Abstract
In recent years, short-chain fatty acids (SCFAs) have been reported to play an important role in maintaining human health. Fecal SCFA concentrations correlate well with colonic SCFA status and gut microbiota composition. However, the associations with the gut microbiota functional pathway, dietary intake, blood SCFAs, and fecal SCFAs remain uncertain. To clarify these relationships, we collected fecal samples, blood samples, and dietary habit data from 12 healthy adults aged 22–51 years. The relative abundance of several SCFA-producing bacteria, gut microbiota diversity, and functional pathways related to SCFA biosynthesis were positively associated with fecal SCFAs even after adjusting for age and sex. Furthermore, fecal acetate was likely to be positively associated with serum acetate. By contrast, dietary intake was not associated with fecal SCFAs. Overall, the present study highlights the potential usefulness of fecal SCFAs as an indicator of the gut microbiota ecosystem and dynamics of SCFAs in the human body.
Keywords: gut microbiota, fecal short-chain fatty acids, serum short-chain fatty acids, dietary intake, human
INTRODUCTION
Short-chain fatty acids (SCFAs) are the products of gut microbial fermentation of host-derived carbohydrates, dietary fiber, protein, and resistant starch [1,2,3]. The colon is responsible for the absorption of most SCFAs produced by gut microbiota into the body [4, 5]. Acetate, propionate, and butyrate are the major SCFA subtypes and play important roles in maintaining human health by acting as substrates for cholesterol synthesis, de novo lipogenesis, and gluconeogenesis in the liver [4, 6, 7] or as primary nutrients for colonocytes [8,9,10]. Fecal SCFAs concentrations are easily determined and correlate well with SCFA status in the colon [11, 12].
Recent epidemiological studies clarified the association between fecal SCFAs and gut microbiota composition [12,13,14,15]. While these previous studies are useful for understanding associations between the gut microbiota and human health in conjunction with SCFAs, they did not account for the functional profiles of the gut microbiota. In addition, consideration of dietary intake may be necessary, as it can affect SCFAs in the colon. Therefore, further research is required to elucidate SCFA metabolism in humans. The aim of the present study was to investigate the associations of gut microbiota composition and functional profiles, as well as dietary intake and fecal SCFAs, in healthy adults. Moreover, we investigated the correlation between fecal SCFAs and serum SCFAs.
MATERIALS AND METHODS
Study participants and sample collection
Participants were recruited among students and employees at our university departments in Sapporo, Hokkaido, Japan, in September 2017. A total of 12 healthy Japanese volunteers (10 males and 2 females) were enrolled in this study. Fasting blood samples were obtained in the morning by cubital venipuncture after overnight fasting from 9:00 p.m. Serum was separated and centrifuged after blood coagulation. Serum samples were stored at −80°C until metabolite analysis. Within 0–7 days of blood collection, participants provided two fresh fecal samples, one which was transferred by the participant into a plastic tube and one collected using a feces collection kit containing guanidine solution (TechnoSuruga Laboratory, Shizuoka, Japan) [16]. Fecal metabolite analysis was performed using the fresh fecal sample soon after collection, while the fecal collection kit sample was refrigerated and sent to TechnoSuruga Laboratory Co. Ltd. (Shizuoka, Japan) for fecal bacterial DNA extraction. Dietary intake one week prior to blood collection was assessed using a brief-type self-administered diet history questionnaire (BDHQ) [17]. Data on dietary intake estimated using the BDHQ have been validated in previous publications [17, 18]. Although the BDHQ is designed to ask about dietary habits in the month prior to the day of the questionnaire, we asked participants to answer about their dietary habits for the week prior to the day of blood collection. This study was approved by the Ethics Committee of the Faculty of Medicine, Hokkaido University (17-007), and written informed consent was obtained from all participants.
Measurement of fecal and serum SCFAs
Fresh fecal samples were homogenized with a 10-fold volume of phosphate buffer solution (50 mM Na2HPO4/NaH2PO4, pH 7.4, and 10% v/v D2O) containing 0.5 mM 3-trimethylsilypropionatel-2,2,3,3,-d4 (TSP-d4) and 0.04% NaN3. The mixtures were vortexed for 30 sec and shaken for 15 min at 4°C. The homogenates were centrifuged at 15,000 rpm for 10 min at 4°C. Serum samples were added to the phosphate buffer solution at the same final concentration as the fecal samples. Supernatants (550 µL) were transferred into 5 mm NMR tubes. All NMR data were obtained using a Bruker AVANCE III spectrometer (Bruker Biospin Inc.) at 600 MHz with a TXI z-gradient probe at 25°C. A NOESY pulse sequence with presaturation for water suppression with a relaxation delay of 4 sec and mixing time of 100 ms was used to record 1H NMR spectra of fecal samples. A Carr-Purcell-Meiboom-Gill sequence with a total time filter of 125 ms was used to record 1H NMR spectra of serum samples. Spectra were collected for each sample with 65,536 data points, a spectral width of 16 ppm, and accumulation of 128 scans. All FIDs were zero-filled, and an exponential line-broadening function of 0.2 Hz was applied before the spectra were Fourier transformed. All spectra were manually phase- and baseline-corrected using Delta 5.0.4 (JEOL RESONANCE Inc.). Metabolites were assigned using the Chenomx NMR Suite 8.2 software (Chenomx Inc.). In this report, total SCFAs were defined as the sum of the subtypes of SCFAs, including acetate, butyrate, and propionate.
Fecal bacterial DNA extraction
Bacterial DNA was extracted from fecal samples according to a previously described method [19]. Briefly, fecal solids, lysing buffer, and lysing matrix—including zirconia beads—were added to a BioPulverizer tube and crushed at 5 m/sec for 2 min using a FastPrep-24 Instrument (MP Biomedicals, Irvine, CA, USA). Bacterial DNA was extracted from 200 µL of fecal suspension using an automatic nucleic acid extractor (Magtration System 12gC, Precision System Science, Matsudo, Japan) with a reagent for extracting nucleic acid (MagDea DNA 200, Precision System Science, Matsudo, Japan).
Fecal bacterial 16S ribosomal RNA (rRNA) library preparation and sequencing
16S rRNA library preparation and sequencing were conducted at Morinaga Milk Industry Co., Ltd. (Zama, Japan). The V3-V4 region of bacterial 16S rRNA was amplified and sequenced using an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) according to a previously described method [20]. After removing sequences consistent with data from Genome Reference Consortium human build 37 (GRCh37) or PhiX 174 from the raw Illumina paired-end reads, the 3′ region of each read with a PHRED quality score less than 17 was trimmed. Trimmed reads less than 150 bp in length with an average quality score less than 25 or those lacking paired reads were also removed. Reads that passed the quality filters were combined using the fastq-join script in ea-utils (version 1.1.2-537) [21].
Bioinformatics analysis
All steps from the trimming of paired-end read FASTQ files, which were obtained by 16S rRNA amplicon sequencing, to gut microbiota diversity analysis were automatically performed using QIIME 1.9.1 [22] according to a previously described method (http://doi.org/10.5281/zenodo.1439555). The operational taxonomic units (OTUs) were clustered against the SILVA 128 reference database [23] at 97% similarity using the USEARCH algorithm [24]. The clustered OTUs were classified into five taxonomic rank categories (phylum, order, class, family, and genus) using the SILVA 128 reference database at 97% similarity. Subsequently, three alpha-diversity indices (Chao 1 index, phylogenetic diversity whole tree, and observed OTUs) were calculated. The representative SCFA-producing bacteria were selected based on previous studies [25, 26]. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) 1.0.0 [27] was used to predict the functional gene compositions based on the 16S rRNA gene sequences against the Greengenes [28] database with the default settings (type of functional predictions: KEGG Orthologs).
Statistical analysis
Data on fecal SCFAs are shown as absolute quantitative values (mM), while data on serum SCFAs are shown as relative concentrations because of the use of a Carr–Purcell–Meiboom–Gill sequence. Relative concentrations of serum SCFAs (no unit provided) represent the ratio of serum SCFA values of a given participant relative to the values of each other participant. To avoid confusion about serum SCFA data, all SCFA data were standardized using mean and standard deviation values of fecal and serum SCFAs among the study participants. The standardization yielded a normal distribution with a mean of zero and a standard deviation of one. The Firmicutes to Bacteroidetes ratio (F:B ratio) was determined by dividing the relative abundance of the phylum Firmicutes (%) by the relative abundance of the phylum Bacteroidetes (%). The Shannon diversity index (H’) [29], Simpson diversity index (1-D), and inverse Simpson index (1/D) [30] were calculated using the following equations at the genus level: H’ = -Σpilnpi and D = Σpi^2, where pi is the relative abundance (%) of genus i in the community. Initially, a linear regression model was used to quantify the association of the relative abundance of each bacterium, each functional pathway of the gut microbiota, or intake of each food and nutrient with fecal SCFAs, with age and sex incorporated as covariates. Next, to verify the correlation between fecal SCFAs and SCFAs in the body, a Pearson’s correlation coefficient (r) was calculated between SCFAs in feces and serum. Furthermore, a similar linear regression model was used to examine the association of fecal SCFAs with serum SCFAs, accounting for age and sex. All statistical analyses were performed using R version 3.4.1. P-values less than 0.05 were considered to be statistically significant.
RESULTS
Characteristics of study participants
All 12 participants (10 males and 2 females) were included in the analysis. Participant age ranged from 22 to 51 years, with a median of 37 years. Four participants collected their feces on the same day as blood collection, and the maximum interval between blood collection and feces collection was 7 days. According to the NMR spectroscopy results, the serum butyrate and propionate values of all the participants were lower than the normal detection limits. Therefore, only serum acetate could be examined among the subtypes of serum SCFAs.
Gut microbiota composition and fecal SCFAs
Table 1 shows the associations of the phylum-level compositions and alpha-diversity indices of the gut microbiota with fecal SCFAs. The relative abundances of phylum Firmicutes, the F:B ratio, Shannon index, Simpson index, and inverse Simpson index were significantly positively associated with fecal butyrate. Table 2 shows the associations of the relative abundances of the representative SCFA-producing bacteria with fecal SCFAs. Of these bacteria, the relative abundances of the representative butyrate-producing bacteria class Clostridia, order Clostridiales, family Ruminococcaceae, genus Subdoligranulum, and family Erysipelotrichaceae were significantly positively associated with fecal butyrate. In addition, the relative abundance of the representative acetate-producing bacteria genus Ruminococcus was significantly positively associated with fecal total SCFAs and fecal acetate, while that of Ruminiclostridium was significantly positively associated with fecal acetate. The relative abundances of the representative propionate-producing bacteria family Porphyromonadaceae, genera Bacteroides, and Ruminococcus were positively associated with fecal propionate, although the relationships were not statistically significant.
Table 1. Associations of phylum-level compositions and alpha-diversity indices of the gut microbiota with fecal SCFAs after adjusting for sex and age.
| Fecal total SCFAs† |
Fecal acetate† |
Fecal butyrate† |
Fecal propionate† |
||||||
|---|---|---|---|---|---|---|---|---|---|
| β | 95%CI‡ | β | 95%CI‡ | β | 95%CI‡ | β | 95%CI‡ | ||
| Phylum (%) | |||||||||
| Actinobacteria | −7.88 | (−23.30, 7.54) | −7.3 | (−22.81, 8.21) | −9.93 | (−26.44, 6.59) | −4.86 | (−20.92, 11.20) | |
| Bacteroidetes | −2.21 | (−10.70, 6.28) | −2.44 | (−10.84, 5.96) | −8.04 | (−14.99, −1.09)* | 4.48 | (−3.30, 12.27) | |
| Firmicutes | 4.11 | (−3.91, 12.12) | 4.17 | (−3.77, 12.11) | 9.73 | (4.39, 15.07)* | −2.65 | (−10.98, 5.68) | |
| Proteobacteria | −0.20 | (−54.23, 53.84) | 1.42 | (−52.3, 55.14) | 29.64 | (−24.71, 83.98) | −30.03 | (−77.62, 17.55) | |
| Alpha-diversity indices | |||||||||
| F:B ratio | 1.3 | (−2.15, 4.76) | 1.11 | (−1.66, 3.87) | 0.42 | (0.1, 0.75)* | −0.23 | (−0.78, 0.32) | |
| Shannon index | 1.08 | (−0.43, 2.59) | 1.1 | (−0.39, 2.59) | 1.82 | (0.6, 3.05)* | −0.18 | (−1.90, 1.55) | |
| Simpson index | 9.06 | (−5.66, 23.77) | 9.63 | (−4.74, 24.00) | 15.13 | (1.85, 28.41)* | −3.36 | (−19.44, 12.71) | |
| Inverse Simpson index | 0.14 | (−0.06, 0.34) | 0.15 | (−0.05, 0.35) | 0.25 | (0.08, 0.41)* | −0.06 | (−0.28, 0.17) | |
| Chao 1 index | 0.003 | (−0.015, 0.020) | 0.002 | (−0.015, 0.02) | 0.009 | (0.01, 0.027) | −0.002 | (−0.020, 0.015) | |
| Observed OTUs | 0.009 | (−0.011, 0.030) | 0.009 | (−0.011, 0.029) | 0.015 | (0.006, 0.035) | 0.002 | (−0.020, 0.023) | |
| PD whole tree | 0.17 | (−0.23, 0.56) | 0.16 | (−0.23, 0.55) | 0.19 | (0.24, 0.62) | 0.09 | (−0.32, 0.49) | |
CI: confidence interval; F:B: Firmicutes to Bacteroidetes; OTUs: operational taxonomic units; PD: phylogenetic diversity; SCFAs: short-chain fatty acids. †Values were standardized. ‡Linear regression analyses, adjusted for age and sex. *p<0.05.
Table 2. Associations of the relative abundances of SCFAs-producing bacteria with fecal SCFAs after adjusting for gender and age.
| Fecal total SCFAs†
|
Fecal acetate†
|
Fecal butyrate†
|
Fecal propionate†
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | 95%CI‡ | β | 95%CI‡ | β | 95%CI‡ | β | 95%CI‡ | |||||
| Phylum Actinobacteria | ||||||||||||
| Bifidobacterium (%) | −8.07 | (−23.83, 7.69) | −7.44 | (−23.30, 8.43) | −9.24 | (−26.47, 7.99) | −5.84 | (−22.07, 10.40) | ||||
| Phylum Bacteroidetes | ||||||||||||
| Porphyromonadaceae (%) | 7.04 | (−5.62, 19.71) | 7.73 | (−4.58, 20.04) | −0.58 | (−15.88, 14.73) | 5.24 | (−7.86, 18.33) | ||||
| Bacteroides (%) | −1.86 | (−7.87, 4.14) | −1.97 | (−7.92, 3.98) | −5.83 | (−10.72, −0.94)* | 2.61 | (−3.15, 8.37) | ||||
| Phylum Firmicutes | ||||||||||||
| Streptococcus (%) | 0.79 | (−105.21, 106.79) | −0.59 | (−105.99, 104.82) | −18.93 | (−134.59, 96.73) | 21.67 | (−81.81, 125.16) | ||||
| Clostridia (%) | 19.38 | (−17.80, 56.57) | 16.29 | (−13.44, 46.02) | 5.15 | (1.94, 8.36)* | −2.05 | (−8.32, 4.21) | ||||
| Clostridiales (%) | 4.04 | (−3.71, 11.79) | 4.19 | (−3.46, 11.84) | 9.03 | (3.4, 14.67)* | −2.64 | (−10.70, 5.42) | ||||
| Lachnospiraceae (%) | 0.41 | (−7.50, 8.31) | 0.76 | (−7.09, 8.60) | 1.59 | (−7.02, 10.21) | −2.45 | (−10.03, 5.13) | ||||
| Anaerostipes (%) | 1.76 | (−63.13, 66.65) | 1.78 | (−62.75, 66.30) | 0.36 | (−71.09, 71.81) | 1.68 | (−62.59, 65.95) | ||||
| Blautia (%) | 24.06 | (−6.40, 54.52) | 23.64 | (−6.79, 54.08) | 10.13 | (−28.88, 49.15) | 22.78 | (−7.92, 53.48) | ||||
| Coprococcus spp. (%) | −6.68 | (−56.06, 42.70) | −9.32 | (−58.13, 39.50) | 20.54 | (−31.52, 72.60) | −9.68 | (−58.25, 38.89) | ||||
| Eubacterium hallii (%) | 1.82 | (−86.04, 89.67) | 1.78 | (−85.58, 89.15) | −2.33 | (−99.03, 94.38) | 4 | (−82.97, 90.96) | ||||
| Eubacterium rectale (%) | −1.12 | (−26.81, 24.58) | 0.38 | (−25.19, 25.94) | 7.01 | (−20.72, 34.73) | −13.90 | (−36.71, 8.91) | ||||
| Roseburia (%) | −53.71 | (−218.41, 110.99) | −35.92 | (−202.80, 131.00) | −32.74 | (−218.4, 152.95) | −127.80 | (−260.58, 5.03) | ||||
| Ruminococcaceae (%) | 5.62 | (−3.32, 14.56) | 5.37 | (−3.60, 14.35) | 9.83 | (2.22, 17.44)* | 0.6 | (−9.34, 10.53) | ||||
| Faecalibacterium (%) | 2.09 | (−14.73, 18.90) | 1.32 | (−15.45, 18.10) | 11.92 | (−3.95, 27.79) | −2.48 | (−19.10, 14.14) | ||||
| Ruminiclostridium spp. (%) | 71.56 | (−1.06, 144.19) | 74.06 | (3.8, 144.31)* | 28.8 | (−71.02, 128.62) | 50.14 | (−32.58, 132.86) | ||||
| Ruminococcus spp. (%) | 19.37 | (2.39, 36.36)* | 19.29 | (2.42, 36.17)* | 18.65 | (−1.86, 39.17) | 9.42 | (−12.24, 31.07) | ||||
| Subdoligranulum (%) | 166.64 | (−139.71, 472.99) | 129.79 | (−118.65, 378.24) | 36.61 | (4.51, 68.71)* | 0.24 | (−53.57, 54.04) | ||||
| Erysipelotrichaceae (%) | 50.51 | (−135.50, 236.53) | 38.2 | (−112.13, 188.52) | 22.09 | (4.87, 39.31)* | −9.78 | (−39.31, 19.76) | ||||
CI: confidence interval; SCFAs: short-chain fatty acids. †Values were standardized. ‡Linear regression analyses, adjusted for age and gender. *p<0.05.
Gut microbiota functional profiles and fecal SCFAs
Table 3 shows the associations of the gut microbiota functional profiles related to the production and metabolism of SCFAs with fecal SCFAs. The functional pathway fatty acid biosynthesis was significantly and positively associated with fecal total SCFAs and fecal acetate. This functional pathway was also positively associated with fecal butyrate and fecal propionate, although the relationship was not statistically significant.
Table 3. Associations of the functional profiles of the gut microbiota with fecal SCFAs adjusting for sex and age.
| Fecal total SCFAs†
|
Fecal acetate†
|
Fecal butyrate†
|
Fecal propionate†
|
|||||
|---|---|---|---|---|---|---|---|---|
| β | 95%CI‡ | β | 95%CI‡ | β | 95%CI‡ | β | 95%CI‡ | |
| Carbohydrate digestion and absorption (%) | 18.71 | (−116.58, 154) | 18.57 | (−115.97, 153.1) | 38.52 | (−108.01, 185.06) | −5.62 | (−140.38, 129.15) |
| Carbohydrate metabolism (%) | 1.81 | (−27.93, 31.56) | 2.93 | (−26.59, 32.45) | −17.78 | (−47.18, 11.62) | 9.56 | (−18.88, 38.01) |
| Fatty acid biosynthesis (%) | 35.56 | (11.63, 59.49)* | 35.16 | (11.16, 59.15)* | 26.22 | (−9.21, 61.65) | 24.38 | (−7.10, 55.87) |
| Fatty acid metabolism (%) | −19.00 | (−85.91, 47.92) | −20.37 | (−86.62, 45.88) | −12.47 | (−87.38, 62.45) | −6.23 | (−74.07, 61.60) |
| Protein digestion and absorption (%) | −32.33 | (−163.11, 98.45) | −35.54 | (−165.00, 93.92) | −90.12 | (−217.28, 37.03) | 44.27 | (−82.84, 171.38) |
| Starch and sucrose metabolism (%) | −1.14 | (−11.82, 9.55) | −1.00 | (−11.64, 9.63) | −7.27 | (−17.48, 2.94) | 3.32 | (−6.95, 13.60) |
CI: confidence interval; SCFAs: short-chain fatty acids. †Values were standardized. ‡Linear regression analyses, adjusted for age and sex. *p<0.05.
Dietary intake and fecal SCFAs
Table 4 shows the associations of dietary intake with fecal SCFAs. Intake of foods containing SCFAs (low fat milk, normal milk, pickled vegetables, soy source, and western confectioneries) and intake of the major sources of SCFAs (protein, animal protein, vegetable protein, carbohydrate, total dietary fiber, water-soluble dietary fiber, insoluble dietary fiber, and butyrate) showed no significant associations with fecal SCFAs.
Table 4. Associations of dietary intake with fecal SCFAs adjusting for sex and age.
| Fecal total SCFAs†
|
Fecal acetate†
|
Fecal butyrate†
|
Fecal propionate†
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β | 95%CI‡ | β | 95%CI‡ | β | 95%CI‡ | β | 95%CI‡ | ||||
| Foods | |||||||||||
| Low-fat milk | (g/day) | −0.004 | (−0.012, 0.004) | −0.003 | (−0.011, 0.005) | −0.002 | (−0.011, 0.007) | −0.005 | (−0.013, 0.002) | ||
| Normal milk | (g/day) | 0.005 | (−0.015, 0.025) | 0.004 | (−0.016, 0.024) | 0.011 | (−0.009, 0.031) | 0.004 | (−0.016, 0.024) | ||
| Pickled vegetables | (g/day) | −0.006 | (−0.062, 0.051) | −0.006 | (−0.063, 0.050) | −0.021 | (−0.082, 0.039) | 0.013 | (−0.043, 0.068) | ||
| Soy sauce | (g/day) | −0.075 | (−2.374, 2.223) | −0.248 | (−2.526, 2.029) | 1.095 | (−1.273, 3.463) | −0.026 | (−2.303, 2.251) | ||
| Western confectionery | (g/day) | −0.002 | (−0.030, 0.026) | −0.001 | (−0.029, 0.027) | −0.012 | (−0.041, 0.017) | 0.001 | (−0.026, 0.029) | ||
| Nutrients | |||||||||||
| Protein | (g/day) | 0.008 | (−0.022, 0.038) | 0.009 | (−0.021, 0.039) | 0.016 | (−0.015, 0.047) | −0.010 | (−0.039, 0.020) | ||
| Animal protein | (g/day) | 0.020 | (−0.022, 0.061) | 0.022 | (−0.019, 0.062) | 0.030 | (−0.012, 0.073) | −0.008 | (−0.052, 0.035) | ||
| Vegetable protein | (g/day) | −0.013 | (−0.097, 0.071) | −0.010 | (−0.093, 0.074) | 0.013 | (−0.079, 0.106) | −0.044 | (−0.120, 0.032) | ||
| Carbohydrates | (g/day) | −0.0004 | (−0.006, 0.006) | −0.0002 | (−0.006, 0.006) | 0.001 | (−0.005, 0.008) | −0.003 | (−0.008, 0.003) | ||
| Total dietary fiber | (g/day) | −0.035 | (−0.211, 0.141) | −0.036 | (−0.211, 0.139) | −0.016 | (−0.211, 0.180) | −0.024 | (−0.199, 0.152) | ||
| Water-soluble dietary fiber | (g/day) | −0.222 | (−0.865, 0.422) | −0.233 | (−0.869, 0.404) | −0.012 | (−0.747, 0.724) | −0.196 | (−0.838, 0.446) | ||
| Insoluble dietary fiber | (g/day) | −0.028 | (−0.293, 0.237) | −0.028 | (−0.291, 0.236) | −0.010 | (−0.303, 0.283) | −0.027 | (−0.290, 0.235) | ||
| Butyrate | (mg/day) | −0.007 | (−0.021, 0.008) | −0.007 | (−0.021, 0.008) | −0.004 | (−0.021, 0.012) | −0.004 | (−0.019, 0.011) | ||
CI: confidence interval; SCFAs: short-chain fatty acids. †Values were standardized. ‡Linear regression analyses, adjusted for age and sex.
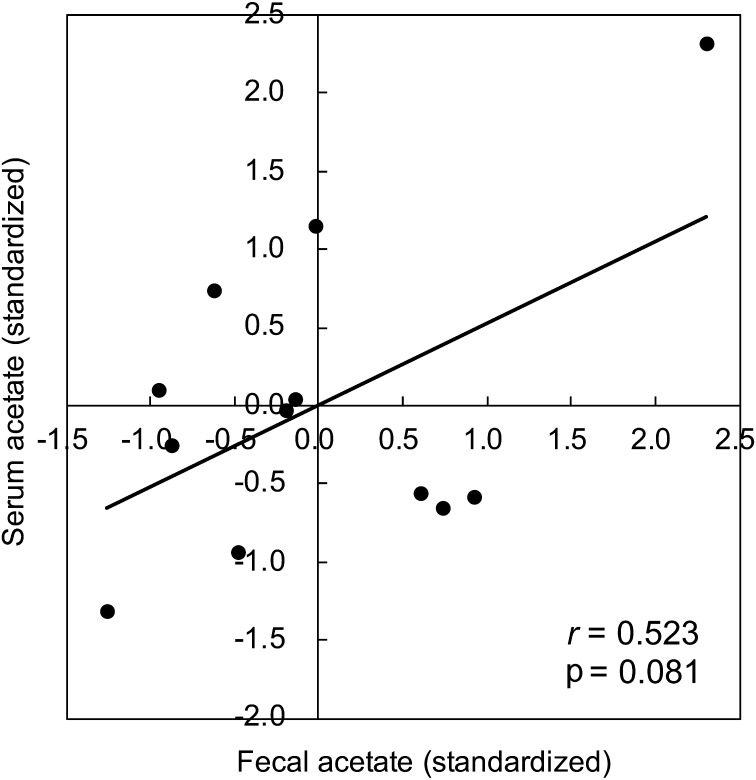
Fecal acetate and serum acetate
Fecal acetate showed a trend toward a positive correlation with serum acetate, although the relationship was not statistically significant (r=0.523, p=0.081; Fig. 1 and Supplementary Fig. 1, illustrated using data on non-standardized fecal and serum acetate values). The possible positive association between fecal and serum acetate was similar even after adjusting for sex and age in the multiple linear regression analysis (β=0.564, 95% CI= −0.123 to 1.251; data not shown).
Fig. 1.
Correlation between fecal acetate (standardized value) and serum acetate (standardized value). Pearson’s correlation coefficients (r) and p-values were calculated to assess the correlation of interest.
DISCUSSION
The present study investigated interrelationships among gut microbiota compositions and functional profiles, dietary intake, and fecal and serum SCFAs in 12 healthy adults. The relative abundance of several SCFA-producing bacteria, gut microbiota diversity, and the functional pathway related to SCFAs biosynthesis were positively associated with fecal SCFAs even after adjusting for age and sex. Furthermore, fecal acetate showed a trend toward a positive association with serum acetate. Taken together, these findings provide evidence that fecal SCFAs positively correlate well with the composition of SCFA-producing bacteria, gut microbiota SCFA productivity, and SCFA status in the body.
Previous epidemiological studies reported that the butyrate-producing phylum Firmicutes and F:B ratio are positively associated with fecal butyrate [12, 14], which is consistent with our results. Moreover, we found that gut microbiota species richness and evenness were positively associated with fecal butyrate. Almost all colonic butyrate is metabolized in colonocytes as primary nutrients [8,9,10]. Our study suggested that the relative abundances of butyrate producers and the diversity of the gut microbiota were associated with the nutritional status of the colonocytes. Likewise, representative acetate-producers and propionate-producers showed a positive association with fecal acetate and propionate, respectively. Fecal acetate and propionate are positively associated with obesity status in humans [12, 14, 31,32,33]. Colonic acetate and propionate are mainly metabolized in the liver as substrates for cholesterol synthesis and lipogenesis [4, 6, 7]. The functional pathway of the gut microbiota related to SCFA biosynthesis was positively associated with all types of fecal SCFAs, especially with total SCFAs and acetate. Because SCFAs are the gut microbial fermentation products of host-derived dietary fiber [1,2,3], it seems reasonable that there were positive associations between the SCFA productivity of the gut microbiota and fecal total SCFAs and acetate. To our knowledge, this study is the first to investigate the association of functional profiles of the gut microbiota with fecal SCFAs. Several previous studies examined whether the presence or absence of SCFA-producing bacteria is associated with fecal SCFAs [12,13,14,15]. Our study offers new insights, finding that the SCFA production capability of the gut microbiota—but not other functions—affects fecal SCFAs. Overall, fecal SCFAs were positively associated with the presence of producers of each SCFA as well as gut microbiota diversity and the SCFA production capability of the gut microbiota.
A previous study of 570 healthy Japanese adults reported that the mean daily intake of SCFAs was 0.37 g [34], which is drastically lower than the 20 g of SCFAs produced by the human gut microbiota each day [35]. In addition, orally administered SCFAs were reported to be rapidly absorbed in the stomach or small intestine and cleared from circulated blood within 1 hr [36, 37]. Based on these reports, trace amounts of SCFAs in the diet may not reach the large intestine and feces, which supports our finding that intake of SCFA-containing food was not associated with fecal SCFAs. Conversely, previous in vitro studies [38, 39] and human intervention studies [40,41,42] have shown positive associations between dietary fiber intake and fecal SCFAs, which are inconsistent with our results. Unlike these previous experiments and intervention studies, we investigated the association between dietary fiber intake and fecal SCFAs in a real-life setting, which may have resulted in the inconsistent finding with respect to the effect of dietary fiber intake on fecal SCFAs.
In humans, fecal acetate correlates well with colonic acetate status [11, 12], even though approximately 95% of colonic acetate is rapidly absorbed by the body [43, 44]. The majority of absorbed acetate is metabolized in the liver, with residual acetate appearing in systemic circulation [45]. Based on these conditions, our study showed that fecal acetate was likely to be positively associated with serum acetate. Regarding fecal acetate, a recent report suggested that serum acetate was positively associated with obesity status in humans [46].
We acknowledge that this study is subject to several important limitations. First, the study participants were middle-aged and comprised of predominantly Japanese males. The generalizability of our results is thus limited. Second, we attempted to investigate interrelationships between dietary intake, gut microbiota, and fecal and blood SCFAs on the day of the survey. However, dietary habits were evaluated for the week prior to blood collection, and some participants provided fecal samples up to one week after blood collection. Third, the 16S rRNA gene sequencing approach we used in this study made it possible to calculate only the relative abundance of the microbiota. Unfortunately, no information is available on the absolute abundance of bacteria. However, the gut microbiota is an ecosystem, and it is important to consider the complex bacteria–bacteria interactions. Therefore, it is valuable to evaluate the relative abundance of each SCFA-producing bacterium in the entire microbiota. Fourth, correlations between fecal and serum SCFAs other than acetate were not assessed due to the detection limits for serum samples in NMR spectroscopy. Acetate is the most abundant SCFA in the human body [47] and constitutes about 60% of fecal total SCFAs and over 90% of serum total SCFAs [48, 49]. Therefore, we believe that the positive association between fecal and serum acetate observed in this study will be helpful for understanding the dynamics of SCFAs in the human body.
In conclusion, our study provided evidence that fecal SCFAs positively correlate well with the presence of SCFA-producing bacteria as well as the diversity and SCFA productivity of the gut microbiota in humans. Moreover, fecal acetate was likely positively associated with serum acetate. These findings highlight the potential usefulness of fecal SCFAs as an indicator of the gut microbiota ecosystem and dynamics of SCFAs in the human body. Recently, in the field of biomedical sciences, there has been increasing interest in gut microbiota as a possible factor linked to human health [50]. For example, recent studies suggest that the gut microbiota is linked with obesity via SCFAs [15, 46]. Therefore, the measurement of fecal SCFAs helps investigate the interrelationships among the gut microbiota SCFAs and obesity in humans. We expect that further epidemiological studies in large populations will yield critical insights that will help elucidate the associations between the gut microbiota and human health in conjunction with SCFAs.
Conflict of interest
No potential conflicts of interest were reported by the authors.
Supplementary
Acknowledgments
This work was supported by the Hitachi Hokkaido University Laboratory, Hitachi, Ltd. The source of funding had no role in this research.
References
- 1.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. 2011. Human nutrition, the gut microbiome and the immune system. Nature 474: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- 3.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. 2016. The role of gut microbiota in the development of obesity and diabetes. Lipids Health Dis 15: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54: 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakobsdottir G, Bjerregaard JH, Skovbjerg H, Nyman M. 2013. Fasting serum concentration of short-chain fatty acids in subjects with microscopic colitis and celiac disease: no difference compared with controls, but between genders. Scand J Gastroenterol 48: 696–701. [DOI] [PubMed] [Google Scholar]
- 6.Bloemen JG, Venema K, van de Poll MC, Olde Damink SW, Buurman WA, Dejong CH. 2009. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin Nutr 28: 657–661. [DOI] [PubMed] [Google Scholar]
- 7.Bloemen JG, Olde Damink SW, Venema K, Buurman WA, Jalan R, Dejong CH. 2010. Short chain fatty acids exchange: Is the cirrhotic, dysfunctional liver still able to clear them? Clin Nutr 29: 365–369. [DOI] [PubMed] [Google Scholar]
- 8.Bugaut M. 1987. Occurrence, absorption and metabolism of short chain fatty acids in the digestive tract of mammals. Comp Biochem Physiol B 86: 439–472. [DOI] [PubMed] [Google Scholar]
- 9.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27: 104–119. [DOI] [PubMed] [Google Scholar]
- 10.Kim YI. 2000. AGA technical review: impact of dietary fiber on colon cancer occurrence. Gastroenterology 118: 1235–1257. [DOI] [PubMed] [Google Scholar]
- 11.Vogt JA, Wolever TM. 2003. Fecal acetate is inversely related to acetate absorption from the human rectum and distal colon. J Nutr 133: 3145–3148. [DOI] [PubMed] [Google Scholar]
- 12.Rahat-Rozenbloom S, Fernandes J, Gloor GB, Wolever TM. 2014. Evidence for greater production of colonic short-chain fatty acids in overweight than lean humans. Int J Obes 38: 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes J, Wang A, Su W, Rozenbloom SR, Taibi A, Comelli EM, Wolever TM. 2013. Age, dietary fiber, breath methane, and fecal short chain fatty acids are interrelated in Archaea-positive humans. J Nutr 143: 1269–1275. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes J, Su W, Rahat-Rozenbloom S, Wolever TM, Comelli EM. 2014. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr Diabetes 4: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, Velásquez-Mejía EP, Sierra JA, Corrales-Agudelo V, Carmona JA, Abad JM, Escobar JS. 2018. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 11: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosomi K, Ohno H, Murakami H, Natsume-Kitatani Y, Tanisawa K, Hirata S, Suzuki H, Nagatake T, Nishino T, Mizuguchi K, Miyachi M, Kunisawa J. 2017. Method for preparing DNA from feces in guanidine thiocyanate solution affects 16S rRNA-based profiling of human microbiota diversity. Sci Rep 7: 4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi S, Murakami K, Sasaki S, Okubo H, Hirota N, Notsu A, Fukui M, Date C. 2011. Comparison of relative validity of food group intakes estimated by comprehensive and brief-type self-administered diet history questionnaires against 16 d dietary records in Japanese adults. Public Health Nutr 14: 1200–1211. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi S, Honda S, Murakami K, Sasaki S, Okubo H, Hirota N, Notsu A, Fukui M, Date C. 2012. Both comprehensive and brief self-administered diet history questionnaires satisfactorily rank nutrient intakes in Japanese adults. J Epidemiol 22: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5: 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato K, Ishida S, Tanaka M, Mitsuyama E, Xiao JZ, Odamaki T. 2018. Association between functional lactase variants and a high abundance of Bifidobacterium in the gut of healthy Japanese people. PLoS One 13: e0206189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aronesty E. 2013. Comparison of sequencing utility programs. Open Bioinforma 7: 1–8. [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- 25.Macfarlane GT, Macfarlane S. 2012. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int 95: 50–60. [DOI] [PubMed] [Google Scholar]
- 26.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165: 1332–1345. [DOI] [PubMed] [Google Scholar]
- 27.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon C. 1948. A mathematical theory of communication. Bell Syst Tech J 27: 379–423. [Google Scholar]
- 30.Simpson EH. 1949. Measurement of diversity. Nature 163: 688–688. [Google Scholar]
- 31.Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. 2010. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18: 190–195. [DOI] [PubMed] [Google Scholar]
- 32.Patil DP, Dhotre DP, Chavan SG, Sultan A, Jain DS, Lanjekar VB, Gangawani J, Shah PS, Todkar JS, Shah S, Ranade DR, Patole MS, Shouche YS. 2012. Molecular analysis of gut microbiota in obesity among Indian individuals. J Biosci 37: 647–657. [DOI] [PubMed] [Google Scholar]
- 33.Teixeira TF, Grześkowiak Ł, Franceschini SC, Bressan J, Ferreira CL, Peluzio MC. 2013. Higher level of faecal SCFA in women correlates with metabolic syndrome risk factors. Br J Nutr 109: 914–919. [DOI] [PubMed] [Google Scholar]
- 34.Otsuka R, Kato Y, Nishita Y, Tange C, Imai T, Ando F, Shimokata H. 2015. Effect of short- and medium-chain fatty acid intake on cognitive score decline over 8 years among community-dwelling elderly. Nippon Eiyo Shokuryo Gakkaishi 68: 101–111. [Google Scholar]
- 35.Hara H. 2002. Physiological effects of short-chain fatty acid produced from prebiotics in the colon. Biosci Microflora 21: 35–42. [Google Scholar]
- 36.Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SE, MacDougall K, Preston T, Tedford C, Finlayson GS, Blundell JE, Bell JD, Thomas EL, Mt-Isa S, Ashby D, Gibson GR, Kolida S, Dhillo WS, Bloom SR, Morley W, Clegg S, Frost G. 2015. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64: 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomare EW, Branch WJ, Cummings JH. 1985. Carbohydrate fermentation in the human colon and its relation to acetate concentrations in venous blood. J Clin Invest 75: 1448–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortensen PB, Nordgaard-Andersen I. 1993. The dependence of the in vitro fermentation of dietary fibre to short-chain fatty acids on the contents of soluble non-starch polysaccharides. Scand J Gastroenterol 28: 418–422. [DOI] [PubMed] [Google Scholar]
- 39.McBurney MI. 1991. Starch malabsorption and stool excretion are influenced by the menstrual cycle in women consuming low-fibre Western diets. Scand J Gastroenterol 26: 880–886. [DOI] [PubMed] [Google Scholar]
- 40.Fredstrom SB, Lampe JW, Jung HJG, Slavin JL. 1994. Apparent fiber digestibility and fecal short-chain fatty acid concentrations with ingestion of two types of dietary fiber. JPEN J Parenter Enteral Nutr 18: 14–19. [DOI] [PubMed] [Google Scholar]
- 41.Noakes M, Clifton PM, Nestel PJ, Le Leu R, McIntosh G. 1996. Effect of high-amylose starch and oat bran on metabolic variables and bowel function in subjects with hypertriglyceridemia. Am J Clin Nutr 64: 944–951. [DOI] [PubMed] [Google Scholar]
- 42.van Munster IP, Tangerman A, Nagengast FM. 1994. Effect of resistant starch on colonic fermentation, bile acid metabolism, and mucosal proliferation. Dig Dis Sci 39: 834–842. [DOI] [PubMed] [Google Scholar]
- 43.Cook SI, Sellin JH. 1998. Review article: short chain fatty acids in health and disease. Aliment Pharmacol Ther 12: 499–507. [DOI] [PubMed] [Google Scholar]
- 44.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. 2006. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40: 235–243. [DOI] [PubMed] [Google Scholar]
- 45.Rombeau JL, Kripke SA. 1990. Metabolic and intestinal effects of short-chain fatty acids. JPEN J Parenter Enteral Nutr 14 Suppl: 181S–185S. [DOI] [PubMed] [Google Scholar]
- 46.Goffredo M, Mass K, Parks EJ, Wagner DA, McClure EA, Graf J, Savoye M, Pierpont B, Cline G, Santoro N. 2016. Role of gut microbiota and short chain fatty acids in modulating energy harvest and fat partitioning in youth. J Clin Endocrinol Metab 101: 4367–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergman EN. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev 70: 567–590. [DOI] [PubMed] [Google Scholar]
- 48.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28: 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binder HJ. 2010. Role of colonic short-chain fatty acid transport in diarrhea. Annu Rev Physiol 72: 297–313. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura K, Sakuragi N, Takakuwa A, Ayabe T. 2016. Paneth cell α-defensins and enteric microbiota in health and disease. Biosci Microbiota Food Health 35: 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.