Abstract
Background
State‐of‐the‐art 3‐dimensional (3D) electron microscopy approaches provide a new standard for the visualization of human platelet ultrastructure. Application of these approaches to platelets rapidly fixed prior to purification to minimize activation should provide new insights into resting platelet ultrastructure.
Objectives
Our goal was to determine the 3D organization of α‐granules, dense granules, mitochondria, and canalicular system in resting human platelets and map their spatial relationships.
Methods
We used serial block face–scanning electron microscopy images to render the 3D ultrastructure of α‐granules, dense granules, mitochondria, canalicular system, and plasma membrane for 30 human platelets, 10 each from 3 donors. α‐Granule compositional data were assessed by sequential, serial section cryo‐immunogold electron microscopy and by immunofluorescence (structured illumination microscopy).
Results and Conclusions
α‐Granule number correlated linearly with platelet size, while dense granule and mitochondria number had little correlation with platelet size. For all subcellular compartments, individual organelle parameters varied considerably and organelle volume fraction had little correlation with platelet size. Three‐dimensional data from 30 platelets indicated only limited spatial intermixing of the different organelle classes. Interestingly, almost 70% of α‐granules came within ≤35 nm of each other, a distance associated in other cell systems with protein‐mediated contact sites. Size and shape analysis of the 1488 α‐granules analyzed revealed no more variation than that expected for a Gaussian distribution. Protein distribution data indicated that all α‐granules likely contained the same major set of proteins, albeit at varying amounts and varying distribution within the granule matrix.
Keywords: electron microscopy, hemostasis, platelet granules, platelet organelles, platelet ultrastructure, super‐resolution light microscopy
Essentials.
State‐of‐the‐art 3D electron microscopy provides a new precision standard for platelet ultrastructural studies.
Human platelets fixed rapidly prior to purification preserve the ultrastructure of resting blood platelets, which are highly sensitive to activation during preparation.
Three‐dimensional ultrastructural characterization of organelle variation in immediately fixed cells provides a needed quantitative model of the resting platelet.
At a snapshot in time, 70% of α‐granules are found within 35 nm of each other, suggesting granule‐specific contact sites in resting human blood platelets.
1. INTRODUCTION
Recent advances in imaging technology have profoundly shifted the state‐of‐the‐art for platelet visualization from 2‐dimensional (2D) to 3‐dimensional 3D at near nanometer resolution. Serial block face–scanning electron microscopy (SBF‐SEM) approaches now permit the routine visualization of full platelet cell volumes for plastic‐embedded samples at a resolution of 5‐7 nm in XY or better.1, 2, 3, 4 Cryo‐electron microscopy (EM) approaches are demonstrating the potential for the visualization of hydrated samples at nanometer resolution.5 This new technology eliminates the inability of earlier 3D ultrastructural methods such as electron tomography6 or scanning transmission electron microscopy (STEM)7 to visualize α‐granule or canalicular system (CS) 3D ultrastructures in full platelet volumes. These advances are yielding new insights into platelet structure and function. For example, SBF‐SEM studies have placed new emphasis on the importance of compound fusion to the platelet release reaction1 and the contribution of direct α‐granule/plasma membrane fusion in α‐granule decondensation,4 while initial cryo‐EM studies point to platelet alterations in ovarian cancer.5 Likewise, the improved resolution of super‐resolution light microscopy, for example, 3D‐structured illumination (3D‐SIM), 2‐fold better in XY and Z,8 is beginning to have an impact on platelet studies.9, 10
To date, however, there has been no systematic study applying these state‐of‐the‐art techniques to resting human platelets and the in situ ultrastructural characterization of their major secretory organelles: α‐granules, dense granules, and CS. Our goal here was to fill that gap by characterizing at near nanometer resolution the 3D ultrastructure of organelles within 30 resting human platelets. This should yield valuable reference information, for example, on the spatial relationship between α‐granules and CS, extent of organelle size/shape dispersity, organelle abundance versus platelet size, and the incidence of putative organelle contact sites. In other cell types, close proximity between organelles is indicative of protein‐dependent contact sites (for reviews, see Saheki and Camilli,11 Wu et al,12 and Scorrano et al13). We chose to use SBF‐SEM of plastic‐embedded, fixed human platelets as the primary 3D structural approach because immediate chemical fixation stabilizes platelet structure without interfering with the subsequent platelet purification necessary for EM visualization.7
As expected, based on previous morphometry (for review, see King and Reed14), our studies showed that α‐granule or CS volume fraction was substantially higher than that of mitochondria or dense granules. For all 4 organelles, volume fraction had little correlation with platelet size. However, we found that α‐granule number correlated most highly with platelet volume with dense granules or mitochondria displaying little, if any, correlation. Spatially, dense granules and mitochondria tended to be the most peripherally located, while α‐granules tended to be somewhat more centrally located. Detailed size and shape analysis of the α‐granule population revealed the dispersion expected of a normal distribution. Interestingly, almost 70% of α‐granules came within ≤35 nm of each other, a distance associated in other cell systems with protein‐mediated physical contact sites. 3D‐SIM protein composition data indicated that all α‐granules likely contained the same major set of proteins, albeit at varying amounts while serial section immunogold labeling at the EM level provided the required resolution to show that von Willebrand factor (VWF), an eccentrically distributed protein within the granule, was present in essentially each and every α‐granule. In sum, our data suggest a single population of α‐granules, many of which are in close contact with one another.
2. MATERIALS AND METHODS
2.1. Blood collection and platelet preparation
For SBF‐SEM, freshly drawn, citrated human blood was immediately fixed with paraformaldehyde (PFA)/glutaraldehyde.4, 7, 15 Purified platelets were then postfixed with glutaraldehyde and heavy metal–stained preembedding4, 15 For immunogold labeling, blood was collected into citrate and was mixed immediately with 2× fixative containing 8% PFA in 0.1 mol/L phosphate buffer (PB) for 2 hours. Platelet‐rich plasma was prepared, transferred to a new tube, and spun to obtain a platelet pellet.7 For immunofluorescence labeling, platelets were fixed, purified, and then postfixed as described.5, 9 All experiments were approved by the University of Arkansas for Medical Sciences Institutional Review Board.
2.2. Analysis of SBF‐SEM Images
The 3D ultrastructure of platelets was obtained using a Zeiss Sigma VP scanning electron microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY, USA), operated in high‐vacuum mode, and equipped with a Gatan 3View SBF system (Gatan Inc., Pleasanton, CA, USA).4, 15 Backscattered electron images were acquired at 2000 by 2000 pixels with a pixel dwell time of 1 μsec and an electron dose of 25 e/nm2. The cutting increment was set to 30 nm. For Donor 1 platelets, block‐face images were acquired at a primary beam energy of 1.8 kV, with x and y pixel size of 6.8 nm. For Donors 2 and 3, block‐face images were acquired at a primary beam energy of 1.1 kV with a 6.9 nm pixel size. The image stacks were aligned using Digital Micrograph (Gatan Inc.) and processed in Amira software (Thermo Fisher Scientific, Waltham, MA, USA). Processing included computer‐assisted segmentation with manual adjustments, 3D rendering, and quantitative analysis.3, 4, 7, 15 Designated data were plotted against a linear fit using KaleidaGraph software (Synergy, Inc., Reading, PA, USA).
2.3. Immunofluorescence Staining and 3D‐SIM Microscopy
For immunofluorescence staining, platelets were manually counted with a Bright‐Line Hemocytometer (Cambridge Instruments Inc., Buffalo, NY, USA) and suspended at a working concentration of 380 000 to 400 000. Staining was performed as described.15, 16 Three‐dimensional–SIM image stacks were taken with a 63×/NA 1.4 or 100×/NA 1.46 objectives with Optovar settings of 1.6× using an Elyra PS.1, inverted, super resolution‐imaging microscope (Carl Zeiss Microscopy). Three‐dimensional–SIM image stacks were visualized with Zen 2 software (Carl Zeiss Microscopy). Representative image slices are displayed using lookup tables adjusted to give minimal incidence of saturated pixels. For each immunostaining pairing, co‐localization was determined on a voxel basis for full platelet volumes using Huygens Professional software (Scientific Volume Imaging, Hilversum, The Netherlands) as previously described.2, 9, 17, 18
2.4. Immunogold Labeling and Immunoelectron Microscopy
The washed platelet pellet was incubated in 0.15% glycine/0.1 mol/L of PB to quench aldehyde groups, washed again, and embedded in 12% gelatin.16 Before being frozen in liquid nitrogen, the gelatin blocks were 2.3 mol/L sucrose infused in 0.1 mol/L PB at 4°C overnight. Frozen samples were sectioned at −120°C with a (Leica UltraCut‐UCT microtome, Leica Mikrosysteme GmbH, A‐1170 Wien, Austria) at 60 or 95 nm thickness and collected onto carbon‐coated, formvar slot grids.
Gold labeling was performed at room temperature by incubating the sections over a series of drops on a Parafilm sheet. Specimens were blocked in 1% bovine serum albumin (BSA)/phosphate buffered saline (PBS), incubated in primary antibodies overnight at 4°C (in a humidified chamber), washed in 0.1% BSA/PBS, incubated in secondary antibodies for 1 hour, and washed in PBS.
Sections were either single or double labeled. For double labeling, grids were incubated with sequential sets of primary antibodies followed by incubation with gold‐labeled secondary antibodies 18. After the first labeling round, grids were rinsed in 0.1% BSA/PBS; bound antibodies were then stabilized by fixation in 1% glutaraldehyde/PBS for 5 minutes and quenched in 0.15 mol/L glycine/PBS.16 The second immune incubation was initiated by blocking in 0.1% BSA/PBS, followed by second primary antibody washes and incubation with different‐sized gold secondary antibodies.
Primary antibodies were diluted 1:50 in PBS containing 1% BSA. Donkey secondary antibodies adsorbed to either 10 nm or 6 nm gold were used at 1:50 dilution in the same buffer. The specificity of immunolabeling was verified using the secondary antibodies alone. Three‐dimensional models were rendered with Amira 6.3 software.3, 4, 7
3. RESULTS
3.1. Immediately fixed, resting human platelets were uniformly discoid in shape
We chose to interrogate platelet ultrastructure in immediately fixed platelets. Our previous work has shown that this population most closely approximates circulating resting platelets,4, 7 and we, therefore, expected that the purified platelets would be discoid in shape. When viewed in block‐face images or in single planes of the 3D data set, the randomly exposed platelets were cut at various angles (Figure 1A‐D). To yield 3D shapes, we performed segmentation and volume rendering of 30 randomly selected platelets, 10 each from 3 donors. As expected,7, 14 almost all of the rendered platelets were highly discoid in shape and had a similar appearance irrespective of donor (Figures 1D and E and 2).
Figure 1.
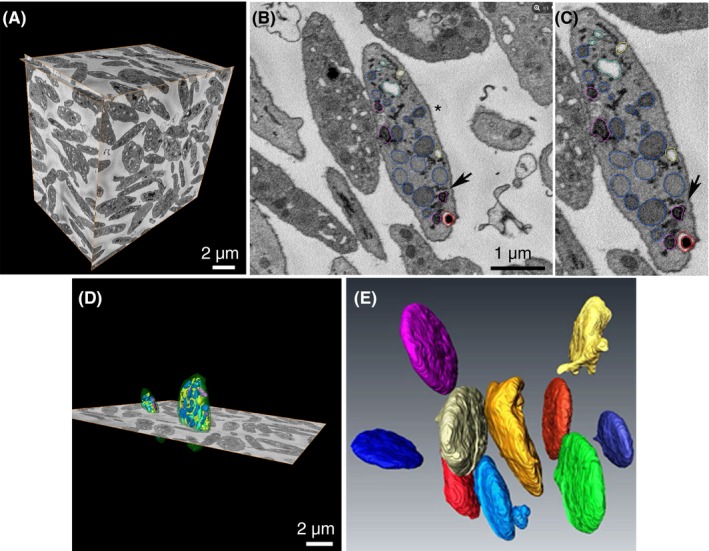
SBF‐SEM of resting human platelets. (A) Three orthogonal slices delineate a complete Donor 1 data set. Multiple platelets can be seen in XY, XZ, and YZ planes. (B) Once validated for total inclusion within the image stack, individual platelets (**) were randomly chosen for segmentation analysis. (C) 1.33× magnification of B to illustrate segmented organelles and the color scheme used: α‐granules (blue), dense granules (red), dense cores (burgundy), mitochondria (magenta), open canalicular system (yellow), closed canalicular system (cyan). Black arrow in B and C points to area of close contact between dark DTS element and α‐granule. (D) Example of 2 platelet surfaces rendered in 3D together with one XY orthoslice. (E) Example surface renderings of 10 different Donor 1 platelets are shown in different colors and display a typical discoid shape, with few of the platelets displaying any pseudopods. Ten platelets each from Donors 2 and 3 were similarly identified, their organelles segmented, and surface rendered. DTS, dense tubular system; SBF‐SEM, serial block face–scanning electron microscopy
Figure 2.
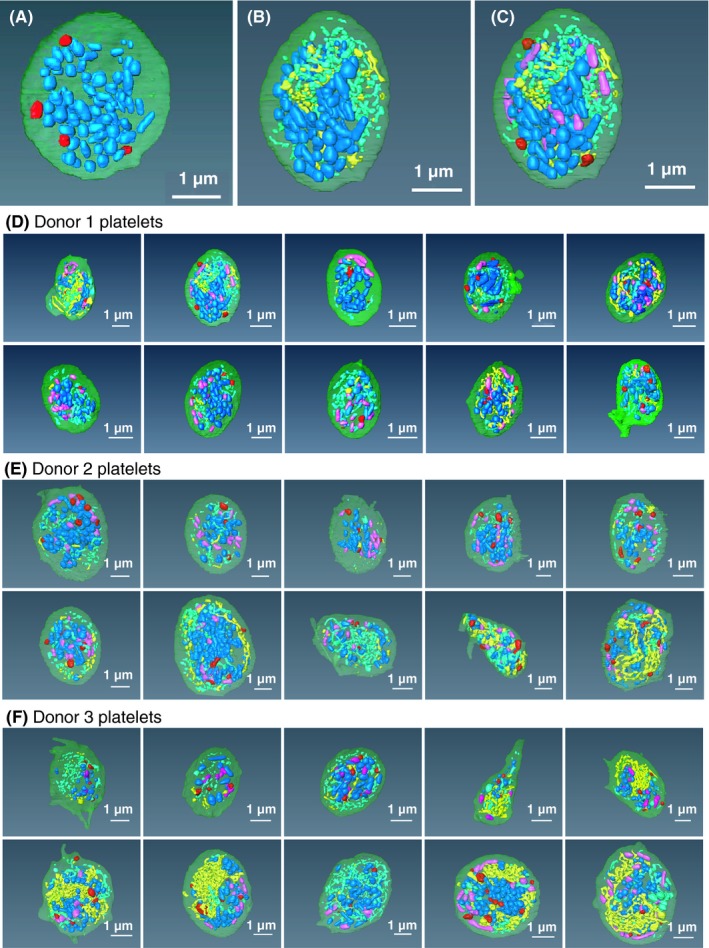
Spatial arrangement of α‐granules (blue) vs. dense granules (red), CS (open and closed, yellow and turquoise), and mitochondria (purple) Example platelet (Donor 1, platelet 2). (A) α‐granules (blue) vs. dense granules (red); (B) α‐granules (blue) vs. CS (open and closed, yellow and turquoise); (C) α‐granules, dense granules, CS versus mitochondria (purple). (B, C, D) Thirty example platelets, 10 each from 3 donors. CS, canalicular system
Platelet organelles were identified morphologically over a series of XY images encompassing the full organelle volume (example XY image, 1B and C). α‐Granules were defined as structures limited by a single membrane, often of a round to ovoid shape that enclosed a matrix devoid of enfolded cristae. As shown, the α‐granule lumen was devoid of nucleoids in these conventionally dehydrated preparations. This was an unexpected result that we attribute to the protective effect of the high concentrations of contrast‐enhancing heavy metals used prior to dehydration and plastic embedding. In the absence of such heavy metal staining, we find nucleoids to be a frequent feature of nonfreeze substitution dehydrated preparations.7 Dense granules were identified as a round, single‐membrane limited organelle with a bullet‐shaped, high‐density core that was typically surrounded by a stain‐free rim. The CS was defined in sequential XY planes as a tubular, single‐membrane limited organelle with a stain‐free interior. We grouped the CS into 2 morphological classes based on whether the membrane element was connected to the cell surface/PM (ie, open CS) or unconnected (ie, closed CS).1, 4, 7 Mitochondria were defined in sequential XY planes as organelles containing frequent membrane infoldings (cisternae) and an electron dark interior/matrix. In addition, elements of the dense tubular system (DTS) marked by an electron dense lumen were present in the block‐face images. We chose to concentrate further segmentation‐driven analysis on α‐granules, dense granules, CS, and mitochondria. In practice, we found DTS extremely difficult to segment in a semiautomated manner using conventional algorithms.
3.2. Quantitative 3D characterization of organelle frequency and volume in resting human platelets
The morphological parameters of the rendered human platelets and the 4 major organelles rendered: α‐granules, dense granules, CS, and mitochondria are summarized numerically in Table 1. When considered as population means, there was little variation across the 3 donors in platelet size. Resting human platelets had a mean short‐axis length of 1.47 μm ± 0.06 μm, a mean long‐axis length of 3.82 μm ± 0.12 μm, and a mean volume of 7.7 μm3 ± 0.5 μm3. Similarly, when averaged and compared across donors, little variation was found in the frequency of α‐granules, dense granules, CS, and mitochondria and/or other organellar‐specific parameters. α‐Granules were the major population, at 50 ± 5 granules per platelet with dense granules, as expected being rare, at 5.5 ± 0.5 per platelet and mitochondria somewhat more numerous, at 10.9 ± 0.9 per platelet. On a volume fraction basis, canalicular system was the most voluminous rendered organelle at 0.113 ± 0.011 (approximately equal portions open, plasma membrane connected and closed CS, no detectable plasma membrane connection1, 7) and showed the most variability from donor to donor with increased open CS abundance in the oldest donor. α‐Granules were somewhat less abundant at a volume fraction of 0.083 ± 0.009, while dense granules and mitochondria at 0.007 ± 001 and 0.019 ± 0.001, respectively, were relatively minor contributors to total platelet cell volume. When compared with previous 3D studies, the α‐granule volume fraction results reported here agree with our previous much smaller scale STEM experiments.7 Moreover, the mean values reported here are similar to those reported by others for washed human platelets imaged by focused ion beam SEM or electron cryotomography.1, 5 The incidence of α‐granules per platelet reported by any of these 3D imaging approaches, 42 to 58 per platelet, is on the low side of the 50 to 80/platelet reported based on older stereology techniques.14 Stereology techniques may overestimate α‐granule number because of possible biases in the statistical extrapolations.
Table 1.
Full platelet structural parameters, 10 platelets each, 3 donors (age stratified): platelets, α‐granules, dense granules, canalicular system, and mitochondria
| Cell | Overall platelet dimensions | α ‐Granule properties | Dense granule properties | Canalicular system propertiesa | Mitochondria properties | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Platelet volume (μm3) | Short axis length (μm) | Long axis length (μm) | α‐Granule number | Average α‐granule short axis (μm) | Average α‐granule long axis (μm) | Mean α‐granule volume (μm³) | α‐Granule volume fraction | Dense granule number | Mean dense granule volume (μm³) | Dense granule volume fraction | Open CS volume (μm³) | Closed CS volume (μm³) | Total CS volume (μm³) | CS volume fraction | Mitochondrial number | Mitochondrial volume fraction | |
| Donor 1, M, 21 y | |||||||||||||||||
| Platelet 1 | 14.80 | 2.09 | 4.71 | 42 | 0.225 | 0.286 | 0.017 | 0.047 | 6 | 0.009 | 0.004 | 1.14 | 0.58 | 1.71 | 0.115 | 18 | 0.016 |
| Platelet 2 | 8.70 | 1.43 | 4.06 | 58 | 0.282 | 0.361 | 0.019 | 0.126 | 5 | 0.011 | 0.006 | 0.34 | 0.78 | 1.12 | 0.128 | 15 | 0.027 |
| Platelet 3 | 3.90 | 1.01 | 3.02 | 30 | 0.296 | 0.338 | 0.017 | 0.129 | 4 | 0.009 | 0.009 | 0.02 | 0.09 | 0.11 | 0.027 | 3 | 0.023 |
| Platelet 4 | 7.40 | 2.06 | 3.65 | 19 | 0.322 | 0.382 | 0.035 | 0.093 | 5 | 0.010 | 0.007 | 0.04 | 0.64 | 0.68 | 0.092 | 3 | 0.011 |
| Platelet 5 | 7.10 | 1.20 | 4.02 | 50 | 0.273 | 0.339 | 0.018 | 0.128 | 3 | 0.009 | 0.004 | 0.44 | 0.23 | 0.67 | 0.094 | 16 | 0.031 |
| Platelet 6 | 6.00 | 1.50 | 3.57 | 41 | 0.206 | 0.255 | 0.009 | 0.063 | 2 | 0.010 | 0.002 | 0.03 | 0.44 | 0.47 | 0.078 | 17 | 0.027 |
| Platelet 7 | 7.40 | 1.21 | 3.75 | 47 | 0.251 | 0.309 | 0.013 | 0.084 | 4 | 0.006 | 0.003 | 0.05 | 0.49 | 0.53 | 0.072 | 12 | 0.020 |
| Platelet 8 | 3.90 | 1.21 | 2.89 | 23 | 0.226 | 0.273 | 0.011 | 0.063 | 3 | 0.005 | 0.004 | 0.03 | 0.37 | 0.40 | 0.103 | 12 | 0.028 |
| Platelet 9 | 5.30 | 1.28 | 3.40 | 40 | 0.246 | 0.285 | 0.011 | 0.081 | 4 | 0.005 | 0.004 | 0.34 | 0.11 | 0.45 | 0.086 | 8 | 0.018 |
| Platelet 10 | 8.10 | 1.92 | 3.50 | 45 | 0.262 | 0.327 | 0.021 | 0.114 | 8 | 0.014 | 0.014 | 0.03 | 0.59 | 0.79 | 0.097 | 6 | 0.013 |
| Donor 1 Mean | 7.30 | 1.49 | 3.66 | 40 | 0.259 | 0.316 | 0.017 | 0.093 | 4.4 | 0.009 | 0.006 | 0.26 | 0.43 | 0.69 | 0.089 | 11.0 | 0.021 |
| Standard error of the mean | 1.00 | 0.12 | 0.17 | 4 | 0.011 | 0.013 | 0.002 | 0.010 | 0.5 | 0.001 | 0.001 | 0.11 | 0.07 | 0.14 | 0.009 | 1.8 | 0.002 |
| Donor 2, F, 37 y | |||||||||||||||||
| Platelet 1 | 9.60 | 1.57 | 4.89 | 89 | 0.277 | 0.321 | 0.013 | 0.118 | 6 | 0.013 | 0.008 | 0.03 | 0.29 | 0.32 | 0.034 | 15 | 0.020 |
| Platelet 2 | 7.00 | 1.49 | 4.26 | 33 | 0.305 | 0.358 | 0.017 | 0.081 | 4 | 0.009 | 0.005 | 0.04 | 0.29 | 0.33 | 0.046 | 12 | 0.018 |
| Platelet 3 | 9.80 | 1.64 | 5.13 | 30 | 0.289 | 0.337 | 0.027 | 0.056 | 6 | 0.009 | 0.006 | 0.09 | 0.14 | 0.22 | 0.023 | 14 | 0.015 |
| Platelet 4 | 8.10 | 1.47 | 4.79 | 45 | 0.307 | 0.368 | 0.021 | 0.117 | 9 | 0.013 | 0.015 | 0.02 | 0.26 | 0.28 | 0.035 | 13 | 0.018 |
| Platelet 5 | 5.30 | 1.24 | 4.03 | 24 | 0.294 | 0.349 | 0.014 | 0.065 | 8 | 0.005 | 0.008 | 0.11 | 0.21 | 0.32 | 0.060 | 13 | 0.017 |
| Platelet 6 | 4.30 | 1.00 | 3.45 | 32 | 0.229 | 0.282 | 0.012 | 0.088 | 4 | 0.012 | 0.011 | 0.14 | 0.29 | 0.43 | 0.099 | 14 | 0.028 |
| Platelet 7 | 13.10 | 1.98 | 4.73 | 141 | 0.259 | 0.312 | 0.013 | 0.141 | 6 | 0.013 | 0.006 | 0.62 | 0.92 | 3.56 | 0.118 | 13 | 0.013 |
| Platelet 8 | 7.80 | 1.55 | 3.87 | 37 | 0.232 | 0.280 | 0.011 | 0.051 | 3 | 0.006 | 0.002 | 0.12 | 0.96 | 1.08 | 0.138 | 8 | 0.011 |
| Platelet 9 | 5.70 | 2.42 | 3.10 | 52 | 0.249 | 0.297 | 0.011 | 0.099 | 8 | 0.016 | 0.023 | 0.67 | 0.74 | 1.41 | 0.248 | 7 | 0.019 |
| Platelet 10 | 9.20 | 1.72 | 3.83 | 77 | 0.228 | 0.266 | 0.008 | 0.065 | 8 | 0.008 | 0.007 | 1.11 | 0.38 | 1.50 | 0.163 | 6 | 0.008 |
| Donor 2 Mean | 8.00 | 1.61 | 4.21 | 56 | 0.267 | 0.317 | 0.015 | 0.088 | 6.2 | 0.011 | 0.009 | 0.29 | 0.45 | 0.74 | 0.097 | 11.5 | 0.017 |
| Standard error of the mean | 0.80 | 0.12 | 0.21 | 12 | 0.010 | 0.011 | 0.002 | 0.010 | 0.6 | 0.001 | 0.002 | 0.12 | 0.10 | 0.18 | 0.023 | 1.0 | 0.002 |
| Donor 3, M, 70 y | |||||||||||||||||
| Platelet 1 | 6.10 | 1.26 | 4.15 | 22 | 0.202 | 0.237 | 0.006 | 0.020 | 5 | 0.004 | 0.003 | 0.08 | 0.39 | 0.46 | 0.076 | 3 | 0.009 |
| Platelet 2 | 4.10 | 0.87 | 3.09 | 9 | 0.284 | 0.317 | 0.023 | 0.049 | 3 | 0.007 | 0.005 | 0.05 | 0.16 | 0.21 | 0.050 | 6 | 0.017 |
| Platelet 3 | 6.10 | 1.28 | 2.98 | 45 | 0.247 | 0.285 | 0.014 | 0.102 | 7 | 0.007 | 0.008 | 0.11 | 0.46 | 0.56 | 0.092 | 11 | 0.028 |
| Platelet 4 | 4.90 | 1.69 | 2.30 | 23 | 0.190 | 0.211 | 0.005 | 0.022 | 4 | 0.003 | 0.003 | 0.47 | 0.41 | 0.88 | 0.178 | 3 | 0.010 |
| Platelet 5 | 8.90 | 1.29 | 3.61 | 44 | 0.249 | 0.280 | 0.011 | 0.057 | 4 | 0.006 | 0.003 | 0.98 | 0.28 | 1.26 | 0.142 | 10 | 0.019 |
| Platelet 6 | 9.20 | 1.63 | 3.92 | 91 | 0.215 | 0.247 | 0.008 | 0.077 | 9 | 0.008 | 0.008 | 1.89 | 0.48 | 2.37 | 0.255 | 7 | 0.011 |
| Platelet 7 | 10.20 | 1.32 | 4.54 | 56 | 0.254 | 0.298 | 0.013 | 0.071 | 6 | 0.021 | 0.013 | 1.60 | 0.28 | 1.88 | 0.184 | 10 | 0.012 |
| Platelet 8 | 9.40 | 1.22 | 3.76 | 64 | 0.243 | 0.284 | 0.015 | 0.104 | 3 | 0.008 | 0.002 | 0.08 | 1.08 | 1.16 | 0.123 | 8 | 0.013 |
| Platelet 9 | 8.40 | 1.27 | 3.57 | 116 | 0.239 | 0.267 | 0.009 | 0.127 | 14 | 0.010 | 0.016 | 1.37 | 0.32 | 1.69 | 0.200 | 17 | 0.030 |
| Platelet 10 | 10.30 | 1.24 | 4.22 | 63 | 0.234 | 0.264 | 0.010 | 0.062 | 4 | 0.004 | 0.002 | 1.44 | 0.73 | 2.17 | 0.211 | 18 | 0.028 |
| Donor 3 Mean | 7.80 | 1.31 | 3.61 | 53 | 0.236 | 0.269 | 0.011 | 0.069 | 5.9 | 0.008 | 0.006 | 0.81 | 0.46 | 1.26 | 0.151 | 9.30 | 0.018 |
| Standard error of the mean | 0.70 | 0.07 | 0.21 | 10 | 0.009 | 0.010 | 0.002 | 0.011 | 1.1 | 0.002 | 0.002 | 0.23 | 0.09 | 0.23 | 0.021 | 1.60 | 0.003 |
| All donor mean | 7.70 | 1.47 | 3.83 | 50.0 | 254 | 301 | 0.014 | 0.083 | 5.5 | 0.009 | 0.007 | 0.46 | 0.44 | 0.90 | 0.113 | 10.6 | 0.019 |
| Standard error of the mean | 0.50 | 0.06 | 0.12 | 5.0 | 6 | 8 | 0.001 | 0.006 | 0.5 | 0.001 | 0.001 | 0.09 | 0.05 | 0.12 | 0.011 | 0.9 | 0.001 |
When considered on an individual platelet basis, variation was more obvious (Figure 3). As might be expected for a linear rather than cubic value, platelet axis lengths clustered more tightly about the mean/average than platelet volumes (Figure 3A). Organelle number per platelet varied least for dense granule and mitochondria and more for α‐granules (Figure 3B). Curve fit analysis showed a distinct linear trendline linking α‐granule number and platelet size/volume and a decidedly less tight relationship for dense granules (DGs) and mitochondria (Figure 3C). Organelle volume fraction per platelet, a value that might be expected to near constant, varied over a 4‐ to >10‐fold range for α‐granules and CS, respectively, and appeared tighter for dense granules and mitochondria (Figure 3D). Importantly, the graphical depiction of variation presented here provided little to no evidence for distinct subsets of platelets, α‐granules, dense granules, CS, and mitochondria. Our analysis in explicitly considering individual dispersity as a factor goes beyond the typical ultrastructural presentation of means/averages and standard errors.
Figure 3.
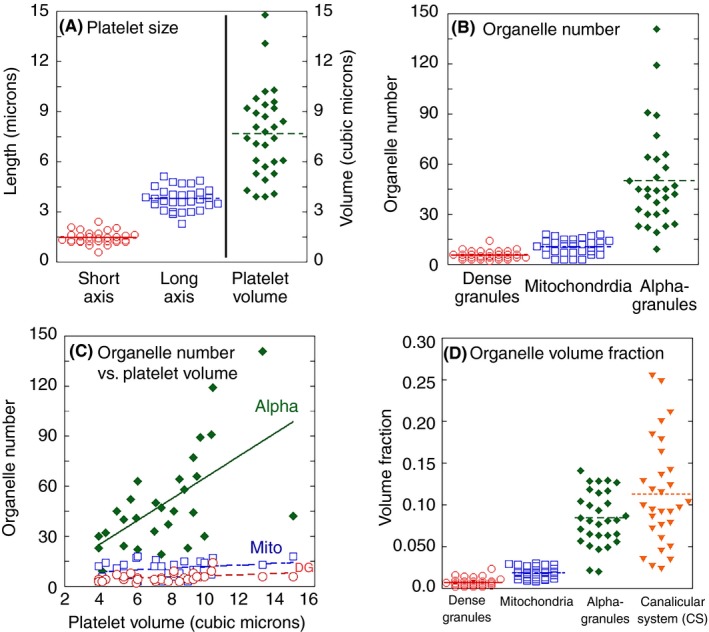
Quantitative platelet parameters summarized as dot and line plots. The data from 30 platelets, 3 donors are grouped together, as little difference was noted between donors. (A) Platelet size; (B) organelle number; (C) organelle number vs. platelet volume; (D) organelle volume fraction. Linear trendlines in C were plotted using Kaleidagraph software. Alpha, α‐granules, R 2 = 0.34; DG, dense granules, R 2 = 0.12; mito, mitochondria, R 2 = 0.04
3.3. 3D spatial arrangement of organelles in resting human platelets
As shown in 2A and Figure S1, dense granules (red) and α‐granules (blue) had different spatial distributions in resting human platelets. Dense granules were relatively peripherally located, while α‐granules were distributed over much of the platelet cell volume (see additional examples, Figure 2D‐F). Quantitatively, almost 25% of dense granules were within 35 nm of the cell surface, while only 7.5% α‐granules were similarly located (Table S1). Comparison of the distributions of CS and α‐granules showed the 2 to be spatially separated, with CS tending to be present in the gaps in α‐granule distribution (Figure 2B and D‐F; Figure S1). In general, there was little interdigitation of CS elements between α‐granules. In total, dense granules, mitochondria, and CS appeared to be spatially separated from α‐granules (Figure 2C‐F; Figure S1). Lack of organelle interdigitation between α‐granules could be a consequence of tight clustering of α‐granules one with another. To test for this possibility, we analyzed the 3D data sets for α‐granule spacing and found that almost 70% of α‐granules were located within ≤35 nm of each other (Table S1). As displayed in Figure S1, these close neighbor distances were over mostly small granule areas. Each may well be analogous to organelle contact sites observed in other cells systems (for review, see Saheki and Camilli,11 Wu et al,12 and Scorrano et al13). Such contact sites mediate various transport processes, for example, lipid transport between organelles, and membrane trafficking events. We note that, as shown in Figure 1B and C, elements of a dense tubular network, the platelet equivalent of endoplasmic reticulum, can be in close contact with α‐granules. Such contacts are also apparent in mouse platelets.4 DTS/α‐granule contacts have not been analyzed further due to difficulties in organelle segmentation.
3.4. Detailed 3D characterization of α‐granule size, shape, and composition
α‐Granules are the major protein storage compartment in platelets and the subject of conflicting reports regarding their homogeneity in size, shape, and composition.6, 9, 17, 19, 20, 21, 22, 23, 24 Because of the importance of this organelle in platelet secretion and to hemostasis and thrombosis in general, we decided to data mine our resting human platelet, 3D SBF‐SEM data sets, to determine the variation in granule size and shape over ~1500 examples. Qualitatively, we saw little apparent variation in α‐granule size, shape, or volume (ie, an ovoid morphology), from platelet to platelet or donor to donor (Figure 2). Quantitatively, the mean granule short‐axis length averaged across 1488 α‐granules was 251 ± 2 nm, and the mean long‐axis length was 297 nm ± 2 nm (standard error of the mean). When displayed as frequency histograms (Figure 4), the data followed a normal distribution with little to no indications of granule subpopulations based on size or shape. As expected for a standard normal distribution, the mean and median granule axis values differed little from one another, 251 nm (short axis) vs. 255 nm and 297.4 nm (long axis) vs. 297.1 nm. Moreover, the skewness in the distributions was small, 0.29 (short axis) and 0.79 (long axis), and the kurtosis values at 2.14 and 3.92 were near the expected 3.0 of a standard normal distribution.25 The mean α‐granule cell volume fraction across the 3 donors showed no significant differences from the mean upon calculating P values. In sum, α‐granules in each of the 3 age‐stratified donors present as a single, major population showing no more than normal Gaussian variation in size and shape. We also examined shape variability in thawed cryosections of human platelets. In agreement with the work of van Nispen et al,6 we detected as a rare event <5% in thin section, putative varied α‐granule forms, that is, vacuoles with tubular extensions or long tubular forms (Figure S2). By immunogold labeling, again in agreement with van Nispen et al, we find that these tubules are enriched in fibrinogen (Figure S2A‐C). Because fibrinogen is endocytosed and delivered through membrane trafficking to α‐granules, we raise the alternative interpretation that the tubules could well be endocytic carriers rather than variant α‐granule structures.
Figure 4.
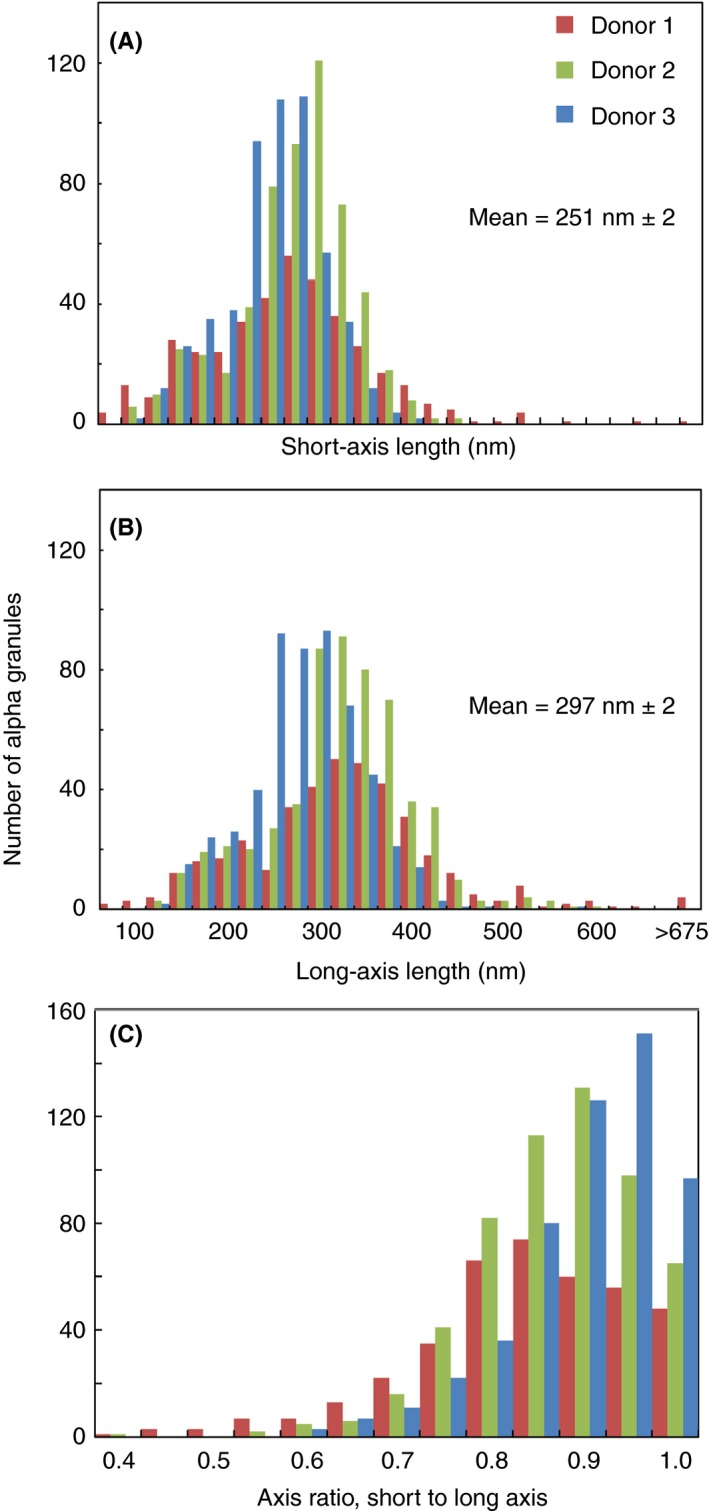
Quantification of the axial shape parameters of 1488 α‐granules from 3 donors, 10 platelets each, revealed a single Gaussian population based on shape and size. Binned short‐axis (A) and long‐axis (B) lengths of each α‐granule for all 3 donors. The α‐granule axis ratio distribution (C) is calculated by dividing the width in A by the length in B and is a measure of the shape of the granules. In C, the x‐axis scale of 1 is defined as a perfect sphere and anything <1 is a spheroid. The short‐ and long‐axis lengths are calculated in Amira software by creating a “Custom Measure” in the “Label Analysis” module after using the “Connected Components” module to assign a unique material to each α‐granule in a platelet
Given the ultrastructural evidence for the resting human α‐granule population showing no more than Gaussian variation in size and shape, we decided to revisit the compositional variation of the population through immunofluorescence labeling and super‐resolution light microscopy and at the electron microscope level through immunogold labeling. At the light microscope level, we used 3D‐SIM, the most commonly used super‐resolution immunofluorescence technique.8 We have shown previously in rare, favorable 3D‐SIM image slices examples of VWF mapping to a subregion of fibrinogen‐positive granules9 that were difficult to impossible to resolve in diffraction‐limited resolution confocal micrographs.9, 17 Here, we mapped the distribution of 5 major cargo proteins versus VAMP‐8, a vesicle‐associated membrane protein that mediates granule fusion with the plasma membrane and CS.1, 4, 26, 27 We also mapped VAMP‐8 distribution relative to a second α‐granule vesicle‐soluble NSF attachment protein receptor,VAMP‐7.28, 29 We found that VAMP‐8 co‐localized highly with VAMP‐7 in resting platelets by 3D‐SIM (Figure S3). In our 3D‐SIM images, neither VAMP‐8 nor VAMP‐7 could be resolved in the ringlike staining pattern expected for a fully resolved granule membrane protein whether lookup tables gave a slight degree of saturation or not (Figure 5, Figure S3, data not shown). As there was no obvious improvement in resolution, we show slightly saturating lookup tables to display a greater range of stained objects in each frame. In either case, the intensity of antibody staining for a membrane protein varied significantly to the eye across platelet slices (Figure 5A and B), indicating individual α‐granule variation in the protein level (see also Figure 5E, VAMP‐8 staining across a Z‐image stack). As expected for the higher resolution achieved in the current study (Figure 5D), the granule object count per platelet in the 3D‐SIM images was higher, at ~60 with a range of ~30 to 120 (2 different VAMP‐8 antibodies, Figure 5F) than that which we reported earlier by confocal microscopy, ~30 in total.17 Three‐dimensional–SIM α‐granule count per platelet and range corresponded remarkably well to those determined by rendering SBF‐SEM micrograph sets (Figure 2B). We note that 3D‐SIM is incapable of providing significant information regarding the shape of objects the size of α‐granules because of its limited Z‐resolution relative to the size of individual α‐granules. This limited resolution is clearly indicated by the inability of 3D‐SIM to resolve VAMP staining as the defining membrane feature of the α‐granule. In comparison, by electron microscopy, a method in which structure is directly visualized at nanometer resolution, the enclosing membrane is a readily recognized feature.
Figure 5.
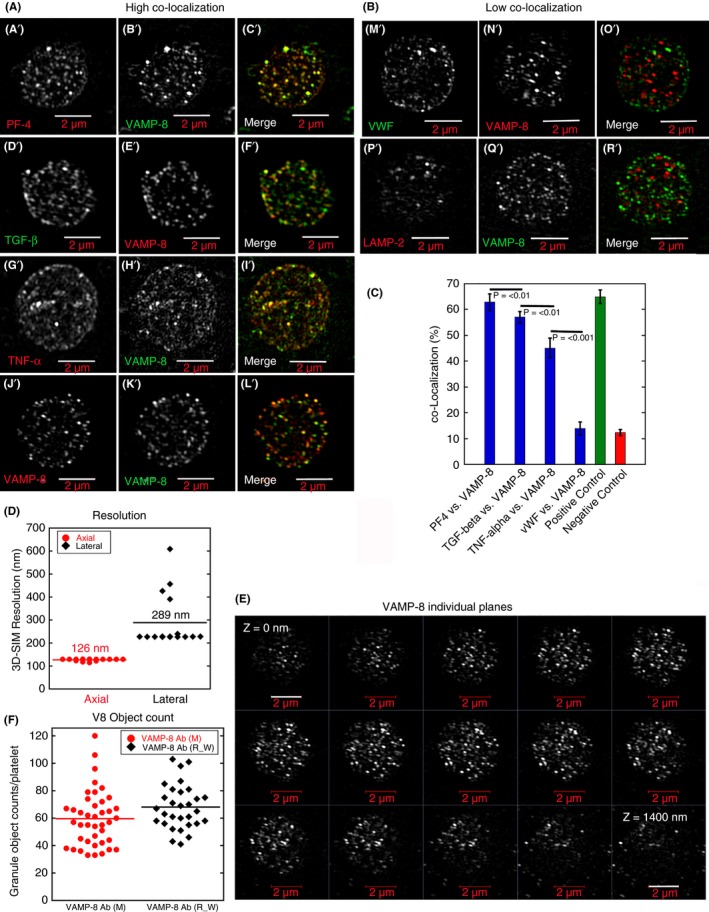
Qualitative (A, B: A’‐R’; E) and quantitative super‐resolution (C,D,F 3D‐SIM) data indicating a high co‐localization of PF4, TGF‐β, TNF‐α (cargo markers, A’‐I’) versus VAMP‐8 (α‐granule membrane marker) in comparison to VWF (M’‐O’). PF4 was detected using goat polyclonal antibodies directed against a peptide sequence present in the N‐terminal portion of human PF4 (Santa Cruz, sc‐23519, Santa Cruz Biotecnology, Inc., Dallas, TX, USA), TGF‐β1/2: mouse monoclonal antibodies directed against full‐length recombinant TGF‐β2 of cow origin (Santa Cruz, sc‐80346, TNF‐alpha: goat polyclonal antibodies directed against recombinant TNF‐α of human origin (R&D Systems, AF‐210‐NA, R&D Systems, Minneapolis, MN, USA), VWF: rabbit polyclonal antibodies directed against human VWF (Dako, A0082, Agilent (Dako), Santa Clara, CA, USA); and LAMP‐2: mouse monoclonal antibodies directed against human LAMP‐2 (Developmental Studies Hybridoma Bank, H4B4, Developmental Studies Hybridoma Bank, Iowa City, IA, USA). For VAMP8 labeling, either rabbit polyclonal VAMP‐8 antibodies (a gift of the S. W. Whiteheart Laboratory, University of Kentucky) or mouse monoclonal antibodies directed against a peptide present in N‐terminal portion of human VAMP‐8 (Santa Cruz, sc‐166820) were used as appropriate for double labeling. The fluorescent donkey secondary antibodies were purchased from (Jackson ImmunoResearch, Inc., West Grove, PA, USA) Positive control (J’‐L’): VAMP‐8 as first antibody stained with 2 different colors of second antibody; negative control (P’‐R’): VAMP‐8 antibody staining versus LAMP‐2 (late endosome/lysosomal membrane marker). Twenty platelets were scored for each pairing. For co‐localization details, see McBride et al,3 Kamykowski et al,9 Sehgal and Storrie,17, and Rhee et al.18 The fluorescence micrographs shown are all single‐plane slices from 3D‐SIM image stacks. Lookup tables were chosen to show a range of intensities, with only a few granules being near intensity saturation. Control data: (D) Calculated resolution values for 3D‐SIM platelet immunofluorescence. (E) Qualitative evidence for 3D‐SIM resolution in Z: VAMP‐8 distribution shown across a series of Z‐planes. (F) The 2 VAMP‐8 antibodies gave equivalent labeling as shown by fluorescent object counting. Micron bars = 2 micron. 3D‐SIM, 3‐dimensional–structured illumination; LAMP‐2, late endosomal/lysosomal membrane protein 2; PF4, platelet factor 4; TGF‐β, transforming growth factor‐β; VAMP‐8, vesicle‐associated membrane protein 8; VWF, von Willebrand factor
We chose cargo markers for immunofluorescence localization against VAMP‐8 that included extremes in the zoned distribution of cargo proteins within the α‐granule. Based on previous electron microscopy: platelet factor 4 (PF4), transforming growth factor‐β (TGF‐β), tumor necrosis factor‐α (TNF‐α), and fibrinogen distribute relatively uniformly through the granule matrix.30, 31, 32, 33 Qualitatively and quantitatively, each of these 4 cargo marker proteins were highly co‐localized with VAMP‐8, as shown in individual images from 3D‐SIM image stacks (5A and C; Figure S4). Quantitatively, 3D co‐localization was nearly as high for each as it was for the positive control, 2 differently labeled secondary antibodies against the VAMP‐8 primary antibody (Figure 5C). We attribute the high degree of co‐localization reported in these studies vs. our previous confocal microscope studies in which few pairings reached more than ~50% co‐localization9 to the 120 nm XY and 290 nm Z resolution (Figure 5D), a 2‐fold improvement in X, Y, and Z relative to confocal microscopy or in total an 8‐fold improvement overall in the tightness of localization achieved.
The fifth content marker, VWF, is known from previous EM to be located eccentrically within individual granule thin sections.6, 33, 34, 35 At the level of light microscopy, the point spread function (PSF) distribution for VWF staining and that of the membrane marker VAMP‐8 would be expected to show no more than limited overlap, a conclusion supported qualitatively by our previous 3D‐SIM studies.9 Although rare examples of side‐by‐side VWF/VAMP‐8 immunofluorescent pairing can be seen in Figure 5B, O’, the 3D‐SIM immunofluorescence analysis of multiple platelets showed little, if any, greater co‐localization of VWF with VAMP‐8 than for the negative control protein, LAMP2 (a late endosomal/lysosomal membrane protein) (Figure 5B and C).
We reasoned that the failure of VWF immunofluorescence to co‐localize extensively with VAMP‐8 or other markers9, 17, 22 (present work) is due to the statistical nature of mapping fluorescence intensity maxima distributions (ie, PSF). We chose an extension of an existing ultrastructural approach, immunoelectron microscopy, to resolve this problem. In previous electron microscopy immunogold labeling in thin sections6, 21, 35 and in our present studies (Figure S2D‐E), a VWF label failed to localize to a significant fraction of morphologically identified α‐granule profiles. We hypothesized that this outcome was due to a sampling problem as the volume of a thin section, a 60‐nm thick slice, is small relative to the size of α‐granules. To sample full granule volumes and hence to ask ultrastructurally whether VWF is present in each individual, morphologically identifiable α‐granule, we used sequential, serial cryo‐sectioning, a technically demanding approach. Serial sections were cut at a thickness of 95 nm to minimize loss between sections. As shown qualitatively in Figure 6A‐D, immunogold labeling for both VWF and the second content marker, fibrinogen, in individual, singly or doubly labeled sections, was concentrated over membrane‐limited α‐granule profiles in the 95‐nm section with, as expected in the case of VWF, some granule profiles unlabeled (Figure 6A, white **). Quantitatively, labeling density for VWF and fibrinogen over α‐granules was 50 and 225 gold particles per μm2, respectively, while cytosolic labeling density was <2 and 6 gold particles per μm2, a difference of ~30‐ fold (Figure 6C and D). When examined in a sequential, serial section set (Figure 6D, purple hoops, granule membrane perimeter in a given sequential section), each granule was positive for VWF (green dots) and fibrinogen (red dots). Significantly, VWF (green dots, arrowheads) was, as expected, concentrated in an eccentric zone within each granule. Fibrinogen was spread across a much larger volume of the granule but not the full granule volume. Fibrinogen itself appeared to be less tightly but, nevertheless, zonally distributed within the granule matrix.
Figure 6.
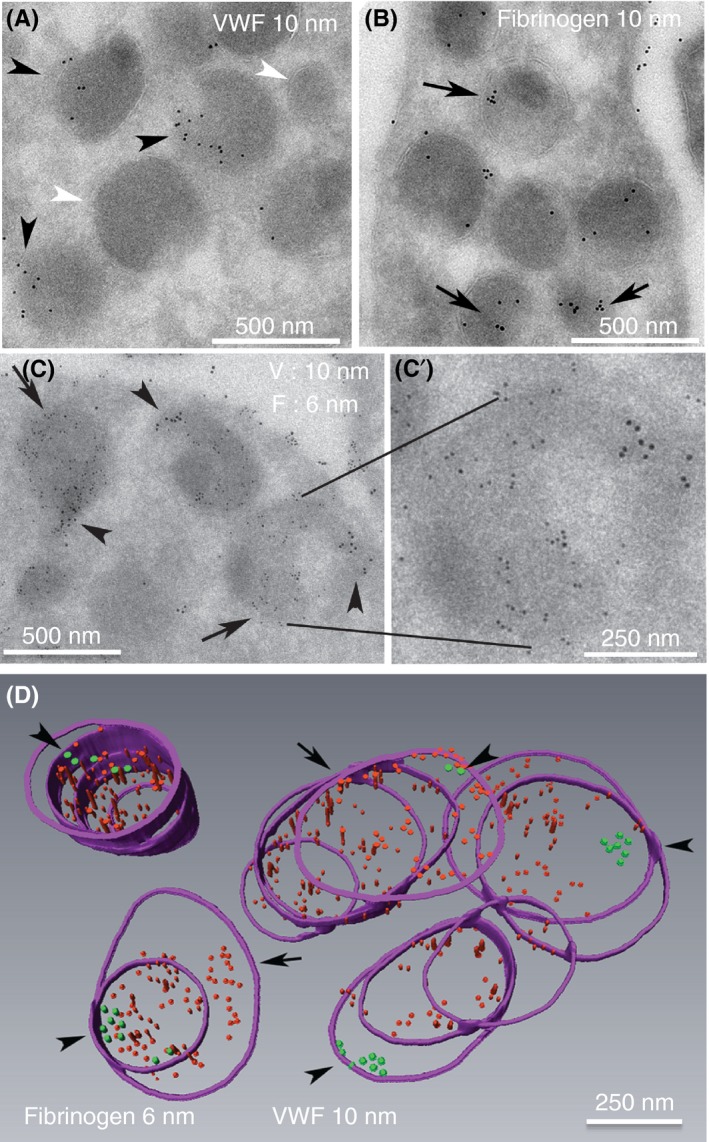
α‐granules in sequential, serial cryosections immunogold labeled for VWF or fibrinogen exhibited a high labeling incidence. (A, B) Representative 95‐nm cryosections, single immunogold‐labeled for VWF (A, green arrowheads, rabbit polyclonal antibodies, Dako, A0082; **, VWF negative granule profiles) or fibrinogen (red arrows, rabbit polyclonal antibodies, Calbiochem, 341552, MilliporeSigma (Calbiochem), Burlington, MA, USA). The fibrinogen labeling was more common across the granule population; 6‐nm gold particles were used to highlight the fibrinogen labeling shown in C and D, and the larger 10‐nm gold was used for VWF. (C,C’) Double‐label immunogold labeling for VWF and fibrinogen in 95‐nm sections. The double label was done sequentially with a fixation step between the first‐round labeling for fibrinogen and the second‐round labeling for VWF (see Materials and Methods for more details); 2× enlarged single representative α‐granule shown in insert demonstrates abundantly labeled fibrinogen and zoned labeling for VWF. (D) Purple hoops mark the membrane perimeter of the α‐granules as seen in the sequential serial sections. Qualitatively, rendered reconstruction of 5 individual α‐granules immunogold labeled for both fibrinogen (red particles) and VWF (green particles, arrowheads) showed that all were positive for both proteins, with the VWF labeling being restricted to a small zone within each granule. Note the lack of intermixing of fibrinogen and VWF labeling in this sequential, fibrinogen, followed by VWF labeling procedure. With respect to any given granule, the angle of cut may vary, and the full granule volume may or may not fall in the 6‐10 sequential sections of a serial section ribbon. Hence, the number of hoops (perimeter outlines) will vary from granule to granule
As shown quantitatively in Table 2, the incidence of α‐granules labeled for fibrinogen and VWF over singly labeled, sequential, serial section sets was high, at ~85%. In doubly labeled, sequential, serial 95‐nm serial cryo‐section sets, the labeling incidence for VWF and fibrinogen labeling was even higher, ~95% for both. The high specificity of that labeling is indicated by the distinct, nonoverlapping zonal immunogold distributions exhibited for each target protein (Figure 6). We conclude that the serial section immunogold labeling procedures provided high‐specificity data and gave a ready explanation for why the fluorescence intensity profiles for VWF and VAMP‐8 do not overlap substantially in our multiplatelet, 3D‐SIM experiments.
Table 2.
Volume sampling through serial sections shows that virtually all α‐granules are positive for both fibrinogen and VWF
| Incidence of granules positive for the respective label | |||
|---|---|---|---|
| Single labeling, sequential serial sections scored | |||
| 95 nm sections | Fibrinogen (10 nm gold) | VWF (10 nm gold) | VWF (6 nm gold) |
| Positive α‐granules, % | 86 ± 17 | 81 ± 15 | 82 ± 9 |
| α‐Granules scored | 111 | 55 | 134 |
| Platelets scored | 8 | 2 | 8 |
| Double‐labeling, sequential serial sections scored | ||
|---|---|---|
| 95‐nm sections | Fibrinogen (6‐nm gold) | VWF (10‐nm gold) |
| Positive α‐granules, % | 99 ± 1 | 93 ± 7 |
| α‐Granules scored | 118 | 118 |
| Platelets scored | 6 | 6 |
Data are presented plus or minus standard error of the mean.
Abbreviation: VWF, von Willebrand factor.
We note in both cryo‐sectioned, immunogold‐ (eg, Figure 6C) or immunofluorescent‐labeled samples that α‐granules whether morphologically or immunologically identified, appear less centrally located than observed in our glutaraldehyde fixed preparations (eg, Figures 1 and 2; Figure S1). We suggest that this outcome is readily explained on the basis of fixative/preparation differences. Glutaraldehyde as a dialdehyde is a strong crosslinking reagent, while formaldehyde/paraformaldehyde as a monoaldehyde is not. The formaldehyde fixation used in the immunolabeling experiments was chosen to preserve antigenicity rather than to give the full structural preservation of a crosslinking fixative. In brief, in our immunolabeling experiments, we have preserved antigenicity and many structural features within the platelet at the cost of poor preservation of the circumferential band of microtubules present in resting platelets. The choices made achieve the overall goal of striking a balance between preserving structure and retaining antigenicity across a range of experiments done at varying resolution levels.
4. DISCUSSION
Our experiments were designed to characterize, in 3D, the key organelle size, shape, and spacing parameters in resting human platelets. A major validation that the immediately fixed human platelets studied were indeed resting platelets came from rendering the morphology of the 30 randomly chosen examples in 3D; 28 of 30 were discoid shaped, a characteristic feature of resting platelets. When we analyzed average organelle sizes in our SBF‐SEM data sets, across 3 donors, we found little evidence for dispersity in the data. However, when analyzed on an individual platelet basis, evidence for significant and interesting dispersity did emerge. We gave an especially detailed consideration to the α‐granule as an important example of a major secretory organelle within platelets. In general, α‐granule parameters such as number were more closely correlated with platelet size than that of dense granules, mitochondria, or CS elements. Importantly, in all cases, the data did not cluster into apparent subsets indicative of organelle subpopulations.
At the level of the individual α‐granule, we collected ultrastructural data on granule size and shape at a resolution of just under 7 nm in XY and 30 nm in Z for 1488 examples over the 30 individual platelets. The dispersity of short‐axis and long‐axis lengths and axis ratios varied in a manner consistent with a single Gaussian population with no evidence for subpopulations. On average, α‐granules were ovoid with a short axis of 250 nm ± 8 nm and a long axis of 300 nm ± 8 nm. These values are somewhat smaller than the 300‐ to 500‐nm dimensions reported from stereological analysis.14 Protein composition data from 3D‐SIM immunofluorescence microscopy indicated co‐localization of 4 different α‐granule cargo proteins with VAMP‐8 or VAMP‐7, SNARE‐class membrane proteins. These data suggest a qualitative similarity between α‐granules as a class. However, staining intensity between granules varied in intensity, suggesting differences in protein copy number per organelle. The expected exception to extensive immunofluorescence co‐localization was VWF, a protein known to cluster eccentrically within the α‐granule lumen.6, 34, 35 By previous thin section immunogold labeling, colabeling is no more than 50%.21 As a final test of compositional similarity within the α‐granule population, we scored morphologically recognizable α‐granules in sequential, serial sections for co–immunogold labeling for VWF and fibrinogen. Nearly 100% of the granules were positive for both, albeit with VWF present in 1 eccentric cluster per α‐granule and fibrinogen being more generally distributed. In sum, the data suggest that α‐granules in resting human platelets consist of a single population with limited quantitative variation in size, shape, and protein copy number.
In conclusion, we draw attention to 2 aspects of our data on the relative spatial distribution of α‐granules, dense granules, mitochondria, and CS in resting human platelets. We found dense granules to be the most peripherally distributed, with almost 25% located within 35 nm of the plasma membrane, while only 7.5% of α‐granules showed a similar location. We suggest that the initial peripheral location of a significant portion of dense granules could be important in facilitating the rapid release of small molecules such as ADP from dense granules in response to platelet activation. Overall, our data indicated little interdigitation of α‐granules, dense granules, mitochondria, and CS with each other in 3D space. We suggest that this may result in part from the close spacing of α‐granules, the most numerous granule population. Approximately 70% of α‐granules were located within 35 nm, a distance that in other cell systems is the result of the formation of specific protein complexes.11, 12, 13 In other cell systems, these organelle contact sites have been implicated in lipid transport and membrane trafficking events.11, 12, 13 We also observed close contacts between DTS and α‐granules. To answer what is the physiological function of such putative contact sites in resting human platelets, further studies are needed.
RELATIONSHIP DISCLOSURE
The authors report nothing to disclose.
AUTHOR CONTRIBUTIONS
IDP was responsible for electron microscope sample preparation, data coordination in the Storrie laboratory, quality control of the platelet rendering done in the Leapman laboratory, and all the serial section immunogold labeling work. SY performed all the 3D‐SIM fluorescence microscopy and the resulting data analysis. AR and EM in the Leapman laboratory did all the serial block‐face SEM image analysis, initial platelet rendering, and resulting data tabulation. JAK and GZ supported sample loading and electron microscope operations. MAA coordinated the experimental work within the Leapman laboratory. RDL and BS were responsible for experimental design, data quality control, and manuscript preparation. BS prepared the first draft and took senior author responsibility for seeing the manuscript through to completion.
Supporting information
ACKNOWLEDGMENTS
The Storrie laboratory was supported in part by NIH grants R01 HL119393 and R56 HL119393. The Leapman laboratory was supported by the intramural program of NIBIB at the National Institutes of Health, Bethesda, Maryland. The Zeiss Elyra microscope used for super‐resolution fluorescence microscopy was funded by NIH grant S10 OD018065 to Brian Storrie. We would like to thank Dr. Sidney W. Whiteheart for his comments during the preparation of this manuscript.
Pokrovskaya ID, Yadav S, Rao A, et al. 3D ultrastructural analysis of α‐granule, dense granule, mitochondria, and canalicular system arrangement in resting human platelets. Res Pract Thromb Haemost. 2020;4:72–85. 10.1002/rth2.12260
REFERENCES
- 1. Eckly A, Rinkel JY, Proamer F, Ulas N, Joshi S, Whiteheart SW, et al. Respective contributions of single and compound granule fusion to secretion by activated platelets. Blood. 2016;128:2538–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yadav S, Williamson JK, Aronova MA, Prince AA, Pokrovskaya ID, Leapman RD, et al. Golgi proteins in circulating human platelets are distributed across non‐stacked, scattered structures. Platelets. 2017;28:400–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McBride EL, Rao A, Zhang G, Hoyne JD, Calco GN, Kuo BC, et al. Comparison of 3D cellular imaging techniques based on scanned electron probes: serial block face SEM vs. axial bright‐field STEM tomography. J Struct Biol. 2018;202:216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pokrovskaya I, Joshi S, Tobin M, Desai R, Aronova MA, Kamykowski JA, et al. SNARE‐dependent membrane fusion initiates α‐granule matrix decondensation in mouse platelets. Blood Advances. 2018;2947–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang R, Stone RL, Kaelber JT, Rochat RH, Nick AM, Vijayan KV, et al. Electron cryotomography reveals ultrastructural alterations in platelets from patients with ovarian cancer. Proc Natl Acad Sci USA. 2015;112:14266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Nispen tot Pannerden H, de Haas F, Gerts W, Posthuma G, van Dijk S, Heijnen HF. The platelet interior revisited: electron tomography reveals tubular alpha‐granule subtypes. Blood. 2010;118:1147–56. [DOI] [PubMed] [Google Scholar]
- 7. Pokrovskaya ID, Aronova MA, Kamykowski JA, Prince AA, Hoyne JD, Calco GN, et al. STEM tomography reveals that the canalicular system and α‐granules remain separate compartments during early secretion stages in blood platelets. J Thromb Haemost. 2016;14:572–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toomre D, Bewersdorf J. A new wave of cellular imaging. Annu Rev Cell Dev Biol. 2010;26:285–314. [DOI] [PubMed] [Google Scholar]
- 9. Kamykowski J, Carlton P, Sehgal S, Storrie B. Quantitative immunofluorescence mapping reveals little functional coclustering of proteins within platelet α‐granules. Blood. 2011;118:1370–3. [DOI] [PubMed] [Google Scholar]
- 10. Bannerjee M, Joshi S, Zhang J, Moncman CL, Yadav S, Bouchard BA, et al. Cellubrevin/vesicle‐associated membrane protein‐3‐mediated endocytosis and trafficking regulate platelet functions. Blood. 2017;130:2872–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saheki Y, de Camilli P. Endoplasmic reticulum plasma‐membrane contact sites. Annu Rev Biochem. 2017;86:659–84. [DOI] [PubMed] [Google Scholar]
- 12. Wu H, Carvalho P, Voeltz GK. Here, there, and everywhere. The importance of ER membrane contact sites. Science. 2018;361(6401):pii: eaan5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scorrano L, de Matteis MAQ, Emr S, Giordano F, Hajnóczky G, Kornmann B, et al. Coming together to define membrane contact sites. Nat Commun. 2019;10(1):1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. King SM, Reed GL. Development of platelet secretory granules. Semin Cell Dev Biol. 2002;13:293–302. [DOI] [PubMed] [Google Scholar]
- 15. Pfeifer CR, Shomorony A, Zhang G, Cai T, Xu H, Notkins AL, et al. Quantitative analysis of mouse pancreatic islet architecture by serial block‐face SEM. J Struct Biol. 2015;189:216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Storrie B, White J, Röttger S, Stelzer EH, Suganuma T, Nilsson T. Recycling of Golgi‐resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole‐induced Golgi scattering. J Cell Biol. 1998;143:1505–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sehgal S, Storrie B. Evidence that differential packaging of the major platelet granule protein von Willebrand factor and fibrinogen can support differential release. J Thromb Haemost. 2007;5:2009–16. [DOI] [PubMed] [Google Scholar]
- 18. Rhee S, Starr T, Forsten‐Williams Storrie B. The steady‐state distribution of glycosyltransferases between the Golgi apparatus and the endoplasmic reticulum is approximately 90:10. Traffic. 2005;6:978–90. [DOI] [PubMed] [Google Scholar]
- 19. Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discov. 2007;6:273–86. [DOI] [PubMed] [Google Scholar]
- 20. White GC 2nd, Rompietti R. Platelet secretion: indiscriminately spewed forth or highly orchestrated? J Thromb Haemost. 2007;5:2006–8. [DOI] [PubMed] [Google Scholar]
- 21. Italiano JE Jr, Richardson JL, Patel‐Hett S, Battinelli E, Zaslavsky A, Short S, et al. Angiogenesis is regulated by a novel mechanism: pro‐and antiangiogenic proteins are organized into separate platelet alpha granules and differentially released. Blood. 2008;111:1227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jonnalagadda D, Izu LT, Whiteheart SW. Platelet secretion is kinetically heterogeneous in an agonist‐responsive manner. Blood. 2012;184:5209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heijnen HF, van der Sluijs P. Platelet secretory behaviour: as diverse as the granules … or not? J Thromb Haemost. 2015;13:2141–51. [DOI] [PubMed] [Google Scholar]
- 24. Yadav S, Storrie B. The cellular basis of platelet secretion: emerging structure/function relationships. Platelets. 2017;28:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. NIST/SEMATECH e‐Handbook of Statistical Methods, Measures of skewness and kurtosis. Available from http://www.itl.nist.gov/div898/handbook/eda/section3/eda35b.htm, 2018. [Accessed 2018 October 31].
- 26. Ren Q, Barber HK, Crawford GL, Karim ZA, Zhao C, Choi W, et al. Endobrevin/VAMP‐8 is the primary v‐SNARE for the platelet release reaction. Mol Biol Cell. 2007;18:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jonnalagadda D, Sunkara M, Morris AJ, Whiteheart SW. Granule‐mediated release of sphingosine‐1‐phosphate by activated platelets. Biochim Biophys Acta. 2014;1831:1581–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peters CG, Michelson AD, Flaumenhaft R. Granule exocytosis is required for platelet spreading: differential sorting of α‐granule expressing VAMP‐7. Blood. 2012;120:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koseoglu S, Peters CG, Fitch‐Tevfik JL, Aisiku O, Danglot L, Galli T, et al. VAMP‐7 links granule exocytosis to actin reorganization during platelet activation. Blood. 2015;126:651–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sander HJ, Slot JW, Bouma BV, Bolhuis PA, Pepper DS, Sixma JJ. Immunocytochemical localization of fibrinogen, platelet factor 4, and beta thromboglobulin in thin frozen sections of human blood platelets. J Clin Invest. 1983;72:1217–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stenberg PE, Shuman MA, Levine SP, Bainton DF. Optimal techniques for the immunocytochemical demonstration of beta‐thromboglobulin, platelet factor 4, and fibrinogen in the alpha granules of unstimulated platelets. Histochem J. 1984;16:983–1001. [DOI] [PubMed] [Google Scholar]
- 32. Sixma JJ, Slot JW, Geuze JH. Immunocytochemical localization of platelet granule proteins. Methods Enzymol. 1989;169:301–11. [DOI] [PubMed] [Google Scholar]
- 33. Harrison P, Savidge GF, Cramer EM. The origin and physiological relevance of alpha‐granule adhesive proteins. Br J Haematol. 1990;74:125–30. [DOI] [PubMed] [Google Scholar]
- 34. Heijnen HFG, Debili N, Vainchenker W, Vainchenker W, Breton‐Gorius J, Geuze HJ, Sixma JJ. Multivesicular bodies are an intermediate stage in the formation of platelet α‐granules. Blood. 1998;91:2313–25. [PubMed] [Google Scholar]
- 35. Cramer EM, Meyer D, le Menn R, Breton‐Gorius J. Eccentric localization of von Willebrand factor in an internal structure of platelet alpha‐granule resembling that of Weibel‐Palade bodies. Blood. 1985;66:710–3. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


