kawamoto holds the patent (Japanese patent application number JP3424126B2) for the Kawamoto’s film method and received royalties from the patent. YM is supported by a fellowship of MERU Foundation, Italy, at The Scripps Research Institute, MERU‐Roon Research Center for Vascular Biology. EW is supported by University of California, San Diego/Rady Children's Hospital Pediatric Hematology/Oncology fellowship. The remaining authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
YM designed and performed the experiments, interpreted the data, and wrote the manuscript. SK and EW performed experiments and revised the manuscript. T Kawamoto helped with experimental design and revised the manuscript. T Kanaji designed the experiments, directed the study, and reviewed the manuscript.
ACKNOWLEDGMENTS
We thank Kersi Pestonjamasp for technical assistance with STORM imaging, and Scott Henderson for CLSM (both are Microscopy core at the Scripps Research Institute).
Morodomi Y, Kanaji S, Won E, Kawamoto T, Kanaji T. Modified application of Kawamoto’s film method for super‐resolution imaging of megakaryocytes in undecalcified bone marrow. Res Pract Thromb Haemost. 2020;4:86–91. 10.1002/rth2.12276
Handling Editor: Dr Yotis Senis
REFERENCES
- 1. Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated‐emission‐depletion fluorescence microscopy. Opt Lett. 1994;19:780–2. [DOI] [PubMed] [Google Scholar]
- 2. Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–8. [DOI] [PubMed] [Google Scholar]
- 3. Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–5. [DOI] [PubMed] [Google Scholar]
- 4. Westmoreland D, Shaw M, Grimes W, Metcalf DJ, Burden JJ, Gomez K, et al. Super‐resolution microscopy as a potential approach to diagnosis of platelet granule disorders. J Thromb Haemost. 2016;14:839–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poulter NS, Pollitt AY, Davies A, Malinova D, Nash GB, Hannon MJ, et al. Platelet actin nodules are podosome‐like structures dependent on Wiskott‐Aldrich syndrome protein and ARP2/3 complex. Nat Commun. 2015;6:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tønnesen J, Inavalli V, Nägerl UV. Super‐resolution imaging of the extracellular space in living brain tissue. Cell. 2018;172:1108–15. [DOI] [PubMed] [Google Scholar]
- 7. Baddeley D, Crossman D, Rossberger S, Cheyne JE, Montgomery JM, Jayasinghe ID, et al. 4D super‐resolution microscopy with conventional fluorophores and single wavelength excitation in optically thick cells and tissues. PLoS ONE. 2011;6:e20645–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ilgen P, Stoldt S, Conradi L‐C, Wurm CA, Rüschoff J, Ghadimi BM, et al. STED super‐resolution microscopy of clinical paraffin‐embedded human rectal cancer tissue. PLoS ONE. 2014;9:e101563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paul DS, Casari C, Wu C, Piatt R, Pasala S, Campbell RA, et al. Deletion of the Arp2/3 complex in megakaryocytes leads to microthrombocytopenia in mice. Blood Adv. 2017;1:1398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guerrero JA, Bennett C, van der Weyden L, McKinney H, Chin M, Nurden P, et al. Gray platelet syndrome: proinflammatory megakaryocytes and alpha‐granule loss cause myelofibrosis and confer metastasis resistance in mice. Blood. 2014;124:3624–35. [DOI] [PubMed] [Google Scholar]
- 11. Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao M, Li L. Dissecting the bone marrow hematopoietic stem cell niches. Cell Res. 2016;26:975–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schrijver W, van der Groep P, Hoefnagel L, ter Hoeve ND, Peeters T, Moelans CB, et al. Influence of decalcification procedures on immunohistochemistry and molecular pathology in breast cancer. Mod Pathol. 2016;29:1460–70. [DOI] [PubMed] [Google Scholar]
- 14. Savi FM, Brierly GI, Baldwin J, Theodoropoulos C, Woodruff MA. Comparison of different decalcification methods using rat mandibles as a model. J Histochem Cytochem. 2017;65:705–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawamoto T, Kawamoto K. Preparation of thin frozen sections from nonfixed and undecalcified hard tissues using Kawamot's film method (2012). Methods Mol Biol. 2014;1130:149–64. [DOI] [PubMed] [Google Scholar]
- 16. Ishimaru T, Nakazono M, Masumura T, Abiko M, San‐oh Y, Nishizawa NK, et al. A method for obtaining high integrity RNA from developing aleurone cells and starchy endosperm in rice (Oryza sativa L.) by laser microdissection. Plant Sci. 2007;173:321–6. [Google Scholar]
- 17. Narisawa S, Yadav MC, Millán JL. In vivo overexpression of tissue‐nonspecific alkaline phosphatase increases skeletal mineralization and affects the phosphorylation status of osteopontin. J Bone Miner Res. 2013;28:1587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujino Y, Minamizaki T, Yoshioka H, Okada M, Yoshiko Y. Imaging and mapping of mouse bone using MALDI‐imaging mass spectrometry. Bone Rep. 2016;5:280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakamura‐Ishizu A, Takubo K, Kobayashi H, Suzuki‐Inoue K, Suda T. CLEC‐2 in megakaryocytes is critical for maintenance of hematopoietic stem cells in the bone marrow. J Exp Med. 2015;212:2133–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang M, Arai A, Udagawa N, Hiraga T, Lijuan Z, Ito S, et al. Osteogenic factor Runx2 marks a subset of leptin receptor‐positive cells that sit atop the bone marrow stromal cell hierarchy. Sci Rep. 2017;7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open‐source platform for biological‐image analysis. Nat Methods. 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM, Henriques R. Fast live‐cell conventional fluorophore nanoscopy with ImageJ through super‐resolution radial fluctuations. Nat Commun. 2016;7:12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olivier N, Keller D, Rajan VS, Gönczy P, Manley S. Simple buffers for 3D STORM microscopy. Biomed Opt Express. 2013;4:885–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang SJ, Berndl M, Michael Ando D, Barch M, Narayanaswamy A, Christiansen E, et al. Assessing microscope image focus quality with deep learning. BMC Bioinformatics. 2018;19:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lambert TJ, Waters JC. Navigating challenges in the application of superresolution microscopy. J Cell Biol. 2017;216:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suraneni PK, Corey SJ, Hession MJ, Ishaq R, Awomolo A, Hasan S, et al. Dynamins 2 and 3 control the migration of human megakaryocytes by regulating CXCR4 surface expression and ITGB1 activity. Blood Adv. 2018;2:3540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nomura M, Sakitani N, Iwasawa H, Kohara Y, Takano S, Wakimoto Y, et al. Thinning of articular cartilage after joint unloading or immobilization. An experimental investigation of the pathogenesis in mice. Osteoarthritis Cartilage. 2017;25:727–36. [DOI] [PubMed] [Google Scholar]
- 28. Oka S, Kanagawa M, Doi Y, Schuster DM, Goodman MM, Yoshimura H. PET tracer 18F‐fluciclovine can detect histologically proven bone metastatic lesions: a preclinical study in rat osteolytic and osteoblastic bone metastasis models. Theranostics. 2017;7:2048–64. [DOI] [PMC free article] [PubMed] [Google Scholar]


